Abstract
BACKGROUND
We have previously described a new rapid approach that relies on monitoring intentionally stressed bacteria in contaminated PCs. This earlier work included human cell lysis with Triton X-100 and filtration as steps in the sample preparation. The present study was undertaken to develop an improved and time saving protocol that enables direct bacterial detection in PCs without lysis and filtration.
STUDY DESIGN AND METHODS
Apheresis- or whole blood-derived PCs were spiked with 17 model bacteria and tested at final concentrations from 103 to 106 CFUs/mL. The contaminated PCs were treated with a chemical compound which induce a stress response in bacteria and monitored using differential impedance sensing to detect and record subtle changes in the dielectric permittivities of the contaminated platelet samples.
RESULTS
No measurable responses from sterile platelet samples were observed during exposure to the compounds used as stressors. In contrast, distinct response profiles were obtained without exception for all 17 bacterial species for all bacterial concentrations tested. Bacterial presence was established within 5–10 minutes for high inoculums (106 and 105 CFUs/mL) while low inoculums (104 and 103 CFUs/mL) were usually detectable within 20 min. The entire testing process routinely took less than 30 minutes from the point of sampling to the time that the final results are available.
CONCLUSION
The results described here demonstrate that monitoring the development of stress in bacteria is a fast and simple way to detect 103 CFUs/mL or more bacteria in complex cellular blood products such as PCs.
Keywords: stress response, differential impedance sensing, dielectric permittivity, platelet contamination, bacterial detection
INTRODUCTION
In a previous report, we described a new approach for detecting bacteria in platelet concentrate (PC) rapidly using differential impedance sensing to monitor the stress developed by bacteria 1. This approach was initially implemented with a protocol that used Triton X-100 to simultaneously lyse all human blood cells while inducing a measurable stress response in any contaminating bacteria. The present study describes significant improvements to our earlier work resulting from the identification of a chemical stressor that induces a stress response in bacteria but not in human cells. The ability to distinguish between bacteria and human cells takes full advantage of the differential sensing approach and enables the direct detection of as few as 103 CFUs/mL stressed bacteria in 1010 platelets/mL using a one-step protocol that does not require lysis and filtration as preparation steps. The data reported here further characterize the potential of exploiting the bacterial stress response as a tool for the routine quality control testing of PC units with a simple and rapid assay.
MATERIALS AND METHODS
Platelet preparations
A total of 141 in-dated and out-dated apheresis-derived single donor and leukocyte-reduced whole blood-derived platelets were obtained from the Rhode Island Blood Center (Providence, RI) and The Blood Connection (Piedmont, SC) over the nearly 2-year period which this study was conducted. Typical in-dated PC units received were 2–3 days old; typical out-dated PC units received were 6–7 days old. All PC preparations were stored at 20–24°C with constant rotation (Orbitron Rotator II, Boekel Scientific, Feasterville, PA). After arrival, the sterility of each PC unit was tested. Bacterial culture medium was inoculated with an aliquot of the platelet sample and incubated at 37°C for up to 3 days. In parallel, 1 mL of the PC sample was plated onto agar plates and incubated at 37°C for 48 hours.
Bacterial species
17 species of bacteria associated with PC transfusion-septic reactions were used in this study. All but one of the bacterial strains were acquired from the bacterial collection of the ATCC (Manassas, VA). The exception, Salmonella typhimurium, was isolated from clinical specimens at the FDA Center for Veterinary Medicine (courtesy of Dr. David G. White). The list of the 17 species used, their respective ATCC numbers, and the cell populations tested are summarized in Table 1. All bacteria were routinely cultured in their recommended broth medium and on agar plates. Aliquots of each strain were subsequently stored frozen in liquid nitrogen.
TABLE 1.
List of bacterial species and concentrations tested.
| Bacterial Species | ATCC# | Tested Concentrations (CFU/mL) | Mean NIR Value |
|---|---|---|---|
| Bacillus cereus | 7064 | CTRL (N=3) 2.7×103 (N=3) 4.9×103(N=3) 2.7×104 (N=3) 1.9×105 (N=3) |
0.9999 ± 0.0001 0.9969 ± 0.0014 0.9955 ± 0.0011 0.9924 ± 0.0030 0.9809 ± 0.0059 |
| Listeria monocytogenes | 19115 | CTRL (N=2) 2.0×102 (N=5) 1.9×103 (N=4) 1.7×104 (N=5) 1.7×105 (N=2) |
1.0001 ± 0.0007 0.9976 ± 0.0018 0.9926 ± 0.0016 0.9848 ± 0.0047 0.9711 ± 0.0018 |
| Propionibacterium acnes | 6919 | CTRL (N=4) 1.6×103 (N=4) 1.6×104 (N=5) 1.6×105 (N=3) 1.6×106 (N=3) |
1.0001 ± 0.0001 0.9983 ± 0.0034 0.9932 ± 0.0034 0.9816 ± 0.0021 0.9532 ± 0.0161 |
| Staphylococcus aureus | 29213 | CTRL (N=3) 2.8×103 (N=3) 2.6×104 (N=3) 2.4×105 (N=3) 2.2×106 (N=3) |
1.0001 ± 0.0007 0.9966 ± 0.0020 0.9930 ± 0.0005 0.9861 ± 0.0054 0.9673 ± 0.0056 |
| Staphylococcus epidermidis | 700567 | CTRL (N=3) 1.3×103 (N=3) 2.8×104 (N=3) 2.7×105 (N=3) 2.8×106 (N=3) |
1.0000 ± 0.00004 0.9938 ± 0.0016 0.9849 ± 0.0053 0.9806 ± 0.0022 0.9689 ± 0.0041 |
| Streptococcus mitis | 49456 | CTRL (N=4) 3.7×103 (N=3) 3.7×104 (N=4) 3.9×105 (N=3) 4.0×106 (N=3) |
1.0000 ± 0.0005 0.9980 ± 0.0005 0.9937 ± 0.0020 0.9837 ± 0.0034 0.9653 ± 0.0126 |
| Streptococcus pneumoniae | 49619 | CTRL (N=4) 2.6×103 (N=7) 2.8×104 (N=4) 3.0×105 (N=4) 2.5×106 (N=3) |
1.0000 ± 0.0005 0.9958 ±0.0012 0.9926 ±0.0020 0.9857 ± 0.0033 0.9735 ±0.0067 |
| Acinetobacter baumannii | 17961 | CTRL(N=3) 1.4×104 (N=3) 1.1×105 (N=3) 1.0×106 (N=3) 1.2×107 (N=3) |
1.0000 ± 0.0027 0.9928 ± 0.0004 0.9897 ± 0.0042 0.9872 ± 0.0006 0.9644 ± 0.0078 |
| Enterococcus faecalis | 19433 | CTRL(N=3) 2.8×104 (N=3) 1.5×105 (N=3) 9.1×105 (N=3) 3.2×106 (N=3) |
1.0001 ± 0.0007 0.9922 ± 0.0005 0.9852 ± 0.0029 0.9806 ± 0.0054 0.9758 ± 0.0016 |
| Escherichia coli | 25922 | CTRL (N=3) 9.8×102 (N=5) 1.1×104 (N=4) 1.3×105 (N=3) 1.0×106 (N=3) |
1.0000 ± 0.0003 0.9964 ± 0.0015 0.9869 ± 0.0022 0.9756 ± 0.0048 0.9508 ± 0.0062 |
| Klebsiella pneumoniae | 13882 | CTRL (N=3) 1.3×103 (N=3) 1.6×104 (N=3) 1.8×105 (N=3) 1.2×106 (N=3) |
1.0005 ± 0.0008 0.9956 ± 0.0004 0.9943 ± 0.0010 0.9917 ± 0.0005 0.9836 ± 0.0010 |
| Proteus mirabilis | 12453 | CTRL (N=3) 3.4×103 (N=4) 2.9×104 (N=3) 2.7×105 (N=4) 2.9×106 (N=3) |
0.9998 ± 0.0004 0.9955 ± 0.0027 0.9835 ± 0.0025 0.9802 ± 0.0027 0.9682 ± 0.0025 |
| Pseudomonas aeruginosa | 15442 | CTRL (N=3) 3.9×103 (N=5) 3.5×104 (N=4) 4.0×105 (N=3) 3.9×106 (N=2) |
0.9999 ± 0.0004 0.9972 ± 0.0011 0.9868 ± 0.0043 0.9749 ± 0.0013 0.9596 ± 0.0188 |
| Pseudomonas fluorescens | 49838 | CTRL (N=3) 5.3×103 (N=5) 5.8×104 (N=4) 5.5×105 (N=4) 6.1×106 (N=2) |
1.0000 ± 0.0002 0.9968 ± 0.0011 0.9874 ± 0.0059 0.9742 ± 0.0044 0.9457 ± 0.0085 |
| Salmonella typhimurium | n/a | CTRL (N=3) 3.0×103 (N=4) 3.8×104 (N=3) 6.5×105 (N=3) 5.6×106 (N=3) |
0.9998 ± 0.0002 0.9968 ± 0.0011 0.9945 ± 0.0011 0.9894 ± 0.0009 0.9868 ± 0.0008 |
| Serratia marcescens | 27143 | CTRL (N=4) 1.9×103 (N=7) 1.9×104 (N=6) 1.9×105 (N=5) 1.9×106 (N=3) |
1.0000 ± 0.0003 0.9966 ± 0.0017 0.9917 ± 0.0026 0.9846 ± 0.0034 0.9679 ± 0.0037 |
| Yersinia enterocolitica | 23715 | CTRL (N=3) 2.2×104 (N=3) 6.6×105 (N=3) 6.2×106 (N=3) 7.3×107 (N=3) |
1.0000 ± 0.0010 0.9940 ± 0.0032 0.9932 ± 0.0032 0.9847 ± 0.0021 0.9726 ± 0.0091 |
Differential impedance measurement platform
All measurements of the impedance and the corresponding dielectric permittivity were made using custom-built instrumentation. The measurement system consisted of a cassette holding the test samples and a temperature-controlled table-top sized device into which the cassette was inserted for analysis. The table-top sized device consisted of thermally controlled platens that surrounded the cassette and maintained a constant temperature with high precision and all sensing electronics. All impedance measurements were made at a single fixed frequency (1000 Hz) and a voltage of 75 mV to minimize any localized heating. The magnitude and angle of the impedance vector for each detection chamber was continuously recorded over the measurement period using circuitry developed in-house and stored in a computer.
All cassettes used in these experiments contained two 100 μL test chambers, each having the same thermo-mechanical properties for optimal differential measurement. Each chamber was defined by a sub-millimeter electrode gap structure that encompassed a sample and sensed its respective electrical properties. The electrode surfaces were made of pure gold deposited onto glass plate (Borofloat™; Schott Glass, AG). A photograph of the apparatus and a test cassette used to obtain the data reported here is shown in Fig. 1.
Fig. 1.
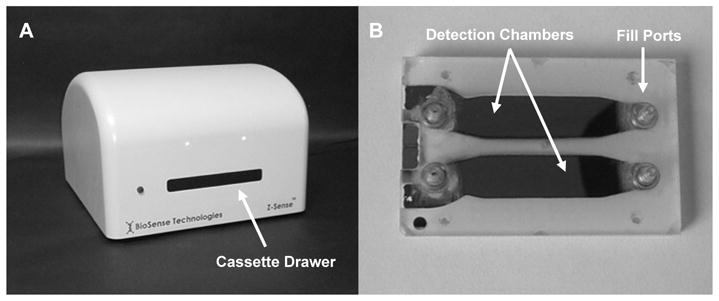
Photograph of the BioSense differential impedance sensing analyzer (A) and two-chamber test cassette (B). The cassette is loaded into a drawer located on the face of the analyzer that extends and retracts and is designed to hold the cassette during measurement.
PC sample preparation
All bacterial species used for spiking experiments were first adapted for growth in PC as previously described 1 to avoid interfering effects from the bactericidal activities of any PC components. The adapted bacteria were aseptically spiked into aliquots of the PCs, allowed to grow to stationary phase, and diluted accordingly to achieve the target measurement concentrations. After each dilution, the spiked PCs were incubated at 20–24°C for 1–2 hours on an orbital shaker (60 rpm) to ensure further adaptation of the bacterial cells and resumption of their growth. In all experiments except those involving P. acnes and S. marcescens, the actual concentrations of bacteria were confirmed by quantitative culture on agar plates. The concentration of P. acnes cells in stationary culture was determined by agar plating followed by viable acridine orange staining using UV microscopy for all subsequent dilutions. For S. marcescens, only bacterial concentration in stationary culture was measured.
Impedance measurements of spiked and non-spiked PC
To prepare a PC sample for measurement (either spiked or sterile), approximately 1 mL of PC was transferred to a sterile vial, mixed with an equal volume of 0.8% glucose (added as an additional bacterial energy source) and EDTA (1 mM), and then divided into two equal parts. As a final step, a fixed amount of the stressor compound was added to one of the parts and mixed.
One chamber of the test cassette was then manually filled with 100 μL of the PC suspension containing the stressor and the second chamber, acting as a reference, was filled with the same volume of PC suspension prepared without the stressor. The filled cassette was then inserted into the analyzer set at 30°C and the impedance signals from each chamber were continuously recorded and analyzed. Each experiment was repeated a minimum of three times including different spikes on different days.
Analysis of the data
For the planar geometry used in the design of these cassettes, the dielectric permittivity is directly proportional to the measured capacitance (obtained from the reactive component of the recorded impedance vector). For all experiments, the dielectric permittivity was calculated according to the equation C = ε (A/d) where ε is the dielectric permittivity of the suspension, A is the electrode area, and d is the separation distance between the electrodes.
The two detection chambers are located in close proximity so that both are subjected to identical thermal/mechanical perturbations. This provides a means for minimizing signal changes that are unrelated to the bacterial stress response. For example, changes in the dielectric properties due to the platelets occurring in the first chamber which are also common in the second chamber are suppressed. Any difference in signal remaining after comparison is quantified by a parameter defined as the “Normalized Impedance Response” (NIR). The value of the NIR is calculated by dividing the value of the capacitance from the chamber containing the stressor-treated PC suspension by that from the adjacent (reference) chamber containing the same but stressor-untreated suspension at each recorded time point. The NIR reflects the changes in the dielectric permittivity due exclusively to the stressed bacteria. The values of the NIR profiles were arbitrarily scaled to a value equal to 1.000 at the start of each experiment for ease in comparison. Otherwise, no other modifications to the NIR values were made.
RESULTS
Impedance response of PC during exposure to stressor
In an initial set of experiments, both sterile apheresis- and whole blood-derived platelet concentrates were treated with varying concentrations of different stressors to evaluate any possible effects of these compounds on the dielectric permittivities of the platelets and any other human cells present in the PC. Extensive impedance measurements of sterile PC treated with varying concentrations of two different stressors were recorded and the corresponding NIR profiles were calculated. In addition, the treated PC suspensions were examined for microscopic visual changes such as aggregation. The choice of stressor was based on the absence of a measurable response from PC, the ability to induce stress in bacterial cells, and chemical stability. A bacterial membrane permeability modifying cationic compound designated as STR-103 was selected as the stressor of first-choice for all subsequent experiments with spiked PCs. This compound is a commercially available derivative of α-chlorotoluene that is non-toxic to work with and stable over time.
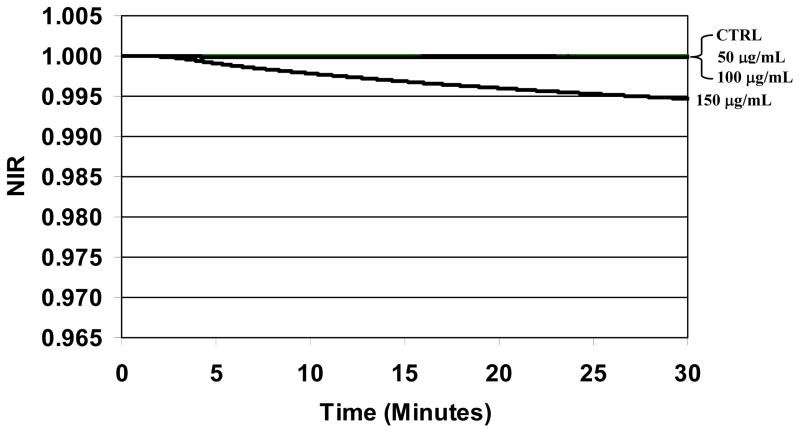
The NIR profiles obtained for sterile whole blood derived PC continuously exposed to STR-103 are plotted in Fig. 2 for the initial 30 minutes of measurement. For this stressor, platelet aggregation was detected in samples treated with 200 μg/mL or greater establishing the threshold for deleterious effects. Consequently, NIR profiles were studied for concentrations less than 200 μg/mL. The NIR profile for PC without the stressor is plotted as a negative control. As expected, because the two detection chambers are filled with the same biological sample, the impedance signals from each chamber are also the same and the resulting NIR value is a constant value equal to one over the entire measurement period. Similar results were obtained for sterile PC treated with STR-103 at the concentrations 50 μg/mL and 100 μg/mL. The corresponding NIR profiles tightly overlap the control curve indicating that the stressor has no measurable effect on the dielectric permittivity of the PC over that time period. However, when the concentration was increased to 150 μg/mL, the NIR value immediately deviated from a constant value and decreased continuously through the end of the measurement period despite the absence of any visually detected aggregation. The magnitude of this response at 30 minutes was commensurate with the responses from the lowest bacterial loads measured in the spiked PCs (Fig. 3). Similar results were also obtained for apheresis-derived PCs (data not shown). Thus, for the stressor STR-103, a concentration of 100 μg/mL was determined to be optimal and used for the spiked PC experiments.
Fig. 2.
Differential impedance response from sterile platelet concentrate treated with STR-103. Three curves corresponding to the control run (“CTRL”; no stressor), PC treated with 50 μg/mL, and 100 μg/mL are plotted together and overlap having the same value. In contrast, the NIR profile for PC treated with 150 μg/mL deviates from these three curves over the 30 minute measurement period.
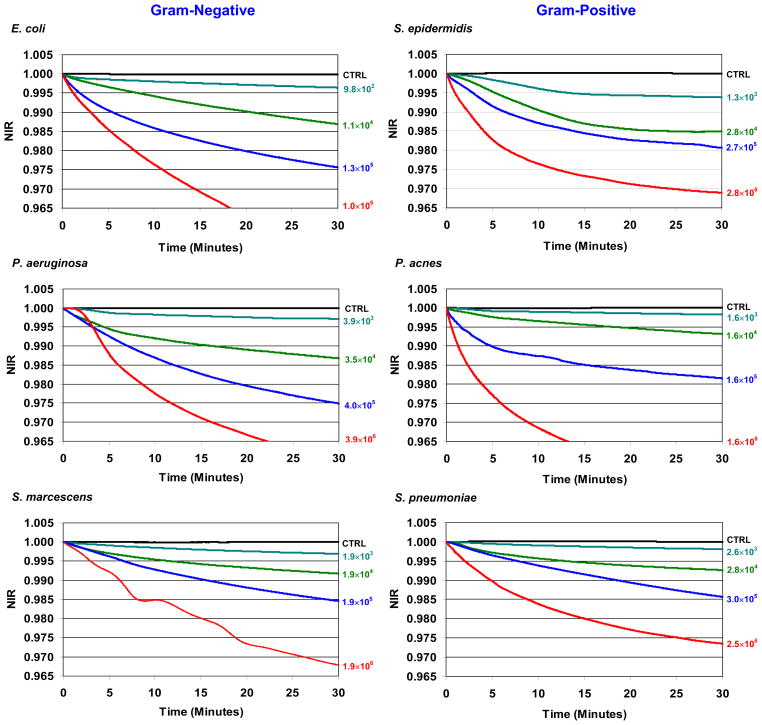
Fig. 3. Normalized Impedance Response (NIR) profiles from PC infected with six representative bacterial species out of 17 tested treated with treated 100 μg/mL of STR-103.
NIR profiles for PC infected with between 103 CFUs/mL to 106 CFUs/mL of the Gram-negative species E. coli, P. aeruginosa, S. marcescens, and Gram-positive species S. epidermidis, P. acnes, and S. pneumoniae. The actual bacterial concentrations tested are displayed next to their corresponding NIR profiles. Sterile but treated PC is plotted as a negative control (CTRL) in all plots. A list of all bacterial species and the respective concentrations tested are found in Table 1.
Bacterial detection in PCs
NIR profiles for spiked PC treated with STR-103 were obtained for 17 bacterial species associated with contaminated PCs. For these experiments, the final bacterial concentrations in the spiked PCs were targeted between 103 and 106 CFUs/mL. A list of all 17 bacterial species, the actual bacterial loads tested, and the corresponding NIR values after 30 minutes are summarized in Table 1.
Graphs of the NIR profiles for 6 bacterial species chosen as representative Gram-negative (E. coli, P. aeruginosa, and S. marcescens), Gram-positive (S. epidermidis, S. pneumoniae) species, and the slow-growing anaerobe P. acnes are presented in Fig. 3. The NIR values plotted for each species are an average of three or more measurements obtained with the same spiked PC unit but with different cassettes and analyzers. The final data sets for each bacterial specie were typically constructed from between one and three different PC units.
For each species, multiple negative controls were obtained. Before spiking, NIR profiles were obtained for every PC unit used during exposure to STR-103 to monitor any potential effects from the stressor on the platelets. NIR profiles were also obtained for spiked but untreated PC to monitor any variability associated with the differential measurement of identical growing bacterial populations in PC. For all bacterial species and PCs tested, the NIR profiles obtained for these controls had constant values equal to one over the measurement period.
In contrast, and without exception, the corresponding NIR profiles for all stressor-treated spiked PCs displayed a characteristic decrease in the NIR values over the same measurement period irrespective of the species. In addition, an increase in the number of contaminant bacteria resulted in a more pronounced NIR profile. In all cases, the treated PC could be delineation from its respective control by visual inspection in significantly less than 30 minutes.
Overall, the NIR profiles for all 17 species are qualitatively similar to each other irrespective of their Gram staining characteristics, culture requirements, or doubling times. These attributes are illustrated by the qualitatively similar NIR profiles for E. coli, S. epidermidis, and P. acnes.
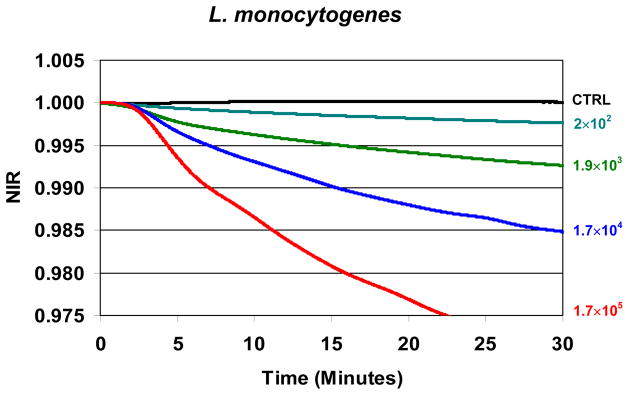
Although no effort was made to investigate systematically the detection of less than 103 CFUs/mL in this study, data at bacterial populations as low as 2 × 102 CFUs/mL were obtained for the PC spiked with L. monocytogenes. These NIR profiles are plotted together in Fig. 4. The NIR values obtained for this low bacterial concentration are the average of 5 replicates using the same PC sample with different cassettes and analyzers. These data were reconfirmed on a different day with different PC inoculated with a low titer bacterial spike. Despite the small NIR values, the profile is readily discernable from its control within 30 minutes by visual inspection.
Fig. 4. Normalized Impedance Response (NIR) from PC infected with L. monocytogenes and treated with treated 100 μg/mL of STR-103.
NIR profiles are plotted for test bacterial concentrations of 2 × 102 CFUs/mL to 1.7 × 106 CFUs/mL and are visually differentiable from the negative control.
Statistical Analysis
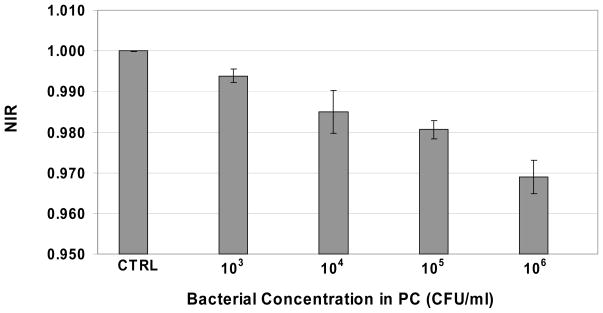
The statistical significance of the NIR profiles for the species E. coli and S. epidermidis was analyzed to investigate the reproducibility of the measurements. Because the NIR profiles were measured continuously, average NIR values were calculated at a single time point (30 minutes). To assess the statistical significance of the different populations with respect to each other, an analysis of variance (ANOVA) was performed. The resulting value was <0.0001 indicating the overall statistical significance of each NIR profile. The results for S. epidermidis are shown graphically in Fig. 5.
Fig. 5. Mean Normalized Impedance Response (NIR) at 30 minutes for PC spiked with different bacterial concentrations of growing S. epidermidis and treated with 100 μg/mL of STR-103.
The mean values and standard deviations of the NIR values at the single time point 30 minutes are plotted for sterile treated PC (“CTRL”) and treated PC infected with S. epidermidis at the concentrations 103, 104, 105, and 106 CFUs/mL. All values are the average of three measurements obtained for the same sample but using different analyzers and cassettes. ANOVA analysis of the mean NIR values revealed a p-value less than 0.0001 indicating their respective statistical significance.
Survival Studies
Survival studies of the bacteria in stressor-treated spiked apheresis- and whole blood-derived PC were undertaken to determine if the corresponding NIR profiles are primarily associated with cell death. PCs containing 106 CFUs/mL logarithmically growing E. coli or S. epidermidis were treated with 100 μg/mL of the stressor STR-103. The corresponding numbers of the stressed bacteria isolated from the PC were determined at ten minute intervals over the initial 60 minutes. Because the preparation and treatment of these PCs were identical to the method used to generate the NIR profiles plotted in Fig. 2, the bacterial survival data can be compared directly with the NIR profiles. For both species studied, the numbers of viable bacteria in the stressor-treated PC remained constant over the initial 30 minute measurement period (irrespective of the type of PC) indicating no significant bacterial cell death.
DISCUSSION
Bacterial Testing of PC
The transmission of bacteria by platelet transfusion and the consequent septic reactions in recipients continues to be a significant clinical problem despite mandatory testing of donor blood and blood products. Septic reactions due to bacterial contamination now are ranked second among the most frequent causes of transfusion-related fatalities in the US 2. Recent surveillance studies of PCs and PC transfusion recipients suggest a bacterial contamination rate of about 1 per 3,000 units, clinical sepsis in about 1 per 20,000 transfusions, and related fatalities in about 1 per 100,000 transfusions 3. As a result, PC transfusion-related transmission of bacteria is now considered to be the most common infectious risk from PC transfusion worldwide 2,4,5. Associated morbidity and mortality most often occur at the end of the PC’s shelf life reflecting higher bacterial loads from the longer storage time of the contaminated unit 6,7. More than 20 different species of Gram-negative and Gram–positive bacteria including anaerobic species have been found to contaminate PC units 8–12. There is unanimous agreement among transfusion scientists and medical professionals that an ideal sterility test should be rapid, sensitive for all bacteria, affordable, simple to perform, and compatible with the workflow at a blood center or hospital.
Automated bacterial blood culturing systems fulfill many of these requirements because a wide range of species can be detected at concentrations of only 1 to 10 CFUs/ml 13. Unfortunately, despite high sensitivity, culture-based techniques are not rapid, not all can detect anaerobic species, and recently published reports indicate an unsettling number of false-positive and false-negative results caused by sampling errors 14,15.
Different approaches for detecting bacteria in PC that have been developed or are under development including the detection of bacterial growth by a variety of methods 16–19. Molecular techniques based on detecting bacterial nucleic acids have also been developed 20. Sensitivities as low as 30 CFUs/ml have been reported using real-time PCR 21. Recently, Verax Biomedical has received FDA approval for an assay intended for point-of-issue (POI) use to detect bacteria in both pooled and single-donor platelets units by detecting bacterial cell wall antigens common for Gram-positive and -negative organisms. With the availability of a FDA approved rapid method for pooled whole blood derived platelets, test procedures known to have poor sensitivity including platelet swirling and measurements of pH or glucose levels22 are no longer considered to satisfy AABB requirements 23. Even when routine quality control (QC) testing is used with all of these methods, septic reactions and fatalities linked to contaminated platelets continue to occur 15.
While pathogen inactivation holds the promise of a complete solution for sterilizing PCs, no such system has been approved for use in the US to date. Despite the INTERCEPT Blood System for platelets 24 (Cerus Corp.) having received CE Mark approval in Europe, the FDA continues to express concerns about the possible detrimental effects on the hemostatic properties of treated platelets 25 along with issues of patient safety. Consequently, in the US, the safety of platelet transfusion continues to rely exclusively on effective testing.
Direct Detection of Stressed Bacteria in Platelet Concentrate
It is well-documented that certain chemical compounds which affect the bacterial cell wall, translational/transcriptional machinery, or the cellular genome trigger a measurable stress response in susceptible bacterial cells 26,27. Because the activation of the stress response does not depend on the growth rate of the organism, bacterial detection can be obtained in real-time with the availability of an appropriate sensing modality to monitor the response of intentionally stressed bacterial cells. In our previous report, we described a new approach for detecting bacteria in PC by monitoring the development of stress in bacteria during exposure to Triton X-100 using differential impedance sensing 1. The method employed Triton X-100 to lyse all human cells in PC as well as stress bacterial cells and required a filtration step to remove cellular fragments while passing not filtering out the bacterial cells. Data were presented showing that concentrations as low as 103 CFUs/mL of the representative Gram-negative and Gram-positive aerobic and anaerobic species E. coli, S. epidermidis, and P. acnes growing in PC could be detected within 30 minutes.
In this latest work we have simplified the sample preparation and eliminated the need for any lysis or filtration steps reducing the sample preparation time to less than 3 minutes. In the current assay, the PC sample is combined with pre-mixed reagents prior to measurement significantly reducing the complexity and number of preparation steps needed and is amenable to simple automated operation.
The overall detection method is based on two discoveries. We have found that the bacterial response to stress causes specific changes to the dielectric permittivity of a suspension. These changes can be easily monitored using differential impedance sensing enabling the ubiquity and immediacy of the stress response to be exploited. The further inclusion of differential sensing ensures that the resulting dielectric permittivities are restricted to bacterial stress-specific signals only and hence, their detection in complex platelet samples.
The dielectric permittivity is a measure of the overall polarizability of both inorganic and organic molecules within the cell suspension. Cellular activities affecting polarizability include a decrease in regular protein synthesis and an increase in the synthesis of stress response proteins, changes in the cell membrane potential and associated ion concentrations, inhibition of DNA replication, changes to DNA and protein conformational structures, and other processes associated with the conservation of energy and the restoration of damages in the cells quest to survive. However, the exact contribution to the dielectric properties of the multitude of intra- and extra-cellular biochemical interactions remains an open question.
Second, we have identified a chemical stressor to which bacteria growing in PC are susceptible and produce a significant change in dielectric permittivity while human cells show no measurable response over relevant experiment times. The identification of a discriminating compound enables the full capabilities of differential sensing to be utilized and the detection of clinically relevant numbers of contaminant bacteria directly in PC without the need for a lysis step.
To date, as many as 50 different sterile PC units (both whole-blood derived and apheresis) have been monitored during exposure to 100 uL/mL of STR-103 without detecting any measurable response. Although these data are consistent, the small number of PC units used in this study is inadequate to establish a corresponding false-positive rate because of limitations in both sample diversity and statistics and a larger study is needed.
All PC units infected with as low as 103 CFUs/mL of the 17 bacterial species tested could be distinguished from sterile control PC without concentrating the sample in well under 30 minutes. The ability to detect these low numbers of bacteria without a concentration step eliminates the need for centrifugation and a potential obstacle to the practical implementation of the rapid methods in blood centers or hospitals.
The clinical significance of an assay capable of detection 103 CFUs/mL or more is the potential avoidance of all serious reactions and 95% probability of avoiding of all possible reactions encountered from transfusing PC contaminated at these bacterial burdens 28.
We note that in a clinical setting, the negative control for all tests would be inferred from a database of previous measurements of sterile PC and continuously updated by the overwhelming number of negative test results. As a practical matter, the development of a standard for routine use as a positive control would be prudent to ensure the proper daily operation.
While test concentrations of 103 CFUs/mL were targeted for all species, uncertainty in the growth of the different bacterial species in the different PC units resulted in actual test concentrations that sometimes varied by as much as a factor of ten as seen in Table 1. Although this study was not designed to determine the limits of detection, the NIR profiles for L. monocytogenes suggest that the overall limit is less than 103 CFUs/mL. Future experiments are planned to investigate if this detection threshold is specific to the susceptibility of L. monocytogenes or is characteristic of the overall sensitivity inherent in the technical approach.
Summary and conclusions
The development of stress is an immediate event in cellular life, contrary to the slow process of population growth. Because the activation of the stress response does not depend on the growth rate, Gram straining characteristics, or culturing requirements of a bacterium, there is significant potential for its use as a truly rapid diagnostic tool in applications where a broad range of bacterial species is encountered. The use of differential impedance sensing, a simple physical method capable of registering the development of bacterial stress response and precisely separating the signals produced by stressed bacteria from other signals in a complex biological system, is an ideal sensing modality for detecting contaminant bacteria in platelets. When combined with the use of a stressor compound that discriminates between human and bacterial cells, the approach is rapid, operationally simple to implement, and capable of detecting all bacteria below clinically relevant concentrations needed for point-of-issue testing.28
Acknowledgments
Sources of Support:
National Heart, Lung, and Blood Institute: Grant 9R44HL090636
Funding for this work was provided by a grant from the National Heart, Lung, and Blood Institute at the National Institutes of Health (9R44HL090636).
Footnotes
Both Dr. Ronald Rieder and Dr. Boris Zavizion are stockholders in BioSense Technologies, Inc.; Dr. Zhihui Zhao and Aphakorn Nittayajarn have no conflicts of interest to report.
References
- 1.Rieder RJ, Zavizion B. Monitoring the physiological stress response: a novel biophysical approach for the rapid detection of bacteria in platelet concentrates. Transfusion. 2008;48:2596–605. doi: 10.1111/j.1537-2995.2008.01880.x. [DOI] [PubMed] [Google Scholar]
- 2.Rao P, Strausbaugh L, Liedtke L, et al. Bacterial infections associated with blood transfusion: experience and perspective of infectious diseases consultants. Transfusion. 2007;47:1206–11. doi: 10.1111/j.1537-2995.2007.01269.x. [DOI] [PubMed] [Google Scholar]
- 3.Walther-Wenke G. Incidence of bacterial transmission and transfusion reactions by blood components. Clin Chem Lab Med. 2008;46:919–25. doi: 10.1515/CCLM.2008.151. [DOI] [PubMed] [Google Scholar]
- 4.Ezuki S, Kawabata K, Kanno T, Ohto H. Culture-based bacterial detection systems for platelets: the effect of time prior to sampling and duration of incubation required for detection with aerobic culture. Transfusion. 2007;47:2044–9. doi: 10.1111/j.1537-2995.2007.01428.x. [DOI] [PubMed] [Google Scholar]
- 5.Mohr H, Lambrecht B, Bayer A, et al. Basics of flow cytometry-based sterility testing of platelet concentrates. Transfusion. 2006;46:41–9. doi: 10.1111/j.1537-2995.2005.00668.x. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs M, Good C, Lazarus H, Yomtovian R. Relationship between bacterial load, species virulence, and transfusion reaction with transfusion of bacterially contaminated platelets. Cli Infect Dis. 2008;46:1214–20. doi: 10.1086/529143. [DOI] [PubMed] [Google Scholar]
- 7.Lee J. Workshop on Bacterial Contamination of Platelets; CBER, FDA; Sep 24, 1999. http://www.fda.gov/cber/minutes/workshop-min.htm. [Google Scholar]
- 8.Kuehnert M, Roth V, Haley N, et al. Transfusion-transmitted infection in the United States, 1998 through 2000. Transfusion. 2001;41:1493–9. doi: 10.1046/j.1537-2995.2001.41121493.x. [DOI] [PubMed] [Google Scholar]
- 9.Wagner S, Friedman L, Dodd R. Transfusion-associated bacterial sepsis. Clin Microbiol Rev. 1994;7:290–302. doi: 10.1128/cmr.7.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrezenmeier H, Walther-Wenke G, Müller T, et al. Bacterial contamination of platelet concentrates: results of a prospective multicenter study comparing pooled whole blood-derived platelets and apheresis platelets. Transfusion. 2007;47:644–52. doi: 10.1111/j.1537-2995.2007.01166.x. [DOI] [PubMed] [Google Scholar]
- 11.Boekhorst P, Beckers E, Vos M, et al. Clinical significance of bacteriologic screening in platelet concentrates. Transfusion. 2005;45:514–9. doi: 10.1111/j.0041-1132.2005.04270.x. [DOI] [PubMed] [Google Scholar]
- 12.Sazama K. Bacteria in blood for transfusion. Arch Pathol Lab Med. 1994;118:350–65. [PubMed] [Google Scholar]
- 13.Brecher M, Means N, Jere C, et al. Evaluation of an automated culture system for detecting bacterial contamination of platelets: an analysis of 15 contaminating organisms. Transfusion. 2001;41:477–82. doi: 10.1046/j.1537-2995.2001.41040477.x. [DOI] [PubMed] [Google Scholar]
- 14.Benjamin R, Wagner S. The residual risk of sepsis: modeling the effect of concentration on bacterial detection in two-bottle culture system and an estimation of false-negative culture rates. Transfusion. 2007;47:1381–9. doi: 10.1111/j.1537-2995.2007.01326.x. [DOI] [PubMed] [Google Scholar]
- 15.Eder A, Kennedy J, Dy B, et al. Bacterial screening of apheresis platelets and the residual risk of septic transfusion reactions: the American Red Cross experience (2004–2006) Transfusion. 2007;47:1134–42. doi: 10.1111/j.1537-2995.2007.01248.x. [DOI] [PubMed] [Google Scholar]
- 16.Depcik-Smith ND, Hay S, Brecher M. Bacterial contamination of blood products: factors, options, and insights. J Clin Apheresis. 2001;16:192–201. doi: 10.1002/jca.10004. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell K, Brecher M. Approaches to the detection of bacterial contamination in cellular blood products. Transfus Med Rev. 1999;13:132–44. doi: 10.1016/s0887-7963(99)80008-x. [DOI] [PubMed] [Google Scholar]
- 18.Wagner S, Friedman L, Dodd R. Transfusion associated bacterial sepsis. Clin Microbiol Rev. 1994;7:290–302. doi: 10.1128/cmr.7.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yomtovian R. Novel methods for detection of platelet bacterial contamination. Vox Sang. 2002;38:129–31. doi: 10.1111/j.1423-0410.2002.tb05285.x. [DOI] [PubMed] [Google Scholar]
- 20.Petershofen E, Fislage R, Faber R, et al. Detection of nucleic acid sequences from bacterial species with molecular genetic methods. Transfus Sci. 2000;23:21–7. doi: 10.1016/s0955-3886(00)00051-5. [DOI] [PubMed] [Google Scholar]
- 21.Sen K. Rapid identification of Yersinia enterocolitica in blood by the 5′ nuclease PCR assay. J Clin Microbiol. 2000;38:1953–8. doi: 10.1128/jcm.38.5.1953-1958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner S, Robinette D. Evaluation of swirling, pH, and glucose tests for the detection of bacterial contamination in platelet concentrates. Transfusion. 1996;36:989–93. doi: 10.1046/j.1537-2995.1996.36111297091744.x. [DOI] [PubMed] [Google Scholar]
- 23.Association bulletin 10-05 (August 19, 2010) Bethesda, MD: AABB; 2010. Suggested Options For Transfusion Services and Blood Collectors to Facilitate Implementation of BB/TS Interim Standard 5.1.5.1.1. [Available at < http://www.aabb.org/resources/publications/bulletins/Pages/ab09-04.aspx>] [Google Scholar]
- 24.Knutson F, Alfonso R, Dupuis K, et al. Photochemical inactivation of bacteria and HIV in buffy-coat derived platelet concentrates under conditions that preserve in vitro platelet function. Vox Sang. 2000;78:209–16. doi: 10.1159/000031183. [DOI] [PubMed] [Google Scholar]
- 25.Picker S, Schneider V, Gathof B. Platelet function assessed by shear-induced deposition of split triple-dose apheresis concentrates treated with pathogen reduction technologies. Transfusion. 2009;49:1224–32. doi: 10.1111/j.1537-2995.2009.02092.x. [DOI] [PubMed] [Google Scholar]
- 26.Kannan G, Wilks J, Fitzgerald D, et al. Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol. 2008;8:37–50. doi: 10.1186/1471-2180-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominguez-Cuevas P, Gonzalez-Pastor J, Marques S, et al. Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J Biol Chem. 2006;281:11981–91. doi: 10.1074/jbc.M509848200. [DOI] [PubMed] [Google Scholar]
- 28.Yomtovian R, Palavecino E, Dykstra A, et al. Evolution of surveillance methods for detection of bacterial contamination of platelets in a university hospital, 1991–2004. Transfusion. 2006;46:719–30. doi: 10.1111/j.1537-2995.2006.00790.x. [DOI] [PubMed] [Google Scholar]