Abstract
Hydroxylated metabolites of polychlorinated biphenyls (OHPCBs) interact with rat sulfotransferase 1A1 (rSULT1A1) as substrates and inhibitors. Previous studies have shown that there are complex and incompletely understood structure-activity relationships governing the interaction of rSULT1A1 with these molecules. Furthermore, modification of the enzyme with glutathione disulfide (GSSG) results in the conversion of some OHPCBs from inhibitors to substrates. We have now examined estimated values for the acid-dissociation constant (Ka) and the octanol-water distribution coefficient (D), as well as experimentally determined dissociation constants for enzyme complexes, to assist in the prediction of interactions of OHPCBs with rSULT1A1. Under reducing conditions, initial velocities for rSULT1A1-catalyzed sulfation exhibited a positive correlation with pKa and a negative correlation with log D of the OHPCBs. IC50 values of inhibitory OHPCBs decreased with decreasing pKa values for both the glutathione (GSH)-pretreated and GSSG-pretreated forms of rSULT1A1. Comparison of GSH- and GSSG-pretreated forms of rSULT1A1 with respect to binding of OHPCB in the presence and absence of adenosine 3’,5’-diphosphate (PAP) revealed that the dissociation constants with the two redox states of the enzyme were similar for each OHPCB. Thus, pKa and log D values are useful in predicting the binding of OHPCBs to the two redox forms of rSULT1A1 as well as the rates of sulfation of those OHPCBs that are substrates. However, the differences in substrate specificity for OHPCBs that are seen with changes in redox status of the enzyme are not directly related to specific structural effects of individual OHPCBs within inhibitory enzyme-PAP-OHPCB complexes.
Keywords: log D, OHPCB, glutathione disulfide, PCB, pKa, sulfotransferase
Introduction
The mammalian cytosolic sulfotransferases (SULTs) constitute a superfamily of enzymes that are critical to many physiologic, pharmacologic and toxicologic processes [1–6]. Among the various isoforms of these enzymes, rat SULT1A1 (rSULT1A1) has been extensively used as a model for mechanistic studies on the interaction of various compounds with SULTs [2]. Among the molecules examined with rSULT1A1 are the hydroxylated metabolites of polychlorinated biphenyls (OHPCBs) [7], and these provide a unique series of substrates and inhibitors with which to examine the molecular basis for specificity of the enzyme under both reduced and partially oxidized conditions.
Polychlorinated biphenyls (PCBs) are world-wide environmental contaminants with varied toxicities and associated health effects [8–10]. The OHPCBs can be formed metabolically through cytochrome P450-catalyzed reactions [10–13], although there is also evidence for abiotic sources [14]. OHPCBs are present in humans [15–19], and have been shown to interact with several isoforms of SULTs. For example, OHPCBs have been observed to potently inhibit human estrogen sulfotransferase (hSULT1E1), and this has been proposed to be a mechanism for the estrogen-like effect of PCBs in humans and other animals due to the role of this SULT in the inactivation of estrogens via sulfation [20]. Inhibition of hSULT1A1-, hSULT1A3-, and rSULT1C1-catalyzed sulfation of thyroid hormones by OHPCBs has also been suggested to play a part in PCB-mediated changes in thyroid hormone levels [21]. Studies on the human hydroxysteroid sulfotransferase, hSULT2A1, also indicate that OHPCBs may be involved in the inhibition of the sulfation of hydroxysteroids, bile acids, and xenobiotics catalyzed by this enzyme [7, 22]. Although the highly chlorinated congeners of PCBs are especially persistent in the environment and in mammals, the lower chlorinated PCBs are volatile components of both indoor and outdoor air [23–25], and it is increasingly apparent that these are readily metabolized in mammals to OHPCBs. Our current studies are focused on these lower chlorinated PCBs (e.g., those containing from one to four chlorine atoms) due to the likelihood of airborne exposure, metabolism to OHPCBs, and the increasing discovery of toxicities associated with the oxidized metabolites.
We have recently reported that OHPCBs either inhibit or undergo sulfation in reactions catalyzed by rSULT1A1 [7]. These results provided some initial structure-activity relationships for OHPCBs with rSULT1A1, and also showed that, after treatment of the enzyme with glutathione disulfide (GSSG), the specificity of the enzyme for some OHPCBs was altered, with some inhibitors for the reduced enzyme becoming substrates for the oxidized rSULT1A1[7]. A more detailed understanding of the fundamental structure-activity relationships for interactions of OHPCBs with rSULT1A1 is the subject of the current investigation.
As is common with other family 1 SULTs, rSULT1A1 displays substantial inhibition with increasing concentrations of many substrates, including OHPCBs [7, 26, 27]. This substrate inhibition is due to the formation of dead end complexes between substrate, enzyme and the co-product of sulfation, adenosine-3’,5’-diphosphate (PAP) [26, 27]. The release of PAP from these dead-end complexes becomes a rate-limiting component of the rSULT1A1-catalyzed reaction [27], and changes in the conformation at the PAP/PAPS-binding site of the enzyme may affect catalytic function and inhibition through facilitation of PAP-release [2, 27]. Thus the present studies are focused on molecular characteristics of OHPCB-substrates that influence the rate of reaction, the use of calculated estimates of physicochemical parameters in the prediction of interactions of OHPCBs as substrates and inhibitors, and the potential role of the redox status of the enzyme in determining its binding interactions with OHPCBs and PAP.
2. Materials and Methods
2.1. Chemicals
The synthesis and characterization of 4’-OH PCB 3, 4’-OH PCB 6, 4-OH PCB 8, 4’-OH PCB 9, 4’-OH PCB 12, 4-OH PCB 14, 4’-OH PCB 25, 4’-OH PCB 33, 4-OH PCB 34, 4’-OH PCB 35, 6’-OH PCB 35, 4-OH PCB 36, 4’-OH PCB 36, 4’-OH PCB 68, and 4’-OH PCB 79 have been reported previously [7, 28]. Adenosine 3’,5’-diphosphate (PAP), PAP-agarose, glutathione, and glutathione disulfide were obtained from Sigma-Aldrich (St. Louis, MO). Adenosine 3’-phosphate 5’-phosphosulfate (PAPS) was also obtained from Sigma-Aldrich and further purified by a published procedure [29] to a purity greater than 98% as determined by HPLC. The ammonium salt of 8-anilinonaphthalene-1-sulfonic acid (ANS), from Fluka (Steinheim, Germany), and 2'-(or-3')-O-(trinitrophenyl)adenosine 5'-monophosphate (TNP-AMP), from Invitrogen (Eugene, OR), were used without further purification. All other chemicals and reagents were of the highest purity commercially available.
2.2. Expression and purification of recombinant rat SULT1A1
Rat SULT1A1 was expressed in recombinant Escherichia coli BL 21 (DE3) cells by using a pET-3c vector as previously described [30]. Cells were grown, cell extract was prepared, and the enzyme was purified by using PAP-agarose affinity column chromatography as recently reported [7]. The protein obtained was homogeneous by SDS-PAGE upon staining with Coomassie brilliant blue. Protein concentrations were determined by the modified Lowry procedure [31], with bovine serum albumin as standard.
2.3. Sulfation of OHPCBs catalyzed by rSULT1A1
Reactions were conducted at 37°C in a total volume of 30 µl containing 0.25 M potassium phosphate buffer at pH 7.0, 200 µM PAPS, 7.5 mM 2-mercaptoethanol, and 10 µM of the stated OHPCB. The OHPCB was added to the reaction mixture using acetone as co-solvent, with a final acetone concentration of 3.3% (v/v). Purified rSULT1A1 (0.75 µg) was added to initiate the reaction. After 6 min incubation, the reaction was stopped by addition of 30 µl of methanol, and the rate of sulfation was determined by HPLC analysis of the substrate-dependent formation of PAP as described previously [32, 33].
2.4. Pretreatment of rSULT1A1 with GSSG and GSH for analysis of the binding of OHPCBs and PAP
Prior to incubations with GSSG and GSH, dithiothreitol (DTT) in the purified rSULT1A1 preparations was removed by PD-10 gel filtration column chromatography. The eluted protein was concentrated by a factor of 5–10 using a 10-ml Amicon stirred cell with a PM-10 membrane (Millipore Corporation, Billerica, MA) [7]. The residual concentration of DTT was below 0.01 mM, as determined by a standard assay for thiols using 5,5’-dithiobis(2-nitrobenzoic acid) [34]. After removal of DTT, rSULT1A1 was incubated with either 1 mM GSH or 1 mM GSSG at 25 °C for 1 h under argon (to prevent auto-oxidation of the enzyme) [7]. The resulting enzyme preparations were then used for determination of dissociation constants as described below. Previous studies have confirmed that treatment of the enzyme with GSH following removal of DTT by this procedure results in the maintenance of all cysteine residues in the thiol-form [23].
2.5. Determination of dissociation constants (Kd) for OHPCBs and PAP
Values for Kd were determined with a PerkinElmer LS-55 luminescence spectrometer (PerkinElmer Life and Analytical Sciences, Waltham, MA) by measuring the displacement of ANS from the enzyme. ANS binds to the sulfuryl acceptor site of rSULT1A1, and the change in its fluorescence intensity upon displacement from the enzyme due to binding of a OHPCB was utilized to determine Kd values as previously described for other substrates of rSULT1A1 [26]. rSULT1A1 at a concentration of 0.34 µM was pre-incubated in 0.25 M potassium phosphate buffer (pH 7.0) (1.0 ml total volume) at 37 °C for 2 min, under argon. The solution was then transferred to a quartz cuvette of 1-cm excitation path length and 0.4 cm emission path length, and ANS (10 µM final concentration) was added. Fluorescence emission was measured at 465 nm (excitation at 380 nm) following the addition of an aliquot of each OHPCB (0.2–1.0 µL aliquots of each OHPCB in absolute ethanol). In titrations of the enzyme-PAP complex, the appropriate concentration of PAP was added after addition of 8-anilinonaphthalene-1-sulfonic acid (ANS) and before titration with an OHPCB. After each addition to the reaction mixture, the solution was mixed by inverting the closed cuvette three times and then placing it in the fluorimeter for 10 seconds before determining the fluorescence. Each binding experiment was performed in duplicate at 37°C. Since the total dilution of the assay during titration was always less than 0.8%, no corrections were made for this dilution upon addition of OHPCBs. The fluorimeter shutters were closed unless spectra were being recorded in order to minimize the time of exposure to the excitation beam.
Binding of PAP to rSULT1A1 was determined in the presence or absence of OHPCB by monitoring the displacement of 2'-(or-3')-O-(trinitrophenyl)adenosine-5'-monophosphate (TNP-AMP). Fluorescence of the TNP-AMP was determined (excitation at 435 nm; emission at 545 nm) in a quartz cuvette of 1-cm excitation path length and 0.4 cm emission path length at 37°C in potassium phosphate buffer at pH 7.0 with 0.34 µM rSULT1A1 present. The effect of each OHPCB on the binding of PAP to rSULT1A1 was determined following pretreatment of the enzyme with either GSSG or GSH, as described above. A saturating concentration of TNP-AMP (30 µM) and a 10 µM concentration of each OHPCB were added before titration with PAP. Triplicate determinations were analyzed for each OHPCB. The means and standard errors of Kd values were calculated by fit of the data to a single-site binding equation corrected for non-specific binding (SigmaPlot 11.0; Systat Software; Chicago, IL).
2.6. Calculation of pKa and log D
For each of the OHPCBs examined, calculated estimates of pKa, and log D (the partition coefficient between octanol and water) at pH 7.0 were obtained by using the ACD /I-Lab Web service (with program ACD/pKa 8.03, and ACD/log D 8.02), Advanced Chemistry Development Inc. (Ontario, Canada).
3. Results
3.1. Relationships between initial velocity of the sulfation reaction and pKa and log D for OHPCBs that were substrates for rSULT1A1
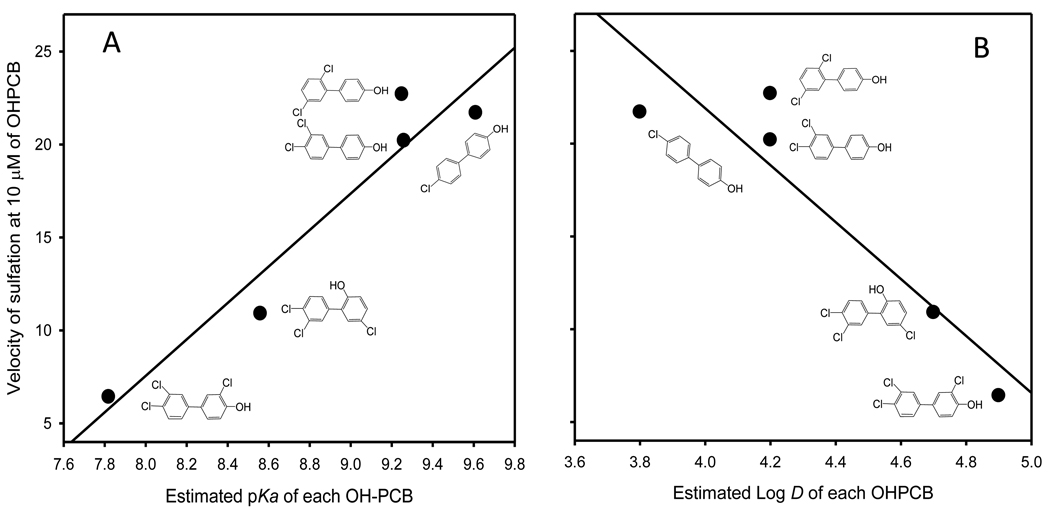
As shown in Figure 1, the estimated pKa values of five OHPCBs that were substrates for rSULT1A1 were positively correlated with their velocity of sulfation by rSULT1A1 (panel A), and the estimated log D values for these OHPCBs were negatively correlated with their velocity of sulfation (panel B). Sulfation of each OHPCB was determined under fully reduced conditions (in the presence of 7.5 mM 2-mercaptoethanol) at a substrate concentration of 10 µM, a concentration that did not cause significant substrate inhibition of the enzyme for these OHPCBs. This substrate concentration was chosen for comparison of rates due to the difficulties inherent in calculation of kinetic parameters for these OHPCBs, where there are varying degrees of substrate inhibition as well as a lack of solubility for some of the substrates at sufficiently high concentrations to unambiguously determine Km and Vmax values.
Figure 1.
Relationships between the rate of sulfation of OHPCBs catalyzed by rSULT1A1 and the estimated values for pKa and log D. The velocity of the sulfation reaction under fully reduced conditions at a OHPCB concentration of 10 µM is correlated with pKa (A) and log D (B). Values for r2 were 0.92 and 0.86, respectively.
3.2. Relationship between the pKa’s of OHPCBs and IC50 values for inhibition of rSULT1A1
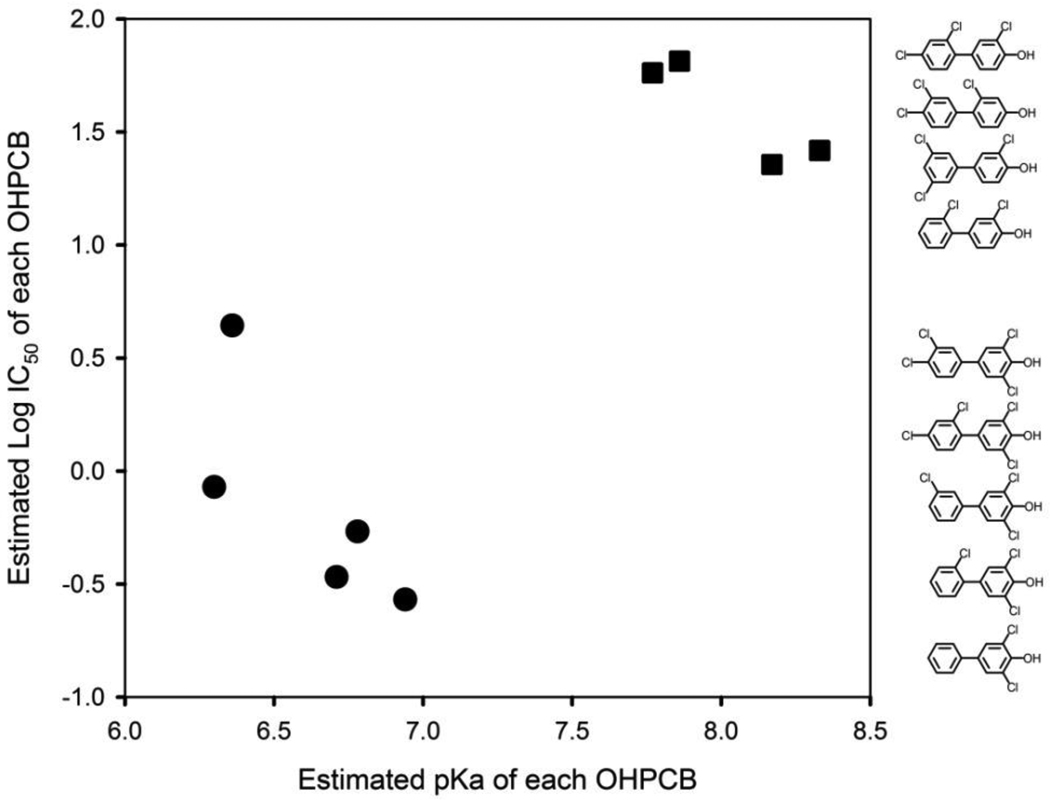
The log IC50 values for nine OHPCBs that inhibit the reduced form of rSULT1A1 [7] were analyzed as a function of their estimated pKa values, and the result is seen in Figure 2. In general, these inhibitory OHPCBs were distributed into two distinct groups in relation to their pKa and IC50 values (p = 3.4 × 10−6, Hotelling’s T-square test). The 4-hydroxy-3,5-dichloro-substituted OHPCBs demonstrated both lower log IC50 and lower pKa values (lower left quadrant of Figure 2), while the other four OHPCBs containing 4-hydroxy-2 (or 3)-chloro-substitutions had both higher IC50 and higher pKa values (upper right quadrant of Figure 2). There were no corresponding correlations observed between log D values (pH 7.0) and IC50 values for these OHPCBs (data not shown).
Figure 2.
Relationship between the estimated pKa values of OHPCBs and their inhibition of rSULT1A1. Log IC50 values for rSULT1A1 (with all cysteines in the thiol form) were determined previously [7]. The OHPCBs were divided into two groups, i.e., 4-OH-3,5-dichloro-PCBs (solid circles) and 4-OH-2(or 3)-chloro-PCBs (solid squares). The bivariate mean vectors (pKa and log IC50) in the two groups were significantly different from each other (Hotelling’s T-square test, p = 3.4 × 10−6). The chemical structures of the OHPCBs are shown in the order of their corresponding IC50 values.
3.3. Dissociation constants for OHPCBs from the PAP-bound enzyme complex for both GSH-and GSSG-pretreated rSULT1A1
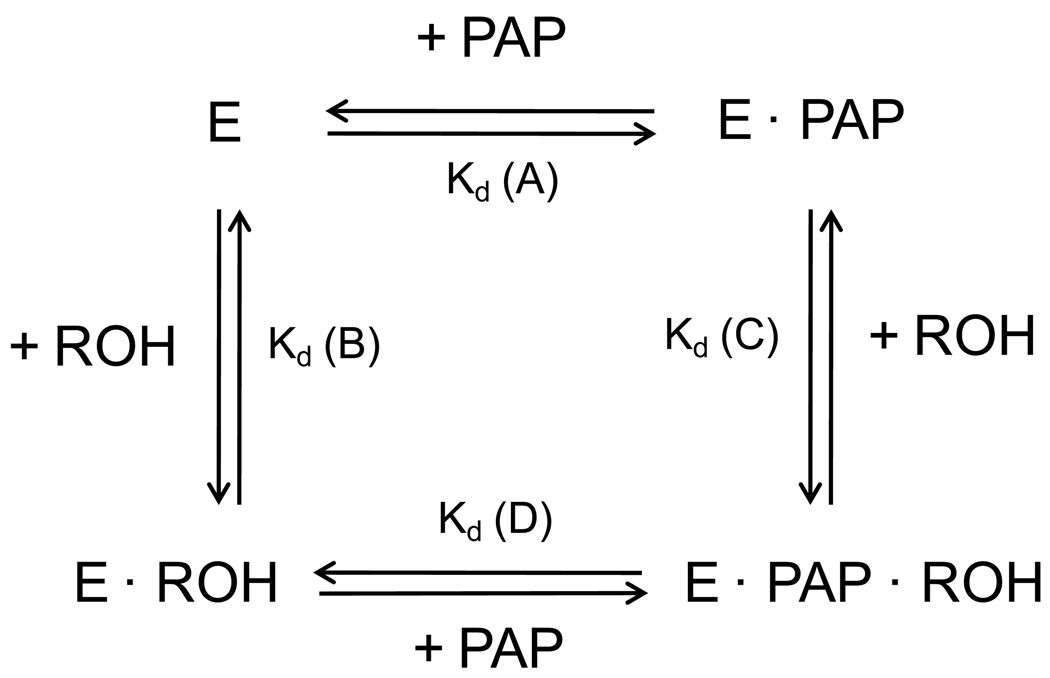
The formation of binary and ternary complexes among rSULT1A1, PAP, and sulfuryl acceptors has been previously shown to be important in determining both substrate inhibition and substrate specificity [27]. Therefore, we have examined the various dissociation constants outlined in Figure 3 for any differences that would be dependent upon the structure of the OHPCB. As indicated in Table 1, 4’-OH PCB 9, 4’-OH PCB 12, 6’-OH PCB 35 and 4’-OH PCB 35, as substrates for rSULT1A1 under standard assay conditions, showed higher Kd values with rSULT1A1 (for both GSH- and GSSG-pretreated enzyme in the absence of PAP or PAPS) than 4’-OH PCB 68 and 4-OH PCB 34 (inhibitors of the enzyme under standard assay conditions and non-substrates for GSSG-pretreated enzyme). In the presence of 0.27 mM PAP, a concentration 10-fold greater than the previously observed Kd for PAP with GSSG-pretreated rSULT1A1[26]), the dissociation constants for those OHPCBs that served as substrates for the reduced enzyme either remained approximately the same or were decreased. Specifically, for 4’- OH PCB 12 and 4’-OH PCB 35, the Kd values for the GSH-pretreated rSULT1A1 and the GSH-pretreated rSULT1A1-PAP complex remained similar, while Kd values for 6’-OH PCB 35 and 4’-OH PCB 9 decreased upon formation of the rSULT1A1-PAP complex with the GSH-pretreated enzyme. It is, however, noteworthy that these Kd values were still 4–10 times higher than observed for the OHPCBs with 3,5-dichloro-4-hydroxy substituents (e.g., 4-OH PCB 14, 4-OH PCB 34 and 4’-OH PCB 68). For those OHPCBs that have been previously shown [7] to be converted from inhibitors to substrates by GSSG-pretreatment of rSULT1A1 (i.e., 4’-OH PCB 6, 4-OH PCB 14, and 4’-OH PCB 33), Kd values with rSULT1A1 and with the rSULT1A1-PAP complex exhibited no consistent differences from Kd values for those OHPCBs that were substrates for the GSH-pretreated enzyme. When pretreatment with GSH was compared with pretreatment of the enzyme with GSSG, no significant alterations in Kd values were observed for any of the nine OHPCBs, either with the free enzyme or the enzyme-PAP complex.
Figure 3.
Formation and dissociation of enzyme complexes comprising rSULT1A1 (E), OHPCB (ROH), and PAP. Kd (A), (B), (C), and (D) indicate the dissociation constants for PAP binding to rSULT1A1, OHPCB binding to rSULT1A1, OHPCB binding to the enzyme-PAP complex, and PAP binding to an enzyme-OHPCB complex, respectively.
Table 1.
Dissociation constants for OHPCBs with rSULT1A1 that had been pretreated with either GSH or GSSG
| OHPCBs | Chemical structure |
IC50a (µM) |
Kd values for OHPCBs with rSULT1A1 (µM)b | |||
|---|---|---|---|---|---|---|
| Without PAP | With 0.27 mM PAP | |||||
| GSH | GSSG | GSH | GSSG | |||
| 4’-OH PCB 9 |  |
—c | 12 ± 4 | 7.1 ± 1.3 | 3.1 ± 1.5 | 3.3 ± 0.1 |
| 4’-OH PCB 12 | —c | 4.0 ± 0.8e | 2.5 ± 0.7e | 3.1 ± 0.01 | 4.6 ± 1.2e | |
| 6’-OH PCB 35 |
|
—c | 65 ± 32 | 117 ± 36 | 3.7 ± 0.01e | 3.6 ± 1.3e |
| 4’-OH PCB 35 |
|
— c | 3.6 ± 0.9 | 1.6 ± 0.4 | 1.6 ± 0.1 | 1.7 ± 0.1 |
| 4’-OH PCB 6 |
|
23d | 0.66 ± 0.2 | 0.50 ± 0.1 | 0.88 ± 0.03 | 0.72 ± 0.01 |
| 4’-OH PCB 33 |
|
26 d | 2.0 ± 0.1 | 2.5 ± 0.4 | 2.0 ± 0.01 | 2.4 ± 0.04 |
| 4-OH PCB 14 |
|
0.27 d | 0.26 ± 0.03 | 0.22 ± 0.02 | 0.40 ± 0.05f | 0.29 ± 0.04f |
| 4’-OH PCB 68 |
|
0.85 | 0.67 ± 0.1 | 0.53 ± 0.1 | 0.47 ± 0.02 | 0.40 ± 0.04 |
| 4-OH PCB 34 |
|
0.34 | 0.37 ± 0.01 | 0.23 ± 0.01e | 0.49g | 0.54 ± 0.01 |
IC50 values with rSULT1A1 (all cysteines in the thiol form) were determined previously with 2-naphthol as the substrate [7].
Kd values were determined utilizing rSULT1A1 that had been pretreated with either 1 mM GSH or 1 mM GSSG as described in section 2.4, and, unless otherwise indicated, data were fit to a one-site saturation equation with correction for non-specific binding of the ANS probe. Values are means and S.E. of two determinations unless otherwise indicated.
This OHPCB is a substrate for rSULT1A1.
This OHPCB is an inhibitor for the enzyme pretreated with GSH, but it is a substrate for rSULT1A1 pretreated with 1 mM GSSG.
The Kd value shown is the first Kd when the data were fit to a two-site equation; the second Kd was over 35 times greater than the first Kd.
Three determinations were performed.
A single determination was performed.
3.4. Dissociation of PAP from OHPCB-bound rSULT1A1 complexes following pretreatment of the enzyme with either GSH or GSSG
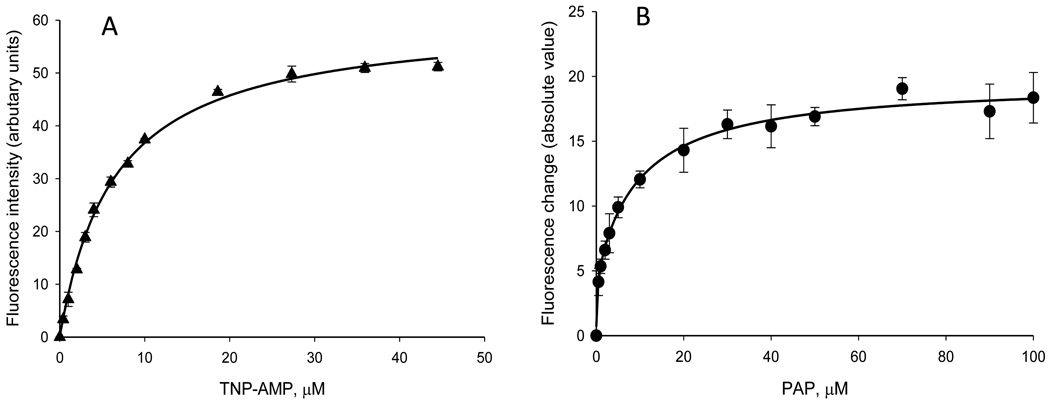
Trinitrophenyl derivatives of AMP, ADP, and ATP (i.e., TNP-AMP, TNP-ADP, and TNP-ATP) have been previously employed as fluorescent probes for adenosine nucleoside- and nucleotide-requiring enzymes [35]. For example, TNP-AMP has been used to analyze ligand binding to proteins such as Ca2+-ATPase [36] and fructose 1,6-bisphosphatase [37]. Due to its structural similarity to PAP, we investigated TNP-AMP for its suitability as a fluorescent probe to determine PAP-binding in the present study. Thus, we titrated 0.34 µM rSULT1A1 in potassium phosphate buffer (pH 7.0) at 37°C with TNP-AMP and determined the fluorescence at 545 nm (excitation at 435 nm). As seen in Figure 4, TNP-AMP bound to the enzyme with a Kd of 6.5 µM and saturation occurred at concentrations greater than 20 µM (Figure 4A). As seen in Figure 4B, PAP displaced the TNP-AMP bound to rSULT1A1 with a Kd value of 2.6 µM.
Figure 4.
Use of TNP-AMP in determination of Kd values for PAP. (A) Binding of TNP-AMP to rSULT1A1 as indicated by an increase in fluorescence intensity at 545 nm (excitation at 435 nm). (B) Displacement of TNP-AMP from the enzyme upon titration with PAP as indicated by the absolute value of the decrease in fluorescence at 545 nm.
As indicated in Table 2, Kd values for the binding of PAP to GSH- versus GSSG-pretreated rSULT1A1 were significantly different, as previously described [26]. When the Kd values for PAP were determined for OHPCBs complexed with either GSH- or GSSG-pretreated rSULT1A1, the result was that there was no statistically significant difference for any individual OHPCB between enzyme pretreated with GSH and GSSG (p>0.05 by Student’s t test). However, the binding of PAP to an enzyme-OHPCB complex was significantly different from PAP-binding in the absence of OHPCB. This was true for all OHPCBs examined and was independent of the redox status of the rSULT1A1.
Table 2.
Binding of PAP to rSULT1A1 and to rSULT1A1-OHPCB complexes following pretreatment of the enzyme with GSH or GSSG
| OHPCB (10 µM) | Kd for binding of PAP with rSULT1A1a | |
|---|---|---|
| GSH | GSSG | |
| None | 4.01 ± 0.06b | 5.10 ± 0.40b |
| 4’-OH PCB 35 | 1.29 ± 0.33 c | 1.07 ± 0.21 c |
| 4-OH PCB 34 | 2.64 ± 1.00 c | 1.24 ± 0.25 c |
| 4-OH PCB 14 | 1.14 ± 0.28 c | 2.67 ± 0.76 c |
| 4’-OH PCB 6 | 0.86 ± 0.24 c | 1.99 ± 0.50 c |
Kd values were determined utilizing rSULT1A1 that had been pretreated with either 1 mM GSH or 1 mM GSSG as described in section 2.4. Data are means and S.E. of three replicates unless otherwise specified.
Data are the means and S.E. of two determinations. The Kd values for PAP-binding in the absence of OHPCB were significantly different from those seen in the presence of a OHPCB (p<0.05: Student’s t test). The Kd value for PAP-binding in the presence of each OHPCB was significantly lower than that observed in the absence of any OHPCB, and this was seen with both GSH- and GSSG-pretreated enzyme (p < 0.05; Student’s t test).
Data are the means and S.E. of three determinations. There were no significant differences between rSULT1A1 pretreated with GSH and the enzyme pretreated with GSSG for any individual OHPCB (p > 0.05 by Student’s t test).
3.5 Relationships between pKa values of OHPCBs and their dissociation constants for binding to GSH-pretreated and GSSG-pretreated rSULT1A1
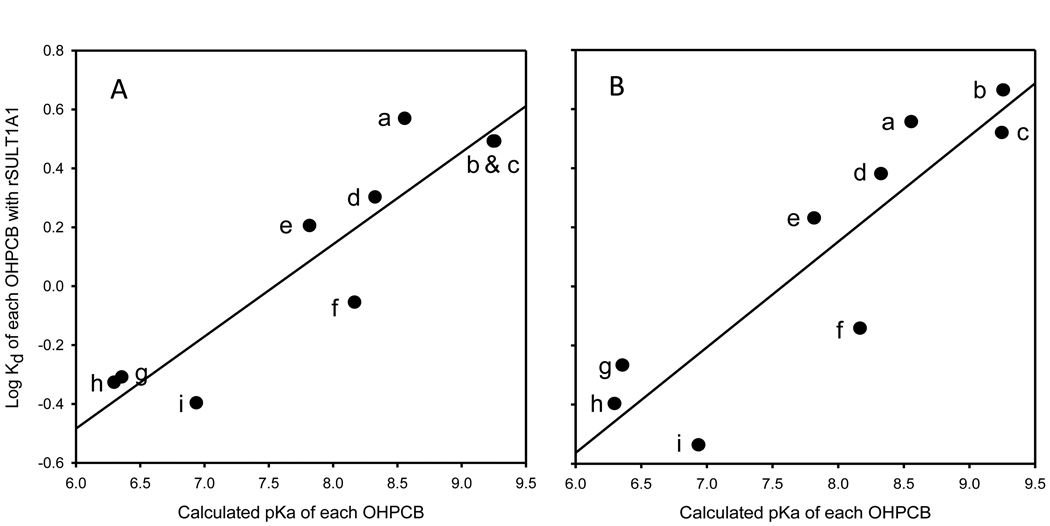
As shown in Figure 5, the estimated pKa values of the nine OHPCBs examined showed a positive correlation with their Kd values with rSULT1A1. Similar relationships for the individual OHPCBs with respect to their pKa and Kd values were observed for both the enzyme that had been pretreated with GSH (panel A) and for the GSSG-pretreated enzyme (panel B) (r2 values were 0.83 and 0.79, respectively). It is also of note that there was no correlation seen between log D and Kd values for the OHPCB with either GSH- or GSSG-pretreated rSULT1A1 (data not shown).
Figure 5.
Relationships between pKa values of OHPCBs and their Kd values for binding to rSULT1A1 under different redox environments. (A) rSULT1A1 pretreated with 1 mM GSH. (B) rSULT1A1 pretreated with 1 mM GSSG. The y axis for panel B is the same as for panel A. The corresponding correlation coefficients were 0.83 and 0.79, respectively. Labeling of data points in both panels: a, 6’-OH PCB 35; b, 4’-OH PCB 12; c, 4’-OH PCB 9; d, 4’-OH PCB 33; e, 4’-OH PCB 35; f, 4’-OH PCB 6; g, 4-OH PCB 34; h, 4’-OH PCB 68; i, 4-OH PCB 14
4. Discussion
The OHPCBs are persistent metabolites of PCBs, and they are of increasing interest due to their biological activities [20, 38–42] and their presence in the serum and tissues of humans [15– 19, 43] and other animals [44–47]. This is particularly true of the lower chlorinated PCBs, where exposure from airborne sources occurs [23–25], and metabolic reactions are prevalent [9–13]. Sulfation represents one possible metabolic reaction for OHPCBs, although it is not yet known whether or not this biotransformation results in detoxication of these molecules. Inhibition of the sulfotransferases catalyzing sulfation of key physiological molecules is also a potential mechanism for biological responses to OHPCBs. Previous work has shown that various isoforms of sulfotransferase display distinct specificities for OHPCBs as substrates and inhibitors [7, 20, 21, 48, 49]. When one considers the large number of OHPCBs that might be derived from PCB congeners, it becomes important to establish predictive relationships that are applicable to understanding their interactions with sulfotransferases. Toward this goal, we have investigated the use of calculated estimates of pKa and log D for several lower chlorinated OHPCBs to aid in predicting the ability of these compounds to serve as substrates and inhibitors of a model aryl (phenol) sulfotransferase, rSULT1A1.
Our present studies indicate that the velocity of sulfation of OHPCBs catalyzed by rSULT1A1, at low substrate concentrations and under reducing conditions, correlates with the estimated pKa values of the OHPCBs (Figure 1A). These findings with the OHPCBs are consistent with a previous report on fluoronitrophenols as substrates for rSULT1A1, in which the log of the apparent second-order rate constant (Kcat/Km) of each compound paralleled the corresponding pKa value [50]. Moreover, our examination of the relationship between the pKa values for OHPCBs and their log IC50 values for inhibition of rSULT1A1 shown in Figure 2 revealed a relationship wherein the most potent inhibitors (i.e., those with IC50 values lower than 4.4 µM) had lower pKa values, while the less potent inhibitors (i.e., those with IC50 values greater than 26 µM) had higher pKa values. These effects provide a useful initial structure-activity relationship, but they also suggest that future studies on the interactions of each of these OHPCBs with the enzyme at varied pH will provide further mechanistic details about the catalytic specificity and inhibition of rSULT1A1.
While one might hope that relationships among rates of reaction, IC50, and pKa would be directly helpful in predicting whether a specific OHPCB is either a substrate or an inhibitor, there was no distinct pKa value that could be used to discriminate among OHPCBs as either substrates or inhibitors for rSULT1A1. This was indicated by the overlapping pKa values of 4’-OH PCB 35 (the weakest substrate for rSULT1A1), 4’-OH PCB 36 and 4’-OH PCB 33 (both being relatively weak inhibitors for reduced rSULT1A1 and substrates for oxidized rSULT1A1), and 4’-OH PCB 25 (the weakest inhibitor of rSULT1A1 observed). While a quantitative division between substrates and inhibitors based on pKa is not evident, it is noteworthy that OHPCBs serving as substrates for rSULT1A1 generally had higher pKa values than those that were inhibitors. This would be consistent with structural studies on the mechanism of a sulfotransferase-catalyzed reaction where the catalytic histidine is involved in orientation of a substrate and abstraction of the phenolic proton during an in-line sulfuryl transfer [51]. Likewise, this is consistent with linear free-energy analyses [50] and kinetic isotope studies [52] that indicate a relatively loose, dissociative transition state in the transfer of the sulfuryl group in the mechanisms of sulfotransferases. Those OHPCBs with very low pKa values, such as the 4-OH-3,5-chloro-substituted PCBs, bind to the enzyme well (as seen by their log Kd values), however, they may not be in the correct orientation for sulfuryl transfer to occur. Additional effects, such as steric effects of the adjacent chlorine atoms, difference in hydrophobic characteristics, and altered stabilization of the transition state are also possible contributors to the inhibition seen by these OHPCBs. Indeed, log D values exhibited an inverse correlation with the velocity of sulfation, and this may be indicative of differences in binding and orientation of the OHPCB in a catalytically competent position at the active site.
An additional component of predicting the interactions of OHPCBs with the sulfotransferase is a more complete understanding of the role that the thiol:disulfide status plays in altering the specificity of the enzyme for substrates and inhibitors. Previous studies on the mechanism of rSULT1A1 have shown the importance of dead-end ternary complexes comprising the enzyme, PAP, and a phenolic substrate in regulation of the rate of the reaction [26, 27]. Our discovery that some OHPCBs were substrates for the rSULT1A1 after the enzyme was incubated with GSSG, but were solely inhibitors of rSULT1A1 when the thiols in the enzyme were maintained in a reduced state [7], led us to investigate whether these redox effects were due to differences in binding of the nucleotide (PAPS and PAP) or the OHPCB in a ternary complex.
Treatment of rSULT1A1 with GSSG prior to assay of the catalytic activity did not result in any significant effects on Kd of each OHPCB (compared with GSH-pretreated enzyme), and this was true even in the presence of a saturating concentration of PAP (Table 1). Similarly, the Kd values for PAP with the GSSG-pretreated enzyme that had been subjected to a saturating concentration of each of five OHPCBs were not statistically different from those with GSH-pretreated enzyme (Table 2). Similar to previous studies [26], however, the dissociation constants for PAP binding to rSULT1A1 differed when the GSH-pretreated and the GSSG-pretreated enzyme were compared. Thus, for those OHPCBs that are inhibitors of reduced rSULT1A1 and substrates for enzyme after GSSG-pretreatment, differences in the decomposition of the dead-end ternary complex (Enzyme-PAP-OHPCB) due to structural differences in the OHPCB are not likely to be responsible for this change in specificity.
Since structural differences among the OHPCBs do not determine specificity solely through differences in the stability of the dead-end ternary complexes under different redox conditions, we must consider the potential for structural differences in OHPCBs providing alterations in the rates of transfer of the sulfuryl group in the transition state of the reaction. It has been previously determined with studies on substituted phenols that electron-withdrawing substituents (e.g., chlorine atoms) increase the rate of reaction for the GSSG-treated rSULT1A1 [27], and electron-withdrawing substituents decrease the rate of reaction in the fully reduced enzyme [27, 53]. This has been interpreted as a shift in rate-determining step of the reaction as a function of the thiol:disulfide status of the enzyme. That is, the rate-determining step for the reduced enzyme was proposed to be the dissociation of the enzyme-PAP-phenol complex, while the transfer of the sulfuryl group was the rate-determining step for the GSSG-treated enzyme [2, 27]. Our results with the OHPCBs examined in the current studies, however, indicate that the rate-determining step of the rSULT1A1-catalyzed reaction for this group of substrates may involve the sulfuryl group transfer in both the GSH-treated and GSSG-treated enzyme forms. There would still be differences in the release of PAP from the enzyme-PAP complex that are dependent upon the redox status of the enzyme, however, the dissociation constant for PAP in an enzyme-PAP-OHPCB complex is not dependent upon either the redox state of the enzyme or the structures of the OHPCBs examined.
Structural characteristics of the OHPCBs would, however, affect the orientation of initial binding to the active site of the enzyme as well as the transition state of the sulfuryl transfer. Structural features of the OHPCB could result in efficient binding and orientation of the phenolic hydroxyl, but insufficient stabilization of the transition state of the reaction might then cause the OHPCB to be an inhibitor due to the lack of transfer of the sulfuryl group. Conformational changes in rSULT1A1 resulting from redox changes in the protein might alter one or more of these critical components for catalysis and thus change interactions that would be specific to the structure of the OHPCB. Further investigation will be needed to elucidate the complex interactions between the redox states of the enzyme and those structural features of the OHPCB that affect the sulfuryl transfer step of the enzyme mechanism.
It is also important to recognize that the number of OHPCBs studied to date represent a small sample of potential OHPCB metabolites. While the structure-activity relationships seen with this set of molecules may provide useful predictive information for other lower chlorinated OHPCBs, it remains to be determined if these relationships are modified for highly chlorinated OHPCB congeners.
Acknowledgements
The authors express their appreciation to Professor Kai Wang for assistance with the statistical analysis of the data in Figure 2. This work was supported by the National Institutes of Health through research grants R01 CA038683 from the National Cancer Institute and P42 ES 013661 from the National Institute of Environmental Health Sciences. We also acknowledge programmatic support through the University of Iowa Environmental Health Sciences Research Center (NIEHS/NIH P30 ES05605). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare that there are no conflicts of interest.
References
- 1.Duffel MW. Sulfotransferases. In: McQueen CA, Guengerich FP, editors. Comprehensive Toxicology, Vol. 4 Biotransformation. Elsevier; Oxford: 2010. pp. 367–384. (Vol. Ed.) [Google Scholar]
- 2.Duffel MW, Marshall AD, McPhie P, Sharma V, Jakoby WB. Enzymatic aspects of the phenol (aryl) sulfotransferases. Drug Metab. Rev. 2001;33:369–395. doi: 10.1081/dmr-120001394. [DOI] [PubMed] [Google Scholar]
- 3.Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 4.Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 5.Glatt H, Boeing H, Engelke CE, Ma L, Kuhlow A, Pabel U, Pomplun D, Teubner W, Meinl W. Human cytosolic sulphotransferases: genetics, characteristics, toxicological aspects. Mutat. Res. 2001;482:27–40. doi: 10.1016/s0027-5107(01)00207-x. [DOI] [PubMed] [Google Scholar]
- 6.Pacifici GM, Coughtrie MW, editors. Human cytosolic sulfotransferases. Boca Raton, FL: CRC Press; 2005. [Google Scholar]
- 7.Liu Y, Smart JT, Song Y, Lehmler HJ, Robertson LW, Duffel MW. Structure-activity relationships for hydroxylated polychlorinated biphenyls as substrates and inhibitors of rat sulfotransferases and modification of these relationships by changes in thiol status. Drug Metab. Dispos. 2009;37:1065–1072. doi: 10.1124/dmd.108.026021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen LG. Stepping backward to improve assessment of PCB congener toxicities, Environ. Health Perspect. 1998;106 Suppl. 1:171–189. doi: 10.1289/ehp.98106s1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson LW, Hansen LG. PCBs: Recent Advances in Environmental Toxicology and Health Effects. Lexington: University of Kentucky Press; 2001. [Google Scholar]
- 10.Safe SH. Polychlorinated biphenyls (PCBs): environmental impact, biochemical and toxic responses, and implications for risk assessment. Crit. Rev. Toxicol. 1994;24:87–149. doi: 10.3109/10408449409049308. [DOI] [PubMed] [Google Scholar]
- 11.Kaminsky LS, Kennedy MW, Adams SM, Guengerich FP. Metabolism of dichlorobiphenyls by highly purified isozymes of rat liver cytochrome P-450. Biochemistry. 1981;20:7379–7384. doi: 10.1021/bi00529a009. [DOI] [PubMed] [Google Scholar]
- 12.Ludewig G, Esch H, Robertson LW. Polyhalogenierte Bi- und Terphenyle. Chapter 20. Wiley-VCH Weinheim; 2007. [Google Scholar]
- 13.McLean MR, Bauer U, Amaro AR, Robertson LW. Identification of catechol and hydroquinone metabolites of 4-monochlorobiphenyl. Chem. Res. Toxicol. 1996;9:158–164. doi: 10.1021/tx950083a. [DOI] [PubMed] [Google Scholar]
- 14.Ueno D, Darling C, Alaee M, Campbell L, Pacepavicius G, Teixeira C, Muir D. Detection of hydroxylated polychlorinated biphenyls (OH-PCBs) in the abiotic environment: surface water and precipitation from Ontario, Canada. Environ. Sci. Technol. 2007;41:1841–1848. doi: 10.1021/es061539l. [DOI] [PubMed] [Google Scholar]
- 15.Bergman A, Klasson-Wehler E, Kuroki H. Selective retention of hydroxylated PCB metabolites in blood. Environ. Health Perspect. 1994;102:464–469. doi: 10.1289/ehp.94102464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dirtu AC, Jaspers VL, Cernat R, Neels H, Covaci A. Distribution of PCBs, their hydroxylated metabolites, and other phenolic contaminants in human serum from two European countries. Environ. Sci. Technol. 2010;44:2876–2883. doi: 10.1021/es902149b. [DOI] [PubMed] [Google Scholar]
- 17.Fängström B, Athanasiadou M, Grandjean P, Weihe P, Bergman Å. Hydroxylated PCB metabolites and PCBs in serum from pregnant Faroese women. Environ. Health Perspect. 2002;110:895–899. doi: 10.1289/ehp.110-1240989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park JS, Bergman A, Linderholm L, Athanasiadou M, Kocan A, Petrik J, Drobna B, Trnovec T, Charles MJ, Hertz-Picciotto I. Placental transfer of polychlorinated biphenyls, their hydroxylated metabolites and pentachlorophenol in pregnant women from eastern Slovakia. Chemosphere. 2008;70:1676–1684. doi: 10.1016/j.chemosphere.2007.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandau CD, Ayotte P, Dewailly E, Duffe J, Norstrom RJ. Analysis of hydroxylated metabolites of PCBs (OH-PCBs) and other chlorinated phenolic compounds in whole blood from Canadian inuit. Environ. Health Perspect. 2000;108:611–616. doi: 10.1289/ehp.00108611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kester MH, Bulduk S, Tibboel D, Meinl W, Glatt H, Falany CN, Coughtrie MW, Bergman A, Safe SH, Kuiper GG, Schuur AG, Brouwer A, Visser TJ. Potent inhibition of estrogen sulfotransferase by hydroxylated PCB metabolites: a novel pathway explaining the estrogenic activity of PCBs. Endocrinology. 2000;141:1897–1900. doi: 10.1210/endo.141.5.7530. [DOI] [PubMed] [Google Scholar]
- 21.Schuur AG, van Leeuwen-Bol I, Jong WM, Bergman A, Coughtrie MW, Brouwer A, Visser TJ. In vitro inhibition of thyroid hormone sulfation by polychlorobiphenylols: isozyme specificity and inhibition kinetics. Toxicol. Sci. 1998;45:188–194. doi: 10.1006/toxs.1998.2504. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Apak TI, Lehmler HJ, Robertson LW, Duffel MW. Hydroxylated polychlorinated biphenyls are substrates and inhibitors of human hydroxysteroid sulfotransferase SULT2A1. Chem. Res. Toxicol. 2006;19:1420–1425. doi: 10.1021/tx060160+. [DOI] [PubMed] [Google Scholar]
- 23.Harrad S, Hazrati S, Ibarra C. Concentrations of polychlorinated biphenyls in indoor air and polybrominated diphenyl ethers in indoor air and dust in Birmingham, United Kingdom: implications for human exposure. Environ Sci Technol. 2006;40:4633–4638. doi: 10.1021/es0609147. [DOI] [PubMed] [Google Scholar]
- 24.Hu D, Martinez A, Hornbuckle KC. Discovery of non-aroclor PCB (3,3'-dichlorobiphenyl) in Chicago air. Environ Sci Technol. 2008;42:7873–7877. doi: 10.1021/es801823r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persoon C, Peters TM, Kumar N, Hornbuckle KC. Spatial distribution of airborne polychlorinated biphenyls in Cleveland Ohio and Chicago, Illinois. Environ Sci Technol. 2010;44:2797–2802. doi: 10.1021/es901691s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall AD, Darbyshire JF, Hunter AP, McPhie P, Jakoby WB. Control of activity through oxidative modification at the conserved residue Cys66 of aryl sulfotransferase IV. J. Biol. Chem. 1997;272:9153–9160. doi: 10.1074/jbc.272.14.9153. [DOI] [PubMed] [Google Scholar]
- 27.Marshall AD, McPhie P, Jakoby WB. Redox control of aryl sulfotransferase specificity. Arch. Biochem. Biophys. 2000;382:95–104. doi: 10.1006/abbi.2000.2020. [DOI] [PubMed] [Google Scholar]
- 28.Lehmler HJ, Robertson LW. Synthesis of hydroxylated PCB metabolites with the Suzuki coupling. Chemosphere. 2001;45:1119–1127. doi: 10.1016/s0045-6535(01)00052-2. [DOI] [PubMed] [Google Scholar]
- 29.Sekura RD. Adenosine 3'-phosphate 5'-phosphosulfate. Methods Enzymol. 1981;77:413–415. [Google Scholar]
- 30.Chen X, Yang Y-S, Zheng Y, Martin BM, Duffel MW, Jakoby WB. Tyrosine-ester sulfotransferase from rat liver: bacterial expression and identification. Protein Expr. Purif. 1992;3:421–426. doi: 10.1016/s1046-5928(05)80045-2. [DOI] [PubMed] [Google Scholar]
- 31.Bensadoun A, Weinstein D. Assay of proteins in the presence of interfering materials. Anal. Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 32.Duffel MW, Binder TP, Rao SI. Assay of purified aryl sulfotransferase suitable for reactions yielding unstable sulfuric acid esters. Anal. Biochem. 1989;183:320–324. doi: 10.1016/0003-2697(89)90486-7. [DOI] [PubMed] [Google Scholar]
- 33.Sheng J, Sharma V, Duffel MW. Current Protocols in Toxicology. New York: John Wiley & Sons, Inc.; 2001. Measurement of aryl and alcohol sulfotransferase activity; pp. 4.5.1–4.5.9. [DOI] [PubMed] [Google Scholar]
- 34.Jocelyn PC. Spectrophotometric assay of thiols. Methods Enzymol. 1987;143:44–66. doi: 10.1016/0076-6879(87)43013-9. [DOI] [PubMed] [Google Scholar]
- 35.Hiratsuka T. Biological activities and spectroscopic properties of chromophoric and fluorescent analogs of adenine nucleoside and nucleotides, 2',3'-O-(2,4,6-trinitrocyclohexadienylidene) adenosine derivatives. Biochim. Biophys. Acta. 1982;719:509–517. doi: 10.1016/0304-4165(82)90240-9. [DOI] [PubMed] [Google Scholar]
- 36.Nakamoto RK, Inesi G. Studies of the interactions of 2',3'-O-(2,4,6-trinitrocyclohexyldienylidine)adenosine nucleotides with the sarcoplasmic reticulum (Ca2+ + Mg2+)-ATPase active site. J. Biol. Chem. 1984;259:2961–2970. [PubMed] [Google Scholar]
- 37.Rakus D, Maciaszczyk E, Wawrzycka D, Ulaszewski S, Eschrich K, Dzugaj A. The origin of the high sensitivity of muscle fructose 1,6-bisphosphatase towards AMP. FEBS Lett. 2005;579:5577–5581. doi: 10.1016/j.febslet.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 38.Schuur AG, Brouwer A, Bergman A, Coughtrie MW, Visser TJ. Inhibition of thyroid hormone sulfation by hydroxylated metabolites of polychlorinated biphenyls. Chem. Biol. Interact. 1998;109:293–297. doi: 10.1016/s0009-2797(97)00140-3. [DOI] [PubMed] [Google Scholar]
- 39.Martinez JM, Stephens LC, Jones LA. Long-term effects of neonatal exposure to hydroxylated polychlorinated biphenyls in the BALB/cCrgl mouse. Environ. Health Perspect. 2005;113:1022–1026. doi: 10.1289/ehp.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Espandiari P, Glauert HP, Lehmler HJ, Lee EY, Srinivasan C, Robertson LW. Initiating activity of 4-chlorobiphenyl metabolites in the resistant hepatocyte model. Toxicol. Sci. 2004;79:41–46. doi: 10.1093/toxsci/kfh097. [DOI] [PubMed] [Google Scholar]
- 41.Machala M, Blaha L, Lehmler HJ, Pliskova M, Majkova Z, Kapplova P, Sovadinova I, Vondracek J, Malmberg T, Robertson LW. Toxicity of hydroxylated and quinoid PCB metabolites: inhibition of gap junctional intercellular communication and activation of aryl hydrocarbon and estrogen receptors in hepatic and mammary cells. Chem. Res. Toxicol. 2004;17:340–347. doi: 10.1021/tx030034v. [DOI] [PubMed] [Google Scholar]
- 42.Twaroski TP, O'Brien ML, Larmonier N, Glauert HP, Robertson LW. Polychlorinated biphenyl-induced effects on metabolic enzymes, AP-1 binding, vitamin E, and oxidative stress in the rat liver. Toxicol. Appl. Pharmacol. 2001;171:85–93. doi: 10.1006/taap.2000.9114. [DOI] [PubMed] [Google Scholar]
- 43.Guvenius DM, Hassanzadeh P, Bergman A, Noren K. Metabolites of polychlorinated biphenyls in human liver and adipose tissue. Environ. Toxicol. Chem. 2002;21:2264–2269. [PubMed] [Google Scholar]
- 44.Buckman AH, Wong CS, Chow EA, Brown SB, Solomon KR, Fisk AT. Biotransformation of polychlorinated biphenyls (PCBs) and bioformation of hydroxylated PCBs in fish. Aquat. Toxicol. 2006;78:176–185. doi: 10.1016/j.aquatox.2006.02.033. [DOI] [PubMed] [Google Scholar]
- 45.Fangstrom B, Athanasiadou M, Athanassiadis I, Weihe P, Bergman A. Hydroxylated PCB metabolites in nonhatched fulmar eggs from the Faroe Islands. Ambio. 2005;34:184–187. [PubMed] [Google Scholar]
- 46.Kato Y, Ikushiro S, Haraguchi K, Yamazaki T, Ito Y, Suzuki H, Kimura R, Yamada S, Inoue T, Degawa M. A possible mechanism for decrease in serum thyroxine level by polychlorinated biphenyls in Wistar and Gunn rats. Toxicol. Sci. 2004;81:309–315. doi: 10.1093/toxsci/kfh225. [DOI] [PubMed] [Google Scholar]
- 47.Nomiyama K, Murata S, Kunisue T, Yamada TK, Mizukawa H, Takahashi S, Tanabe S. Polychlorinated biphenyls and their hydroxylated metabolites (OH-PCBs) in the blood of toothed and baleen whales stranded along Japanese coastal waters. Environ. Sci. Technol. 2010;44:3732–3738. doi: 10.1021/es1003928. [DOI] [PubMed] [Google Scholar]
- 48.Wang LQ, Lehmler HJ, Robertson LW, James MO. Polychlorobiphenylols are selective inhibitors of human phenol sulfotransferase 1A1 with 4-nitrophenol as a substrate. Chem.-Biol. Interact. 2006;159:235–246. doi: 10.1016/j.cbi.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 49.Wang LQ, Lehmler HJ, Robertson LW, Falany CN, James MO. In vitro inhibition of human hepatic and cDNA-expressed sulfotransferase activity with 3-hydroxybenzo[a]pyrene by polychlorobiphenylols. Environ. Health Perspect. 2005;113:680–687. doi: 10.1289/ehp.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chapman E, Bryan MC, Wong CH. Mechanistic studies of beta-arylsulfotransferase IV. Proc. Natl. Acad. Sci. USA. 2003;100:910–915. doi: 10.1073/pnas.0337638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kakuta Y, Petrotchenko EV, Pedersen LC, Negishi M. The sulfuryl transfer mechanism. Crystal structure of a vanadate complex of estrogen sulfotransferase and mutational analysis. J. Biol. Chem. 1998;273:27325–27330. doi: 10.1074/jbc.273.42.27325. [DOI] [PubMed] [Google Scholar]
- 52.Hoff RH, Czyryca PG, Sun M, Leyh TS, Hengge AC. Transition state of the sulfuryl transfer reaction of estrogen sulfotransferase. J. Biol. Chem. 2006;281:30645–30649. doi: 10.1074/jbc.M604205200. [DOI] [PubMed] [Google Scholar]
- 53.Duffel MW, Jakoby WB. On the mechanism of aryl sulfotransferase. J. Biol. Chem. 1981;256:11123–11127. [PubMed] [Google Scholar]