Abstract
Alkaliphilic Bacillus species grow at pH values up to approximately 11. Motile alkaliphilic Bacillus use electrochemical gradients of Na+ (sodium-motive force) to power ion-coupled, flagella-mediated motility as opposed to the electrochemical gradients of H+ (proton-motive force) used by most neutralophilic bacteria. Membrane-embedded stators of bacterial flagella contain ion channels through which either H+ or Na+ flow to energize flagellar rotation. Stators of the major H+-coupled type, MotAB, are distinguishable from Na+-coupled stators, PomAB of marine bacteria and MotPS of alkaliphilic Bacillus. Dual ion-coupling capacity is found in neutralophilic Bacillus strains with both MotAB and MotPS. There is also a MotAB variant that uses both coupling ions, switching as a function of pH. Chemotaxis of alkaliphilic Bacillus depends upon flagellar motility but also requires a distinct voltage-gated NaChBac-type channel. The two alkaliphile Na+ channels provide new vistas on the diverse adaptations of sensory responses in bacteria.
Keywords: alkaliphile, Bacillus, chemotaxis, flagella, motility, MotPS, NaChBac, Na+ cycle, sodium-dependent, voltage-gated, Na+ channel
Extremophilic bacteria are of great utility because of the natural products they provide and the insights their adaptations provide into the plasticity of biological systems [1-3]. In addition, studies of the adaptations to their specific environmental challenges often lead to discoveries that apply more broadly to nonextremophilic bacteria, including pathogenic strains. Studies of extremely alkaliphilic Bacillus species have led to insights into ion coupling to ATP synthesis and alkaline pH homeostasis that are now applied way beyond alkaliphiles [3-6], and recent studies of flagellar-based motility and chemotaxis in these extremophiles have identified participating components and raised questions that extend broadly [7-11]. Thus, the alkaliphile as a model system of bioenergetic work has shed light on swimming, which underpins chemotaxis, the ability to move away from detrimental conditions and toward favorable ones, and the ability to inhabit niches in animal hosts as well as environmental settings or perform complex behaviors [12,13].
Structure of bacterial flagella
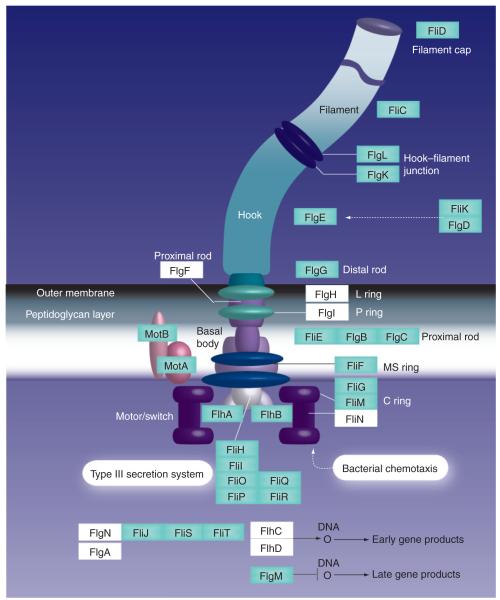
Like most other motile bacteria, alkaliphilic Bacillus species use filamentous propeller-like flagella for swimming (Figure 1). Those alkaliphiles studied to date have peritrichous flagella, in other words, flagella that are distributed around the cell surface as opposed to being confined to one or both cell poles [9,11]. The flagellum is a complex rotary nanomachine that is composed of multiple copies of at least 25 different proteins (Figure 1) [14,15]. The structure is composed of a helical filament, a basal body that is embedded in the cytoplasmic membrane and a hook connecting the filament to the basal body. The flagellar motor in the basal body consists of a rotor and stator that function comparably to those of nonbiological motors. The rotor encompasses a switch complex that is composed of the proteins FliG, FliM and FliN, and is involved in the generation of the torque for flagellar rotation as well as in the control of the rotational direction, either counterclockwise or clockwise, and in the assembly of the flagella [16-25]. The membrane-embedded stators form a ring around the base of the flagella [26] and support conversion of the energy from transmembrane electrochemical ion gradients, in the form of a proton-motive force (PMF) or sodium-motive force (SMF), into mechanical energy [27]. MotA and MotB proteins were identified as the protein components of the stator of proton-driven motors of Gram-negative Salmonella enterica serovar Typhimurium and Escherichia coli in which the most extensive studies of bacterial motility and chemotaxis have been conducted [28-30]. MotA has four transmembrane helices (TM1–4). A large cytoplasmic domain between TM2 and TM3 is involved in generation of torque by its interaction with the C-terminal region of FliG [31]. MotB has one TM and a large periplasmic domain that binds to the peptidoglycan and acts as the anchor [32]. MotB also possesses a conserved and essential aspartate residue that is thought to play a critical role in proton movement [33,34]. The functional stoichiometry of each stator complex is thought to be 4A:2B [33,35]. The typical number of such complexes surrounding the basal motor appears to be at least 11 [26,36,37]. The torque, which is required for rotating the flagellar filament, is generated by the interaction between the rotor and the stator in the basal body.
Figure 1. Flagellum in the Gram-negative bacteria Escherichia coli.
Gram-positive species such as Bacillus lack the LP-ring assembly. Protein names, shown in green squares, are components that are conserved in many Bacillus species including alkaliphilic Bacillus. In Bacillus species, there is an FlhO protein that has been reported to substitute for FlgF, FlgI and FlgH [112]. An additional flagellar assembly factor, FliW, was reported in Bacillus subtilis and Campylobacter jejuni [113]. B. subtilis has the orthologs FliM and FliG, but instead of FliN this species has FliY, which is only homologous to FliN in its C-terminus [114]. B. subtilis has two kinds of flagellar stator complex, MotAB and MotPS. Some alkaliphilic Bacillus species only have a sodium-coupled, MotPS-type stator instead of a proton-coupled, MotAB-type one.
C: Cytoplasmic; L: Lipopolysaccharide; MS: Membrane and supramembranous; P: Peptidoglycan. Reproduced with permission from [201].
Na+-coupled stators
Initial studies of motility in Gram-positive bacteria showed that the rotational energy for the flagellar motor is provided by the electrochemical ion gradients across the cytoplasmic membrane, as had been established for Gram-negative bacteria [38]. In these studies, motility of Streptococcus spp. [39] and of Bacillus subtilis [40,41] was shown to be powered by the PMF as had been shown for E. coli and S. Typhimurium. Additional cations, such as sodium or potassium, were not required for motility (Figure 2).
Figure 2. An unrooted tree shows relationships among the stator subunits MotB, MotS and PomB from Escherichia coli, Salmonella enterica serovar Typhimurium, Bacillus and Vibrio species.
Multiple sequence alignment by ClustalW [202] was used for calculating a multiple alignment and for generating an unrooted phylogenetic tree. Branch lengths are proportional to the sequence divergence and can be measured relative to the bar shown (bar = 0.1 substitutions per amino acid site).
*Proteins from alkaliphilic bacteria.
As the energetic profile of aerobic alkaliphilic Bacillus strains was revealed through studies by several laboratories, motility was observed at alkaline pH values from 9 to 10.5 [42-44]. However, it was also clear that in this range of pH, the PMF available to energize proton-coupled bioenergetic work such as motility decreases progressively [43,45]. Alkaliphilic Bacillus species were found to have sizeable ‘reversed’ chemical gradients of H+, in other words, cytoplasmic pH values that are markedly lower than high external pH values during optimal growth [6,43,46-48]. In carefully pH-controlled studies in continuous cultures, alkaliphilic Bacillus pseudofirmus OF4 was shown to grow at pH values above 11. This alkaliphile sustains a cytoplasmic pH 2.3 units lower than the external pH of 10.5, resulting in the low bulk PMF of −44 mV, while growing logarithmically with a generation time of approximately 37 min on a semidefined, malate-containing medium and exhibiting a clear capacity for motility [43,47]. It was therefore anticipated that motility in such extremely alkaliphilic Bacillus strains would be energized by the larger SMF that exists under these conditions, as had earlier been found to be the case for ion-coupled solute uptake [49]. That Na+ coupling was indeed used for motility was first demonstrated by Hirota et al., who showed that the energy source for the rotation of flagellar motor of extremely alkaliphilic Bacillus species is the SMF and not the PMF [42,50]. However, the Na+-dependent stator of the alkaliphilic Bacillus species was not the first SMF-driven Mot-type system to be identified. Rather, it was discovered that marine Vibrio species also use the SMF rather than PMF to power motility of their polar flagellum [51], and the Mot-type stator-force generator was designated PomA/PomB, a MotAB-like pair of proteins that required additional proteins, MotX and MotY, for torque generation in Vibrio alginolyticus [8,52-55]. Recently, Paulick et al. showed polar flagellar localization of both MotB and PomB in Shewanella oneidensis MR-1 and demonstrated the coexistence of the two sets of stator units in a single flagellar motor [56].
The MotPS that functions as the stator of the flagellar motor of alkaliphilic Bacillus species were first identified in B. pseudofirmus OF4 based on homology of the predicted products of a set of putative mot genes to MotAB [8]. The motPmotS genes are universally located downstream of the ccpA gene, which encodes a central regulator of carbon metabolism in Bacillus species and other Gram-positive organisms [57,58]. In extremely alkaliphilic Bacillus halodurans C-125 [59] and B. pseudofirmus OF4 [8], Na+-dependent MotPS is the only stator and the flagellar motor is driven by SMF. By contrast, the more moderately alkaliphilic Oceanobacillus iheyensis, an alkaliphilic marine bacterium, has both sodium-coupled MotPS and proton-coupled MotAB (Figure 2) [60]. There are no reports of motility assays for this bacterium but studies described below for neutralophilic B. subtilis, which also has both MotAB and MotPS, suggest that both stators contribute to the torque required for flagellar rotation, with different relative contributions under different conditions.
Why is there so little motility of Na+-dependent extremely alkaliphilic strains at near-neutral pH?
Bacillus pseudofirmus OF4 grows optimally up to pH 10.6 on semidefined malate-containing medium and the low end of its pH range under these conditions is 7.5 [43,61]. Sturr et al. reported that the motility of B. pseudofirmus OF4 declined at the low end of the pH range for growth and that no motility was observed at pH 7.5 [43]. This raised the possibility that in extreme alkaliphiles, which are adapted to growth at high pH, sodium-coupled processes are particularly sensitive to inhibition by protons. This has been hypothesized to be the case for sodium-coupled solute transporters of alkaliphilic B. pseudofirmus OF4 [62]. After swimming assays confirmed that motility of B. pseudofirmus OF4 is sodium dependent and the number of flagella per cell were shown to be comparable at pH 7.5 and 10.3, the possibility that competitive inhibition by protons was responsible for poor swimming at near-neutral pH was tested [63]. Indeed, motility was observed at pH values as low as 7 when the concentration of Na+ was raised sufficiently and in the range of pH 8–10, swimming speed increased with increasing [Na+] up to 230 mM.
A lesson from the alkaliphile: MotPS is a second stator in neutralophilic B. subtilis
Once MotPS was recognized as the functional, sodium-dependent stator for motility of extreme alkaliphiles, it was of interest to investigate whether the homolog that had earlier been described in B. subtilis, but for which no function had been demonstrated [64], might be a second functional stator that responded to Na+ and elevated pH in some neutralophiles [8]. Studies revealed that in B. subtilis, MotPS functioned as a Na+-dependent stator that made a contribution to swimming of wild-type B. subtilis, especially at elevated pH, sodium and viscosity [8,9]. Hybrid forms were active and provided information about the contributions of the two stator components to different stator properties; this included the finding that the MotB and MotS proteins were the major determinants of ion specificity [9]. Up-motile variants of B. subtilis mutants lacking functional MotAB were described and found to have a mutation in the intergenic region between ccpA and motPS that resulted in more cotranscription of the latter two genes with the upstream ccpA [65]. This finding suggests that under appropriate selective pressure, MotPS could play a major role in motility of some B. subtilis strains. There have been increasing reports of dual flagellar systems in different bacterial species [66], with this one being the first example of two distinct stators that interact with a single rotor [8]. Other neutralophilic Bacillus strains also have both motAB and motPS loci, as indicated by genomic data (Figure 2). Recent studies have shown, using the two B. subtilis stators, that each of the stators with different single ion-coupling preferences can be modified by mutagenesis of the motB or motS gene to forms that can use both Na+ and H+, bringing it closer to the different mode of dual ion coupling that is described below for the single Bacillus clausii stator [11]. Additional studies will be required to determine how the ratio of MotPS to MotAB stators is determined in a given strain and to test the suggestion that a given flagellum is surrounded by a mixture of the two types. These are issues that are of general interest in the field [67].
One stator that couples to two different ions: flagellar stator & motility of B. clausii KSM-K16
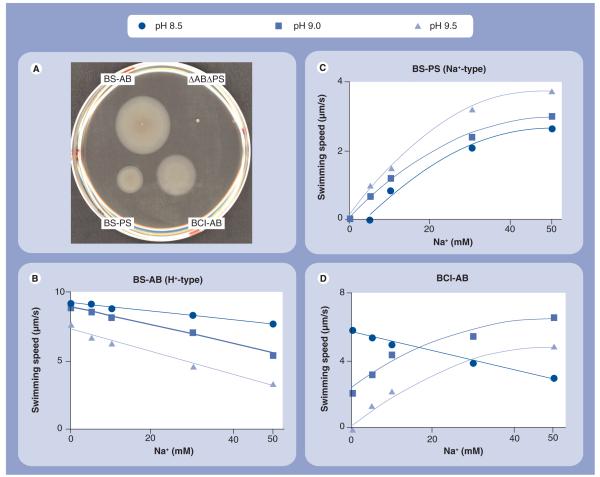
Bacillus clausii strains are alkaliphilic bacteria that are noted for useful biological products such as alkaline proteases [68,69]. The genome sequence of B. clausii KSM-K16 was completed in 2005 and revealed only a single set of genes encoding a potential stator for this motile alkaliphile. These genes were predicted to encode a MotAB-like pair of proteins (GenBank accession no. AP006627) (Figure 2). The B. clausii stator proteins were closely related to the MotA and MotB that compose the proton-coupled stator in B. subtilis (identity: 52 and 45%; similarity: 73 and 64%, respectively), and were much less closely related to the MotP and MotS that compose the sodium-coupled stator in B. subtilis (identity: 32 and 28%; similarity: 57 and 55%, respectively). This raised the possibility that either B. clausii was only poorly motile in the alkaline edge of its pH range or that it had a novel type of MotAB stator that could couple to both sodium and protons. Studies of this question first showed that the organism was very motile over a range of pH that extended from pH 7.0 to 11 [11]. Therefore, the properties of the B. clausii MotAB (BCl-MotAB) were examined to test the hypothesis of a single, native stator with dual ion-coupling capacity. The BCl-MotAB was introduced into a stator-less (ΔmotAmotBΔmotPmotS) mutant strain of B. subtilis, because B. clausii KSM-K16 is not yet genetically accessible. The heterologous BCl-MotAB functioned in the B. subtilis setting, as shown in Figure 3, making it possible to compare its properties with those of the two single stators of the host, MotAB and MotPS of B. subtilis, introduced into the statorless mutant in an identical fashion. The studies assessed ion dependence at different pH values as well as the pH-dependent effects of a sodium-channel inhibitor, 5-(N-ethyl-N-isopropyl)-amiloride (EIPA), an amiloride analog, and the protonophore cyanide m-chlorophenyl hydra-zone (CCCP), which dissipates electrochemical proton gradients. The results strongly supported the hypothesis that BCl-MotAB is a bifunctional stator that uses protons at near-neutral pH and sodium at high pH and probably uses both at intermediate pH values.
Figure 3. The motility of a stator-less Bacillus subtilis mutant (ΔABΔPS) expressing either B. subtilis motAB, B. subtilis motPS or Bacillus clausii motAB.
(A) Motility in a soft agar plate assay. Fresh cells were inoculated into Luria-Bertani medium (pH 7.0) soft agar and incubated at 37°C for 16 h. (B–D) Motility in liquid medium. BS-AB (B), BS-PS (C) and BCl-AB (D) cells were grown at 37°C in Luria-Bertani medium (pH 7.0) and the swimming speed was measured at various concentrations of sodium in 1/20 Tryptone Yeast Extract (TY) medium. 1/20 TY medium contained 0–1 mM sodium. The pH values of 1/20 TY medium used in the swimming speed assays of B. subtilis strains are indicated by the key.
Bcl-AB: B. clausii motAB; BS-AB: B. subtilis motAB; BS-PS: B. subtilis motPS; ΔABΔPS: Deletion strain of the motAB and motPS genes.
Reproduced with permission from [11].
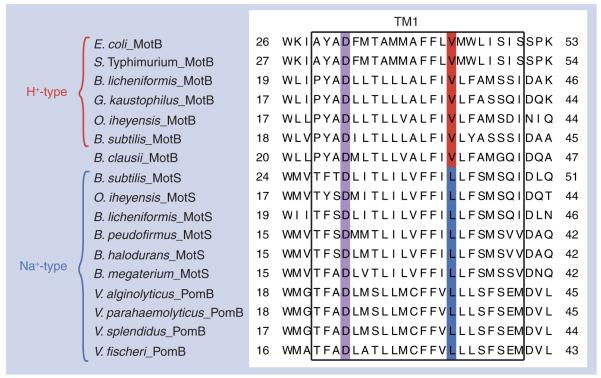
It was further possible to introduce mutations into BCl-MotB that made the BCl-MotAB much more single-ion dependent, either for sodium or for protons. The strategy was developed from alignments of BCl-MotB with MotB and MotS. These alignments supported inferences with respect to which residues were of potential importance in ion specificity (Figure 4). A triple mutant of BCl-MotB with G42S, Q43S and Q46A mutations was constructed on this basis and behaved like the MotB subunit of a proton-coupled stator. A second triple mutant with V37L, A40S and G42S mutations behaved like the MotS subunit of the sodium-coupled stator. Further studies of single and double mutants showed that the Q43S mutation was critical for loss of sodium coupling and had to be introduced together with either the G42S or Q46A mutation to produce a proton-coupled form. The V37L mutation was critical for sodium selectivity and a combination of the V37L mutation and either the A40S or the G42S mutation was required for production of the BCl-MotB form that exhibits sodium coupling at low pH [11]. Thus, the novel bifunctional stator of B. clausii yielded insights into the determinants of ion-coupling specificity that will be useful in identifying other candidates in bacteria for dual function. When high-resolution structures of flagellar stators are achieved, such information will also play a role in modeling the cation sites.
Figure 4. Alignment of the region containing the single transmembrane segment of MotB, MotS and PomB from Escherichia coli, Salmonella enterica serovar Typhimurium, Bacillus and Vibrio species.
The amino acid residues that are important for proton or sodium-ion selectivity are highlighted with red and blue, respectively. A conserved aspartic acid residue is critical for flagellar rotation and is highlighted with violet [33-34].
Reproduced with permission from [11].
Chemotaxis of Bacillus species
Motile bacteria can respond to stimulation by chemical and physical effectors in the external environment [12,27,70,71]. The temporal shift of temperature, pH and oxygen concentration can be effectors (attractants or repellents), as can diverse nutrient or inhibitory chemical compounds. The most intensively studied bacterial chemotaxis system is that of E. coli, which is thought to involve a less complex set of networks than that of B. subtilis chemotaxis [71-73], but still continues to provide novel information [74,75]. B. subtilis provides one of the other chemotaxis systems for which a great amount of information is available and is by far the most studied among Bacillus species, revealing commonalities with E. coli chemotaxis but also significant differences [71,72,76-78]. In both model systems, the bacteria swim in straight runs (with counterclockwise rotation of the flagellar bundle) with occasional tumbles (initiated by clockwise rotation) that increase the likelihood of a change in direction [71]. Therefore, movement toward attractants is favored by smooth runs in response to an attractant and movement away from repellents is favored by increased tumbling. Chemoreceptor proteins are usually transmembrane signaling proteins that bind external signaling molecules and/or respond to physical effectors in a manner that also initiates a change that is transmitted to the cytoplasmic domain. The chemoreceptors are called methylated chemotaxis proteins (MCPs) because of an adaptation feature of chemotaxis that involves a methylation/demethylation of sites on the MCPs. Clusters of MCPs have been found at the cell poles of rod-shaped bacteria [79-81]. The response of the MCPs is transmitted via the CheA histidine kinase that is complexed, together with adaptor protein CheW, on the cytoplasmic side. The resulting state of autophosphorylation of CheA determines the phosphorylation state of the response regulator CheY, which interacts with the switch domain of the rotor and determines the direction of rotation and, hence, the frequency of tumbling as opposed to runs. The response of the CheA kinase activity to binding of chemo-effectors to the MCPs is different in E. coli than in B. subtilis, and the Bacillus has distinct and more complex layers of adaptive capacity that correlate with the presence of chemotaxis proteins that are not found in E. coli (Table 1) [71,76]. B. subtilis also has redundancies in several chemotaxis proteins, which is not observed in E. coli and is thought to increase robustness of B. subtilis chemotaxis (Table 1) [12,70,77].
Table 1.
Comparison of the chemotaxis protein homologs of alkaliphilic Bacillus species, Bacillus subtilis and Escherichia coli.
| Protein | Functions | Escherichia coli |
Bacillus subtilis |
Bacillus halodurans |
Bacillus clausii |
Oceanobacillus iheyensis |
Ref. |
|---|---|---|---|---|---|---|---|
| CheW | Adaptor protein | P0A964 | P39802 | Q9K8N6 | Q5WEU4 |
Q8CXH0, Q8CX96 |
[102] |
| CheV | Adaptor protein | – | P37599 |
Q9KAM5, Q9KC62 |
Q5WG28 | – | [77,102] |
| CheA | Histidine kinase | P07363 | P29072 | Q9K8N5 | Q5WEU3 | Q8ENE1* | [103] |
| CheY | Response regulator | P0AE67 | P24072 |
Q9KA46, Q9K8N7 |
Q5WFR9 | Q8EQW9 | [104] |
| CheR | Methyltransferase | P07364 | P31105 | Q9KCB8 | Q5WGS9 | Q8EQB5 | [105,106] |
| CheB | Deamidase/methylesterase | P07330 | Q05522 | Q9KA55 | Q5WFS3 | Q8EQW0 | [79,107] |
| CheC | Phosphatase | – | P40403 | Q9KDX3 | Q5WFS4 | Q8EQV9 | [77-78,108,109] |
| CheD | Deamidase/methylesterase | – | P40404 | Q9KA57 | Q5WFS5 | Q8EQV8 | [77,109] |
| CheX | Phosphatase | – | – | Q9KDX3 | – | – | [109,110] |
| FliY | Phosphatase | – | P24073 | Q9KA45 | Q5WFR8 | Q8EQX0 | [78,109] |
| CheZ | Phosphatase | P0A9H9 | – | – | – | – | [111] |
The GenBank accession numbers are indicated.
Indicates the protein that was newly annotated in this research.
Alkaliphilic Bacillus species show similar chemotaxis ‘toolkits’ to B. subtilis (Table 1) and are expected to share many of the same chemoeffectors as well as the general design principles of the signal transduction mechanism, since both of those show a high degree of conservation among even more distantly related bacteria [70-71]. There is as yet no information in alkaliphilic Bacillus species about the involvement of particular MCPs or other chemotaxis elements in responses to pH or PMF, as there are for E. coli [82-84]. However, evidence has emerged for a dependence of chemotaxis in alkaliphilic B. pseudofirmus OF4 on a voltage-gated sodium channel (see below). This incompletely understood layer of adaptation in alkaliphile chemotaxis may be distinct in these organisms, perhaps reflecting the challenge of alkaliphily and its dependence on a sodium cycle. Quite possibly, though, the involvement of a channel in alkaliphile chemotaxis is a particularly pronounced extremophile version of a role for an ion channel that will turn up in other bacteria and whose identification and study will be facilitated by the extremophile model.
Channel involvement in chemotaxis
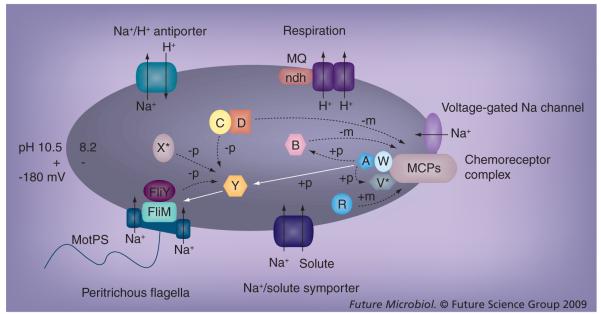
In the aerobic, alkaliphilic Bacillus species, the respiratory chain pumps protons outward during electron transport and secondary sodium/proton antiporters (exchangers) convert the PMF produced by this proton pumping to a SMF and a ‘reversed’ pH gradient, in other words, they achieve the required acidification of the cytoplasm relative to the outside pH (Figure 5) [3,45]. Although alkaliphilic Bacillus species all have multiple sodium/proton antiporters encoded in their genomes, there is a specific antiporter, the multisubunit Mrp-type antiporter, which is essential for alkaliphily of aerobic B. halodurans C-125 and B. pseudofirmus OF4. It is the only bacterial antiporter that has been definitively shown to be irreplaceable by others of the organism’s antiporter complement for alkaline pH homeostasis [85-88]. The same family of antiporters is found widely in bacterial pathogens [88]. The continuous antiporter-mediated efflux of sodium, which is coupled to cytoplasmic proton accumulation during respiration, necessitates continuous re-entry of sodium from the outside. It was expected that the MotPS sodium channel would be a major mediator of such sodium re-entry, especially under conditions in which the medium or environment was not rich in nutrients and other solutes that are taken up together with Na+ [89]. Mutant studies showed that MotPS plays a modest role in sodium circulation in support of pH homeostasis under conditions in which both the concentrations of Na+ and of solutes that enter with sodium are low. A distinct channel, the voltage-gated Na+ channel called NaChBac in B. halodurans C-125 and NaVBP in B. pseudofirmus OF4, has a greater role in cytoplasmic pH homeostasis, as shown in mutants of the latter alkaliphile strain with defects in the ncbA gene encoding the channel [10,90].
Figure 5. Relationship between Na+-dependent motility, chemotaxis and the Na+ cycle of extremely alkaliphilic Bacillus species.
White arrows show signal transduction in support of chemotaxis. Dotted arrows show signal removal and adaptation systems. Solid black arrows show transport of ions and solutes. Alkaliphilic Bacillus species use a sodium-motive force for flagellar motility and solute symport [115]. Methyl-accepting chemotaxis proteins (chemoreceptors, MCPs), W, A, Y, B, R and FliM are chemotaxis components that are found in all bacteria that have flagellar motility [71]. V, C, D and FliY are not present in Escherichia coli but are found in many Bacillus species including alkaliphilic Bacillus (see Table 1). CheX is also present in several Bacillus species (e.g. Bacillus halodurans C-125). The asterisks show the proteins that are not conserved in alkaliphilic Bacillus species. Bacterial voltage-gated Na channels are found in alkaliphilic Bacillus species (e.g., Bacillus halodurans, Bacillus pseudofirmus, Bacillus clausii and Oceanobacillus iheyensis). The voltage-gated Na channel of B. pseudofirmus OF4 (NaVBP) is required for normal chemotaxis of this alkaliphile and it colocalizes with putative chemoreceptors (McpX) at cell poles [7,10].
−m: Demethylation; +m: methylation; −p: Dephosphorylation; +p: Phosphorylation.
A: CheA; B: CheB; C: CheC; D: CheD; MCP: Methylated chemotaxis protein; MQ: Menaquinone; ndh: NADH dehydrogenase; R: CheR; V: CheV; W: CheW; Y: CheY.
The NaChBac–NaVBP type of channel is also found in other alkaliphilic Bacillus strains, as well as in other bacteria that face a spectrum of salt/alkali challenges [91]. The NaChBac family is of great general interest because of its homology with voltage-gated sodium channels of eukaryotes, which are profoundly important in various disease processes [90]. The smaller, bacterial version provides an excellent experimental model for structural studies [92,93]. It was also of great interest with respect to the physiology of bacterial motility and chemotaxis since the finding that B. pseudofirmus OF4 mutants lacking a functional NaVBP exhibited a striking chemotaxis abnormality, in other words, a ‘tumbly’ phenotype accompanied by ‘inverse chemotaxis’, wherein the bacteria moved away from nutrient attractants, such as malate, and toward repellent conditions, such as pH 10.5, in the absence of an energy source to support cell energetics [10]. The inverse chemotactic phenotype was not secondary to a problem with pH homeostasis because the inverted response to nutrients was observed under conditions of pH and sodium in which pH homeostasis was normal. This suggests that the NaVBP channel has a direct role of some kind in chemotaxis of the alkaliphilic Bacillus. Additional support for this suggestion was the observation that nifedipine, a sodium-channel blocker, caused inverse chemotaxis in the wild-type alkaliphile [10]. A role for ion channels in bacterial chemotaxis has been suggested multiple times, but had not previously been associated with a specific channel [94-96].
When properties of the NaVBP channel shown in electrophysiological studies were compared with the conditions existing in its native setting, the results indicated that NaVBP function may depend upon modulation by interaction with other proteins that provide ‘additional triggers’ for the channel [10]. Hypothetically, an interaction between the channel and the chemotaxis machinery might serve the dual purposes of activating the channel in some way that coordinates with chemotactic signaling and serves pH homeostasis simultaneously. This idea needs to be studied further, but thus far it has been shown that NaVBP of B. pseudofirmus OF4 colocalizes at the cell poles of its native host with one of the MCPs, as indicated by its cross-reaction with antibodies against a B. subtilis MCP (Figure 5) [7].
Future perspective
Alkaliphilic bacteria are model organisms for study of sodium cycles that support motility, solute transport as well as pathogenicity in bacteria including Pseudomonas aeruginosa and Vibrio cholerae [97-99], and is implicated in many others [97]. The number of examples and distinct modes of sodium-dependent or dual ion-dependent motility are likely to continue to increase. This will be of general ecological and physiological interest and the findings are likely to include bacteria in which the motility system will be a useful target for inhibition of pathogenicity.
There also remain many basic questions that are yet to be answered, some of which have been mentioned. It will be important to unravel whether the stator rings around individual flagella contain multiple types of stators in organisms with single rotors and dual stator types. The controls that determine the ratio of the two types need to be resolved. It will also be of interest to determine the distribution of ratios in ‘wild’ rather than only of long-time laboratory-dwelling strains. Earlier studies suggested that the sodium-dependent MotPS channels have multiple Na+-binding sites [9]; this merits further investigation into the nature and roles of such sites. A related issue is the relatively poor function of the alkaliphile MotPS stator at very high concentrations of Na+ [9]. A similar phenomenon has been reported in Vibrio alginolyticus [100], and the possibility has been raised that there is a Na+ site somewhere in the motor that acts as a brake.
The recent findings of diversity in the ion-coupling patterns of different bacteria should encourage those with interests in ‘sodium energetics’ to move beyond the coupling patterns of the ATP synthase as a major sentinel of whether a bacterium is primarily in the camp of sodium energetics as opposed to proton energetics (for example, see [101]). The findings reviewed here indicate that different bacteria display: proton-only coupling to motility, sodium-only coupling, and dual proton and sodium coupling motility using several different strategies, for example shared rotors and multiple stators; separate flagellar systems entirely; and a single stator that switches ions as a function of pH. The correlation of these different modes with degrees of overall use of sodium versus proton energetics would enrich the analyses greatly.
In the chemotaxis area, the basis for the effect of NaVBP on alkaliphile chemotaxis needs to be elucidated. It may offer clues for identifying (perhaps more subtle) roles of channels in chemotaxis of nonextremophilic bacteria. Conversely, the possibility of interactions between the channel and chemotaxis proteins at the cell poles requires further probing. Further study of NaVBP or other NaChBac forms is expected to continue to contribute to mechanistic details about voltage-gated sodium channels.
Executive summary.
Structure of bacterial flagella
The bacterial flagellum is a complex structure that functions as a rigid propeller that turns as part of a rotary nanomachine with broad roles in physiology and pathogenicity.
Stators of the flagellar motor encompass ion channels that transduce energy of the sodium- or proton-motive forces into mechanical energy.
Na+-coupled stators
A conserved pair of stator proteins, MotPS, that are homologs of MotAB mediate Na+-coupled motility of extreme alkaliphiles.
One of two stators interacting with a single motor in other Bacillus species.
Why is there so little motility of Na+-dependent extremely alkaliphilic strains at near-neutral pH?
Poor motility supported by alkaliphile MotPS at low external pH values results from competitive inhibition by protons that is reversible by high [Na+].
Use of a motor that is inhibited by protons is compatible with the hard-wired adaptation of extreme alkaliphiles to life at high pH.
A lesson from the alkaliphile: MotPS is a second stator in neutralophilic Bacillus subtilis
Neutralophilic Bacillus strains, including Bacillus subtilis, exhibit dual ion-coupling capacity using MotPS and MotAB as stators for a single rotor.
One stator that couples to two different ions: flagellar stator & motility of Bacillus clausii KSM-K16
A strain of the probiotic alkaliphile B. clausii has a novel form of dual ion coupling to motility in which a single MotAB stator uses both sodium and protons at different ranges of pH.
Deductions from studies of alignments of the MotB and MotS subunits of sodium- and proton-coupled motility systems facilitated conversion of single ion-coupled stators to dual use and the B. clausii dual-ion stator to single-ion use (either sodium or protons).
Additional examples of this type of bifunctional stator may now be recognized.
Chemotaxis of Bacillus species
Alkaliphile genomes are predicted to encode an array of chemotaxis proteins that is comparable to the ‘toolkit’ of B. subtilis.
Channel involvement in chemotaxis
A voltage-gated sodium channel with homology to the eukaryotic channels that have major roles in physiology and pathophysiology plays a role in alkaliphile pH homeostasis.
The NaVBP sodium channel of Bacillus pseudofirmus OF4 is required for normal chemotaxis of this alkaliphile and it colocalizes with chemoreceptors at cell poles.
Mutational loss of the NavBP voltage-gated Na+ channel of B. pseudofirmus OF4 results in ‘inversed’ chemotaxis.
Future perspective
A sodium cycle is central to the lifestyle of alkaliphiles and many nonalkaliphilic and pathogenic bacteria.
Further investigation of dual coupling systems will resolve the controls of the ratios of sodium- to proton-coupled stators in motility as well as the mechanism of the channel interaction with the chemotaxis system in alkaliphiles and, perhaps, beyond alkaliphiles.
The multiple ion-coupling patterns found among different bacteria present a new sentinel for assessments of the engagement of different wild and clinical strains in sodium energetics versus proton energetics.
Acknowledgments
Financial & competing interests disclosure This work was supported by a research grant GM28454 from the National Institute of General Medical Sciences (to Terry Ann Krulwich) and grants from the Kurata Memorial Foundation for Promoting Science and Special Research, from Toyo University and from the 21st Century Center of Excellence program of the Ministry of Education, Culture, Sports, Science and Technology of Japan (to Masahiro Ito) and a JSPS Research Fellowships for Young Scientists (to Naoya Terahara).
Footnotes
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Shun Fujinami, NITE Bioresource Information Center, Department of Biotechnology, National Institute of Technology and Evaluation, 2-10-49 Nishihara, Shibuya-ku, Tokyo 151-0066, Japan Tel.: +81 334 811 972 Fax: +81 334 818 424 fujinami-shun@nite.go.jp.
Naoya Terahara, Bio-nano Electronics Research Center, Toyo University, Kawagoe, Saitama, 350-8585, Japan Tel.: +81 276 82 9231 Fax: +81 276 82 9231 dx0600027@toyonet.toyo.ac.jp.
Terry Ann Krulwich, Department of Pharmacology & Systems Therapeutics, Mount Sinai School of Medicine, Box 1603, One Gustave L. Levy Place, New York, NY 10029, USA Tel.: +1 212 241 7280 Fax: +1 212 996 7214 terry.krulwich@mssm.edu.
Masahiro Ito, Graduate School of Life Sciences, Toyo University, Oura-gun, Gunma 374-0193, Japan and Bio-nano Electronics Research Center, Toyo University, Kawagoe, Saitama, 350-8585, Japan Tel.: +81 276 829 202 Fax: +81 276 829 202 ito1107@toyonet.toyo.ac.jp.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Horikoshi K. Alkaliphiles: some applications of their products for biotechnology. Microbiol. Mol. Biol. Rev. 1999;63:735–750. doi: 10.1128/mmbr.63.4.735-750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horikoshi K. Past, present and future of extremophiles. Extremophiles. 2008;12:1–2. doi: 10.1007/s00792-007-0127-5. [DOI] [PubMed] [Google Scholar]
- 3.Krulwich TA, Hicks DB, Swartz TH, Ito M. Bioenergetic adaptations that support alkaliphily. In: Gerday C, Glansdorff N, editors. Physiology and Biochemistry of Extremophiles. ASM Press; Washington, DC, USA: 2007. pp. 311–329. [Google Scholar]
- 4.Liu J, Fujisawa M, Hicks DB, Krulwich TA. Characterization of the functionally critical AXAXAXA and PXXEXXP motifs of the ATP synthase C-subunit from an alkaliphilic Bacillus. J. Biol. Chem. 2009;284:8714–8725. doi: 10.1074/jbc.M808738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padan E, Bibi E, Ito M, Krulwich TA. Alkaline pH homeostasis in bacteria: new insights. Biochim. Biophys. Acta. 2005;1717:67–88. doi: 10.1016/j.bbamem.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yumoto I. Environmental and taxonomic biodiversities of Gram-positive alkaliphiles. In: Gerday C, Glansdorff N, editors. Physiology and Biochemistry of Extremophiles. ASM Press; Washington, DC, USA: 2007. pp. 295–310. [Google Scholar]
- 7.Fujinami S, Sato T, Trimmer JS, et al. The voltage-gated Na+ channel NaVBP colocalizes with methyl-accepting chemotaxis protein at cell poles of alkaliphilic Bacillus pseudofirmus OF4. Microbiology. 2007;153:4027–4038. doi: 10.1099/mic.0.2007/012070-0. ▪▪Demonstrates colocalization of the voltage-gated sodium channel of alkaliphilic Bacillus with methylated chemotaxis proteins.
- 8.Ito M, Hicks DB, Henki TM, et al. MotPS is the stator-force generator for motility of alkaliphilic Bacillus, and its homologue is a second functional Mot in Bacillus subtilis. Mol. Microbiol. 2004;53:1035–1049. doi: 10.1111/j.1365-2958.2004.04173.x. ▪ Identified MotPS as the sodium-dependent stator that is required for motility of extremely alkaliphilic Bacillus strains and is a second functional stator that confers dual ion-coupling capacity on motility of Bacillus subtilis upon the same flagellar motor.
- 9.Ito M, Terahara N, Fujinami S, Krulwich TA. Properties of motility in Bacillus subtilis powered by the H+-coupled MotAB flagellar stator, Na+-coupled MotPS or hybrid stators MotAS or MotPB. J. Mol. Biol. 2005;352:396–408. doi: 10.1016/j.jmb.2005.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito M, Xu H, Guffanti AA, et al. The voltage-gated Na+ channel NaVBP has a role in motility, chemotaxis, and pH homeostasis of an alkaliphilic Bacillus. Proc. Natl Acad. Sci. USA. 2004;101:10566–10571. doi: 10.1073/pnas.0402692101. ▪ Demonstration of a role for a bacterial voltage-gated sodium channel in both alkaline pH homeostasis and chemotaxis of an alkaliphilic Bacillus.
- 11.Terahara N, Krulwich TA, Ito M. Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on Bacillus subtilis motors. Proc. Natl Acad. Sci. USA. 2008;105:14359–14364. doi: 10.1073/pnas.0802106105. ▪▪ Identified the first bacterium in which a single stator–rotor uses both protons and sodium ions for ion coupling, depending upon the pH, demonstrated mutations that convert the bifunctional stator to each single stator type and applied the same approach to confer dual ion use on the two single ion-use stators of B. subtilis.
- 12.Miller LD, Russell MH, Alexandre G. Diversity in bacterial chemotactic responses and niche adaptation. Adv. Appl. Microbiol. 2009;66:53–75. doi: 10.1016/S0065-2164(08)00803-4. ▪▪ Reviews the adaptive value of diverse chemotaxis reponses among different bacteria.
- 13.Ottemann KM, Miller JF. Roles for motility in bacterial–host interactions. Mol. Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 14.MacNab RM. How bacteria assemble flagella. Annu. Rev. Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 15.Rajagopala SV, Titz B, Goll J, et al. The protein network of bacterial motility. Mol. Systems Biol. 2007;3:128. doi: 10.1038/msb4100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi S, Fujita H, Ishihara A, Aizawa S, MacNab RM. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 1986;166:187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 1992;226:433–446. doi: 10.1016/0022-2836(92)90958-m. [DOI] [PubMed] [Google Scholar]
- 18.Zhao R, Pathak N, Jaffe H, Reese TS, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- 19.Lloyd SA, Blair DF. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 1997;266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 20.Lloyd SA, Whitby FG, Blair DF, Hill CP. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature. 1999;400:472–475. doi: 10.1038/22794. [DOI] [PubMed] [Google Scholar]
- 21.Brown PN, Hill CP, Blair DF. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 2002;21:3225–3234. doi: 10.1093/emboj/cdf332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown PN, Mathews MA, Joss LA, Hill CP, Blair DF. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 2005;187:2890–2902. doi: 10.1128/JB.187.8.2890-2902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berg HC. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. ▪▪ Review of properties and mechanistic insights into bacterial flagellar structure and function.
- 24.Park SY, Lowder B, Bilwes AM, Blair DF, Crane BR. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc. Natl Acad. Sci. USA. 2006;103:11886–11891. doi: 10.1073/pnas.0602811103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sowa Y, Berry RM. Bacterial flagellar motor. Q. Rev. Biophys. 2008;41:103–132. doi: 10.1017/S0033583508004691. ▪▪ Principles of flagellar motor function.
- 26.Khan S. Gene to ultrastructure: the case of the flagellar basal body. J. Bacteriol. 1993;175:2169–2174. doi: 10.1128/jb.175.8.2169-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blair DF. How bacteria sense and swim. Annu. Rev. Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 28.Stocker BA. Transduction of flagellar characters in Salmonella. J. Gen. Microbiol. 1953;9:410–433. doi: 10.1099/00221287-9-3-410. [DOI] [PubMed] [Google Scholar]
- 29.Ridgway HG, Silverman M, Simon MI. Localization of proteins controlling motility and chemotaxis in Escherichia coli. J. Bacteriol. 1977;132:657–665. doi: 10.1128/jb.132.2.657-665.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silverman M, Matsumura P, Simon M. The identification of the mot gene product with Escherichia coli–λ hybrids. Proc. Natl Acad. Sci. USA. 1976;73:3126–3130. doi: 10.1073/pnas.73.9.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Lloyd SA, Blair DF. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl Acad. Sci. USA. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chun SY, Parkinson JS. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988;239:276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- 33.Kojima S, Blair DF. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry. 2004;43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, Sharp LL, Tang HL, et al. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blair DF, Berg HC. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 36.Block SM, Berg HC. Successive incorporation of force-generating units in the bacterial rotary motor. Nature. 1984;309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- 37.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl Acad. Sci. USA. 2006;103:8066–8071. doi: 10.1073/pnas.0509932103. ▪ Moving toward the number and diversity among stators surrounding single flagellar rotors.
- 38.Larsen SH, Adler J, Gargus JJ, Hogg RW. Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc. Natl Acad. Sci. USA. 1974;71:1239–1243. doi: 10.1073/pnas.71.4.1239. ▪▪ Identification of electrochemical gradients as the energy source for bacterial motility and chemotaxis.
- 39.Manson MD, Tedesco P, Berg HC, Harold FM, Van der Drift C. A protonmotive force drives bacterial flagella. Proc. Natl Acad. Sci. USA. 1977;74:3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuura S, Shioi J, Imae Y. Motility in Bacillus subtilis driven by an artificial protonmotive force. FEBS Lett. 1977;82:187–190. doi: 10.1016/0014-5793(77)80581-4. [DOI] [PubMed] [Google Scholar]
- 41.Matsuura S, Shioi JI, Imae Y, Iida S. Characterization of the Bacillus subtilis motile system driven by an artificially created proton motive force. J. Bacteriol. 1979;140:28–36. doi: 10.1128/jb.140.1.28-36.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirota N, Kitada M, Imae Y. Flagellar motors of alkalophilic Bacillus are powered by an electrochemical potential gradient of Na+ FEBS Lett. 1981;132:278–280. [Google Scholar]
- 43.Sturr MG, Guffanti AA, Krulwich TA. Growth and bioenergetics of alkaliphilic Bacillus firmus OF4 in continuous culture at high pH. J. Bacteriol. 1994;176:3111–3116. doi: 10.1128/jb.176.11.3111-3116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohta K, Kiyomiya A, Koyama N, Nosoh Y. The basis of the alkalophilic property of a species of Bacillus. J. Gen. Microbiol. 1975;86:259–266. doi: 10.1099/00221287-86-2-259. [DOI] [PubMed] [Google Scholar]
- 45.Krulwich TA. Alkaliphiles: ‘basic’ molecular problems of pH tolerance and bioenergetics. Mol. Microbiol. 1995;15:403–410. doi: 10.1111/j.1365-2958.1995.tb02253.x. [DOI] [PubMed] [Google Scholar]
- 46.Goto T, Matsuno T, Hishinuma-Narisawa M, et al. Cytochrome c and bioenergetic hypothetical model of alkaliphilic Bacillus sp. J. Biosci. Bioeng. 2005;100:365–379. doi: 10.1263/jbb.100.365. [DOI] [PubMed] [Google Scholar]
- 47.Guffanti AA, Hicks DB. Molar growth yields and bioenergetic parameters of extremely alkaliphilic Bacillus species in batch cultures, and growth in a chemostat at pH 10.5. J. Gen. Microbiol. 1991;137:2375–2379. doi: 10.1099/00221287-137-10-2375. [DOI] [PubMed] [Google Scholar]
- 48.Guffanti AA, Susman P, Blanco R, Krulwich TA. The protonmotive force and α-aminoisobutyric acid transport in an obligately alkalophilic bacterium. J. Biol. Chem. 1978;253:708–715. [PubMed] [Google Scholar]
- 49.Krulwich TA, Guffanti AA. The Na+ cycle of extreme alkalophiles: a secondary Na+/H+ antiporter and Na+/solute symporters. J. Bioenerg. Biomembr. 1989;21:663–677. doi: 10.1007/BF00762685. [DOI] [PubMed] [Google Scholar]
- 50.Hirota N, Imae Y. Na+-driven flagellar motors of an alkalophilic Bacillus strain YN-1. J. Biol. Chem. 1983;258:10577–10581. [PubMed] [Google Scholar]
- 51.Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992;355:182–184. doi: 10.1038/355182a0. ▪▪ Demonstrates dual ion coupling in Vibrio.
- 52.Sato K, Homma M. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 2000;275:5718–5722. doi: 10.1074/jbc.275.8.5718. [DOI] [PubMed] [Google Scholar]
- 53.Asai Y, Kojima S, Kato H, Nishioka N, Kawagishi I, Homma M. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J. Bacteriol. 1997;179:5104–5110. doi: 10.1128/jb.179.16.5104-5110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okabe M, Yakushi T, Asai Y, Homma M. Cloning and characterization of motX, a Vibrio alginolyticus sodium-driven flagellar motor gene. J. Biochem. 2001;130:879–884. doi: 10.1093/oxfordjournals.jbchem.a003061. [DOI] [PubMed] [Google Scholar]
- 55.Okunishi I, Kawagishi I, Homma M. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 1996;178:2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paulick A, Koerdt A, Lassak J, et al. Two different stator systems drive a single polar flagellum in Shewanella oneidensis mr-1. Mol. Microbiol. 2009;71(4):836–850. doi: 10.1111/j.1365-2958.2008.06570.x. [DOI] [PubMed] [Google Scholar]
- 57.Henkin TM, Grundy FJ, Nicholson WL, Chambliss GH. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol. Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 58.Moreno MS, Schneider BL, Maile RR, Weyler W, Saier MH., Jr Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 2001;39:1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x. [DOI] [PubMed] [Google Scholar]
- 59.Takami H, Nakasone K, Takaki Y, et al. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takami H, Takaki Y, Uchiyama I. Genome sequence of Oceanobacillus iheyensis isolated from the Iheya Ridge and its unexpected adaptive capabilities to extreme environments. Nucleic Acids Res. 2002;30:3927–3935. doi: 10.1093/nar/gkf526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guffanti AA, Finkelthal O, Hicks DB, et al. Isolation and characterization of new facultatively alkalophilic strains of Bacillus species. J. Bacteriol. 1986;167:766–773. doi: 10.1128/jb.167.3.766-773.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gilmour R, Messner P, Guffanti AA, et al. Two-dimensional gel electrophoresis analyses of pH-dependent protein expression in facultatively alkaliphilic Bacillus pseudofirmus OF4 lead to characterization of an S-layer protein with a role in alkaliphily. J. Bacteriol. 2000;182:5969–5981. doi: 10.1128/jb.182.21.5969-5981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujinami S, Terahara N, Lee S, Ito M. Na+ and flagella-dependent swimming of alkaliphilic Bacillus pseudofirmus OF4: a basis for poor motility at low pH and enhancement in viscous media in an “up-motile” variant. Arch. Microbiol. 2007;187:239–247. doi: 10.1007/s00203-006-0192-7. [DOI] [PubMed] [Google Scholar]
- 64.Grundy FJ, Waters DA, Takova YY, Henkin TM. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol. Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 65.Terahara N, Fujisawa M, Powers B, Henkin TM, Krulwich TA, Ito M. An intergenic stem-loop mutation in the Bacillus subtilis ccpA–motPS operon increases motPS transcription and the MotPS contribution to motility. J. Bacteriol. 2006;188:2701–2705. doi: 10.1128/JB.188.7.2701-2705.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCarter LL. Dual flagellar systems enable motility under different circumstances. J. Mol. Microbiol. Biotechnol. 2004;7:18–29. doi: 10.1159/000077866. ▪ Reviews the utility of dual ion-coupling capacity.
- 67.Delalez N, Armitage JP. Parts exchange: tuning the flagellar motor to fit the conditions. Mol. Microbiol. 2008;71:807–810. doi: 10.1111/j.1365-2958.2008.06573.x. [DOI] [PubMed] [Google Scholar]
- 68.Kobayashi T, Hakamada Y, Adachi S, et al. Purification and properties of an alkaline protease from alkalophilic Bacillus sp. KSM-K16. Appl. Microbiol. Biotechnol. 1995;43:473–481. doi: 10.1007/BF00218452. [DOI] [PubMed] [Google Scholar]
- 69.Kageyama Y, Takaki Y, Shimamura S, et al. Intragenomic diversity of the V1 regions of 16S rRNA genes in high-alkaline protease-producing Bacillus clausii spp. Extremophiles. 2007;11:597–603. doi: 10.1007/s00792-007-0074-1. [DOI] [PubMed] [Google Scholar]
- 70.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell. Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. ▪ Review of the circuitry of bacterial chemotaxis.
- 71.Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. ▪ Review of differences in components and complexity of different bacterial chemotaxis systems.
- 72.Garrity LF, Ordal GW. Chemotaxis in Bacillus subtilis: how bacteria monitor environmental signals. Pharmacol. Ther. 1995;68:87–104. doi: 10.1016/0163-7258(95)00027-5. [DOI] [PubMed] [Google Scholar]
- 73.Kirby JR. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu. Rev. Microbiol. 2009;63:45–59. doi: 10.1146/annurev.micro.091208.073221. [DOI] [PubMed] [Google Scholar]
- 74.Eisenbach M. A hitchhiker’s guide through advances and conceptual changes in chemotaxis. J. Cell. Physiol. 2007;213:574–580. doi: 10.1002/jcp.21238. [DOI] [PubMed] [Google Scholar]
- 75.McCarter LL. Multiple modes of motility: a second flagellar system in Escherichia coli. J. Bacteriol. 2005;187:1207–1209. doi: 10.1128/JB.187.4.1207-1209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muff TJ, Ordal GW. The diverse CheC-type phosphatases: chemotaxis and beyond. Mol. Microbiol. 2008;70:1054–1061. doi: 10.1111/j.1365-2958.2008.06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rao CV, Glekas GD, Ordal GW. The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol. 2008;16:480–487. doi: 10.1016/j.tim.2008.07.003. ▪ Review of recent finding of multiple chemotaxis adaptation systems in Bacillus.
- 78.Szurmant H, Muff TJ, Ordal GW. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J. Biol. Chem. 2004;279:21787–21792. doi: 10.1074/jbc.M311497200. [DOI] [PubMed] [Google Scholar]
- 79.Kirby JR, Niewold TB, Maloy S, Ordal GW. CheB is required for behavioural responses to negative stimuli during chemotaxis in Bacillus subtilis. Mol. Microbiol. 2000;35:44–57. doi: 10.1046/j.1365-2958.2000.01676.x. [DOI] [PubMed] [Google Scholar]
- 80.Lamanna AC, Ordal GW, Kiessling LL. Large increases in attractant concentration disrupt the polar localization of bacterial chemoreceptors. Mol. Microbiol. 2005;57:774–785. doi: 10.1111/j.1365-2958.2005.04728.x. [DOI] [PubMed] [Google Scholar]
- 81.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. ▪▪ Polar localization of bacterial chemoreceptors.
- 82.Krikos A, Conley MP, Boyd A, Berg HC, Simon MI. Chimeric chemosensory transducers of Escherichia coli. Proc. Natl Acad. Sci. USA. 1985;82:1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamamoto K, MacNab RM, Imae Y. Repellent response functions of the Trg and Tap chemoreceptors of Escherichia coli. J. Bacteriol. 1990;172:383–388. doi: 10.1128/jb.172.1.383-388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Umemura T, Matsumoto Y, Ohnishi K, Homma M, Kawagishi I. Sensing of cytoplasmic pH by bacterial chemoreceptors involves the linker region that connects the membrane-spanning and the signal-modulating helices. J. Biol. Chem. 2002;277:1593–1598. doi: 10.1074/jbc.M109930200. [DOI] [PubMed] [Google Scholar]
- 85.Hamamoto T, Hashimoto M, Hino M, et al. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol. Microbiol. 1994;14:939–946. doi: 10.1111/j.1365-2958.1994.tb01329.x. [DOI] [PubMed] [Google Scholar]
- 86.Kajiyama Y, Otagiri M, Sekiguchi J, Kosono S, Kudo T. Complex formation by the mrpABCDEFG gene products, which constitute a principal Na+/H+ antiporter in Bacillus subtilis. J. Bacteriol. 2007;189:7511–7514. doi: 10.1128/JB.00968-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morino M, Natsui S, Swartz TH, Krulwich TA, Ito M. Single gene deletions of mrpA to mrpG and mrpE point mutations affect activity of the Mrp Na+/H+ antiporter of alkaliphilic Bacillus and formation of hetero-oligomeric Mrp complexes. J. Bacteriol. 2008;190:4162–4172. doi: 10.1128/JB.00294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Swartz TH, Ikewada S, Ishikawa O, Ito M, Krulwich TA. The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles. 2005;9:345–354. doi: 10.1007/s00792-005-0451-6. [DOI] [PubMed] [Google Scholar]
- 89.Sugiyama S. Na+-driven re-entry pathway in alkaliphilic bacteria. Mol. Microbiol. 1995;15:592. doi: 10.1111/j.1365-2958.1995.tb02273.x. [DOI] [PubMed] [Google Scholar]
- 90.Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. ▪▪ Identification and characterization of the first bacterial voltage-gated sodium channel.
- 91.Koishi R, Xu H, Ren D, et al. A superfamily of voltage-gated sodium channels in bacteria. J. Biol. Chem. 2004;279:9532–9538. doi: 10.1074/jbc.M313100200. [DOI] [PubMed] [Google Scholar]
- 92.Ouyang W, Jih TY, Zhang TT, Correa AM, Hemmings HC., Jr Isoflurane inhibits NaChBac, a prokaryotic voltage-gated sodium channel. J. Pharmacol. Exp. Ther. 2007;322:1076–1083. doi: 10.1124/jpet.107.122929. [DOI] [PubMed] [Google Scholar]
- 93.Shafrir Y, Durell SR, Guy HR. Models of the structure and gating mechanisms of the pore domain of the NaChBac ion channel. Biophys. J. 2008;95:3650–3662. doi: 10.1529/biophysj.108.135327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goulbourne EA, Jr, Greenberg EP. Inhibition of Spirochaeta aurantia chemotaxis by neurotoxins. J. Bacteriol. 1983;155:1443–1445. doi: 10.1128/jb.155.3.1443-1445.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tisa LS, Olivera BM, Adler J. Inhibition of Escherichia coli chemotaxis by omega-conotoxin, a calcium ion channel blocker. J. Bacteriol. 1993;175:1235–1238. doi: 10.1128/jb.175.5.1235-1238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tisa LS, Sekelsky JJ, Adler J. Effects of organic antagonists of Ca2+, Na+, and K+ on chemotaxis and motility of Escherichia coli. J. Bacteriol. 2000;182:4856–4861. doi: 10.1128/jb.182.17.4856-4861.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hase CC, Fedorova ND, Galperin MY, Dibrov P. Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. 2001;65:353–370. doi: 10.1128/MMBR.65.3.353-370.2001. ▪ Review of bacterial sodium cycles in the context of bacterial pathogenesis.
- 98.Kosono S, Haga K, Tomizawa R, et al. Characterization of a multigene-encoded sodium/hydrogen antiporter (sha) from Pseudomonas aeruginosa: its involvement in pathogenesis. J. Bacteriol. 2005;187:5242–5248. doi: 10.1128/JB.187.15.5242-5248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vimont S, Berche P. NhaA, an Na+/H+ antiporter involved in environmental survival of Vibrio cholerae. J. Bacteriol. 2000;182:2937–2944. doi: 10.1128/jb.182.10.2937-2944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yoshida S, Sugiyama S, Hojo Y, Tokuda H, Imae Y. Intracellular Na+ kinetically interferes with the rotation of the Na+-driven flagellar motors of Vibrio alginolyticus. J. Biol. Chem. 1990;265:20346–20350. [PubMed] [Google Scholar]
- 101.Mulkidjanian AY, Dibrov P, Galperin MY. The past and present of sodium energetics: may the sodium-motive force be with you. Biochim. Biophys. Acta. 2008;177:985–992. doi: 10.1016/j.bbabio.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rosario MM, Fredrick KL, Ordal GW, Helmann JD. Chemotaxis in Bacillus subtilis requires either of two functionally redundant CheW homologs. J. Bacteriol. 1994;176:2736–2739. doi: 10.1128/jb.176.9.2736-2739.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bischoff DS, Bourret RB, Kirsch ML, Ordal GW. Purification and characterization of Bacillus subtilis CheY. Biochemistry. 1993;32:9256–9261. doi: 10.1021/bi00086a035. [DOI] [PubMed] [Google Scholar]
- 104.Fuhrer DK, Ordal GW. Bacillus subtilis CheN, a homolog of CheA, the centrl regulator of chemotaxis in Escherichia coli. J. Bacteriol. 1991;173:7443–7448. doi: 10.1128/jb.173.23.7443-7448.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Burgess-Cassler A, Ordal GW. Functional homology of Bacillus subtilis methyltransferase II and Escherichia coli cheR protein. J. Biol. Chem. 1982;257:12835–12838. [PubMed] [Google Scholar]
- 106.Burgess-Cassler A, Ullah AH, Ordal GW. Purification and characterization of Bacillus subtilis methyl-accepting chemotaxis protein methyltransferase II. J. Biol. Chem. 1982;257:8412–8417. [PubMed] [Google Scholar]
- 107.Kirsch ML, Zuberi AR, Henner D, Peters PD, Yazdi MA, Ordal GW. Chemotactic methyltransferase promotes adaptation to repellents in Bacillus subtilis. J. Biol. Chem. 1993;268:25350–25356. [PubMed] [Google Scholar]
- 108.Muff TJ, Ordal GW. The CheC phosphatase regulates chemotactic adaptation through CheD. J. Biol. Chem. 2007;282:34120–34128. doi: 10.1074/jbc.M706432200. [DOI] [PubMed] [Google Scholar]
- 109.Muff TJ, Ordal GW. Assays for CheC, FliY, and CheX as representatives of response regulator phosphatases. Methods Enzymol. 2007;423:336–348. doi: 10.1016/S0076-6879(07)23015-0. [DOI] [PubMed] [Google Scholar]
- 110.Muff TJ, Foster RM, Liu PJ, Ordal GW. CheX in the three-phosphatase system of bacterial chemotaxis. J. Bacteriol. 2007;189:7007–7013. doi: 10.1128/JB.00896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kuo SC, Koshland DE., Jr Roles of cheY and cheZ gene products in controlling flagellar rotation in bacterial chemotaxis of Escherichia coli. J. Bacteriol. 1987;169:1307–1314. doi: 10.1128/jb.169.3.1307-1314.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zuberi AR, Ying C, Bischoff DS, Ordal GW. Gene–protein relationships in the flagellar hook–basal body complex of Bacillus subtilis: sequences of the flgb, flgc, flgg, flie and flif genes. Gene. 1991;101:23–31. doi: 10.1016/0378-1119(91)90220-6. [DOI] [PubMed] [Google Scholar]
- 113.Titz B, Rajagopala SV, Ester C, Hauser R, Uetz P. Novel conserved assembly factor of the bacterial flagellum. J. Bacteriol. 2006;188:7700–7706. doi: 10.1128/JB.00820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bischoff DS, Ordal GW. Bacillus subtilis chemotaxis: a deviation from the Escherichia coli paradigm. Mol. Microbiol. 1992;6:23–28. doi: 10.1111/j.1365-2958.1992.tb00833.x. [DOI] [PubMed] [Google Scholar]
- 115.Krulwich TA, Ito M, Guffanti AA. The Na+-dependence of alkaliphily in Bacillus. Biochim. Biophys. Acta. 2001;1505:158–168. doi: 10.1016/s0005-2728(00)00285-1. [DOI] [PubMed] [Google Scholar]
- 201.KEGG A bioinformatics resource for linking genomes to life and the environment. www.genome.jp/kegg/pathway/ko/ko02040.html.
- 202.ClustalW Multiple Sequence Alignment. http://clustalw.genome.jp.