Abstract
Conformational changes in the substrate access channel have been observed for several forms of cytochrome P450, but the extent of conformational plasticity exhibited by a given isozyme has not been completely characterized. Here we present crystal structures of P450cam bound to a library of 12 active site probes containing a substrate analog tethered to a variable linker. The structures provide a unique view of the range of protein conformations accessible during substrate binding. Principal component analysis of a total of 30 structures reveals three discrete clusters of conformations; closed (P450cam-C), intermediate (P450cam-I) and fully open (P450cam-O). Relative to P450cam-C, the P450cam-I state results predominantly from a retraction of the F-helix, while both F and G helices move in concert to reach the fully open P450cam-O state. Both P450cam-C and P450cam-I are well defined states, while P450cam-O shows evidence for a somewhat broader distribution of conformations, and includes the open form recently seen in the absence of substrate. The observed clustering of protein conformations over a wide range of ligand variants suggests a multi-step closure of the enzyme around the substrate that begins by conformational selection from an ensemble of open conformations and proceeds through a well defined intermediate, P450cam-I, before full closure to the P450cam-C state in the presence of small substrates. This multi-step pathway may have significant implications for a full understanding of substrate specificity, kinetics and coupling of substrate binding to P450 function.
Substrate recognition by cytochrome P450s has been intensively studied for many decades, and while much is known about how structural variations of these enzymes result in their wide ranging specificity, less is understood about the role played by protein dynamics and active site motion in this process. P450s are an important family of monooxygenases with over 10,000 members widely distributed in living systems from bacteria to humans (1). They use O2 and a cysteine coordinated heme to catalyze the oxidation of a vast array of substrates in reactions as diverse as steroid biosynthesis and xenobiotic metabolism (2). Even distantly related P450s share an overall conserved protein fold, first described for P450cam, CYP101A1, a camphor metabolizing P450 from Pseudomonas putida (3, 4). Despite this overall similarity, significant differences in the sequence, structure and membrane location of individual P450s have been characterized. In addition, exactly how these differences allow some P450s to catalyze the stereo-specific hydroxylation of a single well defined substrate, while others are able to oxidize a wide range of compounds remains incompletely understood (5, 6).
The diversity of substrate specificities of P450s is believed to result largely from differences in the structure, flexibility and/or dynamics of the substrate binding channel which connects the protein surface to the deeply buried heme center. The substrate binding channel is defined by the anti-parallel F and G helices, the intervening F-G loop and segments of the B′ helix, which fold over and around the heme and the I helix to enclose the substrate and position it for attack by the reactive Compound I center of the heme (4, 7). There are significant differences in the sequence of these structural elements between various forms of P450 (8), and in consequence, the structure of the substrate binding channel varies considerably from one form to another (9). Thus, the small, closed substrate access channels seen in many prokaryotic P450s (4, 10–12) are in marked contrast to the larger, more open channels seen for mammalian microsomal enzymes involved in drug metabolism (5, 13, 14).
Conformational change clearly plays an important role in substrate recognition for many if not all P450s, but only recently have specific details become available about the conformational space sampled by a given enzyme (15–18). For a number of P450s, most notably P450cam, the closed conformation observed for the camphor bound state must undergo significant conformational change to allow binding of substrate and release of product (3, 4, 11). Importantly, reports have shown several examples of bacterial P450s that exist both in an open conformation in the absence of substrate, and in a closed conformation in the presence of substrate or ligand (13, 16, 19–24). This now includes P450cam, which was previously observed in the closed conformation even in the absence of substrate (25), as we have recently reported an open conformation of P450cam when crystallized in the absence of substrate that is essentially identical to that observed in the presence of large tethered substrates (18). This suggests that the open conformation does not result from being held open by the tethered substrate, but is instead dynamically sampled in the substrate-free form. While channel closure is likely to influence regio- and/or stereospecificity of oxidation in these enzymes, it is unclear at present whether or how the functional activity of the enzyme is coupled to conformational state.
The role of active site flexibility in P450 substrate recognition is of considerable importance for mammalian P450s. Among the diverse and promiscuous drug metabolizing microsomal enzymes, evidence exists both for and against conformational change associated with substrate binding. For example, CYP2B4 has been observed to undergo significant conformational change upon binding substrates or inhibitors (13, 14, 17), while CYP3A4 has been observed in both the substrate-bound and -free forms with relatively small changes in structure (26). For the highly specific mitochondrial CYP24A1, which is responsible for the specific hydroxylation of 1α,25-dihydroxyvitamin D3, an open conformation was observed without bound substrate, and the importance of channel closure upon substrate binding remains unknown (27). Thus, a general concept has emerged that highly specific biosynthetic P450s may require a more closed and tight fit around the substrate, while the more promiscuous drug metabolizing enzymes make use of a larger, more open, non-specific active site channel.
Substrate recognition by P450s could be realized in a number of ways, ranging from a rigid lock-and-key interaction which matches a particular substrate to a complementary but static binding site, to induced-fit models, in which substrate binding induces a protein conformational change that only exists upon interaction with substrate. Alternatively, conformational selection implies that substrate binds a subset of conformations from a pre-existing ensemble of states, causing a shift in the equilibrium of populated states as the energy landscape changes upon substrate binding (28). Recent results for P450cam (18), EryK (15) and PikC (16) and CYP119 (29), showing population of both open and closed conformations in the absence of substrates strongly suggest that these enzymes make use of conformational selection by dynamically visiting alternate conformations. While these results show how substrates can gain access to the buried active sites, the full range of conformational states sampled by a given P450 or the details about the specific trajectories or conformational intermediates remains undefined. As these features are likely to be critically important for substrate recognition and specificity in this broad class of enzymes, it would be important to characterize the range of conformational states available to a given structure.
In this study we report the structural characterization of P450cam bound to a family of related tethered substrates and describe the extent to which the protein responds to variations in the probe. The results described here provide direct observation of at least three distinct clusters of conformational sub-states referred to as P450cam-C (closed), P450cam-I (intermediate) and P450cam-O (open), and suggest a stepwise pathway for the inter-conversion from the closed to the open state through a well defined intermediate.
EXPERIMENTAL PROCEDURES
Protein preparation
The full-length P450cam containing the C334A mutation was purified as described previously (18). C334A has been shown to prevent intermolecular dimerization and increase protein stability, but does not affect protein activity (30). Purified protein (A417/A280 > 1.45) was exchanged into 50 mM KPi, pH 6.0, 1 mM camphor, 30 mM β-mercaptoethanol, and frozen at −80 °C for further experiments.
Synthesis of substrate analogs
Reagents and solvents were purchased from commercial sources and used without further purification. Anhydrous N,N-diisopropylethylamine and DMF were purchased from Acros Organics. NMR spectra were obtained on a Varian Mercury 400 MHz spectrometer. The 1H chemical shifts are expressed relative to the residual solvent signal. Mass spectra were obtained at the Scripps Research Institute Mass Spectrometry Facility. tert-butyl-6-aminohexylcarbamate, tert-butyl-8-aminooctylcarbamate and tert-butyl N-{2-[2-(2-aminoethoxy)ethoxy]ethyl}-carbamate were synthesized according to the procedure of Jaramillo et al. (31). The synthesis of AdaC1-C8-Dans was reported previously (32).
All Boc-protected adamantane tethered substrate analogues were synthesized by the following general procedure (tert-butyl-8-aminooctylcarbamate and adamantyl carbonyl chloride are used as an illustrative example). tert-butyl-8-aminooctylcarbamate (0.200 g, 0.822 mmol), adamantane-1-carbonyl chloride (0.196 g, 0.989 mmol) and 1 mL N,N-diisopropylethylamine were dissolved in dry DMF (10 mL) under argon and stirred at room temperature overnight. The solvent was removed under reduced pressure, and the crude product purified via flash chromatography (SiO2) using a solvent gradient from 100% CH2Cl2 to CH2Cl2:MeOH (9/1) to give the product as a white solid. For the other adamantyl derivatives the procedure was modified as follows: HOBt (hydroxybenzotriazole) (1.1 eq) and EDC (1-ethyl-3-(3′-dimethylaminopropyl) carbodiimide) (1.1 eq) were added to the reaction mixture.
The Dansyl group was added to the adamantyl tethered substrate analogues following deprotection of the Boc group using the following general procedure (1-(8-tert-butyl-8-aminooctylcarbamate) amidoadamantane is used as an illustrative example). 1-(8-tert-Butyl-8-aminooctylcarbamate) amidoadamantane (0.100 g, 0.247 mmol) was dissolved in CH2Cl2/TFA (9:1) and stirred for 1 hour at room temperature. The solvents were removed under reduced pressure, and the crude product was then dissolved in dry DMF (5 mL). N,N-Diisopropylethylamine (0.5 mL) was added, followed by dansyl chloride (0.100 g, 0.371 mmol). The reaction mixture was stirred under argon at room temperature overnight. The solvent was removed under reduced pressure, and the crude product was purified via flash chromatography (SiO2) using gradient elution from CH2Cl2 (100%) to CH2Cl2:MeOH:Et3N (8/1/1) as eluent to give the product as a pale yellow solid.
Crystallization, data collection and crystal structure determination
Camphor was removed from protein stock solutions by buffer exchange into 20 mM Hepes, pH 6.5 by gel filtration through two sequential PD-10 columns (GE healthcare). 1 mM P450cam was mixed with tethered substrate analogs (Table 1) at a 1:1 ratio. Crystals of P450cam bound to tethered substrates were grown by sitting-drop vapor diffusion at 6 °C from 100 mM cacodylic acid, pH 6.5, 12–22% polyethylene glycol 8000 and 200 mM K+. The crystals were transferred to cryoprotectant buffer consisting of 100 mM cacodylic acid, pH 6.5, 16–18% polyethylene glycol 8000, 25% polyethylene glycol 600 and 200 mM K+, mounted on nylon loops and flash frozen at 77K.
Table 1.
Structural analysis of P450cam conformers.
| Substratea | PDB | PC1b | PC2b | F helix shiftc (Å, deg) | G helix shiftc (Å, deg) | FG helix angle (deg) | Axial Water | Missing B′ helix | Ref |
|---|---|---|---|---|---|---|---|---|---|
| P450CAM-C | |||||||||
| Camphor |
2CPP | 25.7 | 2.9 | — | — | −148.9 | No | — | (4) |
| 1YRC | 29.4 | 4.9 | 0.28, 2.7 | 0.25, 2.8 | −149.4 | (42) | |||
| 5CP4 | 24.8 | 3.7 | 0.22, 2.7 | 0.40, 1.8 | −150.1 | (43) | |||
| 2ZAX | 28.7 | 5.7 | 0.23, 3.1 | 0.28, 2.5 | −148.7 | (44) | |||
| 1DZ4 | 26.7 | 6.5 | 0.11, 1.2 | 0.55, 0.9 | −150.1 | (45) | |||
| 1S-camphor |
1AKD | 23.9 | 6.1 | 0.41, 2.7 | 0.48, 1.4 | −149.0 | No | — | (46) |
| Imidazole |
2H7Q | 24.1 | 4.9 | 0.26, 2.2 | 0.54, 1.5 | −151.4 | No | — | (47) |
Nicotine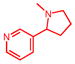
|
1P2Y | 25.9 | 5.6 | 0.08, 5.2 | 0.36, 0.5 | −148.2 | No | — | (48) |
Metyrapone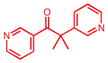
|
1PHG | 25.6 | 3.2 | 0.07, 1.4 | 0.12, 0.5 | −148.6 | No | — | (49) |
| P450CAM-I | |||||||||
| AdaC1-C8-Dans |
1RE9 | 6.4 | −9.9 | 3.30, 17.8 | 1.00, 5.9 | −156.6 | No | — | (66) |
| 1LWL | 4.4 | −11.5 | 3.59, 17.4 | 1.33, 5.4 | −156.5 | (32) | |||
| 3P6M | 2.7 | −11.2 | 3.77, 15.5 | 1.52, 5.4 | −156.7 | This study | |||
| 3P6N | 2.2 | −11.1 | 3.75, 16.0 | 1.50, 6.2 | −157.2 | This study | |||
| AdaC1-C8EtgGlu-Bio |
3OIA | 3.0 | −11.6 | 3.71, 15.8 | 1.27, 7.3 | −153.2 | No | — | Unpublished |
| AdaC1-Etg-Dans |
3P6O | 3.7 | −10.6 | 3.73, 16.7 | 1.42, 6.0 | −153.5 | No | — | This study |
| AdaC1-C6-Bio |
3P6P | 2.5 | −10.3 | 3.91, 15.2 | 1.37, 6.6 | −157.8 | No | — | This study |
| 3OH-AdaC1-C8-Dans |
3OL5 | 1.1 | −10.5 | 3.88, 14.9 | 1.96, 5.1 | −155.8 | Yes | — | Unpublished |
| P450CAM-O | |||||||||
| AdaC1-C4-Dans |
1RF9 | −9.9 | 0.7 | 3.94, 15.9 | 3.68, 10.8 | −149.7 | Yes | — | (66) |
| AdaC2-Etg-Boc |
3P6Q | −15.6 | 3.2 | 4.51, 17.2 | 3.95, 15.2 | — | Yes | 92–95 | This study |
3OH-AdaC1-Etg-Boc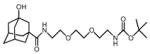
|
3P6R | −16.3 | 4.1 | 4.84, 16.6 | 4.57, 15.2 | −151.1 | No | 90–96 | This study |
| AdaC2-C8-Dansd |
3P6S | −17.3 | 2.3 | 4.59, 17.1 | 4.46, 14.9 | −150.2 | No | — | This study |
| 3P6T | −15.5 | 0.7 | 4.24, 16.8 | 4.28, 15.7 | −150.6 | This study | |||
| AdaC3-C6-Dans |
3P6U | −21.0 | 3.8 | 4.95, 17.8 | 4.83, 17.4 | −150.6 | Yes | 93–94 | This study |
3Et-AdaC1-Etg-Boc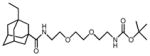
|
3P6V | −20.4 | 4.6 | 5.06, 17.4 | 5.02, 16.0 | −150.7 | No | 90–96 | This study |
| AdaC3-Etg-Boc |
3P6W | −20.5 | 3.8 | 5.04, 17.1 | 4.85, 17.4 | −151.0 | No | 91–95 | This study |
| AdaC3-C8-Dans |
3P6X | −22.2 | 3.9 | 5.07, 18.7 | 4.86, 18.1 | −150.4 | Yes | 90–94 | This study |
AdaC1-perfluorobiphenyl-Ru(bipy)3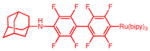
|
1K2O | −22.7 | 3.8 | 5.54, 21.0 | 5.14, 18.8 | −151.6 | Yes | — | (65) |
| AdaC1-C8-Ru(bipy)3 |
1QMQ | −16.0 | 3.3 | 4.43, 16.0 | 4.28, 14.7 | −149.0 | Yes | — | (64) |
| Substrate-free | 3L61 | −24.9 | 4.3 | 5.27, 19.3 | 5.09, 17.9 | −150.2 | Yes | 90–97 | (18) |
| 3L62 | −21.5 | 3.6 | 4.93, 18.2 | 4.73, 16.9 | −150.2 | 91–94 | (18) | ||
Well ordered electron density included for portions of the model shown in red.
PC1 and PC2 are first two principal components in the PCA of crystal structures described in this table.
Helix shift is calculated relative to the camphor-bound closed conformation (PDB entry 2CPP) at the center of the F helix (Lys178) and G helix (Lys197).
The entire substrate was only resolved in the structure of PDB entry 3P6T.
X-ray diffraction data were collected at 100 K using beamline 7-1, 9-1 or 11-1 at the Stanford Synchrotron Radiation Laboratory. Data were processed using Scala (33). Molecular replacement was done using Molrep (34) using the previously determined tethered substrate-bound P450cam structures (PDB entries 1RE9 and 1RF9). Model fitting and refinement were done with Coot (35) and Refmac5 (36) respectively. The final models were validated using the programs Procheck (37), Sfcheck (38), Molprobity (39) and the PDB validation server. Statistics for data collection and refinement are shown in Table S1. All structural graphics were generated using the Pymol Molecular Graphics System (40).
Structure analysis
Structural superposition and principal component analysis were done with the program bio3d package (41). A total of 30 crystal structures of P450cam bound to small substrates or tethered substrate analogs were aligned and the coordinates of the Cα atoms were used as input for principal component analysis. Bound small substrates include camphor (PDB entries 2CPP (4), 1YRC (42), 5CP4 (43), 2ZAX (44) and 1DZ4 (45)), 1S-camphor (PDB entry 1AKD (46)), imidazole (PDB entry 2H7Q (47)), (S)-(−)-nicotine (PDB entry 1P2Y (48)) and metyrapone (PDB entry 1PHG (49)). Chemical structures of all tethered substrate analogs used in this study and previously reported are summarized in the Table 1. Helical displacement relative to the closed conformation (PDB entry 2CPP) was calculated based on the coordinate difference of Cα atoms at the helix center: Lys178 (F helix) and Lys197 (G helix). Rotation of the helix axes relative to the closed conformation (PDB entry 2CPP) was determined by calculation of the angle between the helix axes in the superimposed structures either at Lys178 or at Lys197. Helix axis parameters were determined using the algorithm implemented in the program HELO (50). The helix packing angle and the distance between F and G helices were determined using the program C-HELIX (51).
RESULTS
Structures of P450cam bound to tethered substrates
The use of tethered substrate analogs allowed the trapping of a range of conformational states of the P450cam substrate access channel. X-ray crystal structures were solved for P450cam crystallized in the presence of each member of the library of tethered substrates shown in Table 1. These molecules contain adamantane substrate analogs tethered to surface exposed groups by linkers of variable length and composition. In each case, the substrate binding channel was observed in an open conformation, in contrast to the closed camphor bound state. For six of the new structures, the electron density was very well defined for the complete length of the tethered substrate, and the well ordered portions are shown in red in Table 1. In other cases, additional electron density was apparent within the substrate channel compared with the solvent-filled open state of substrate-free enzyme (18), and in addition, the axial water was absent from the distal heme face (Table 1). This suggests partial occupancy of the channel by these tethered substrates. In only two cases, AdaC2-Etg-Boc and AdaC3-C6-Dans, where the axial water was present and the electron density was not well-resolved, was it not possible to verify occupation of the channel by the ligand. The most significant specific interaction observed between the tethered substrates and the protein is a hydrogen bond between Tyr-96 and the carbonyl oxygen for those analogs containing the AdaC1- and AdaC2- linkage. The loss of this hydrogen bond appears related to the disorder of B′ helix, as residues in the B′ helix were not found in the electron density map of the most fully open structures (Table 1). Further analysis of the tethered substrate conformation for these structures and its effects on the enzyme active site are described in a separate manuscript.
The axial heme bound water is absent in some tethered substrate bound structures, but is present in others (Table 1), and this variation is attributed to differences in protein conformation and the location and occupancy of substrates in the active site. For example, electron density for the water was not evident in the structures of P450cam-I with the exception of 3OH-AdaC1-C8-Dans, in which the hydroxyl of the tethered substrate forms a hydrogen bond with the axial water. On the other hand, the distal water was observed for most of the P450cam-O structures where the electron density for the substrates is less well defined. This is consistent with previous observations that the axial water is displaced by tight-binding substrates (52). However, we cannot exclude the possibility that variation of distal heme water occupation results from partial X-ray induced heme reduction for some structures. Such effects are unlikely be the source of changes in conformational state for the following reasons. While redox dependent changes in backbone dynamics (53) and hydrogen-deuterium exchange rates (54) have been observed, these changes are most prevalent in the B′ and C helices rather than the F and G helices. In addition, the camphor-bound ferrous P450cam structure has been shown to very similar to the ferric form (45), and our structures of substrate bound ferric enzyme are in good agreement with these closed conformations (18). Finally, the structure at 2.0 Å resolution for the dithionite reduced P450cam in complex with AdaC1-C8-Dans is identical to the ferric enzyme within an RMSD of 0.2 Å (unpublished data).
In order to determine whether crystal packing influences the resultant structures, protein-protein interfaces in the crystal were analyzed for all structures reported in the current study. PISA analysis (55) verifies that the protein exists as a monomer in solution and that the largest packing interface in each conformational state varies between 500–700 Å2, suggesting no extensive interface is present. The residues around the substrate channel involved in packing interactions with the neighboring molecules do not differ significantly (Figure S1). In addition, the same unit cell (P212121) accommodates both P450cam-I and P450cam-O structures (Table S2) and the exception to this unit cell (P21, PDB entry 1K2O) indicates that the P450cam-O conformation is not determined by the P212121 crystal form. Finally, the P450cam-C exists in a variety of crystal forms (Table S2), but the alternative lattices do not affect the protein conformation. All of these observations suggest the different structures in P450cam-O, -I and –C do not result in differences in packing interactions.
Clusters of protein conformational states
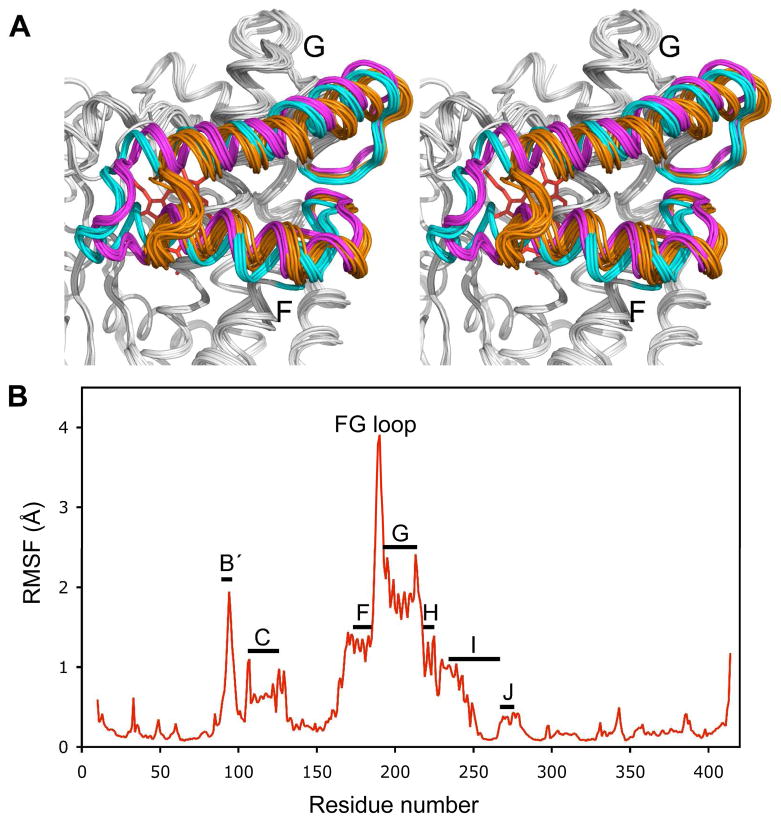
The family of protein structures described above and solved previously displayed a distribution of conformations that range between the completely closed camphor bound state (3) and the fully open form recently reported for P450cam crystallized in the absence of camphor (18). Overlaid in Figure 1A are 30 structures, including the 12 structures reported here (Table S1) and 18 previously reported complexes. A range of protein conformational states are evident, with the most prominent RMS fluctuations from the average structure occurring in the B′, F and G helices and the F-G loop and to a lesser degree in the C, H and I helices (Figure 1B). It is also apparent that the variations in the substrate access channel appear to cluster into distinct families of conformations as indicated by the color scheme of Figure 1. As also indicated in Figure 1B, a number of regions adjacent to, or in contact with the F and G helices appear to undergo minimal fluctuations within this family of structures, suggesting that the conformational variations are highly localized to the elements defining the substrate binding channel. One exception is the C helix which is a part of the proposed putidaredoxin (Pdx) binding site (56).
Figure 1.
Crystal structures of P450cam bound to substrates found in Table 1. (A) Stereo view of superimposed structures of the Cα backbone. (B) Average pair-wise RMSD per residue (RMSF) along the sequence for these structures.
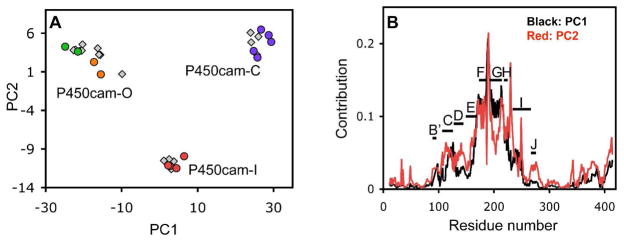
Principal component analysis (PCA) of the combined set of structures shown in Figure 1 was used to characterize the range of observed conformational sub-states. PCA has been used to characterize differences within sets of both experimentally derived structures (57, 58) and those traversed in simulated molecular dynamics trajectories (59, 60). By reducing the number of correlated variables in multiple sets of structural coordinates to those that contribute most to the covariance, PCA has been useful in identifying the number and nature of the primary modes of protein dynamics. PCA analysis of the combined set of P450cam structures described above shows that the first two principal components account for 93% of the covariance (Figure S2) and thus provide a useful description of the conformational space sampled by the structural data. Figure 2A shows a plot of each of these structures projected onto the plane of the first two principal components (PC1 and PC2). The data are clearly clustered into three distinct groups, which we hereafter refer to as P450cam-C (closed), P450cam-I (intermediate) and P450cam-O (open). The lack of additional clustering along the third principal component axis (Figure S2) also indicates that the range of structural differences is well described by only the first two principal components. The P450cam-C cluster includes the previously well characterized closed conformation of the enzyme as typified by complexes with small molecule inhibitors and substrates such as camphor (3, 46–49). At the other end of the range, at the left end of the P450cam-O cluster of Figure 2A, is the recently described open conformation seen in the absence of any substrate (18). It is particularly striking that the conformations for the library of tethered substrates are not evenly distributed across these extremes, but instead fall into three distinct clusters of states.
Figure 2.
Principal component analysis of P450cam crystal structures. (A) 2D plot of the first two principal components for each structure. Data points representing repeat structures are shown in different colors: camphor (purple), AdaC1-C8-Dans (red), AdaC2-C8-Dans (orange) and substrate-free open conformations (green). (B) The contribution of each residue in the sequence to the first (black) and the second (red) principal components.
PCA analysis also provides insight into the nature of the structural differences between these three conformational sub-states. Shown in Figure 2B are the per-residue contributions to the structural changes for the first two principal components. Principal component 1 (PC1) and principal component 2 (PC2) involve similar contributions across several regions of the protein. The F-G loop and the F helix make the largest contribution to both PC1 and PC2, indicating a highly co-variational movement of these structural elements for either of these components. However, Figure 2B also shows distinct differences between PC1 and PC2, in which the G helix makes a larger contribution to PC1 than it does to PC2. In addition, PC2 contains a larger contribution than PC1 for the H-I surface loop centered near Gly230 and a narrow region near Asp251 at the position of the bulge in the center of the I helix. As seen in Figure 2A, the differences between the P450cam-C and P450cam-O conformational groups occur almost entirely with a change in PC1, while the P450cam-I states involve contributions from both PC1 and PC2. Thus, the difference between the fully closed P450cam-C and the partially open P450cam-I conformations can be seen as arising from a complex coordinated movement involving the F helix, the F-G and H-I surface loops and the I helix bulge, while the G helix remains relatively unchanged. In contrast, the largest differences between P450cam-C and the fully open P450cam-O conformations involve a change in both the F and G helices and the intervening F-G loop.
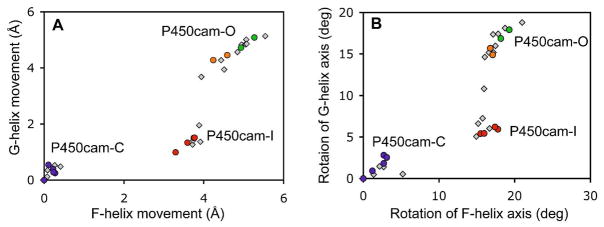
Specific displacements and relative orientations for the F and G helices were examined for these structures to obtain a physical picture of the changes associated with the inter-conversion of the P450cam-C, P45cam-I and P450cam-O states (Graphical description of these parameters are shown in Figure S3). The mean helix displacement and orientation, measured at the helix midpoint, were calculated relative to the closed camphor bound state (PDB entry 2CPP) and plotted in Figure 3. As with PCA analysis, the structures appear distributed into three clusters of conformations. Relative to the P450cam-C closed conformation, the P450cam-I intermediate results from an average retraction of the F helix by 3.7 Å, while the G helix moves in the same direction by only 1.5 Å. However, the difference between P450cam-C and P450cam-O results from a larger and almost equal displacement of both the F and G helices of 4.8 Å and 4.9 Å respectively. As shown in Figure 3B, a similar pattern is observed for the relative helix orientations. Because of distinct movements of the F and G helices in the transition from P450cam-C to the P450cam-I and the P450cam-O states, the F-G loop arrangement is different in the two open conformations. In P450cam-C, the F-G loop is bent toward the solvent accessible surface of the F and G helices and hydrophilic interactions between residues in the F helix and the F-G loop appear to stabilize this conformation (Figure S4). However, in the P450cam-I state, the F helix movement leaves the F-G loop behind (Figure 1), and hydrophilic contacts are lost (Figure S4). Despite significant changes in local contacts, backbone φ and ψ angles around the F-G loop are very similar in the three conformational states (Figure S5). Additional movement of the G helix in the P450cam-O state eventually accompanies movement of the F-G loop with recovery of contacts between the F helix and the F-G loop (Figure S4). These observations indicate that the F-G loop moves as a unit, and as part of the G helix.
Figure 3.
Geometric analysis of the F and G helical movements of P450cam structures relative to that of the camphor bound state (PDB entry 2CPP). Data points from repeat structures are colored as described in Figure 2. (A) Translational movement of the F and G helices. (B) Rotation of the F and G helical axis.
The scatter of structural coordinates for each cluster suggests that both P450cam-C and P450cam-I are well defined states with a small distribution from the average, while P450cam-O represents a more widely distributed set of open conformations. In several cases, redundant structures were determined from separate crystals of the same protein/ligand combination, and these redundant structures are shown in the same color in Figures 2 and 3. The observed average standard deviation for F and G helix displacement within the P450cam-C, P450cam-I and P450cam-O clusters was 0.16 Å, 0.24 Å and 0.46 Å respectively, while that for the redundant structures was 0.18 Å. This suggests that P450cam-C and P450cam-I represent well defined conformations in which individual structures do not vary by more than that seen for crystal-to-crystal variation, while the P450cam-O cluster is more broadly distributed over a range of open conformations. One previously reported ligand, AdaC1-C4-Dans appeared at the boundary between P450cam-I and P450cam-O in the plot of rotations of F and G helices (Figure 3B).
Changes in intra-protein interactions
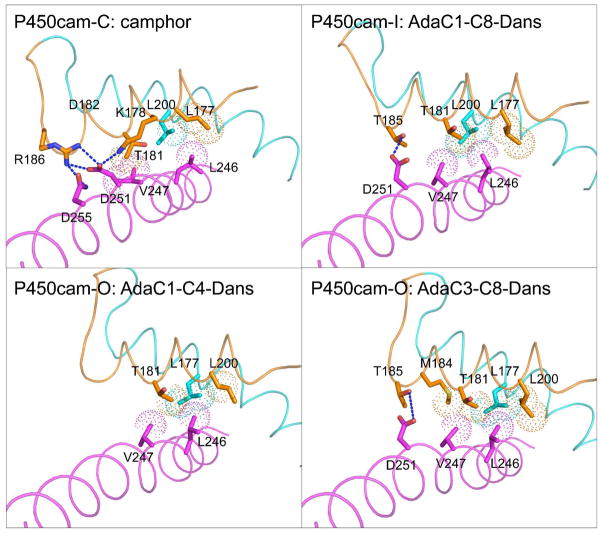
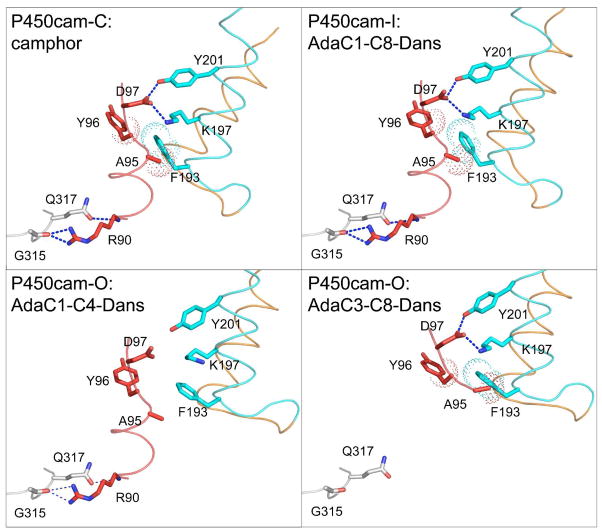
The distinct conformational states seen for P450cam result in a range of altered interactions between protein secondary structural elements. As the structure moves from the closed conformation to either of the open forms, a critical interaction between the F helix and the I helix bulge is disrupted. As shown in Figure 4, P450cam-C contains a bifurcated salt-bridge between Lys178 and Arg186 in the F helix and Asp251 in the I helix. This salt-bridge is lost in both the P450cam-I and P450cam-O conformations, resulting in the retraction of the F helix for both of these open conformations. Additional hydrophobic interactions between Leu177, Leu200 and Leu246 of the F, G and I helices are maintained in all three conformations, as they are near the point at which the F helix pivots over the I helix.
Figure 4.
Inter-helical contacts between F (orange), G (cyan) and I (magenta) helices are shown for P450cam bound to camphor (P450cam-C), AdaC1-C8-Dans (P450cam-I), AdaC1-C4-Dans (P450cam-O), and AdaC3-C8-Dans (P450cam-O). Salt bridge and hydrogen bonding interactions are shown in blue dash, while hydrophobic contacts are shown in as dotted spheres.
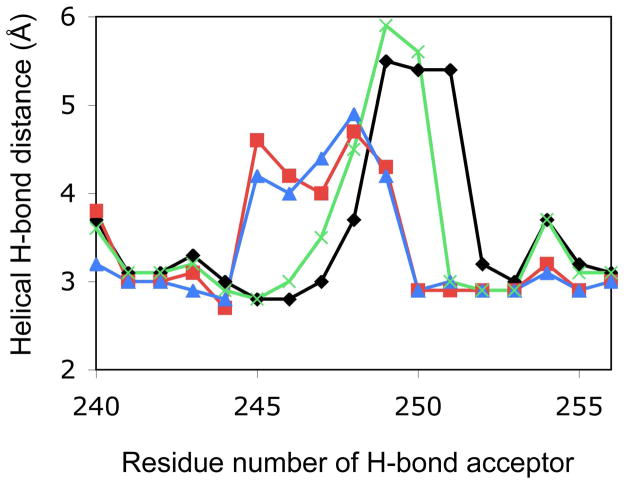
The loss of the salt-bridge between the F and I helix also accompanies changes in the I helix bulge, which provides a pocket for O2 and enables its hydrogen bonding to the catalytically important side chain of Thr252 (Figure 5) (61). In the P450cam-C state, the I helix bulge results from loss of normal hydrogen bond acceptor (residues 249-251) and donor (residues 253-255) interactions (Figure 5). Because the bulge is shorter than one turn of the α-helix, the I-helix appears kinked in the middle (Figure S6). When the F helix retracts in the open conformations, the α-helical bulge is shifted towards the N-terminus of the I helix and becomes distributed over almost two helical turns, and the distance between α-helical hydrogen donors and acceptors becomes shorter, but not enough to form hydrogen bonds. This shift results in a straightening of the I helix (Figure S6). Interestingly, because of this bulge shift, the distance between the normal Gly248 hydrogen bond acceptor and Thr252 donor increases from 3.7 Å (PDB entry 2CPP; P450cam-C) to 4.9 Å (PDB entry 3L62; P450cam-O). This, in turn, widens the α-helical groove near the active site and makes room for a “catalytic water” as is seen in the ferrous dioxygen-bound P450cam structure (45) (Figure S6). Irrespective of the wider α-helical groove around Gly248 and Thr252, the unusual hydrogen bond between Gly248 backbone carboxyl and the Thr252 side chain hydroxyl, which has been thought to be responsible for the I helix bulge, was still preserved in each of the P450cam-I and P450cam-O conformations.
Figure 5.
Geometrical analysis of the I helix bulge. Distance between the hydrogen bond acceptor (i residue) and donor (i + 4 residue) in the I helix is plotted for P450cam-C (black, PDB entry 2CPP), P450cam-I (red, PDB entry 1RE9), P450cam-O (blue, PDB entry 3L62) and oxyferrous P450cam (green, PDB entry 1DZ8).
The G helix is seen to make a distinct switch in its interactions with other secondary structural elements in the protein as the channel converts from closed P450cam-C state through the P450cam-I intermediate to the fully open P450cam-O state. As the F helix initially retracts from P450cam-C to P450cam-I, the G helix remains anchored by contacts with the B′ helix as shown in Figure 6. These contacts include a bifurcated hydrogen bond between Asp97 and two residues of the G helix, Tyr201 and Lys197, and also by hydrophobic interactions between Ala95 and Phe193 (Figure 6). These interactions appear to prevent the G helix from following the F helix as it retracts during the P450cam-C to P450cam-I transition. As the channel opens further during the P450cam-I to P450cam-O conversion, the G helix finally retracts. However, as shown in Figure 6, in order for the interactions between the G and B′ helices to be maintained, the B′ helix itself unwinds, loses its K+ binding site and becomes disordered, as recently described for the substrate-free open conformation (18). One structure, the complex with AdaC1-C4-Dans, which appears at the boundary between the P450cam-I and P450cam-O conformations, retains an ordered B′ helix as the G helix retracts, but it does this at the expense of breaking its interactions with the G helix. As a result of the separate movements of the F and G helix, the inter-helical angle undergoes a small shift as one proceeds from P450cam-C to P450cam-I, but this angle is restored for the P450cam-O cluster (Figure S7). Figure S7 also shows that the F and G helices for P450cam-C and P450cam-O pack against each other in a classic anti-parallel ridge-into-groove interaction with an inter-helical angle of −150° (51). However, for the group of P450cam-I structures, the inter-helical angle is reduced slightly to approximately −158°. As this inter-helical angle changes during the P450cam-C to P450cam-I transition, several hydrophobic interactions between the F and G helices are initially disrupted, but are restored in P450cam-O (Figure S8).
Figure 6.
Interactions between the B′ and G helices shown for P450cam bound to camphor (P450cam-C), AdaC1-C8-Dans (P450cam-I), AdaC1-C4-Dans (P450cam-O), and AdaC3-C8-Dans (P450cam-O). B′, F and G helices are colored in red, orange and cyan, respectively. Residues from the β3 strand is colored in white. Electrostatic interactions are shown in blue dash, while hydrophobic contacts are shown in as dotted spheres.
DISCUSSION
Conformational clusters in P450cam
The process by which diverse P450s bind substrates and control the specificity and functional coupling of catalysis have been extensively studied (11, 62, 63). While some forms such as EryK (15) and PikC (16) have been shown to exist in both closed and open states in the absence of substrates, the extent of structural changes traversed by a given enzyme during substrate binding is not well understood for any P450. Such changes are clearly necessary for some forms, most notably P450cam, which binds substrate in a completely closed solvent inaccessible conformation (3, 4). The open conformation observed for P450cam in the presence of large tethered substrates has demonstrated its ability to exist in open states (64–66), but only recently has this open conformation been observed in the absence of substrate (18). The open conformations of the substrate-free and the most open of the tethered substrate-bound forms are very similar (RMSD of 0.5 Å), suggesting the open conformation observed for the tethered-substrate bound forms are not induced by the tethered substrates, but are already pre-existing in the conformational space of P450cam. Thus, examination of the range of structures exhibited while bound to a library of tethered substrate analogs may be very useful in revealing the distribution of conformational states sampled by the protein during substrate recognition and catalysis.
The 12 crystal structures presented here combined with analysis of 18 previously determined structures show how a wide variety of compounds are accommodated at the active site of P450cam. In response to variation in the composition over the length of the tethered substrate, the protein displays a significant variation in the structure of the substrate access channel. However, the structures are not evenly distributed within this conformational space, as might be expected for a highly plastic substrate binding site, but are instead highly clustered into three families of states, P450cam-C, P450cam-I, and P450cam-O, indicating closed, intermediate and fully open conformations. Both P450cam-C and P450cam-I appear to be well defined states that show a narrow structural distribution that is not larger than that seen in repeat determinations of the same structure, while P450cam-O states are spread over a broader distribution, suggesting an ensemble of open conformations. The three conformations differ largely by movements of secondary structural elements that surround the substrate access channel, including the B′, F, G, and I helices. One exception to this is the small movement of the C helix which forms part of the proposed Pdx binding site, suggesting that the enigmatic role of Pdx as a functional effector may be related to the conformational shifts involving the open to closed transition. Recent studies have suggested that Pdx binding forces selection of a closed conformational transition that prevents escape of substrate (67).
Each of the P450cam-O, P450cam-I and P450cam-C conformational states are observed in the presence of multiple ligands (Table 1). For example, the P450cam-O conformation, which is observed for substrate-free P450cam, lies within 0.4 Å RMSD of that bound to the Ada-C2-Etg-Dans probe. In another example, the well defined P450cam-I state is observed with a variety of ligands that vary over the substrate, linker and surface resident positions (Table 1). Finally, a very narrow distribution is observed for all of the reported P450cam-C conformations despite accommodating a range of small substrates and inhibitors. On the other hand, minor differences in the tethered substrate composition are observed to cause significant changes in protein conformation. For example, AdaC1-C8-Dans and AdaC2-C8-Dans differ by only a single-bond shift in the position of the amide linkage, yet results in a significant shift from the P450cam-I to the P450cam-O state (Table 1). Finally, the axial water ligand, associated with the spin-state shift upon conversion to the camphor bound state is absent from each of the P450cam-C forms but is sometimes present and sometimes missing in both P450cam-I and P450cam-O (Table 1). These results show that a particular protein-substrate interaction does not always lead to a unique conformational outcome, and that each of the conformational states is able to accommodate a variety of probe molecules. Our analysis suggests that the protein displays an innate preference for a small number of pre-existing conformational sub-states, and each of these states can accommodate a range of ligands without undergoing a continuous plastic response of the active site structure. This suggests that substrate recognition by P450cam occurs largely by conformational selection, and is consistent with the conclusions of a recent study of CYP119 (29) and EryK (15).
Multi-step pathway for channel closure
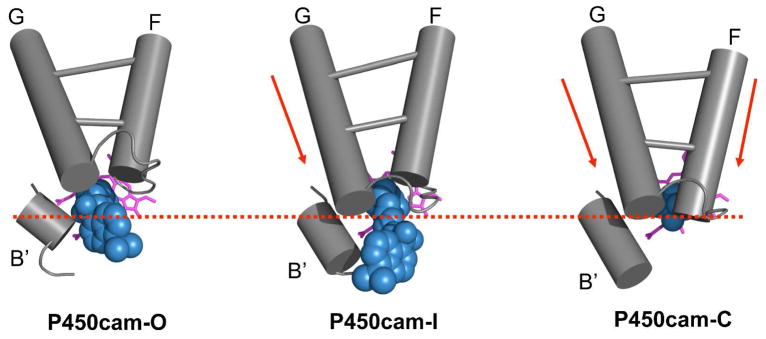
Analysis of the library of structures presented here provides the basis for a proposed multi-step pathway for closure of the substrate access channel around substrate. When each of the structures is displayed in a molecular animation ordered by the principal component displacement, the resulting movie (Supporting Information) gives a clear impression of a coordinated two-step trajectory involving a distinct half-open intermediate. As shown in cartoon model of Figure 7, the P450cam-I state represents a distinct sequential intermediate in the stepwise closure of the channel, P450cam-O → P450cam-I → P450-C. Conversion of P450cam-O to P450cam-I occurs by closure of the G helix over the active site, allowing the ordering of the B′ helix and creation of the K+ binding site, which has been shown to enhance camphor binding by 10 fold (68). Final conversion from P450cam-I to P450cam-C involves further closure of the F helix and formation of the salt bridge between Arg186 and Asp251. This later interaction is crucial for the formation of the well known I helix bulge which alters positioning of Thr-252 and a catalytic water at the active site (61). The fact that the I helix bulge is only formed upon final closure from P450cam-I to P450cam-C may have significant mechanistic implications that will be explored in a separate publication.
Figure 7.
Model for the multi-step closure of P450cam. Helices are represented by cylinders, tethered substrates are shown as a CPK model in blue: camphor (P450cam-C), AdaC1-C8-Dans (P450cam-I) and AdaC1-C4-Dans (P450cam-O), and heme is shown in magenta. Red dashed line was drawn to help track changes between conformational states.
Recent results have shown that the full range of conformations observed in this study is also sampled by adaptive accelerated molecular dynamics (69). Notably, beginning with P450cam-C, the simulations visit both P450cam-I and P450cam-O as distinct states during the molecular dynamics trajectory. This provides strong support that the experimentally determined family of structures described here represent snapshots of the conformational dynamics seen in this enzyme.
In conclusion, the structures presented here show that P450cam samples at least three distinct conformational states and suggests that substrate recognition occurs by a process involving an ordered multi-step closure of the open P450cam-O conformation around the substrate by way of a well defined intermediate structure, P450cam-I. At present it is not clear if the P450cam-I or P450cam-O conformations are catalytically competent. However, the discrete conformational states observed for the 30 P450cam structures analyzed in this study suggests that the initial interaction of camphor with P450cam-O results in a shift in equilibrium from P450cam-O to P450cam-I by conformational selection. Subsequent conversion to P450cam-C could also involve selection of substrate orientation within the channel, which may guide the protein over conformational barriers between pre-exsiting conformations.
Supplementary Material
Acknowledgments
The authors thank Dr. A. Annalora for valuable discussions. We also thank the staff at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Lightsource is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Abbreviations
- RMSD
root mean square deviation
- PCA
principal component analysis
- P450cam-C
P450cam in the closed conformation
- P450cam-I
P450cam in the intermediate open conformation
- P450cam-O
P450cam in the open conformation
- Pdx
putidaredoxin
Footnotes
This work was supported by grant GM41049 from the NIH.
The atomic coordinate and structures factors presented in this word were deposited in the Protein Data Bank under the codes 3P6M, 3P6N, 3P6O, 3P6P, 3P6Q, 3P6R, 3P6S, 3P6T, 3P6U, 3P6V, 3P6W and 3P6X.
SUPPORTING INFORMATION AVAILABLE: Table S1, X-ray crystallographic statistics for all structures reported in this study; Figure S1, crystal packing interface around the substrate channel; Figure S2, 2D plots of first three principal components and the magnitude of covariance from each principal component; Figure S3, graphical description of geometrical parameters analyzed in this study; Figure S4, analysis of contacts around the F-G loop; Figure S5, backbone angles around the F-G loop; Figure S6, geometry of the I-helix bulge; Figure S7, analysis of packing angles and distances between the F and G helices in P450cam structures; Figure S8, analysis of contacts between the F and G helices for P450cam-C, P450cam-I and P450cam-O; Animated video showing a proposed model of the substrate channel opening. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Nelson DR. The cytochrome p450 homepage. Hum Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guengerich FP. Human cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P-450: Structure, Mechanism, and Biochemistry. 3. Kluwer Academics/Plenum; New York: 2005. pp. 377–530. [Google Scholar]
- 3.Poulos TL, Finzel BC, Gunsalus IC, Wagner GC, Kraut J. The 2.6-A crystal structure of Pseudomonas putida cytochrome P-450. J Biol Chem. 1985;260:16122–16130. [PubMed] [Google Scholar]
- 4.Poulos TL, Finzel BC, Howard AJ. High-resolution crystal structure of cytochrome P450cam. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 5.Johnson EF, Stout CD. Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem Biophys Res Commun. 2005;338:331–336. doi: 10.1016/j.bbrc.2005.08.190. [DOI] [PubMed] [Google Scholar]
- 6.Poulos TL, Johnson EF. Structures of cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P-450: Structure, Mechanism, and Biochemistry. 3. Kluwer Academics/Plenum; New York: 2005. pp. 87–114. [Google Scholar]
- 7.Groves JT. Key elements of the chemistry of cytochrome-P-450 - The oxygen rebound mechanism. J Chem Educ. 1985;62:928–931. [Google Scholar]
- 8.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 9.Pochapsky TC, Kazanis S, Dang M. Conformational plasticity and structure/function relationships in cytochromes P450. Antioxid Redox Signal. 2010;13:1273–1296. doi: 10.1089/ars.2010.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meharenna YT, Li H, Hawkes DB, Pearson AG, De Voss J, Poulos TL. Crystal structure of P450cin in a complex with its substrate, 1,8-cineole, a close structural homologue to D-camphor, the substrate for P450cam. Biochemistry. 2004;43:9487–9494. doi: 10.1021/bi049293p. [DOI] [PubMed] [Google Scholar]
- 11.Pylypenko O, Schlichting I. Structural aspects of ligand binding to and electron transfer in bacterial and fungal P450s. Annu Rev Biochem. 2004;73:991–1018. doi: 10.1146/annurev.biochem.73.011303.073711. [DOI] [PubMed] [Google Scholar]
- 12.Poulos TL, Cupp-Vickery J, Li H. Structural studies on prokaryotic cytochromes P450. In: Ortiz de Montellano PR, editor. Cytochrome P-450: Structure, Mechanism, and Biochemistry. 2. Plenum Press; New York: 1995. pp. 125–150. [Google Scholar]
- 13.Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Proc Natl Acad Sci USA. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-A resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J Biol Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 15.Savino C, Montemiglio LC, Sciara G, Miele AE, Kendrew SG, Jemth P, Gianni S, Vallone B. Investigating the structural plasticity of a cytochrome P450: Three dimensional structures of P450 EryK and binding to its physiological substrate. J Biol Chem. 2009;284:29170–29179. doi: 10.1074/jbc.M109.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherman DH, Li S, Yermalitskaya LV, Kim Y, Smith JA, Waterman MR, Podust LM. The structural basis for substrate anchoring, active site selectivity, and product formation by P450 PikC from Streptomyces venezuelae. J Biol Chem. 2006;281:26289–26297. doi: 10.1074/jbc.M605478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao YH, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. Structure of microsomal cytochrome P4502B4 complexed with the antifungal drug bifonazole - Insight into p450 conformational plasticity and membrane interaction. J Biol Chem. 2006:5973–5981. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- 18.Lee Y-T, Wilson RF, Rupniewski I, Goodin DB. P450cam visits an open conformation in the absence of substrate. Biochemistry. 2010;49:3412–3419. doi: 10.1021/bi100183g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Poulos TL. The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat Struct Biol. 1997;4:140–146. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- 20.Zhao B, Guengerich FP, Bellamine A, Lamb DC, Izumikawa M, Lei L, Podust LM, Sundaramoorthy M, Kalaitzis JA, Reddy LM, Kelly SL, Moore BS, Stec D, Voehler M, Falck JR, Shimada T, Waterman MR. Binding of two flaviolin substrate molecules, oxidative coupling, and crystal structure of Streptomyces coelicolor A3(2) cytochrome P450 158A2. J Biol Chem. 2005;280:11599–11607. doi: 10.1074/jbc.M410933200. [DOI] [PubMed] [Google Scholar]
- 21.Park SY, Yamane K, Adachi S, Shiro Y, Weiss KE, Maves SA, Sligar SG. Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties. J Inorg Biochem. 2002;91:491–501. doi: 10.1016/s0162-0134(02)00446-4. [DOI] [PubMed] [Google Scholar]
- 22.Xu LH, Fushinobu S, Ikeda H, Wakagi T, Shoun H. Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state. J Bacteriol. 2009;191:1211–1219. doi: 10.1128/JB.01276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouellet H, Podust LM, de Montellano PRO. Mycobacterium tuberculosis CYP130: crystal structure, biophysical characterization, and interactions with antifungal azole drugs. J Biol Chem. 2008;283:5069–5080. doi: 10.1074/jbc.M708734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zerbe K, Pylypenko O, Vitali F, Zhang W, Rouset S, Heck M, Vrijbloed JW, Bischoff D, Bister B, Süssmuth RD, Pelzer S, Wohlleben W, Robinson JA, Schlichting I. Crystal structure of OxyB, a cytochrome P450 implicated in an oxidative phenol coupling reaction during vancomycin biosynthesis. J Biol Chem. 2002;277:47476–47485. doi: 10.1074/jbc.M206342200. [DOI] [PubMed] [Google Scholar]
- 25.Poulos TL, Finzel BC, Howard AJ. Crystal structure of substrate-free Pseudomonas putida cytochrome P-450. Biochemistry. 1986;25:5314–5322. doi: 10.1021/bi00366a049. [DOI] [PubMed] [Google Scholar]
- 26.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 27.Annalora AJ, Goodin DB, Hong W, Zhang Q, Johnson EF, Stout CD. The crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism. J Mol Biol. 2010;396:441–451. doi: 10.1016/j.jmb.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lampe JN, Brandman R, Sivaramakrishnan S, Ortiz de Montellano PR. 2D NMR and All-atom molecular dynamics of cytochrome P450 CYP119 reveal hidden conformational substates. J Biol Chem. 2010;285:9594–9603. doi: 10.1074/jbc.M109.087593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nickerson DP, Wong LL. The dimerization of Pseudomonas putida cytochrome P450cam: practical consequences and engineering of a monomeric enzyme. Protein Eng. 1997;10:1357–1361. doi: 10.1093/protein/10.12.1357. [DOI] [PubMed] [Google Scholar]
- 31.Jaramillo D, Wheate NJ, Ralph SF, Howard WA, Tor Y, Aldrich-Wright JR. Polyamide platinum anticancer complexes designed to target specific DNA sequences. Inorg Chem. 2006;45:6004–6013. doi: 10.1021/ic060383n. [DOI] [PubMed] [Google Scholar]
- 32.Dunn AR, Hays AM, Goodin DB, Stout CD, Chiu R, Winkler JR, Gray HB. Fluorescent probes for cytochrome p450 structural characterization and inhibitor screening. J Am Chem Soc. 2002;124:10254–10255. doi: 10.1021/ja0271678. [DOI] [PubMed] [Google Scholar]
- 33.Evans PR. Data reduction. CCP4 Study Weekend Proceedings DL/SCI/R34. 1993:114–122. [Google Scholar]
- 34.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 38.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 39.Lovell SC, Davis IW, Adrendall WB, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by C alpha geometry: phi,psi and C beta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 40.DeLano WL. The PyMOL Molecular Graphics System. 2002 www.pymol.org.
- 41.Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 42.Meilleur F, Dauvergne MT, Schlichting I, Myles DA. Production and X-ray crystallographic analysis of fully deuterated cytochrome P450cam. Acta Crystallogr D Biol Crystallogr. 2005;61:539–544. doi: 10.1107/S0907444905003872. [DOI] [PubMed] [Google Scholar]
- 43.Vidakovic M, Sligar SG, Li H, Poulos TL. Understanding the role of the essential Asp251 in cytochrome p450cam using site-directed mutagenesis, crystallography, and kinetic solvent isotope effect. Biochemistry. 1998;37:9211–9219. doi: 10.1021/bi980189f. [DOI] [PubMed] [Google Scholar]
- 44.Harada K, Sakurai K, Ikemura K, Ogura T, Hirota S, Shimada H, Hayashi T. Evaluation of the functional role of the heme-6-propionate side chain in cytochrome P450cam. J Am Chem Soc. 2008;130:432–433. doi: 10.1021/ja077902l. [DOI] [PubMed] [Google Scholar]
- 45.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. The catalytic pathway of cytochrome p450cam at atomic resolution. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 46.Schlichting I, Jung C, Schulze H. Crystal structure of cytochrome P-450cam complexed with the (1S)-camphor enantiomer. FEBS Lett. 1997;415:253–257. doi: 10.1016/s0014-5793(97)01135-6. [DOI] [PubMed] [Google Scholar]
- 47.Verras A, Alian A, de Montellano PR. Cytochrome P450 active site plasticity: attenuation of imidazole binding in cytochrome P450(cam) by an L244A mutation. Protein Eng Des Sel. 2006;19:491–496. doi: 10.1093/protein/gzl035. [DOI] [PubMed] [Google Scholar]
- 48.Strickler M, Goldstein BM, Maxfield K, Shireman L, Kim G, Matteson DS, Jones JP. Crystallographic studies on the complex behavior of nicotine binding to P450cam (CYP101) Biochemistry. 2003;42:11943–11950. doi: 10.1021/bi034833o. [DOI] [PubMed] [Google Scholar]
- 49.Poulos TL, Howard AJ. Crystal structures of metyrapone- and phenylimidazole-inhibited complexes of cytochrome P-450cam. Biochemistry. 1987;26:8165–8174. doi: 10.1021/bi00399a022. [DOI] [PubMed] [Google Scholar]
- 50.Tatulian SA. Determination of helix orientations in proteins. Comput Biol Chem. 2008;32:370–374. doi: 10.1016/j.compbiolchem.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Dalton JA, Michalopoulos I, Westhead DR. Calculation of helix packing angles in protein structures. Bioinformatics. 2003;19:1298–1299. doi: 10.1093/bioinformatics/btg141. [DOI] [PubMed] [Google Scholar]
- 52.Raag R, Poulos TL. Crystal structures of cytochrome P-450CAM complexed with camphane, thiocamphor, and adamantane: factors controlling P-450 substrate hydroxylation. Biochemistry. 1991;30:2674–2684. doi: 10.1021/bi00224a016. [DOI] [PubMed] [Google Scholar]
- 53.Pochapsky SS, Dang M, Ouyang B, Simorellis AK, Pochapsky TC. Redox-Dependent Dynamics in Cytochrome P450(cam) Biochemistry. 2009;48:4254–4261. doi: 10.1021/bi900002k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamuro Y, Molnar KS, Coales SJ, OuYang B, Simorellis AK, Pochapsky TC. Hydrogen-deuterium exchange mass spectrometry for investigation of backbone dynamics of oxidized and reduced cytochrome P450cam. J Inorg Biochem. 2008;102:364–370. doi: 10.1016/j.jinorgbio.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 56.Pochapsky TC, Lyons TA, Kazanis S, Arakaki T, Ratnaswamy G. A structure-based model for cytochrome P450cam-putidaredoxin interactions. Biochimie. 1996;78:723–733. doi: 10.1016/s0300-9084(97)82530-8. [DOI] [PubMed] [Google Scholar]
- 57.Grant BJ, McCammon JA, Caves LS, Cross RA. Multivariate analysis of conserved sequence-structure relationships in kinesins: coupling of the active site and a tubulin-binding sub-domain. J Mol Biol. 2007;368:1231–1248. doi: 10.1016/j.jmb.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 58.Teodoro ML, Phillips GN, Kavraki LE. Understanding protein flexibility through dimensionality reduction. J Comput Biol. 2003;10:617–634. doi: 10.1089/10665270360688228. [DOI] [PubMed] [Google Scholar]
- 59.Amadei A, Linssen AB, Berendsen HJ. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 60.Amadei A, Linssen AB, de Groot BL, van Aalten DM, Berendsen HJ. An efficient method for sampling the essential subspace of proteins. J Biomol Struct Dyn. 1996;13:615–625. doi: 10.1080/07391102.1996.10508874. [DOI] [PubMed] [Google Scholar]
- 61.Nagano S, Poulos TL. Crystallographic study on the dioxygen complex of wild-type and mutant cytochrome P450cam. Implications for the dioxygen activation mechanism. J Biol Chem. 2005;280:31659–31663. doi: 10.1074/jbc.M505261200. [DOI] [PubMed] [Google Scholar]
- 62.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and chemistry of cytochrome P450. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 63.Isin EM, Guengerich FP. Substrate binding to cytochromes P450. Anal Bioanal Chem. 2008;392:1019–1030. doi: 10.1007/s00216-008-2244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dmochowski IJ, Crane BR, Wilker JJ, Winkler JR, Gray HB. Optical detection of cytochrome P450 by sensitizer-linked substrates. Proc Natl Acad Sci USA. 1999;96:12987–12990. doi: 10.1073/pnas.96.23.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunn AR, Dmochowski IJ, Bilwes AM, Gray HB, Crane BR. Probing the open state of cytochrome P450cam with ruthenium-linker substrates. Proc Natl Acad Sci USA. 2001;98:12420–12425. doi: 10.1073/pnas.221297998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hays AMA, Dunn AR, Chiu R, Gray HB, Stout CD, Goodin DB. Conformational states of cytochrome P450cam revealed by trapping of synthetic molecular wires. J Mol Biol. 2004;344:455–469. doi: 10.1016/j.jmb.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 67.Asciutto EK, Madura JD, Pochapsky SS, Ouyang B, Pochapsky TC. Structural and Dynamic Implications of an Effector-Induced Backbone Amide cis-trans Isomerization in Cytochrome P450(cam) J Mol Biol. 2009;388:801–814. doi: 10.1016/j.jmb.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Di Primo C, Hui Bon Hoa G, Douzou P, Sligar S. Mutagenesis of a single hydrogen bond in cytochrome P-450 alters cation binding and heme solvation. J Biol Chem. 1990;265:5361–5363. [PubMed] [Google Scholar]
- 69.Markwick PR, Pierce LC, Goodin DB, McCammon JA. Adaptive accelerated molecular dynamics (Ad-AMD) reveals the molecular plasticity of P450cam. doi: 10.1021/jz101462n. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.