Abstract
Helicases catalytically unwind structured nucleic acids in a nucleoside-triphosphate-dependent and directionally specific manner, and are essential for virtually all aspects of nucleic acid metabolism. ATPase-driven helicases which translocate along nucleic acids play a role in damage recognition or unwinding of a DNA tract containing the lesion. Although classical biochemical experiments provided evidence that bulky covalent adducts inhibit DNA unwinding catalyzed by certain DNA helicases in a strand-specific manner (i.e. , block to DNA unwinding restricted to adduct residence in the strand the helicase translocates), recent studies suggest more complex arrangements that may depend on the helicase under study, its assembly in a protein complex, and the type of structural DNA perturbation. Moreover, base and sugar phosphate backbone modifications exert effects on DNA helicases that suggest specialized tracking mechanisms. As a component of the replication stress response, the single-stranded DNA binding protein Replication Protein A (RPA) may serve to enable eukaryotic DNA helicases to overcome certain base lesions. Helicases play important roles in DNA damage signaling which also involve their partnership with RPA. In this review, we will discuss our current understanding of mechanistic and biological aspects of helicase action on damaged DNA.
Keywords: helicase, DNA damage, DNA repair, replication, Werner syndrome, FANCJ, Fanconi Anemia, Replication Protein A, Xeroderma pigmentosum, nucleotide excision repair
INTRODUCTION
Helicases are molecular motor proteins that couple the hydrolysis of nucleoside triphosphate to nucleic acid unwinding (for review, please see refs. 1–4). Enzymes of this class function coordinately with other proteins as complex machines and play essential roles in pathways of DNA metabolism that include replication, DNA repair, recombination, transcription, and chromosome segregation. Despite considerable efforts to understand biochemical, structural, and genetic aspects of helicase function, the precise mechanisms by which helicases catalyze strand separation and perform their biological roles remain to be fully understood. The growing number of DNA helicases implicated in human disease suggests that these enzymes have vital specialized roles in cellular pathways important for the maintenance of genome stability.
An important aspect of helicase function in a biological context is its interactions with damaged DNA molecules. Literally all DNA metabolic processes are affected by DNA damage. For example, both DNA replication and transcription can be blocked or their fidelity compromised by certain forms of DNA damage. 5–8 DNA damage evokes a cellular response by a genome surveillance system that senses DNA structural perturbation at the site of the lesion and elicits an appropriate reply that may involve direct repair of the lesion, stabilization of the replication fork, or induction of apoptosis. Sophisticated mechanisms and pathways have evolved to handle DNA damage. Indeed, a variety of DNA repair proteins have the ability to recognize poor base stacking induced by a spectrum of DNA lesions. 9 The unique ability of helicases to catalytically separate DNA strands from one another may be utilized to tolerate or repair damaged DNA. Helicases from simple bacteria to mammalian cells participate in the DNA damage response and DNA repair to maintain genome homeostasis and proper cellular function.
In this review, we will discuss the interactions of helicases with DNA damage to provide the reader a sense of how this information has been used to better understand helicase mechanism, structure-function relationships, and their pathway functions. Some specific examples of the roles of helicases in DNA damage recognition that provide a model for future studies will be discussed. We will address studies on the ATPase/helicase subunits of the TFIIH particle that are implicated in DNA repair of bulky adducts which suggest a complex mechanism of DNA damage recognition and creation of an opened DNA damage substrate. The effects of structurally defined intercalating covalent polycyclic aromatic hydrocarbons on the DNA unwinding function catalyzed by human RecQ helicases will be discussed. The importance of base contacts versus interactions with the sugar phosphate backbone for Superfamily (SF)1 and SF2 helicases will be presented. Effects of base damage on helicase function will be discussed and a mechanism for stimulation of helicase activity past a helicase blocking lesion by a protein partner is proposed. Lastly, the importance of helicases in early checkpoint signaling when a replication fork encounters a blocking lesion will be discussed. Recent work in these areas suggests that helicases have unique functions in DNA damage detection, signaling, and processing.
DNA Damage Recognition by the UvrB Helicase of the UvrABC Nucleotide Excision Repair Protein Complex
In prokaryotes, the DNA damage recognition and incision steps of nucleotide excision repair (NER) are mediated by the proteins UvrA, UvrB, and UvrC. A second helicase known as UvrD is responsible for unwinding the 12 nt tract containing the lesion after the incisions have occurred. Several excellent reviews (see refs. 10–12) have summarized the details of the UvrABC NER system. UvrA in the form of an A2B (or A2B2) complex first scans DNA to recognize altered base pair conformations, enabling UvrB to search for the precise site of damage. UvrB searches for the exact position of the lesion. Once DNA damage recognition is verified by UvrB in a reaction dependent on UvrB ATP hydrolysis, UvrA departs and UvrB becomes stably bound to the lesion with the DNA wrapped around UvrB, 13 facilitating localized melting around the site of DNA damage. In this complex the C-terminal domain of UvrB is readily exposed and can interact with UvrC. The resulting UvrBC-complex is capable of performing dual incision, first on the 3’-side and next on the 5’-side of the lesion.
UvrB plays a critical role in the discrimination of the lesion. Elegant structural studies and mutational analyses have provided tremendous insight at the molecular level of how DNA damage recognition is mediated by UvrB (for a detailed and comprehensive review, see ref. 12). It is generally believed that the NER system uses the UvrAB complex to catalyze ATP-dependent strand opening activity which is instrumental for the DNA damage search. Mutational studies showed that the flexible β-hairpin of UvrB, which extrudes from domain 1a (involved in ATP binding) and contacts domain 1b (implicated in DNA binding), contains several highly conserved hydrophobic residues, and these β-hairpin hydrophobic residues are important for the damage recognition. The aromatic residues in the tip of the β-hairpin are important for the strand separating activity of UvrB. 14 Once the DNA damage is located by the UvrAB complex, UvrB uses its ATPase activity to place the DNA in a strained conformation that can be subsequently recognized and incised by UvrC. The role of UvrB in opening damaged DNA may serve as an excellent model for understanding the role of XPD in eukaryotic NER 12 (discussed below).
In recent work, insight into the structural basis for DNA recognition and processing by UvrB was obtained from UvrB complexed with DNA. 15;16 The β-hairpin of UvrB inserts between the strands of the double helix, providing a mechanism to lock down one of the two strands. A model was proposed in which during UvrB translocation, each consecutive nucleotide of one strand is flipped behind the β-hairpin into a small hydrophobic pocket. Base flipping by UvrB plays an important role in optimal 3’ incision by UvrC. 17;18 Modeling of the UvrB interaction with damaged DNA based on studies with benzo[a]pyrene-derived lesions is consistent with a common recognition mechanism that involves UvrB base flipping. 19
Strand-Specific Effects of Bulky DNA Damage on Helicase-Catalyzed DNA Unwinding
Strand-specific inhibition of unwinding activity by DNA lesions has been observed for DNA helicases involved in DNA replication (for a review, see ref. 20). The 1–2 intrastrand d(GpG) cross-link inducedby the antitumor drug cis-diamminedichloroplatinum(II) significantly reduced the unwinding activity of the Herpes simplex virus type1-replicative helicase UL9 only when it was present on the strand along which the protein translocates. 21 The gene 4 protein that is essential for T7 viral replication translocates 5’–3’ along single-stranded DNA, and this movement is blocked by the bulky DNA adducts derived from benzo[a]pyrene (BaP). 22 BaPDNA adducts only inhibited gene 4 helicase activity if they resided in the strand on which the helicase translocates. 23 T7 gene 4 was sequestered by the translocating strand lesion and formed a stable complex with the modified DNA.
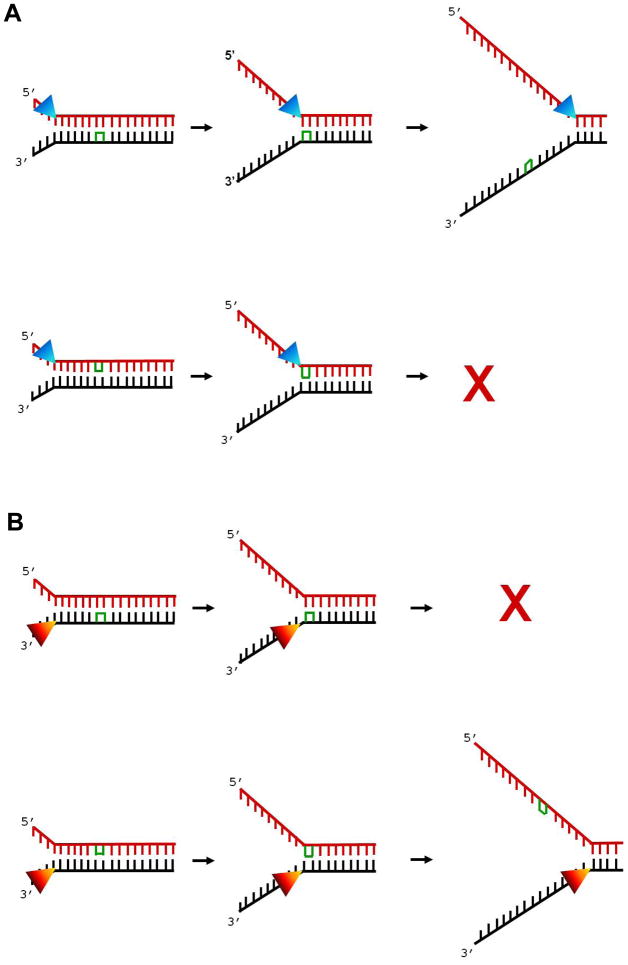
Some experiments with DNA helicases involved in DNA repair have suggested strand-specific inhibition of unwinding activity. Calf thymus DNA helicase E could displace a primer containing a GpG intrastrand cross-link; however, its unwinding was blocked if the lesion resided in the translocating strand in the single-stranded template but not if it was located in the annealed duplex region. 24 DNA unwinding by the purified RecB helicase, a subunit of the E. coli RecBCD complex implicated in double strand break repair by homologous recombination, was inhibited by cisplatin (Pt-d(GpG)) damage introduced on the translocating strand of a partial duplex substrate. 25 RecB helicase activity was not inhibited by the cisplatin damage introduced on the non-translocating strand. These in vitro studies suggest that for a number of helicases, a bulky DNA lesion such as an intrastrand cross-link within a single strand compromises unwinding activity by preventing protein translocation on the strand containing the adduct (Fig. 1).
Fig. 1. Strand-specific inhibition of DNA helicase activity.
A DNA lesion can impede unwinding of double-stranded DNA by a helicase in a strand-specific manner such that inhibition is only observed when the lesion resides in the strand that the helicase translocates. In Panel A, DNA unwinding by a 5’ to 3’ helicase is only impeded by a lesion (intrastrand cross-link) in the top strand. In Panel B, DNA unwinding by a 3’ to 5’ helicase is only impeded by a lesion in the bottom strand. Although this is the case for certain helicases and DNA lesions, there are exceptions to the rule. See text for details. Intrastrand cross-link indicated in green; helicase indicated by triangle.
The RAD3 encodes a DNA-dependent ATPase and 5’ to 3’ DNA helicase of the TFIIH complex required for NER in S. cerevisiae. 26;27 In a classic series of experiments from the Friedberg lab, it was demonstrated that the DNA helicase and ATPase activities of purified yeast Rad3 protein were inhibited in a strand-specific manner by UV-induced photoproducts, cisplatin-induced bulky adducts, or abasic sites. 28–30 Biochemical studies demonstrated that Rad3 was trapped at the sites of these lesions and formed stable complexes with the damaged DNA. It was proposed that the inhibition of Rad3 helicase activity by damaged DNA might be important for damage-specific incision of DNA during NER. Strand-specific inhibition of Rad3 helicase activity by a bulky DNA adduct was proposed to contribute to protein-DNA recognition of the lesion. However, molecular analyses of the XPB and XPD helicase-like proteins which are components of the mammalian TFIIH complex suggest an even greater level of sophistication for opening damaged DNA (see next section).
Analysis of XPB and XPD Implicated in Opening Damaged DNA During Nucleotide Excision Repair
During the initial DNA damage recognition steps of NER, the damaged DNA must be locally unwound or opened to set up the subsequent steps of nucleolytic incisions in the ssDNA tract containing the lesion upstream and downstream of the damage. In human NER, the helicase-like proteins of the TFIIH complex responsible for opening damaged DNA are XPB and XPD. Mutations in either XPB or XPD are genetically linked to the skin cancer disorder Xeroderma pigmentosum (XP) which is defective in NER of UV-induced DNA photoproducts. However, XPB or XPD as components of the TFIIH complex are also important in promoter opening during transcription initiation, and mutations in either helicase are genetically linked to diseases with transcription defects: XP combined with Cockayne syndrome (XP/CS), or the brittle hair disease Trichothiodystrophy (TTD). 31;32
There has been much interest in understanding the precise molecular roles of XPB and XPD in mammalian NER. How these helicases scan or verify DNA damage and permit the subsequent steps of NER is an active area of investigation. Interestingly, the two helicases display opposite directionality with XPB translocating in the 3’ to 5’ direction whereas XPD translocates in the 5’ to 3’ direction. 33;34 Presumably, these helicases must coordinate their translocase activities since they reside within the same TFIIH protein complex. Mutation of the highly conserved Walker A box that abolishes both ATPase and helicase activity of yeast XPB (RAD25) or XPD (RAD3) results in defective NER, 35;36 consistent with the hypothesis that these proteins are involved in creating a bubble around the lesion site in an ATPase-dependent manner. In an elegant set of experiments, Coin and colleagues demonstrated distinct roles of the XPB and XPD helicases in opening damaged DNA during NER. 33 They discovered that XPB helicase activity, but not ATPase activity, was dispensable for NER, whereas XPD helicase activity is required for NER. Thus, XPB does not behave as a conventional DNA unwinding enzyme (helicase) in NER. Rather, the authors propose that XPB causes local melting of the double-stranded DNA around the lesion, enabling XPD to anchor at the site and unwind the DNA to create a bubble of ~27 nt for incision of the damaged strand. XPB may also operate to help locally melt dsDNA around the promoter during transcription initiation. 37 Alternatively, it was proposed that XPB helicase activity may be important for promoter escape in transcription; 38 however, this remains to be experimentally determined. Curiously, the helicase function of XPD is required for DNA damage opening during NER, but is not required for transcription. 33;39;40
The Hanaoka lab demonstrated that NER has the potential to search for lesions over a distance of at least 150 bases from the site where the XPC complex initially detects unpaired bases in the vicinity of the lesion. 41 DNA damage verification involves scanning the DNA from the initial detection site. This search appears to be conducted with a 5’-to-3’ directionality, suggesting a scanning mechanism along a specific DNA strand. The authors proposed that XPD helicase as a component of TFIIH is mainly responsible for this damage search by translocating 5’ to 3’ with respect to the strand that it is bound. 41 The efficiency of NER incision can be greatly affected by the binding orientation of XPC with respect to the lesion, which dictates the strand that is searched for DNA lesions. A two–step model for the DNA damage recognition phase of NER was proposed: 1) directional binding of the XPC complex with unpaired bases in the undamaged strand; and 2) loading of TFIIH and scanning of the damaged strand by XPD in the 5’ to 3’ direction. 41
An Exception to the Rule of Strand-Specific Inhibition of Helicase Activity by a DNA Adduct
The DNA substrate specificity of purified recombinant S. acidocaldarius XPD and its action on damaged DNA substrates was recently investigated by the White laboratory. 42 A key finding of the study was that helicase activity catalyzed by purified XPD was remarkably insensitive to the presence of DNA modifications, such as abasic sites, bulky extrahelical fluorescein adducts, or a cyclobutane pyrimidine dimer (CPD), regardless of whether these modifications were placed on either the displaced or translocated strands. These observations challenge the dogma that DNA lesions repaired by NER are recognized and acted upon in part by their ability to block DNA translocation and unwinding by the XPD helicase. However, it is conceivable that the effect of a given lesion recognized by NER machinery on purified XPD helicase may be distinct from the effect it has on XPD helicase existing within the TFIIH particle. Inhibition of XPD helicase activity by DNA binding proteins (e. g. , Alba1)42 led the authors to suggest a model in which further translocation by XPD may be blocked by XPC complex bound to the DNA on the 3’ side of the lesion. Alternatively, XPD helicase operating within the context of the TFIIH particle may halt its translocation once XPB is anchored on the opposite side of the lesion. Either of these models is consistent with the observations of the White lab that purified XPD can traverse past a lesion recognized by NER.
Can Helicases with Opposite Translocation Polarities Move Together in the Same Direction on the DNA Duplex to Scan for Damage?
Although the opposite translocation polarities of XPB and XPD may suggest a model in which they move in an ATP-driven manner on the same strand in opposite directions to open the duplex around the lesion, it is conceivable that XPB and XPD helicases translocate in the same direction on opposite strands during DNA strand scanning and opening of the damaged DNA duplex. This model of dual helicases moving together is reminiscent of the E. coli RecBCD complex in which helicases (RecB, RecD) of opposite translocation polarities move together on opposite strands of the DNA duplex. 43 This bipolar DNA translocation mechanism facilitates a process in which the RecBCD enzyme operates as a helicase-nuclease to initiate the repair of double-stranded DNA breaks by homologous recombination. It would be highly informative to determine if the XPB and XPD helicases within the TFIIH complex operate in an analogous manner during the damaged DNA recognition/opening steps of NER or during initiation of transcription. On a broader level, the coordinate action of two helicases with opposite translocation polarities may provide a unique mechanism for processing steps of DNA repair or a genome surveillance mechanism in which the protein complex can simultaneously scan both strands of duplex DNA for lesions (Fig. 2). A protein complex containing the 3’ to 5’ Bloom’s syndrome helicase (BLM) and Fanconi anemia (FA) proteins was identified, 44 and the FANCM ATP-dependent translocase mediates the interaction between the BLM and FA core complexes; 45 however, a biochemically functional interaction between BLM helicase and a 5’ to 3’ translocase/helicase is yet to be shown.
Fig. 2. DNA damage surveillance by a dual motor helicase complex.
A hypothetical model for how two helicases with opposite directionalities of translocation might function together to scan double-stranded DNA for damage in either strand. See text for details. Red oval, 5’ to 3’ helicase; green oval, 3’ to 5’ helicase; DNA damage indicated by yellow sunbursts.
Insight to DNA Damage Recognition by Crystal Structures of XPB and XPD
Recently, three groups solved the XPD crystal structure from archaea Sulfolobus acidocaldarius (SaXPD), 46 Sulfolobus tokodaii (StXPD), 47 and Thermoplasma acidophilum (TaXPD). 48 The XPD crystal structures revealed two highly conserved RecA domains with a central interface for ATP binding and hydrolysis, an Iron-Sulfur domain that plays a structural role to stabilize the enzyme and serve as a wedge to physically separate the DNA duplex strands, and an Arch domain that together with the other domains forms an enclosed tunnel. A model was proposed in which DNA damage is recognized by passing one of the two strands through a central hole where a narrow pocket in the wall of the central hole excludes the bulky lesion. Details of the readout mechanism for DNA damage recognition require further study. Nonetheless, the XPD crystal structures have provoked a renewed interest in deciphering the mechanism whereby a helicase might use its structural domains to catalyze DNA unwinding or even facilitate DNA damage recognition. This area of investigation is likely to blossom with the determination of crystal structures of DNA lesions with helicases as occurred in the DNA polymerase field. 49;50 Intriguingly, superimposing the positions of the clinically relevant XPD mutations on the XPD structure has provided some insight to the genotype-phenotype relationships. 46;51
The Archaeoglobus fulgidus (Af) XPB crystal structure revealed several domains of interest. 52 An N-terminal DNA damage recognition domain (DRD) that displays the highest structural similarity with the mismatch recognition domain (MRD) of the bacterial mismatch recognition protein MutS was identified. Helicase domains 1 and 2 (HD1, HD2) retained a RecA topology. An XPB-unique motif resides adjacent to motif III in the helicase core with RED residues in the motif conserved in XPB homologs from archaea to human. Mutagenesis studies demonstrated that the integrity of the RED motif is important for AfXPB helicase activity. Lastly, a positively charged Thumb Motif was identified in AfXPB which possesses structural similarities with the thumb domain of T7 DNA polymerase and Taq polymerase that are proposed to be important for DNA binding.
Biochemical studies demonstrated that purified AfXPB specifically binds dsDNA containing a single tetrahydrofuran as an abasic nucleotide. 52 Furthermore, it was shown that AfXPB preferentially unwinds a blunt duplex DNA molecule containing a single cyclobutane pyrimidine dimer (CPD) or (6–4) photoproduct compared to undamaged duplex DNA. It was proposed that the binding of damaged DNA by the DRD is important for XPB to open the 30 nucleotide bubble required for NER. As suggested by the authors, it is conceivable that the preferential binding of XPB to damaged DNA plays a role in switching TFIIH from transcription to NER, and/or may play a role in the incision of the damaged DNA strand during NER.
A Role for XPB to Recruit TFIIH Complex to Damaged DNA
To better understand the molecular functions of XPB and XPD helicases for the role of TFIIH in nucleotide excision repair, Oksenych and co-workers further examined the importance of ATP hydrolysis catalyzed by XPB or XPD. 53 Using genetically reconstituted cell lines with site-specific mutations in the ATPase Walker A motif, it was observed that a TFIIH complex defective in XPB ATPase activity was not recruited to sites of DNA damage, whereas a TFIIH complex defective in XPD ATPase was recruited. Interestingly, TFIIH recruitment to damaged DNA did not require XPB helicase activity but did depend on the integrity of the RED motif and Thumb motif (mentioned in the previous section), and these motifs were essential for DNA-stimulated ATPase activity of XPB. Foot printing experiments revealed that the RED and Thumb motifs play an important role in opening the DNA around the lesion. The authors proposed that XPB functions as an ATP-dependent hook that uses the ATPase, RED and Thumb motifs to anchor TFIIH to sites of DNA damage during repair.
Collectively, these findings are significant because they provide experimental evidence that the XPB motor ATPase function, uncoupled from helicase activity, plays an important role in DNA repair. An important unanswered question is how the appropriate strand, the damaged one, is acted upon by the NER endonucleases, and what role the XPB and XPD helicases have in conferring strand specificity. It is also debatable if the XPB translocase activity is required for its role in NER or transcription since it is not well understood if the XPB mutant with an intact ATPase activity and proper function in NER 33 is able to efficiently translocate on DNA. A translocase-defective version of XPB is likely to be impaired in its ability to scan damaged DNA; however, this particular type of XPB mutant to our knowledge has not been reported. Although there is not extensive literature on helicase mutants which are specifically defective in coupling ATP hydrolysis to translocase activity, there is one report that a helicase core domain mutation in SF1 PcrA helicase uncoupled ssDNA translocation from helicase activity. 54 An XPB mutant that fails to couple ATP hydrolysis to translocation would be useful to further dissect its role in NER.
The molecular analyses of XPB helicase suggest that certain helicases have roles in nucleic acid metabolism that are strictly dependent on their motor ATPase function, and not necessarily classic DNA unwinding. The ability of certain helicases to use their ATPase function to scan damaged DNA and/or locally melt damaged DNA and recruit protein complexes requires further investigation. As summarized nicely in a recent review, 38 the functional roles of XPB and XPD are regulated by their interactions with proteins that reside within the TFIIH particle as well as proteins extrinsic to the TFIIH complex. Clearly, as the field progresses, there will be an even greater appreciation of the motor ATPase-driven functions of helicase proteins that do not involve classic processive DNA unwinding in the form of strand separation. Moreover, advances in understanding how DNA opening or unwinding by the TFIIH helicases is tightly regulated by protein interactions will provide models for the studies of other DNA helicases, such as those of the RecQ family that are implicated in diseases of premature aging and cancer. 55
Orientation and Stereochemistry of Polycyclic Aromatic Hydrocarbons Influence WRN Helicase Inhibition
In this section, we will describe some biochemical studies of DNA helicases that suggest for certain forms of DNA damage, a lesion can have an effect on helicase-catalyzed DNA unwinding that is dependent on its stereochemistry and orientation of projection within the DNA double helix. This information is pertinent to not only mechanistic aspects of how helicases track along DNA during translocation and the unwinding reaction but also to the biological roles of DNA helicases in the DNA damage response and DNA repair.
Polycyclic aromatic hydrocarbons such as benzo[c]phenanthrene (BcPh) or benzo[a]pyrene (BaP) diol epoxide (DE) adducts that are abundant in our diet and the environment are both mutagenic and carcinogenic. 56 Understanding the basis for the adverse effects of polycyclic aromatic hydrocarbons on DNA metabolic processes has been of general interest in the field. These and other bulky DNA adducts potentially interfere with normal DNA replication by perturbing the functions of DNA polymerases and nucleic acid processing enzymes such as helicases. Work from the Romano lab characterized the ability of T7 gene 4 helicase to unwind DNA substrates prepared from oligonucleotides that had been modified in vitro by incubating with BaP DE. 22;23 These studies demonstrated strand-specific inhibition of gene 4 translocation and unwinding by BaP adducts.
We examined the effects of polycyclic aromatic hydrocarbons on helicase activity catalyzed by the human Werner syndrome (WS) protein (WRN). WS is a premature aging disorder that is defective in a protein with both helicase and exonuclease activities. 57 Thus the premature aging phenotypes of WS is likely to be a consequence of defective DNA metabolism, whereas other progeria syndromes such as Hutchinson-Gilford or Restrictive Dermopathy arise from mutations affecting lamin A production, which is required for normal nuclear and chromatin formation. 58 Indeed, WRN plays an important role in replication fork progression after DNA damage or replication fork arrest. 59 In addition, recent work suggests that WRN has a role in telomere metabolism, 60;61 gene expression, 62;63 and genomic stability through its interactions with topoisomerase(s). 64;65
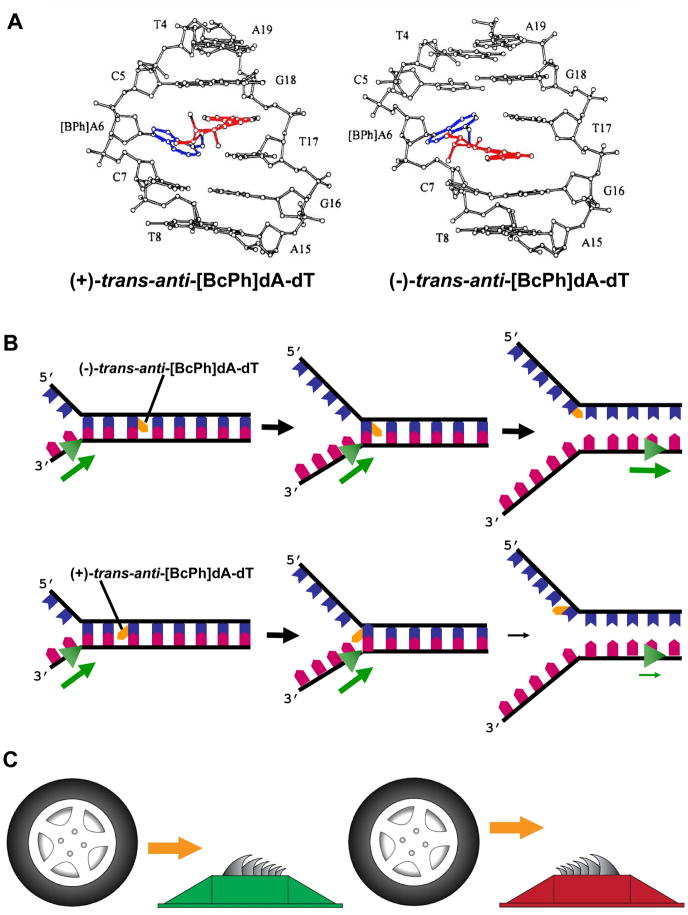
To address the effects of polycyclic aromatic hydrocarbons on WRN helicase, DNA substrates composed of oligonucleotides containing diastereomerically pure cis-and trans-opened BaP or BcPh DE adducts were used. 66;67 Results from helicase assays with the purified recombinant WRN helicase and forked duplex DNA substrates harboring a single BcPh dA adduct indicate that the orientation and stereochemistry of the intercalated adduct influenced its ability to inhibit WRN helicase activity. 67 The hydrocarbon portions of BcPh adducts intercalate into DNA on the 3’ or the 5’ side of the adducted deoxyadenosine for the (−)- and (+)-adducts, respectively (Fig. 3A). In the strand along which WRN translocates, cis-opened adducts were significantly more effective inhibitors than trans-opened isomers, indicating that WRN unwinding is sensitive to adduct stereochemistry. WRN helicase activity was also inhibited but to a lesser extent by cis-opened BcPh DE adducts in the displaced strand independent of their direction of intercalation, whereas inhibition by the trans-opened stereoisomers in the displaced strand depended on their orientation, such that only adducts oriented toward the advancing helicase inhibited WRN activity (see cartoon in Fig. 3B). The effect of trans-opened BcPh DE adducts in the displaced strand of the DNA substrate on WRN helicase activity is analogous to traffic spikes that permit an automobile to traverse from one direction, but not the other (Fig. 3C). Thus, WRN helicase activity is sensitive not only to the strand and stereochemistry of intercalated BcPh adducts, but also their orientation with respect to the strand they reside and the approach of the helicase.
Fig. 3. Benzo[c]phenathrenyl ring orientation in duplex DNA influences its ability to inhibit DNA unwinding by the advancing 3’ to 5’ WRN helicase.
Panel A, View looking into the major groove and normal to the helix axis of the solution structure of the central d(T4-C5-[BPh]A6-C7-T8)-d(A15-G16-T17-G18-A19) segment of Left, the (+)-trans-anti-[BcPh]dA-dT 11-mer duplex, and Right, the (−)-trans-anti-[BcPh]dA-dT 11-mer duplex. The benzo[c]phenanthrenyl moiety, shown in red bonds, is intercalating in the 5’-side direction of the lesion site between d(C5-[BPh]A6)-d(T17-G18) base pairs in the (+)-trans-anti isomer (Left) and in the 3’-side direction between d([BPh]A6-C7-d(G16-T17) base pairs in the (−)-trans-anti isomer (Right). The bonds of the adenine ring are shown in blue. Adapted from a figure that is reproduced by copyright permission from Biochemistry 1995, 34: 1295–1307. Panel B, model depicting effect of trans-opened BcPh dA stereoisomers in the displaced strand on WRN helicase activity that is dependent on their orientation, such that only adducts oriented toward the advancing helicase inhibited WRN unwinding activity. In figure, bold black arrow indicates efficient DNA unwinding past the BcPh lesion (orange polygon). Panel C, Traffic spikes used to control traffic provide a useful analogy for the orientation-specific effects of trans-opened BcPh dA stereoisomers on WRN helicase activity. If the tiger teeth are orientated toward the advancing automobile, they seriously deter further advance past the tire shredders. However, if the tiger teeth are orientated away from the advancing automobile, they allow access. See ref. 67 and text for details.
WRN helicase activity was also tested on DNA substrates that contained a single BaP DE-dG adduct within the duplex portion of the DNA substrate. 66 Depending on their stereochemistry, BaP DE-dG adducts can either occupy the minor groove without significant perturbation of DNA structure (trans adducts), or cause base displacement at the adduct site (cis adducts). 68;69 In contrast to what was observed for BcPh-dA adducts, 67 WRN helicase activity was found to be inhibited in a strand-specific manner by the BaP DE-dG adduct. Similar to WRN, human RECQ1 helicase was profoundly inhibited by a BaP DE-dG adduct in a strand-specific manner, suggesting that RecQ helicases may behave similarly upon encountering other types of intercalating DNA damage. A possible mechanism for the profound inhibition of WRN or RECQ1 helicase activity by the minor groove BaP DE-dG adduct is suggested by a conserved helix-turn-helix motif found in RecQ helicases that was shown to mediate minor groove binding in the human DNA repair protein O6-alkylguanine-DNA alkyltransferase. 70
Consistent with the idea that WRN helicase activity is particularly susceptible to minor groove perturbation, we demonstrated previously that WRN-catalyzed DNA unwinding is potently inhibited by the minor groove-binding drugs distamycin A and netropsin as compared with other DNA-binding drugs. 71 The BLM helicase mutated in the cancer disorder Bloom’s syndrome is also inhibited by minor groove binding drugs. 71 The adverse effects of biologically relevant DNA modifications, either covalent or noncovalent, on the catalytic activities of WRN and other RecQ helicases may be relevant to the genetic damage and cell transformation induced by the adducts and/or the mechanism of action of chemotherapeutic drugs such as distamycin analogs that position themselves in the minor groove.
WRN helicase activity was only mildly affected by intercalating BaP DE-dA adducts that locally perturb DNA double helical structure. 66 Partial unwinding of the duplex at BaP DE-dA adduct sites 72 may make such adducted DNAs more susceptible to the action of WRN helicase than DNA containing the corresponding BcPh DE-dA adducts, which cause little or no destabilization of duplex DNA. 73 Adverse effects of polycyclic aromatic hydrocarbons on WRN and other helicases could contribute to chromosomal damage and carcinogenesis induced by these DEs.
Disruption of Sugar Phosphate Backbone Integrity Negatively Affects DNA Unwinding by Superfamily 2 DNA Helicases
Helicases can be classified according to primary structure analyses which have revealed sequence similarities according to superfamilies or families of putative or bonafide ATP-dependent DNA unwinding enzymes (for review, see ref. 4). The two largest groups, SF1 and SF2, contain signature amino acid clusters or ‘motifs’ that comprise the helicase core domain. For SF1 helicases, the major contacts between the protein and DNA are with the bases; however, electrostatic interactions with the sugar phosphate backbone also exist. An unwinding mechanism known as the ‘Mexican Wave’ has been described for a SF1 helicase PcrA in which DNA bases are flipped out as they make hydrophobic contacts with aromatic amino acids of the helicase. 4 For SF2 helicases, the major contacts are thought to be with the phosphodiester backbone through ionic interactions with the helicase protein.
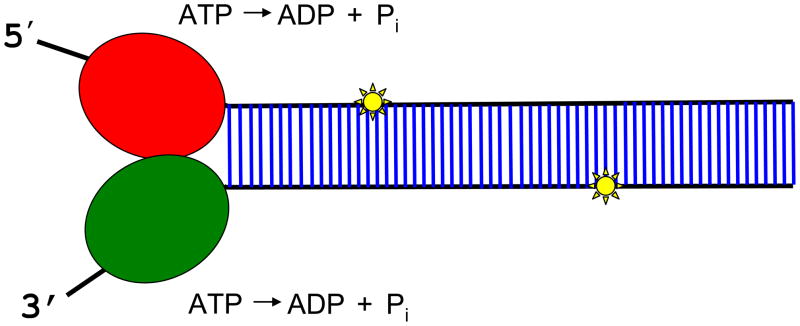
Biochemical studies have examined the effects of sugar phosphate backbone modifications such as polyglycol or polyvinyl synthetic linkages within either the single-stranded loading region or the duplex region of the DNA (or RNA) substrate on the unwinding activities catalyzed by SF1 and SF2 helicases. For example, the 18-atom polyglycol linker estimated to maximally extend up to 6 nt in length for ssDNA or 3 base pairs of duplex DNA has been an effective tool to examine the effect of disrupted backbone discontinuity on helicase mechanism, either the initiation 74;75 or elongation 76 phases of the unwinding reaction. The SF1 DNA helicase Rep efficiently unwound a DNA substrate harboring a polyglycol linkage in the single strand loadingregion adjacent to the duplex, suggesting a mechanism for initiation of DNA unwinding that is not passive and is consistent with a rolling mechanism during which Rep binds to ssDNA and dsDNA simultaneously. 74 In contrast, the SF2 5’ to 3’ helicase FANCJ (BACH1/BRIP1) was strongly inhibited by a polyglycol linkage in the 5’ ssDNA arm residing 6 nt away from the single-stranded: double stranded junction of a simple 5’ tailed DNA substrate or a forked duplex substrate. 77 Thus, FANCJ requires backbone continuity in the 5’ single stranded tail to efficiently translocate during initiation of unwinding.
Placement of the polyglycol linkage within either strand of the duplex region of the DNA substrate strongly inhibited FANCJ helicase activity, suggesting that the enzyme has a unique ability to sense backbone continuity in both strands of the DNA duplex during the unwinding reaction. 78 FANCJ partially unwound the backbone modified DNA substrates and became sequestered, indicating that FANCJ was not able to effectively unwind past the obstacle. However, by increasing the length of the 5′ ssDNA tail used for helicase loading, FANCJ was able to efficiently unwind the backbone-modified DNA substrates. This may be a consequence of the loading of numerous functionally active FANCJ helicase molecules on the DNA substrate with a longer 5′ ssDNA tail under multiple turnover conditions. Surprisingly, an increased motor ATPase of a FANCJ helicase domain variant (M299I) associated with breast cancer also enabled the helicase to unwind the backbone-modified DNA substrate in a more proficient manner. 78
The sensitivity of FANCJ to sugar phosphate backbone discontinuity in either strand of the duplex distinguishes this helicase from a number of other helicases (vaccina virus NPH-II 76, bacteriophage T4 Dda, 79;80, and T7 gene 4 81) that are inhibited by synthetic backbone linkers in a strand-specific manner, i.e. , only the strand the helicase is presumed to translocate. The human 3’ to 5’ helicase RECQ1 is also inhibited by the polyglycol linkage in a strand-specific manner (our unpublished data). In contrast, the hepatitis C virus NS3 RNA helicase (SF2) readily unwound RNA duplexes that contained long stretches of polyglycol linkages in either translocating or non-translocating strands 82, suggesting that the kinetic step size (18 bp) 83 may enable the RNA helicase to ‘step over’ the polyglycol lesions in its track.
Biochemical studies of the NS3 helicase were performed with duplex DNA substrates containing a peptide nucleic acid (PNA) as the displaced strand. 84 PNAs contain normal purine and pyrimidine bases linked together with a neutral N-(2-aminoethyl) glycine backbone and are able to base pair with the complementary strand. The PNA-DNA substrate was unwound by NS3 at a much slower rate despite the fact that the PNA strand was not the helicase translocating strand. In contrast, DNA substrates containing a displaced strand consisting of a morpholino oligomer that is neutral or phosphorothioates which contain a sulfur in place of the oxygen in the phosphate backbone were unwound efficiently by NS3. As suggested by the authors of the study, the lack of a polyanionic backbone is not responsible for the inhibition of NS3 helicase activity observed for the PNA-DNA substrate. It is possible that the greater steric bulk of the morphilino backbone compared to the PNA backbone may contribute to the observed differences.
Vinylphosphonate internucleotide linkages which restrict rotational DNA backbone flexibility inhibited the helicase activity of the SF2 BLM and WRN helicases only when they resided in the translocating strand; however, the inhibitory effect was not as great as that observed for the SF1 PcrA helicase which unwinds DNA as a monomer. 85
The Raney lab examined the importance of electrostatic interactions between the SF1 Dda helicase and the negatively charged sugar phosphate backbone. 79 They tested Dda to unwind a DNA substrate in which one of the backbone phosphate groups within the duplex was replaced with a methylphosphonate (MeP) moiety which substitutes a phosphoryl oxygen with a methyl group, resulting in the loss of one negative charge to the strand that the MeP resides. The MeP modification profoundly inhibited Dda helicase under pre-steady state or excess enzyme conditions. Thus, electrostatic interactions are important for Dda helicase activity, suggesting this may be the case for other SF1 helicases. In contrast, the presence of an abasic site had a significantly reduced effect on Dda helicase activity. 79 Abasic sites in the helicase substrate were also tolerated by the SF2 RNA helicase NPH-II 76 and FANCJ helicase. 78
In summary, these studies suggest that DNA helicases display diversity in DNA substrate requirements (e. g. , phosphodiester backbone continuity) for efficient unwinding. Presumably, these differences reflect distinctions in unwinding mechanisms that are a function of helicase interactions with the translocating and non-translocating strands, assembly state, cooperativity between helicase molecules, efficiency of coupling motor ATPase to strand separation, etc. Although the DNA substrates with backbone disruption tested are due to synthetic linkages, biologically relevant forms of backbone damage exist. Phosphate oxygens are sites for alkylation leading to DNA phosphotriester adducts, which are abundant, persistent, and may contribute to genotoxicity. 86 Moreover, anti-sense technologies may be enhanced with oligonucleotides bearing alkyltriester linkages 87 and other modifications. It will be of interest to explore the effects of such backbone modifications which can occur in nature on the biochemical and cellular functions of various helicases and nucleic acid translocases.
Effects of Chemically Modified DNA Bases on Helicase-Catalyzed DNA Unwinding
Chemical damage to the DNA bases is implicated in mutagenesis and cell lethality, as well as carcinogenesis, aging, and neurological disorders (see ref. 88 and cited references). Various processes including oxidation and alkylation from endogenous or exogenous sources can impose base damage and also sugar modifications, strand breaks, and abasic sites. Cellular repair systems can remove or correct these forms of DNA damage and preserve the informational content and integrity of the genome. Although the DNA repair pathways are finely tuned to recognize DNA damage, recognition of base damage among billions of normal base pairs is formidable. Since helicases may reside in protein complexes that are important for DNA damage recognition, it is of interest to study the interactions of DNA helicases with base lesions and other forms of DNA modifications.
In an effort to understand the interaction of the FANCJ helicase that is defective in Fanconi anemia 89 with damaged DNA, the effects of two naturally occurring DNA base modifications (thymine glycol, 8-oxoguanine) on FANCJ unwinding of a simple forked duplex DNA substrate were examined. 90 Thymine glycol induces a significant, localized structural change with the thymine glycol largely extrahelical, 91 whereas 8-oxoguanine exerts only a mild perturbation of the duplex (for review see ref. 8). A thymine glycol is considered a lethal form of base damage because it blocks cellular DNA replication or transcription, whereas 8-oxoguanine is mutagenic because it causes replication errors but does not pose an immediate block to either replication or transcription. FANCJ was sensitive to a single thymine glycol base damage regardless if thymine glycol resided in the helicase translocating or non-translocating strand. 90 The sensitivity of FANCJ helicase activity to the thymine glycol was unique since other DNA helicases tested were either insensitive to the thymine glycol or only inhibited in a strand-specific manner (Table 1).
Table 1.
Effects of Thymine Glycol on DNA Unwinding by Various Helicasesa
| Helicase | Organism | Directionality | Helicase Inhibition by TG | |
|---|---|---|---|---|
| Translocating Strand | Non-translocating Strand | |||
| FANCJ | Human | 5’ to 3’ | Yes | Yes |
| DinG | E. coli | 5’ to 3’ | No | No |
| DnaB | E. coli | 5’ to 3’ | No | No |
| UvrD | E. coli | 3’ to 5’ | No | No |
| BLM | Human | 3’ to 5’ | Yes | No |
| WRN | Human | 3’ to 5’ | Yes | No |
| RECQ1 | Human | 3’ to 5’ | Yes | No |
See text and reference 81 for details.
In contrast to the observed effect of thymine glycol on FANCJ helicase activity, FANCJ was insensitive to the 8-oxoguanine lesion positioned in either strand of the duplex region of the helicase substrate. 90 Thus, the inhibition of FANCJ helicase activity by the thymine glycol damage was specific and not a general effect exerted by a different form of oxidative base modification, namely the 8-oxoguanine lesion. The observed differences between the effects of thymine glycol and 8-oxoguanine on FANCJ helicase activity may reflect the degree of structural distortion to the DNA double helix. Recently, the unwinding capacities of WRN, BLM and RecQ5 helicases were observed to be two-fold greater on telomeric D-loop substrates harboring an 8-oxoguanine, 92 suggesting these helicases may have a role in recognition of certain forms of oxidative DNA damage.
Strand-Specific Stimulation of FANCJ Helicase Activity on the Thymine Glycol DNA Substrate by RPA
The inhibition of FANCJ helicase activity by a single thymine glycol adduct raised the question if FANCJ might have a mechanism to efficiently unwind past the base damage through an interaction with its protein partner RPA. Previously, it was reported that RPA physically and functionally interacts with FANCJ 93 and other human DNA helicases (RECQ1, WRN, and BLM). 57 Kinetic analyses of initial helicase-catalyzed rates revealed over a 10-fold increase in unwinding for the nontranslocating strand thymine glycol substrate when RPA was present in the FANCJ helicase reaction compared with the reaction containing FANCJ alone. 90 In contrast, RPA failed to stimulate FANCJ helicase activity on the DNA substrate with thymine glycol in the strand that FANCJ translocates. Stimulation of FANCJ helicase activity by RPA was specific since E. coli Single-stranded DNA Binding Protein (SSB) did not have an effect on the DNA unwinding reactions catalyzed by FANCJ on the substrates with thymine glycol in either the nontranslocating or translocating strands. Moreover, RPA was not able to stimulate E. coli DinG helicase activity on DNA substrates with thymine glycol in either strand, indicating a specific interaction between the human RPA and FANCJ.
RPA, which physically and functionally interacts with human RECQ1, 94 was able to stimulate DNA unwinding by the helicase in a strand-specific manner on the thymine glycol DNA substrate. 90 RPA was only able to stimulate RECQ1 helicase activity on the DNA substrate in which the thymine glycol was positioned in the opposite strand to the one that RECQ1 translocates. RPA was discovered to bind with a much higher (17-fold) affinity to single-stranded DNA containing a single thymine glycol compared to undamaged ssDNA, 90 suggesting that the enhanced binding of RPA to the unwound single-stranded DNA tract containing the thymine glycol plays an element in the RPA stimulation of helicase activity past the helicase-blocking thymine glycol lesion. RPA heterotrimers with RPA70 missense mutations characterized by DNA binding defects failed to stimulate FANCJ helicase activity, consistent with the notion that the high affinity binding of RPA for ssDNA plays a role in the strand-specific stimulation of helicase-catalyzed DNA unwinding.
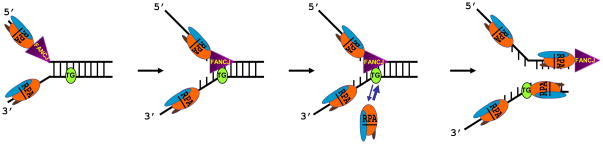
Why does RPA only stimulate FANCJ (or RECQ1) helicase activity on the substrate with the thymine glycol positioned in the helicase-nontranslocating strand but not the helicase-translocating strand? A plausible model for the strand-specific stimulation of FANCJ helicase activity by RPA is shown in Fig. 4. As FANCJ proceeds to translocate on the strand opposite the thymine glycol, the lesion is exposed in its single-stranded state whereby it is bound tightly by RPA. The helicase then proceeds with its unwinding reaction. When the thymine glycol is positioned in the strand that FANCJ translocates, the helicase is blocked in such a manner that thymine glycol remains within the duplex and RPA does not gain access to bind it with high affinity. Since FANCJ fails to interact favorably with RPA at the site of unwinding, the helicase eventually dissociates from the helicase substrate and would have to attempt to unwind it again. Given that RPA is a highly abundant protein in the eukaryotic cell (100,000 proteins per cell), 95 the high affinity binding of RPA to thymine glycol and perhaps other forms of oxidative base damage is physiologically important.
Fig. 4. Strand-specific stimulation of FANCJ helicase activity past a thymine glycol base lesion by RPA.
The model depicts the ability of the single-stranded DNA binding protein RPA to stimulate FANCJ helicase activity when the lesion resides in the strand opposite to the one that FANCJ translocates. RPA binds tightly to the partially unwound ssDNA containing the thymine glycol in the helicase non-translocating strand, enabling the helicase to proceed unwinding past the lesion. See text and ref. 90 for details. RPA, Replication Protein A heterotrimer; FANCJ helicase, triangle; TG oval, thymine glycol base damage.
Roles of Eukaryotic DNA Helicases at Stalled Replication Forks and in DNA Damage Signaling
When the replication fork machinery is stalled by a DNA lesion, DNA synthesis is arrested resulting in the halt of cell cycle progression and the activation of DNA damage checkpoints. Activation of two protein kinases known as ATR and Chk1 is an important step in checkpoint activation after replication fork stalling. 96;97 Uncoupling of DNA unwinding by mini-chromosome maintenance (MCM) helicase at the replication fork and continuation of DNA synthesis by a DNA polymerase leads to the accumulation of single-stranded DNA which is bound by RPA. 98 Presumably, MCM helicase is able to efficiently unwind past DNA lesions that block progression of the replicative polymerase. Although some progress has been made in understanding the unwinding mechanism of the MCM helicase, 99 to our knowledge there is no published information on the effects of DNA damage on MCM helicase activity. The assembly of a multi-protein complex that contains ATR-ATRIP, TopBP1, and Rad9/Hus1/Rad1 (9-1-1) at stalled replication forks together with RPA coating the ssDNA is critical for ATR activation, which is subsequently followed by Chk1 activation. 100–103
Very recently, evidence was presented that the FANCJ (BACH1) helicase plays an early role in DNA replication checkpoint control that is mediated by its DNA unwinding activity and its interaction with TopBP1. 104 FANCJ was shown to be required for loading of RPA on to chromatin during replicational stress induced by the replication inhibitor hydroxyurea, which in turn is required for ATR-dependent phosphorylation events in response to replicational stress. Thus the FANCJ helicase is not only important for the appropriate response to agents that induce interstrand DNA cross-links, but also more generally as a helicase to promote S phase progression, 105 resolve alternate DNA structures such as G-quadruplexes, 106 and control DNA replication checkpoint. Other DNA helicases also have specialized roles at stalled replication forks. A good example is the RecQ family of DNA helicases that are components of the human replication complex 107 or are believed to operate when the replication fork encounters a lesion that blocks leading strand synthesis. 57 Cobb et al. provided evidence that DNA polymerase stabilization at stalled replications forks in yeast requires the RecQ helicase Sgs1 and the ATM-related kinase ATM. 108 A model was proposed in which Sgs1 helicase catalytically unwinds secondary structure at stalled replication forks to maintain single-stranded DNA for RPA (which Sgs1 binds), enabling RPA and Mec1 to promote polymerase association at the stalled fork. It will be of interest to determine if and how the physical interaction of RPA with FANCJ 93 and the RecQ helicases 109 is required for checkpoint control or replication fork processing. Further investigation of the functions of DNA helicases during situations of cellular stress imposed by agents that deplete the nucleotide pool or introduce bulky adducts or oxidative base lesions that perturb normal replication is a high priority.
Finally, it is of note that two new single-stranded DNA binding proteins (hSSB1, hSSB2) in addition to RPA have been discovered that are critical for genomic stability and the DNA damage response. 110–112 Therefore, it will be intriguing to unravel the potential interactions of hSSB1 and hSSB2 with DNA helicases in their interactions with damaged DNA during cellular replication, recombination, and DNA repair.
SUMMARY
We have discussed various aspects of how helicases interact with damaged DNA, and how these interactions are important for cellular pathways of the DNA damage response. Clearly, there is still much to learn about how helicases involved in DNA repair or replication checkpoint control perform their functions faithfully in an orchestrated manner with other nuclear proteins. The growing importance of helicases in diseases characterized by genomic instability prompts close attention to this special class of enzymes. Future work in helicase research promises to be exciting and still unpredictable.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging. We wish to thank Drs. Deborah Croteau and Zheng Cao (Laboratory of Molecular Gerontology, NIA-NIH) for comments.
References
- 1.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 2.Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18625–18628. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 3.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 4.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 5.Branzei D, Foiani M. The DNA damage response during DNA replication 1. Curr Opin Cell Biol. 2005;17:568–575. doi: 10.1016/j.ceb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Tornaletti S, Hanawalt PC. Effect of DNA lesions on transcription elongation. Biochimie. 1999;81:139–146. doi: 10.1016/s0300-9084(99)80046-7. [DOI] [PubMed] [Google Scholar]
- 7.Tornaletti S. DNA repair in mammalian cells: Transcription-coupled DNA repair: directing your effort where it’s most needed. Cell Mol Life Sci. 2009;66:1010–1020. doi: 10.1007/s00018-009-8738-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 9.Yang W. Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair (Amst) 2006;5:654–666. doi: 10.1016/j.dnarep.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 11.Truglio JJ, Croteau DL, Van HB, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 12.Van HB, Croteau DL, DellaVecchia MJ, Wang H, Kisker C. ‘Close-fitting sleeves’: DNA damage recognition by the UvrABC nuclease system. Mutat Res. 2005;577:92–117. doi: 10.1016/j.mrfmmm.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Lu M, Tang MS, et al. DNA wrapping is required for DNA damage recognition in the Escherichia coli DNA nucleotide excision repair pathway. Proc Natl Acad Sci U S A. 2009;106:12849–12854. doi: 10.1073/pnas.0902281106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moolenaar GF, Hoglund L, Goosen N. Clue to damage recognition by UvrB: residues in the beta-hairpin structure prevent binding to non-damaged DNA. EMBO J. 2001;20:6140–6149. doi: 10.1093/emboj/20.21.6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truglio JJ, Karakas E, Rhau B, et al. Structural basis for DNA recognition and processing by UvrB. Nat Struct Mol Biol. 2006;13:360–364. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- 16.Waters TR, Eryilmaz J, Geddes S, Barrett TE. Damage detection by the UvrABC pathway: crystal structure of UvrB bound to fluorescein-adducted DNA. FEBS Lett. 2006;580:6423–6427. doi: 10.1016/j.febslet.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 17.Malta E, Moolenaar GF, Goosen N. Base flipping in nucleotide excision repair 1. J Biol Chem. 2006;281:2184–2194. doi: 10.1074/jbc.M508901200. [DOI] [PubMed] [Google Scholar]
- 18.Malta E, Verhagen CP, Moolenaar GF, et al. Functions of base flipping in E. coli nucleotide excision repair. DNA Repair (Amst) 2008;7:1647–1658. doi: 10.1016/j.dnarep.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Jia L, Kropachev K, Ding S, et al. Exploring damage recognition models in prokaryotic nucleotide excision repair with a benzo[a]pyrene-derived lesion in UvrB. Biochemistry. 2009;48:8948–8957. doi: 10.1021/bi9010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villani G, Tanguy LG. Interactions of DNA helicases with damaged DNA: possible biological consequences. J Biol Chem. 2000;275:33185–33188. doi: 10.1074/jbc.R000011200. [DOI] [PubMed] [Google Scholar]
- 21.Villani G, Pillaire MJ, Boehmer PE. Effect of the major DNA adduct of the antitumor drug cis-diamminedichloroplatinum (II) on the activity of a helicase essential for DNA replication, the herpes simplex virus type-1 origin-binding protein. J Biol Chem. 1994;269:21676–21681. [PubMed] [Google Scholar]
- 22.Brown WC, Romano LJ. Benzo[a]pyrene-DNA adducts inhibit translocation by the gene 4 protein of bacteriophage T7. J Biol Chem. 1989;264:6748–6754. [PubMed] [Google Scholar]
- 23.Yong Y, Romano LJ. Benzo[a]pyrene-DNA adducts inhibit the DNA helicase activity of the bacteriophage T7 gene 4 protein. Chem Res Toxicol. 1996;9:179–187. doi: 10.1021/tx950112h. [DOI] [PubMed] [Google Scholar]
- 24.Huang L, Turchi JJ, Wahl AF, Bambara RA. Activity of calf thymus DNA helicase E on cis-diamminedichloroplatinum (II)-damaged DNA 1. J Biol Chem. 1993;268:26731–26737. [PubMed] [Google Scholar]
- 25.Villani G, Cazaux C, Pillaire MJ, Boehmer P. Effects of a single intrastrand d(GpG) platinum adduct on the strand separating activity of the Escherichia coli proteins RecB and RecA. FEBS Lett. 1993;333:89–95. doi: 10.1016/0014-5793(93)80380-d. [DOI] [PubMed] [Google Scholar]
- 26.Sung P, Prakash L, Matson SW, Prakash S. RAD3 protein of Saccharomyces cerevisiae is a DNA helicase. Proc Natl Acad Sci U S A. 1987;84:8951–8955. doi: 10.1073/pnas.84.24.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung P, Prakash L, Weber S, Prakash S. The RAD3 gene of Saccharomyces cerevisiae encodes a DNA-dependent ATPase. Proc Natl Acad Sci U S A. 1987;84:6045–6049. doi: 10.1073/pnas.84.17.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naegeli H, Bardwell L, Friedberg EC. The DNA helicase and adenosine triphosphatase activities of yeast Rad3 protein are inhibited by DNA damage. A potential mechanism for damage-specific recognition. J Biol Chem. 1992;267:392–398. [PubMed] [Google Scholar]
- 29.Naegeli H, Modrich P, Friedberg EC. The DNA helicase activities of Rad3 protein of Saccharomyces cerevisiae and helicase II of Escherichia coli are differentially inhibited by covalent and noncovalent DNA modifications. J Biol Chem. 1993;268:10386–10392. [PubMed] [Google Scholar]
- 30.Naegeli H, Bardwell L, Friedberg EC. Inhibition of Rad3 DNA helicase activity by DNA adducts and abasic sites: implications for the role of a DNA helicase in damage-specific incision of DNA. Biochemistry. 1993;32:613–621. doi: 10.1021/bi00053a029. [DOI] [PubMed] [Google Scholar]
- 31.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Oh KS, Khan SG, Jaspers NG, et al. Phenotypic heterogeneity in the XPB DNA helicase gene (ERCC3): xeroderma pigmentosum without and with Cockayne syndrome. Hum Mutat. 2006;27:1092–1103. doi: 10.1002/humu.20392. [DOI] [PubMed] [Google Scholar]
- 33.Coin F, Oksenych V, Egly JM. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell. 2007;26:245–256. doi: 10.1016/j.molcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Schaeffer L, Moncollin V, Roy R, et al. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J. 1994;13:2388–2392. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzder SN, Sung P, Bailly V, Prakash L, Prakash S. RAD25 is a DNA helicase required for DNA repair and RNA polymerase II transcription. Nature. 1994;369:578–581. doi: 10.1038/369578a0. [DOI] [PubMed] [Google Scholar]
- 36.Sung P, Higgins D, Prakash L, Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988;7:3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin YC, Choi WS, Gralla JD. TFIIH XPB mutants suggest a unified bacterial-like mechanism for promoter opening but not escape. Nat Struct Mol Biol. 2005;12:603–607. doi: 10.1038/nsmb949. [DOI] [PubMed] [Google Scholar]
- 38.Oksenych V, Coin F. The long unwinding road: XPB and XPD helicases in damaged DNA opening. Cell Cycle. 2010;9:90–96. doi: 10.4161/cc.9.1.10267. [DOI] [PubMed] [Google Scholar]
- 39.Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 40.Winkler GS, Araujo SJ, Fiedler U, et al. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem. 2000;275:4258–4266. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
- 41.Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol Cell. 2009;36:642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 42.Rudolf J, Rouillon C, Schwarz-Linek U, White MF. The helicase XPD unwinds bubble structures and is not stalled by DNA lesions removed by the nucleotide excision repair pathway. Nucleic Acids Res. 2009;38:931–941. doi: 10.1093/nar/gkp1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–71. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meetei AR, Sechi S, Wallisch M, et al. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deans AJ, West SC. FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 46.Fan L, Fuss JO, Cheng QJ, et al. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H, Rudolf J, Johnson KA, et al. Structure of the DNA repair helicase XPD. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolski SC, Kuper J, Hanzelmann P, et al. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hogg M, Wallace SS, Doublie S. Bumps in the road: how replicative DNA polymerases see DNA damage. Curr Opin Struct Biol. 2005;15:86–93. doi: 10.1016/j.sbi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 50.Yang W. Damage repair DNA polymerases Y. Curr Opin Struct Biol. 2003;13:23–30. doi: 10.1016/s0959-440x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 51.Lehmann AR. XPD structure reveals its secrets. DNA Repair (Amst) 2008;7:1912–1915. doi: 10.1016/j.dnarep.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 52.Fan L, Arvai AS, Cooper PK, et al. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Mol Cell. 2006;22:27–37. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 53.Oksenych V, de Jesus BB, Zhovmer A, Egly JM, Coin F. Molecular insights into the recruitment of TFIIH to sites of DNA damage. EMBO J. 2009 doi: 10.1038/emboj.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soultanas P, Dillingham MS, Wiley P, Webb MR, Wigley DB. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brosh RM, Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Committee on the Biologic Effects of Atmospheric Pollutants) Particulate Polycyclic Organic Matter. National Academy of Sciences; Washington, DC: 1972. [Google Scholar]
- 57.Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Musich PR, Zou Z. Genomic instability and DNA damage responses in progeria arising from defective maturation of prelamin A. Aging. 2009;1:28–37. doi: 10.18632/aging.100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li B, Reddy S, Comai L. Sequence-specific processing of telomeric 3’ overhangs by the Werner syndrome protein exonuclease activity. Aging. 2009;1:289–302. doi: 10.18632/aging.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Opresko PL, Otterlei M, Graakjaer J, et al. The Werner Syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 62.Kyng KJ, May A, Stevnsner T, et al. Gene expression responses to DNA damage are altered in human aging and in Werner Syndrome. Oncogene. 2005;24:5026–5042. doi: 10.1038/sj.onc.1208692. [DOI] [PubMed] [Google Scholar]
- 63.Turaga RV, Paquet ER, Sild M, et al. The Werner syndrome protein affects the expression of genes involved in adipogenesis and inflammation in addition to cell cycle and DNA damage responses. Cell Cycle. 2009;8:2080–2092. doi: 10.4161/cc.8.13.8925. [DOI] [PubMed] [Google Scholar]
- 64.Aggarwal M, Brosh RM., Jr WRN helicase defective in the premature aging disorder Werner syndrome genetically interacts with topoisomerase 3 and restores the top3 slow growth phenotype of sgs1 top3. Aging. 2009;1:219–233. doi: 10.18632/aging.100020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laine JP, Opresko PL, Indig FE, et al. Werner protein stimulates topoisomerase I DNA relaxation activity. Cancer Res. 2003;63:7136–7146. [PubMed] [Google Scholar]
- 66.Choudhary S, Doherty KM, Handy CJ, et al. Inhibition of Werner Syndrome helicase activity by Benzo[a]pyrene Diol Epoxide adducts can be overcome by Replication Protein A. J Biol Chem. 2006;281:6000–6009. doi: 10.1074/jbc.M510122200. [DOI] [PubMed] [Google Scholar]
- 67.Driscoll HC, Matson SW, Sayer JM, et al. Inhibition of Werner syndrome helicase activity by benzo[c]phenanthrene diol epoxide dA adducts in DNA is both strand-and stereoisomer-dependent. J Biol Chem. 2003;278:41126–41135. doi: 10.1074/jbc.M304798200. [DOI] [PubMed] [Google Scholar]
- 68.Geacintov NE, Cosman M, Hingerty BE, et al. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 69.Volk DE, Thiviyanathan V, Rice JS, et al. Solution structure of a cis-opened (10R)-N6-deoxyadenosine adduct of (9S,10R)-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a DNA duplex. Biochemistry. 2003;42:1410–1420. doi: 10.1021/bi026745u. [DOI] [PubMed] [Google Scholar]
- 70.Daniels DS, Woo TT, Luu KX, et al. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat Struct Mol Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 71.Brosh RM, Jr, Karow JK, White EJ, et al. Potent inhibition of werner and bloom helicases by DNA minor groove binding drugs. Nucleic Acids Res. 2000;28:2420–2430. doi: 10.1093/nar/28.12.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sayer JM, Shah JM, Liang C, et al. Effect of absolute configuration on the optical and NMR properties of oligonucleotides containing benzo[a]pyrene adducts at N6 of deoxyadenosine. Polycyclic Aromatic Hydrocarbons. 1999;17:95–104. [Google Scholar]
- 73.Ruan Q, Kolbanovskiy A, Zhuang P, et al. Synthesis and characterization of site-specific and stereoisomeric fjord dibenzo[a,l]pyrene diol epoxide-N(6)-adenine adducts: unusual thermal stabilization of modified DNA duplexes. Chem Res Toxicol. 2002;15:249–261. doi: 10.1021/tx010157k. [DOI] [PubMed] [Google Scholar]
- 74.Amaratunga M, Lohman TM. Escherichia coli Rep helicase unwinds DNA by an active mechanism. Biochemistry. 1993;32:6815–6820. doi: 10.1021/bi00078a003. [DOI] [PubMed] [Google Scholar]
- 75.Bertram RD, Hayes CJ, Soultanas P. Vinylphosphonate internucleotide linkages inhibit the activity of PcrA DNA helicase. Biochemistry. 2002;41:7725–7731. doi: 10.1021/bi025755s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kawaoka J, Jankowsky E, Pyle AM. Backbone tracking by the SF2 helicase NPH-II. Nat Struct Mol Biol. 2004;11:526–530. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- 77.Gupta R, Sharma S, Sommers JA, et al. Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J Biol Chem. 2005;280:25450–25460. doi: 10.1074/jbc.M501995200. [DOI] [PubMed] [Google Scholar]
- 78.Gupta R, Sharma S, Doherty KM, et al. Inhibition of BACH1 (FANCJ) helicase by backbone discontinuity is overcome by increased motor ATPase or length of loading strand. Nucleic Acids Res. 2006;34:6673–6683. doi: 10.1093/nar/gkl964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eoff RL, Spurling TL, Raney KD. Chemically modified DNA substrates implicate the importance of electrostatic interactions for DNA unwinding by Dda helicase. Biochemistry. 2005;44:666–674. doi: 10.1021/bi0484926. [DOI] [PubMed] [Google Scholar]
- 80.Tackett AJ, Morris PD, Dennis R, Goodwin TE, Raney KD. Unwinding of unnatural substrates by a DNA helicase. Biochemistry. 2001;40:543–548. doi: 10.1021/bi002122+. [DOI] [PubMed] [Google Scholar]
- 81.Jeong YJ, Levin MK, Patel SS. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc Natl Acad Sci U S A. 2004;101:7264–7269. doi: 10.1073/pnas.0400372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beran RK, Bruno MM, Bowers HA, Jankowsky E, Pyle AM. Robust translocation along a molecular monorail: the NS3 helicase from Hepatitis C Virus traverses unusually large disruptions in its track. J Mol Biol. 2006;358:974–982. doi: 10.1016/j.jmb.2006.02.078. [DOI] [PubMed] [Google Scholar]
- 83.Serebrov V, Pyle AM. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- 84.Tackett AJ, Wei L, Cameron CE, Raney KD. Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 2001;29:565–572. doi: 10.1093/nar/29.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garcia PL, Bradley G, Hayes CJ, et al. RPA alleviates the inhibitory effect of vinylphosphonate internucleotide linkages on DNA unwinding by BLM and WRN helicases. Nucleic Acids Res. 2004;32:3771–3778. doi: 10.1093/nar/gkh709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shooter KV. DNA phosphotriesters as indicators of cumulative carcinogen-induced damage. Nature. 1978;274:612–614. doi: 10.1038/274612a0. [DOI] [PubMed] [Google Scholar]
- 87.Marcus-Sekura CJ, Woerner AM, Shinozuka K, Zon G, Quinnan GV., Jr Comparative inhibition of chloramphenicol acetyltransferase gene expression by antisense oligonucleotide analogues having alkyl phosphotriester, methylphosphonate and phosphorothioate linkages. Nucleic Acids Res. 1987;15:5749–5763. doi: 10.1093/nar/15.14.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilson DM, III, Bohr VA, McKinnon PJ. DNA damage, DNA repair, ageing and age-related disease. Mech Ageing Dev. 2008;129:349–352. doi: 10.1016/j.mad.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu Y, Brosh RM., Jr FANCJ helicase operates in the Fanconi Anemia DNA repair pathway and the response to replicational stress. Curr Mol Med. 2009;9:470–482. doi: 10.2174/156652409788167159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suhasini AN, Sommers JA, Mason AC, et al. FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by Replication Protein A to unwind the damaged DNA substrate in a strand-specific manner. J Biol Chem. 2009;284:18458–18470. doi: 10.1074/jbc.M109.012229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kung HC, Bolton PH. Structure of a duplex DNA containing a thymine glycol residue in solution. J Biol Chem. 1997;272:9227–9236. doi: 10.1074/jbc.272.14.9227. [DOI] [PubMed] [Google Scholar]
- 92.Ghosh A, Rossi ML, Aulds J, Croteau D, Bohr VA. Telomeric D-loops containing 8-oxo-2’-deoxyguanosine are preferred substrates for Werner and Bloom syndrome helicases and are bound by POT1. J Biol Chem. 2009;284:31074–31084. doi: 10.1074/jbc.M109.027532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gupta R, Sharma S, Sommers JA, et al. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007;110:2390–2398. doi: 10.1182/blood-2006-11-057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cui S, Arosio D, Doherty KM, et al. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 96.Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair (Amst) 2009;8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 97.Friedel AM, Pike BL, Gasser SM. ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol. 2009;21:237–244. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 98.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burrows AE, Elledge SJ. How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev. 2008;22:1416–1421. doi: 10.1101/gad.1685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 103.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. MOL Cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol Cell Biol. 2007;27:6733–6741. doi: 10.1128/MCB.00961-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu Y, Shin-Ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia Anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thangavel S, Mendoza-Maldonado R, Tissino E, et al. The human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation 1. Mol Cell Biol. 2010 doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Doherty KM, Sommers JA, Gray MD, et al. Physical and functional mapping of the RPA interaction domain of the Werner and Bloom syndrome helicases. J Biol Chem. 2005;280:29494–29505. doi: 10.1074/jbc.M500653200. [DOI] [PubMed] [Google Scholar]
- 110.Li Y, Bolderson E, Kumar R, et al. HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J Biol Chem. 2009;284:23525–23531. doi: 10.1074/jbc.C109.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Richard DJ, Bolderson E, Cubeddu L, et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature. 2008;453:677–681. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]
- 112.Richard DJ, Bolderson E, Khanna KK. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Crit Rev Biochem Mol Biol. 2000;44:98–116. doi: 10.1080/10409230902849180. [DOI] [PubMed] [Google Scholar]