Abstract
The NMR structure of an RNA with a copy of the 5′CUG/3′GUC motif found in the triplet repeating disorder myotonic dystrophy type 1 (DM1) is disclosed. The lowest energy conformation of the UU pair is a single hydrogen bonded structure; however, the UU protons undergo exchange indicating structural dynamics. Molecular dynamics simulations show that the single hydrogen bonded structure is the most populated one but the UU pair interconverts between 0, 1, and 2 hydrogen bonded pairs. These studies have implications for the recognition of the DM1 RNA by small molecules and proteins.
RNA plays essential roles in many biological processes and is thus an important therapeutic target.(1) In particular, recent studies have shown that an RNA gain of function causes myotonic dystrophy types 1 and 2 (DM1 and DM2,1 respectively).(2, 3) DM1 is caused by a trinucleotide CTG repeat in the 3′UTR of the DMPK gene. Once transcribed, expanded CUG repeats fold into an RNA hairpin with repeating 5′CUG/3′GUC motifs and binds CUG binding protein and the RNA splicing regulator muscleblind-like protein 1 (MBNL-1). The downstream effect of these interactions is mis-splicing of the main muscle chloride channel (clc1) and insulin receptor transcripts, for example.(4, 5)
Oligonucleotides (6) and the small molecule pentamidine (7) target the DM1 CUG repeats and correct pre-mRNA splicing defects in animal models of disease. Other small molecules targeting DM1 RNAs have been developed, including triazine-acridine conjugates (8) and compounds selected via dynamic diversity screening strategy.(9) In addition, rational, modular assembly strategies have been developed to provide nanomolar in vitro inhibitors.(10–13). In order to gain insights into how small molecules and proteins bind DM1 RNAs, the refined NMR structure and a molecular dynamics (MD) simulation of an RNA that contains the 5′CUG/3′GUC motif are disclosed herein.
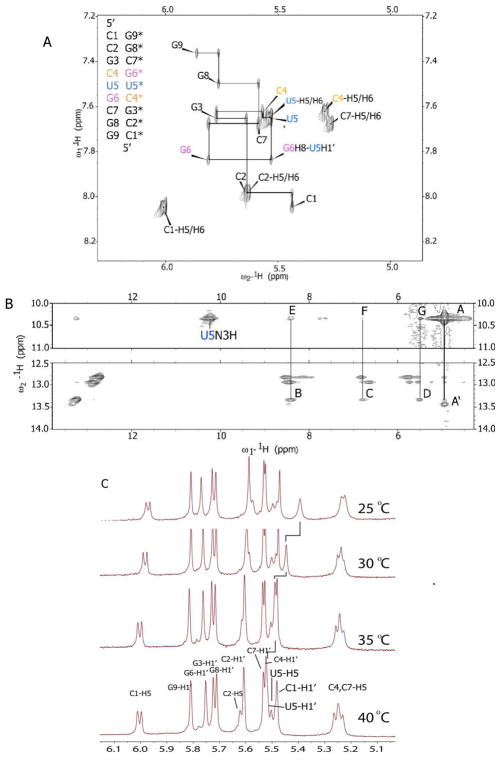
NMR spectra of r(CCGCUGCGG)2 were collected and non-exchangeable protons assigned by analysis of NOESY and DQF-COSY spectra in D2O at 40°C. The base to H1′ NOESY walk (Figure 1) and the observed cross peaks and intensities between inter and intra-nucleotide base to H2′ and base to H3′ are indicative of A-form RNA. NOE connectivities between the C4-H1′ and U5-H6 and between U5-H1′ and G6-H8 are similar to other base to H1′ connectivities in the duplex; this supports a structure where stacking between C4, U5, and G6 is present.
Figure 1.
2D-NMR and 1D-NMR spectra of r(CCGCUGCGG)2. A, 200 ms NOESY spectrum showing base to H1′ connectivities. Labels identify intra-nucleotide base to H1′ NOESY cross peaks. C and U H5 to H6 cross peaks are also identified. B, 100 ms NOESY spectrum collected in 90:10 H2O:D2O at 10°C. Label E identifies cross peaks between the U5-N3H and the G6 imino proton while F and G identify cross peaks between U5 imino proton and the C4 and G6 amino protons, respectively. Strong cross peaks between the U5-N3H and terminal G9 with water are identified in the spectrum as A and A′, respectively. C, temperature-dependent NMR spectra. An upfield shift of ~0.1 ppm in the U5-H1′ peak at lower temperatures indicates dynamic exchange.
Exchangeable proton assignments were made by analysis of a 2D NOESY spectrum recorded in H2O:D2O (9:1) at 10°C. All imino protons are observed. Imino to imino proton, imino to amino proton, and C-H5 proton NOEs are present for the internal GC base pairs. The G3 and G8 imino protons exhibit very little to no exchange with the solvent.
The terminal G9 imino proton (peak A, Figure 1) and the U5 imino proton (peak A′, Figure 1) are broadened by exchange with solvent. The location of the U5 base is best defined by NOEs observed in the water NOESY. The U5 imino proton exhibits an NOE to the G6 imino proton peak (E, Figure 1), and to the C4 and G6 amino protons (F and G, respectively, Figure 1). The large NOE cross peak between the U5 imino proton with water (A, Figure 1) shows that the UU pair, especially at the N3 interface, is dynamic. In order to further confirm dynamic exchange, NMR spectra (C, Figure 1) of the RNA duplex in D2O were acquired at 25°C, 30°C, 35°C and 40°C. Line broadening and an upfield shift in the U5-H1′ resonance with decreased temperature indicates rapid exchange, thus confirming a structurally dynamic UU pair. Also, an upfield shift of U5-H5 was observed as a function of temperature, indicating rapid exchange. (14, 15)
Based on 1H-1H NOE distances and Watson-Crick base pairing restraints, the structure of the duplex was refined using restrained molecular dynamics and Amber 11 (16). There are no NOE distance violations observed at >0.2 Å. Figure 2 shows the superposition of the 25 lowest free energy structures generated. The average RMSD for the pair-wise, all-atom superposition of the structures is 0.78 Å, indicating excellent convergence.
Figure 2.

Overlay of the 25 lowest energy structures for r(CCGCUGGCC)2. Right, view of structures from the major groove. Left, view of the structure from the minor groove.
Globally, the structure adopts a well-defined A-form geometry with a C3′-endo sugar pucker. The helical rise for each base pair step is less than 3 Å. Thus, the overall structure is similar to a standard A-form helix with moderate displacement of the UU pair.
Since NMR spectra indicated that the UU pair was in dynamic exchange as indicated by the NOE between the U5N3 and water (Figure 1), we sought to explore the nature of the dynamics via MD simulations. MD simulations were performed over a 1 ns period using the lowest energy, Amber-refined RNA structure as a starting point. During the course of the simulations, the global RMSD of the structure varied little relative to the initial starting structure indicating equilibration and a stable simulation.
A series of over 4000 discrete frames (structures) were generated during the MD simulation. These structures were analyzed to define the types of UU pairs present in the trajectory. The analysis showed UU pair structures that had 0, 1, or 2 hydrogen bonds (using distance and angle cutoffs of 3.5 Å and 120o, respectively, to define the presence of hydrogen bonds).(17) The relative populations are: 15.9% of the structures have 0 hydrogen bonds; 76.5% have 1 hydrogen bond (U5-N3 to U14-O4); and 7.6% have two hydrogen bonds (U14-N3 to U5-O2 and U5-N3 to U14-O4) (Figure 3).
Figure 3.
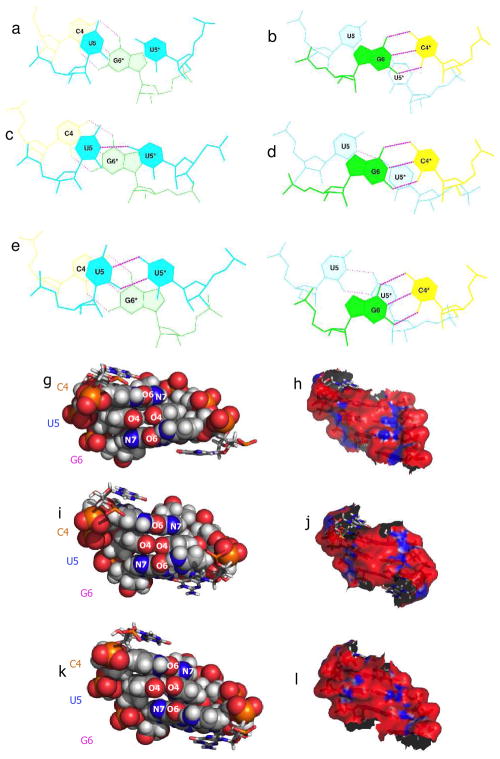
Top view of the stacking interactions for the 0, 1, and 2 hydrogen bonded structures, space filling model views from the minor groove, and the electrostatic potential as viewed from the minor groove. A–F, stacking diagram for the UU loop on the closing GC pairs for the 0 (A and B), 1 (C and D), and 2 (E and F) hydrogen bonded structures, respectively. G–L, view of the major groove in a CPK model and major groove electrostatic calculations for the 0 (G and H), 1 (I and J), and 2 (K and L) hydrogen bonded structures.
In order to understand the features in the different UU pairs and to explain the relative population differences from MD, we studied the geometry of the pairings and their stacking interactions with the loop closing pairs. For the geometry, the inter-strand C1′-C1′ distances were calculated for each type of structure. This analysis showed that UU pairs with 0, 1, and 2 hydrogen bonds have distances of 10.7 Å, 10.6 Å and 8.9 Å, respectively. Since the C1′-C1′ distance in standard A-form RNA helices is ~10.5 Å,(18) the 0 and 1 hydrogen bond structures do not induce a change in standard A-form helical geometry, however, the 2 hydrogen bond structure does. Thus, despite the two hydrogen bonded structure having a bonus in stability from an extra hydrogen bond, it is not the major structure. Evidently, formation of two hydrogen bonds would occur at the expense of introducing a distortion in the RNA helix.
Differences in stacking and electrostatic surface potentials in the three types of structures were also investigated (Figure 3). For all three structures, there is some overlap of the UU pair with the closing GC base pairs; however, there are not large differences in stacking between the three pairing types. Electrostatic potentials (h, j, and l; Figure 3) were calculated using the Poisson-Boltzmann equation.(19) The electrostatic surface potential of the minor groove is largely negative, although there are isolated patches of positive surface area. Akin to stacking interactions, the electrostatic potentials are similar.
Two crystal structures of RNA molecules containing CUG repeats have been reported. (20) In good agreement with these studies with six CUG repeats, the RNA molecule adopts an overall A-form structure. Moreover, there is no distortion in the RNA backbone relative to A-form to accommodate the UU pairs. In contrast to our NMR investigations, the UU pairs in the crystal structure formed water mediated, not direct, hydrogen bonds. A structure of G(CUG)2C was reported by Kiliszek et al.(21) In this structure, the 5′CUG/3′GUC motifs display the one hydrogen bond pattern that was observed by NMR. The difference in the two crystal structures prompted Kiliszek et al. to reanalyze the structure of Mooers et al.; this reanalysis suggested that the Mooers et al. structure is consistent with the 1 hydrogen bond structure. The observation that similar structures are observed with a isolated DM1 motif by NMR compared to crystal structures with multiple DM1 motifs suggests that the structural effects of having multiple DM1 motifs may be negligible.
Recently, an analysis of the structures of 1×1 nucleotide RNA internal loops deposited in the Protein Data Bank (PDB) was disclosed.(22) Several of the structures in this analysis included UU pairs. A variety of structures are observed including 1 and 2 hydrogen bonded structures as well as structures in which the U nucleotides are flipped out of the RNA helix altogether. Taken together with the results reported here, it shows that UU pairs adopt multiple conformations without distorting the global conformation of the surrounding nucleotides. A movie of our MD simulations (Supporting Information) shows that the UU pairs in the 5′CUG/3′GUC motif sample 0, 1, and 2 hydrogen bonded structures without breaking loop closing base pairs.
In summary, NMR spectroscopy and MD simulations show that the 5′CUG/3′GUC motif found in DM1 RNAs is dynamic. The NMR structures suggest that ligand binding may proceed through conformational selection of RNA structures. These investigations provide a foundation to study structures of ligands bound to DM1 RNAs.
Supplementary Material
Acknowledgments
We acknowledge Jon French and Amit Kumar for help with NMR experiments and Doug Kojetin, Anthony Carvalloza, and Andrew Davis for help with Amber.
Footnotes
This work was funded by the National Institutes of Health (3R01GM079235-02S1 and 1R01GM079235-01A2) and Research Corporation. MDD is a Dreyfus New Faculty Awardee, a Dreyfus Teacher-Scholar, and a Cottrell Scholar.
Abbreviations: clc1, Main Chloride Ion Channel; DM1, myotonic dystrophy type 1; DM2, myotonic dystrophy type 2; DMPK, dystrophia myotonia protein kinase; DQF-COSY, Double Quantum Filtered Correlation Spectroscopy; MD, Molecular Dynamics; NOESY, Nuclear Overhauser Effect Correlation Spectroscopy; 3′UTR, 3′ untranslated region.
SUPPORTING INFORMATION AVAILABLE Supporting information includes protocols and a movie of the MD trajectory simulation. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Thomas JR, Hergenrother PJ. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 2.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 3.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Science. 2000;289:1769–1772. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 4.Savkur RS, Philips AV, Cooper TA. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 5.Lueck JD, Mankodi A, Swanson MS, Thornton CA, Dirksen RT. J Gen Physiol. 2007;129:79–94. doi: 10.1085/jgp.200609635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Proc Natl Acad Sci USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. Proc Natl Acad Sci USA. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gareiss PC, Sobczak K, McNaughton BR, Palde PB, Thornton CA, Miller BL. J Am Chem Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MM, Pushechnikov A, Disney MD. ACS Chem Biol. 2009;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. J Am Chem Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Disney MD, Lee MM, Pushechnikov A, Childs-Disney JL. Chembiochem. 2010;11:375–382. doi: 10.1002/cbic.200900716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MM, Childs-Disney JL, Pushechnikov A, French JM, Sobczak K, Thornton CA, Disney MD. J Am Chem Soc. 2009;131:17464–17472. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nonin S, Leroy JL, Gueron M. Biochemistry. 1995;34:10652–10659. doi: 10.1021/bi00033a041. [DOI] [PubMed] [Google Scholar]
- 15.Schroeder KT, Skalicky JJ, Greenbaum NL. RNA. 2005;11:1012–1016. doi: 10.1261/rna.2270205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. J Comp Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, Debolt S, Ferguson D, Seibel G, Kollman P. Comp Phys Commun. 1995;92:1–41. [Google Scholar]
- 18.Limer S. Prog Nucl Acid Res and Mol Biol. 1997;57:1–39. doi: 10.1016/s0079-6603(08)60276-7. [DOI] [PubMed] [Google Scholar]
- 19.Fogolari F, Brigo A, Molinari H. J Mol Recog. 2002;15:377–392. doi: 10.1002/jmr.577. [DOI] [PubMed] [Google Scholar]
- 20.Mooers BH, Logue JS, Berglund JA. Proc Natl Acad Sci USA. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis AR, Kirkpatrick CC, Znosko BM. Nucleic Acids Res. 2010;28:1–14. doi: 10.1093/nar/gkq793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.