Abstract
Daughters of diabetes patients have lower insulin sensitivity than women with no diabetes family history, but increase insulin sensitivity to a greater extent with exercise training. This study aimed to determine whether differences in circulating concentrations of adiponectin and leptin, and adipose tissue expression of their genes and receptors played a role. Women offspring of patients with type 2 diabetes mellitus (n = 34; age, 35.6 ± 7.0 years; body mass index, 28.1 ± 5.1 kg/m2) and matched controls with no diabetes family history (n = 36; age, 33.6 ± 6.1 years; body mass index, 27.3 ± 4.7 kg/m2) participated. Blood and abdominal subcutaneous adipose tissue samples were obtained at baseline and after a controlled 7-week endurance-type exercise intervention (sessions were performed at 65%-80% of maximum heart rate). At baseline, no significant differences were observed between groups in circulating leptin or adiponectin concentrations, or expression of their genes or receptors. In response to exercise, plasma leptin decreased more in offspring than controls (−32.2% vs −7.3%, P = .005 for interaction); and the long isoform of the leptin receptor messenger RNA (mRNA) increased significantly only in the offspring (+39.4%, P = .026 vs +7.7%, P = .892). Leptin mRNA decreased similarly in both groups (−24.7% vs −25.0%, P < .05 for both). Furthermore, changes in plasma leptin (r = −0.432, P < .001) and leptin mRNA (r = −0.298, P = .019) correlated significantly with changes in insulin sensitivity. Plasma adiponectin decreased similarly in both groups (−12.1% vs −15.2%, P < .01 for both), but no significant changes were observed in adiponectin-related gene expression. This work shows that exercise training has differing effects on leptin-related variables between women with and without a diabetes family history and suggests that these molecular differences may contribute to the differential effects of exercise training on insulin sensitivity between these 2 groups.
1. Introduction
The central role of obesity in insulin resistance and type 2 diabetes mellitus suggests an important role for adipose tissue in the development of these conditions. Although liver and skeletal muscle are the central tissues determining “whole-body” insulin sensitivity [1], it is becoming clear that adipose tissue–derived factors have important effects on insulin/glucose metabolism in these organs. Originally, the function of adipose tissue was thought to be as the site of energy storage; however, it has become clear that adipose tissue is in fact the largest endocrine organ in the body, secreting many bioactive substances (adipokines) that are required for normal body function and are found at altered levels in metabolic disease [2]. Two of these, adiponectin [3] and leptin [4], are implicated in insulin resistance and may play a role in the etiology of diabetes.
Offspring of diabetic patients have approximately 3 times the risk of developing diabetes vs those with no family history of diabetes [5,6]; and when matched for body mass index (BMI), they are often still more insulin resistant than control subjects [7-9]. Differences in adipokine concentrations may contribute to this: for example, offspring are reported to have lower circulating adiponectin [10,11] and higher circulating leptin concentrations [12] than matched controls. Although this may be due partly to differences in adipose tissue mass and distribution, differences in the functionality of adipose tissue, in terms of protein production and gene expression per unit mass of adipose tissue, and in the tissue response to altered environments, such as exercise, are also likely to play a role.
Exercise training is an intervention that improves insulin sensitivity. We have recently demonstrated that 7 weeks of moderate-intensity exercise training induced a 3-fold greater improvement in insulin sensitivity in offspring than matched control subjects [13]. Circulating leptin concentrations decreased significantly only in the offspring in response to the intervention and the change correlated significantly with the change in insulin sensitivity, independently of change in body mass [13], suggesting that this change in insulin sensitivity may be mediated through leptin. However, the effect of exercise on circulating adiponectin remains inconclusive, with increases [14,15], decreases [16] and “no change” [17] reported in different studies. Similarly, the effects of exercise training on adipose tissue gene expression of leptin, adiponectin, and their receptors, which would indicate sensitivity to these adipokines, are not known. These data would help to provide a more complete understanding of the mechanisms by which exercise training modulates adiponectin and leptin, and why differential effects are observed in offspring and controls; these would also allow an investigation of the interactions between adipokines, their receptors, and environmental stimuli in different population groups, which may help explain these differential effects.
The purpose of the present study was therefore to determine whether the above factors differed between sedentary offspring and control subjects, whether they correlated with the observed differences in insulin sensitivity, and whether changes in these factors or relationships with insulin sensitivity after exercise training differed between groups.
2. Methods
The participants and study design have previously been described in detail [13]. Briefly, 34 women with at least one parent with type 2 diabetes mellitus (offspring; age, 35.6 ± 7.0 years; BMI, 28.1 ± 5.1 kg/m2) and 36 women with no first- or second-degree relative with type 2 diabetes mellitus, matched for age and BMI with the offspring (control; age, 33.6 ± 6.1 years; BMI, 27.3 ± 4.7 kg/m2), participated. All subjects were sedentary, healthy, normoglycemic, nonsmokers with a regular menstrual cycle and gave written informed consent. Because of dropouts, postintervention data are for 28 offspring and 34 controls. No significant differences at baseline were observed between those who dropped out and those who completed the study (data not shown). The trial was approved by the Research Ethics Committee of the North Glasgow University Hospitals NHS Trust and was registered with ClinicalTrials.gov (Trial identifier: NCT00268541). Subjects underwent adipose tissue biopsy and an oral glucose tolerance test at baseline and after a progressive 7-week endurance-type exercise training program that is described in detail in Barwell et al [13]. Briefly, exercise sessions were performed at 65% to 80% of maximum heart rate beginning with 3 exercise sessions of 30 minutes in week 1 and building to 5 exercise sessions of 60 minutes in weeks 6 and 7. Postexercise testing, including the biopsy, was done between 15 and 24 hours after the final training session. Subjects were asked not to alter their dietary habits during their participation in the study and completed 7-day weighed food diaries before each metabolic testing day that were analyzed using a computerized version of the food composition tables (CompEat Pro; Nutrition Systems, Banbury, United Kingdom).
Body composition and fat distribution measurements were made using dual x-ray absorptiometry scans (LUNAR Prodigy DEXA scanner; GE Healthcare Diagnostic Imaging, Slough, United Kingdom). Blood samples were collected into potassium EDTA on ice, separated within 15 minutes, and stored at −80°C. Adiponectin and leptin concentrations were determined in fasted state blood samples using enzyme-linked immunosorbent assay kits (R&D Systems Europe, Abingdon, United Kingdom). Insulin and glucose were determined in blood samples collected in the fasted state and 30, 60, 90, and 120 minutes after oral glucose tolerance test glucose ingestion by enzyme-linked immunosorbent assay (Mercodia, Uppsala, Sweden) and enzymatic colorimetric reaction (Roche, Welwyn Garden City, United Kingdom) kits, respectively. Within-assay coefficients of variation were 3.9%, 8.0%, 3.9% and 0.8%, respectively. Insulin sensitivity was calculated using the insulin sensitivity index (ISI) [18], which is highly correlated with the rate of whole-body glucose disposal during a euglyemic-hyperinsulinemic clamp [18].
Subcutaneous abdominal adipose tissue biopsies (∼300 mg) were taken with a needle at the level of the umbilicus and had blood and any nonadipose tissue contamination removed with sterile forceps and washing with 0.9% saline solution on a gauze. Each biopsy was split into 2 similarly sized pieces (∼150 mg) before flash-freezing in liquid nitrogen, all within 2 minutes of collection. Total RNA was extracted from one of these pieces using the RNeasy Lipid Tissue Mini Kit (Qiagen, Crawley, United Kingdom) according to the manufacturer's protocol with mechanical homogenization in a RiboLyser (Thermo Scientific, Loughborough, United Kingdom). Complementary DNA was synthesized using the Reverse-iT MAX 1st Strand Synthesis Kit (ABGene, Epsom, United Kingdom) with random hexamers, 500 ng of RNA, and twice the recommended volume of all ingredients, but otherwise according to the manufacturer's protocol. After stopping, reactions were diluted from 40 to 250 μL with distilled water.
Quantitative polymerase chain reaction (PCR) assays were designed for each transcript (Supplementary Table A) using the Universal ProbeLibrary Assay Design Center (http://www.roche-applied-science.com/). Messenger RNA (mRNA) expression levels of transcripts were measured in triplicate using the Universal ProbeLibrary Set (Roche) and ABsolute QPCR Mix (ABGene). Each reaction contained 1.35 μL of each primer (10 μmol/L stock), 0.15 μL probe, and 1 μL complementary DNA in a final volume of 15 μL. Cycle threshold (Ct) outliers were removed using the median absolute deviation method (0.6745 ⁎ absolute difference of data point from median/median) with a maximum acceptable threshold of 3.5. Average amplification efficiency for each assay was determined using version 7.5 of LinReg PCR [19], after exclusion of Ct outlier data points and efficiency values lying greater than ±1.96 standard deviations (SDs) from the mean. Non–Ct-outliers were averaged for each triplicate and corrected for amplification efficiency (Ct ⁎ log [mean efficiency]/log [2]). Each assay required reactions on multiple plates; therefore, control samples were run on each plate with Ct values on subsequent plates corrected to be directly comparable to those on plate 1, using the formula Ct(on plate X) + “average Ct control samples plate 1” − “average Ct control samples plate X”.
Four potential calibrator genes (ACTB, GAPDH, RPLP0, and LRP10) were compared for stability under experimental conditions using NormFinder [20]. RPLP0 was found to be the most stable across all subject groups and intervention conditions. The corrected Ct values for each target gene and RPLP0 were used to calculate the ΔCt, and the average ΔCt of the baseline control group was used to calculate the ΔΔCt. The relative expression values used for statistical analyses and quoted in Table 1 were given by 2−ΔΔCt.
Table 1.
Concentrations of plasma adipokines and gene expression at baseline
| Control (n = 36) | Offspring (n = 34) | P value (unpaired t test) | |
|---|---|---|---|
| ISI | 6.35 (5.27-7.65) | 4.75 (3.85-5.86) | .047 |
| Fat mass (kg) | 27.3 (24.3-30.6) | 29.6 (26.1-33.5) | .359 |
| Plasma adiponectin (mg/mL) | 6.88 (6.04-7.85) | 6.58 (5.65-7.67) | .663 |
| Plasma leptin (pg/mL) | 14.9 (12.1-18.2) | 17.5 (13.5-22.6) | .329 |
| ADIPOQ | 1.00 (0.87-1.14) | 0.92 (0.75-1.13) | .489 |
| ADIPOR1 | 1.00 (0.87-1.15) | 1.03 (0.90-1.18) | .783 |
| ADIPOR2 | 1.00 (0.93-1.08) | 0.97 (0.88-1.06) | .591 |
| LEPTIN | 1.00 (0.83-1.21) | 0.97 (0.76-1.23) | .837 |
| LEPR (long isoform) | 1.00 (0.82-1.21) | 0.99 (0.79-1.25) | .966 |
| LEPR (short isoforms) | 1.00 (0.85-1.17) | 1.03 (0.85-1.25) | .813 |
Statistical analyses were performed on log-transformed data. Values are means and 95% confidence limits back transformed to original units. Gene expression values are relative to RPLP0 expression and the average target to RPLP0 ratio of the control group at baseline. Insulin sensitivity index, fat mass, and plasma leptin have been reported previously [12], but are included here to allow full analyses of combined results.
Data were analyzed using Statistica (version 6.0; StatSoft, Tulsa, OK), Minitab (version 13.1; Minitab, State College, PA), and R (version 2.7.2 [21]; R Foundation for Statistical Computing, Vienna, Austria). Data were tested for normality using the Kolmogorov-Smirnov normality test: plasma adiponectin and leptin were log transformed. Gene expression data were relative and therefore log transformed to ensure normality of errors.
Baseline differences between groups were determined using unpaired t tests. Effects of exercise on the 2 groups were assessed by 2-way analysis of variance with repeated measures and post hoc Tukey tests. Associations between variables were assessed using Pearson correlations.
Adipokine systems were defined as plasma concentration of each adipokine plus subcutaneous adipocyte mRNA expression levels of the adipokine and its receptor genes (Supplementary Table A). Stepwise regression analyses in R identified the best multivariate model to explain the observed data both before (baseline) and in response to (Δ) the exercise intervention. Initial models included all variables plus all possible 2-way interactions. Fat mass correlated well with ISI in both groups at baseline (Table 2) and was therefore included in the modeling to understand if the adipokine “systems” had effects over and above fat mass and if any influence of either “system” was dependent on fat mass. Before modeling, quantitative data were z scored; that is, values for each phenotype were expressed as number of SDs they were away from the mean value for that phenotype. Therefore, the hypothetical average individual would have a baseline score of 0. Similarly, an individual with no change in response to exercise would have a Δ z score of 0. Individuals in the control group were assigned a group value of 0, whereas offspring were assigned a group value of 1. The formulae in Tables 4 and 5 and Supplementary Tables C and D can be interpreted in light of this. The “drop1” and “add1” functions of the “stats” package were used alternately until all terms that remained in the model were either significant (P ≤ .05) or required in the model because of a significant higher-order term. Inclusion of 2-way interactions allowed investigation of whether the influence of certain variables required knowledge of other variables. In biological terms, this allowed us to ask, for example, if the effect of plasma leptin was dependent on the levels of leptin receptor expression. Retrospective calculations of achieved power using G⁎power (v3.0.10; Franz Faul, Universität Kiel, Germany), the full model (ie, the maximum number of predictors), and the achieved adjusted R2 values showed that for adiponectin, leptin, and combined baselines modeling we had 99.7%, 99.8%, and 96.0% power, respectively, to detect effects of the demonstrated size. For modeling of adiponectin and leptin Δs, we had 52.0% and 98.6% power, respectively. Although the power to detect a true effect of adiponectin of the observed size is much lower than we would like, the reduced model produced by the stepwise regression modeling for adiponectin achieved 86.2% power.
Table 2.
Correlations between ISI at baseline with other baseline variables (baseline) and between change in ISI and changes in other variables in response to the exercise intervention (Δ)
| Plasma adipokine (gene) | Combined group |
Control group |
Offspring group |
|||
|---|---|---|---|---|---|---|
| Baseline (N = 70a) | Δ (n = 62a) | Baseline (n = 36a) | Δ (n = 34a) | Baseline (n = 34a) | Δ (n = 28a) | |
| Subject group |
r = −0.239 P = .047 |
r = 0.268 P = .035 |
||||
| Fat mass |
r = −0.540 P < .001 |
r = −0.152 P = .265 |
r = −0.396 P = .023 |
r = 0.181 P = .337 |
r = −0.653 P = < .001 |
r = −0.454 P = .020 |
| Plasma adiponectin |
r = 0.452 P < .001 |
r = 0.068 P = .598 |
r = 0.384 P = .021 |
r = 0.055 P = .757 |
r = 0.512 P = .002 |
r = 0.037 P = .852 |
| ADIPOQ mRNA |
r = 0.203 P = .093 |
r = −0.021 P = .870 |
r = 0.062 P = .720 |
r = −0.015 P = .932 |
r = 0.275 P = .116 |
r = −0.066 P = .739 |
| ADIPOR1 mRNA |
r = 0.129 P = .288 |
r = 0.035 P = .790 |
r = 0.084 P = .625 |
r = −0.094 P = .598 |
r = 0.200 P = .256 |
r = 0.090 P = .648 |
| ADIPOR2 mRNA |
r = 0.137 P = .257 |
r = −0.051 P = .696 |
r = −0.034 P = .846 |
r = −0.155 P = .382 |
r = 0.256 P = .144 |
r = 0.048 P = .808 |
| Plasma leptin |
r = −0.550 P < .001 |
r = −0.432 P < .001 |
r = −0.574 P < .001 |
r = −0.351 P = .042 |
r = −0.516 P < .001 |
r = −0.396 P = .037 |
| Leptin mRNA |
r = −0.217 P = .071 |
r = −0.298 P = .019 |
r = −0.360 P = .031 |
r = −0.297 P = .089 |
r = −0.127 P = .475 |
r = −0.345 P = .072 |
| Leptin receptor (long) mRNA |
r = 0.072 P = .552 |
r = 0.203 P = .113 |
r = 0.063 P = .715 |
r = 0.179 P = .311 |
r = 0.082 P = .646 |
r = 0.136 P = .490 |
| Leptin receptor (short) mRNA |
r = −0.047 P = .702 |
r = 0.105 P = .418 |
r = −0.184 P = .282 |
r = 0.232 P = .187 |
r = 0.077 P = .665 |
r = −0.041 P = .836 |
N values for fat mass were 65 and 56, respectively (combined group); 33 and 30, respectively (control group); and 32 and 26, respectively (offspring group).
Table 4.
Final stepwise regression models for ISI and each adipokine “system” and the 2 “systems” combined at baseline
| Adipokine | Model | |
|---|---|---|
| Adiponectin | ISI = + 0.156 − 0.294*(SG) − 0.593*FM + 0.285*pl.Adipo − 0.762*AdipoQ + 0.495*AdipoR1 − 0.323*AdipoR2 + 1.285*SG:AdipoQ − 1.126*SG:AdipoR1 + 0.621*SG:AdipoR2 + 0.205* AdipoQ:AdipoR2 |
P < .001 Radj2 = 48.0% |
| Leptin | ISI = + 0.125 − 0.339*(SG) + 0.089*(FM) − 0.568*pl.Leptin − 0.124*(Leptin mRNA) + 0.322*LEPRlong − 0.179*(LEPRshort) − 1.282*SG:FM + 1.141*SG:pl.Leptin − 0.335*pl.Leptin:LEPRshort + 0.272*Leptin mRNA:short |
P < .001 Radj2 = 48.6% |
| Combined | ISI = + 0.370 − 0.278*(SG) − 0.490*FM + 0.292*pl.Adipo − 0.039*(AdipoQ) + 0.481*AdipoR1 − 0.630*AdipoR2 + 0.012*(pl.Leptin) − 0.569*Leptin mRNA + 0.102*(LEPRlong) − 0.116*(LEPRshort) − 1.371*SG:AdipoR1 + 1.119*SG:AdipoR2 + 0.440*FM:AdipoQ − 0.481*FM:AdipoR2 + 0.562*FM:pl.Leptin − 0.734*FM:Leptin mRNA + 0.524*pl.Adipo:AdipoQ − 0.448*pl.Adipo:LEPRlong + 0.987*AdipoR1:pl.Leptin − 0.793*AdipoR1:Leptin mRNA − 0.418*AdipoR1:LEPRlong + 0.484*AdipoR1:LEPRshort + 0.439*AdipoR2:Leptin mRNA − 0.443*pl.Leptin:Leptin mRNA − 0.799*pl.Leptin:LEPRlong + 0.635* Leptin mRNA:LEPRlong − 0.310*pl.Adipo:AdipoR1 + 1.191*SG:AdipoQ |
P < .001 Radj2 = 72.4% |
Terms in parentheses are nonsignificant but required in model for higher-order terms. SG indicates subject group; FM, fat mass; pl.Adipo, plasma adiponectin; ADIPOQ, adiponectin mRNA; ADIPOR1, adiponectin receptor 1 mRNA; ADIPOR2, adiponectin receptor 2 mRNA; pl.Leptin, plasma leptin; LEPRlong, the long form of the leptin receptor; LEPRshort, the short forms of the leptin receptor.
Table 5.
Final stepwise regression models for change (Δ) in ISI and changes in each adipokine “system” and the 2 “systems” combined
| Adipokine | Model | |
|---|---|---|
| Adiponectin | ΔISI = + 0.155 − 0.039⁎(SG) − 1.465⁎ΔFM + 0.409⁎(Δpl.Adipo) + 0.059⁎(ΔADIPOR1) − 0.121⁎(ΔADIPOR2) − 1.028⁎SG:Δpl.Adipo − 3.504⁎ΔFM:Δpl.Adipo + 0.203⁎ΔADIPOR1:ΔADIPOR2 |
P = .008 Radj2 = 22.8% |
| Leptin | ΔISI = + 0.081 + 1.405⁎ΔFM − 0.081⁎(Δpl.Leptin) − 0.289⁎ΔLeptin mRNA + 0.001⁎(ΔLEPRlong) + 1.712⁎ΔFM:Δpl.Leptin − 0.244⁎Δpl.Leptin:ΔLEPRlong |
P < .001 Radj2 = 47.4% |
| Combined | Too many possible terms relative to N value to test |
Terms in parentheses are nonsignificant but required in model for higher-order terms.
3. Results
Insulin sensitivity, fat mass, and plasma leptin data have been reported previously [13] but are included in tables to allow interpretation of the gene expression data and circulating adiponectin data in context and consideration of each adipokine “system” as a whole. Further descriptive data about the study participants are also available in that publication [13].
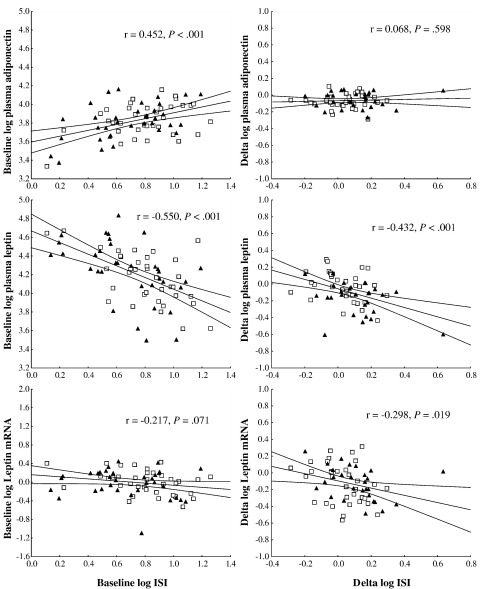
Table 1 describes the baseline data. No significant differences were observed in concentrations of circulating adipokines or in gene expression levels. For all subjects together, ISI correlated positively with circulating adiponectin and negatively with circulating leptin (Fig. 1), explaining 20.4% and 30.3% of the variance, respectively. These relationships with ISI were also evident in the separate offspring and control groups (Table 2). Leptin mRNA levels correlated significantly with ISI in the control group but not the offspring group (Table 2).
Fig. 1.
Correlations between ISI and plasma adipokine or adipocyte mRNA levels at baseline (baseline) and between change in ISI and changes in plasma adipokine or adipocyte mRNA levels (Δ). Black triangles (▲) represent the offspring group, whereas open squares (□) represent the control group. Regression line (central line) and 95% confidence limits (outer lines) are shown.
Changes in variables in response to the exercise intervention are shown in Table 3. In all subjects combined, plasma adiponectin and leptin were significantly reduced by exercise; however, separate consideration of the groups shows that the reduction in leptin was significant only in the offspring group, whereas adiponectin was significantly reduced in both groups. Leptin gene expression was significantly reduced by exercise overall and in separate groups, whereas the long isoform of the leptin receptor (LEPRlong) gene expression was significantly increased overall; but when the groups were considered separately, the increase was only significant in the offspring group. Exercise-induced changes in plasma leptin and leptin gene expression (Fig. 1 and Table 2) correlated with the change in ISI in the combined group, explaining 18.7% and 8.9% of the variance, respectively. Similar correlations were found in each of the subgroups, although they did not reach significance for leptin mRNA in these smaller subgroups. Correlations between the variables and fat mass are shown in Supplementary Table B for information, although the findings of this study were not substantially altered by expressing ISI per unit fat mass (data not shown).
Table 3.
Exercise induced changes (percentage) in ISI, fat mass, plasma adipokines, and gene expression
| Combined group (n = 62) | Pa | Control group (n = 34) | Pb | Offspring group (n = 28) | Pc | Pd | |
|---|---|---|---|---|---|---|---|
| ISI | 16.7 (7.1 to 27.2) | <.001 | 7.3 (−3.0 to 18.7) | .613 | 29.2 (12.4 to 48.5) | .001 | .035 |
| Fat mass | −15.2 (−20.5 to −9.6) | <.001 | −12.1 (−18.7 to −4.8) | .022 | −13.8 (−18 to −9.4) | .004 | .556 |
| Plasma adiponectin | −13.8 (−18.0 to −9.4) | <.001 | −15.2 (−20.5 to −9.6) | <.001 | −12.1 (−18.7 to −4.8) | .007 | .477 |
| Plasma leptin | −19.5 (−28.0 to −10.0) | <.001 | −7.3 (−19.1 to 6.2) | .725 | −32.2 (−42.5 to −20.1) | <.001 | .005 |
| ADIPOQ mRNA | −4.6 (−12.3 to 3.9) | .314 | −6.8 (−16.1 to 3.6) | .633 | −1.8 (−14.6 to 12.8) | .992 | .558 |
| ADIPOR1 mRNA | 7.6 (−1.0 to 17.0) | .083 | 5.4 (−3.9 to 15.5) | .802 | 10.4 (−4.7 to 28.0) | .408 | .585 |
| ADIPOR2 mRNA | 0.6 (−5.8 to 7.5) | .853 | 0.7 (−8.1 to 10.2) | .999 | 0.6 (−8.8 to 11.0) | .999 | .998 |
| Leptin mRNA | −24.9 (−33.7 to −14.9) | <.001 | −25.0 (−37.7 to −9.7) | .008 | −24.7 (−36.1 to −11.3) | .022 | .975 |
| LEPR (long isoform) mRNA | 21.0 (3.8 to 41.0) | .011 | 7.7 (−13.0 to 33.3) | .892 | 39.4 (12.9 to 72.1) | .026 | .101 |
| LEPR (short isoforms) mRNA | 9.1 (−2.5 to 22.1) | .102 | 1.7 (−9.3 to 14.1) | .996 | 18.8 (−3.1 to 45.6) | .186 | .179 |
Statistical analyses were performed on log-transformed data. Values are means and 95% confidence limits. Insulin sensitivity index, fat mass, and plasma leptin have been reported previously [12], but are included here to allow full analyses of combined results. Post hoc tests are Tukey honestly significant difference tests; other P values are from repeated-measures analysis of variance.
Exercise term.
Post hoc exercise term for control group only.
Post hoc exercise term for offspring group only.
Interaction between subject group and exercise term.
At baseline, stepwise regression modeling showed that variation in the leptin “system” could account for 48.6% of the variance in ISI (Table 4). Those in the offspring group had 0.339 SD lower ISI than controls if they had average (ie, 0) values of all other data. More complex terms in the model require evaluation of several variables at once. Control individuals with 1 SD higher than average fat mass had a relatively average ISI (+0.089 SD), whereas offspring individuals had 1.193 SD lower (+0.089 − 1.282) than average ISI. Control individuals with 1 SD higher than average plasma leptin had 0.568 SD lower than average ISI, whereas offspring individuals had a 0.573 SD higher (−0.568 + 1.141) than average ISI. If they also had 1 SD higher short isoform of the leptin receptor (LEPRshort) mRNA expression level, these values would be reduced by 0.514 SD (−0.179 − 0.335). Individuals with a 1 SD higher than average leptin mRNA expression level would have 0.124 SD lower than average ISI. If they also had 1 SD higher than average LEPRshort mRNA expression level, then they would have 0.031 SD lower (−0.124 − 0.179 + 0.272) than average ISI. Those with 1 SD higher than average LEPRlong mRNA expression level had 0.322 SD higher than average ISI.
Baseline variation in the adiponectin “system” accounted for 48.0% of the variance in ISI (Table 4) with individual components quantifiable as above. A similar approach, explaining 72.4% of the ISI variance, can be used to identify important interactions between the adiponectin and leptin “systems.” The influence of ADIPOQ, ADIPOR1, and ADIPOR2 mRNAs were all dependent on subject group, as ADIPOQ and ADIPOR1 mRNAs were for the adiponectin “system” alone. Significant points of interaction between the “systems” were as follows—(1) Plasma adiponectin and LEPRlong mRNA: If 1 SD higher than average for both, ISI was 0.332 SD lower for the offspring group, although it was relatively unchanged (+0.054) in the control group. (2) Plasma leptin and ADIPOR1 mRNA: If 1 SD higher than average for both, ISI was 1.480 SD higher for the control group, although it was relatively unchanged (+0.089) in the offspring group. (3) Leptin mRNA and ADIPOR1 mRNA: If 1 SD higher than average for both, ISI was 0.881 SD lower for the control group and was further reduced to −2.252 in the offspring group. (4) ADIPOR1 mRNA and LEPRlong mRNA: If 1 SD higher than average for both, ISI was slightly higher than average (+0.165) for the control group, but was 1.206 SD lower than average for the offspring group. (5) ADIPOR1 mRNA and LEPRshort mRNA: If 1 SD higher than average for both, ISI was 0.849 SD higher than average for the control group, although it lower than average (−0.522 SD) for the offspring group. (6) ADIPOR2 and leptin mRNAs: If 1 SD higher than average for both, ISI was 0.760 SD lower than average for the control group, but 0.359 SD higher than average for the offspring group.
Variation in the leptin “system” Δs accounted for 47.4% of the variance in Δ ISI (Table 5). None of the changes in the leptin “system” differed by group. The Δ fat mass was an independent predictor of Δ ISI, although paradoxically a reduction in fat mass was correlated with a reduction in ISI (individuals who reduced their fat mass by 1 SD would have reduced their ISI by 1.405 SD if they changed nothing else). Changes in plasma leptin and in LEPRlong mRNA were nonsignificant as individual terms, although they were required in the model for more complex terms. Individuals who reduced their leptin mRNA by 1 SD increased their ISI by 0.289 SD. Individuals who reduced both their fat mass and plasma leptin by 1 SD increased their ISI by 3.117 SD. Those who reduced their plasma leptin and increased their expression of LEPRlong mRNA increased their ISI by 0.326 SD.
Variation in the adiponectin “system” Δs accounted for 22.8% of the variance in Δ ISI (Table 5). Too few individuals completed the study to reliably test for interactions in the Δs of the 2 adipokine “systems.”
4. Discussion
Our results demonstrate that, whereas there were no overt baseline differences in plasma adiponectin, leptin, or related gene expression levels, which could explain the baseline ISI differences between women with and without a family history of diabetes, plasma leptin and LEPRlong mRNA only changed significantly in response to exercise training in the offspring group, and exercise-induced changes in the “leptin system” could explain almost half the exercise-induced change in ISI. Furthermore, whereas adiponectin-related variables did not change differently between the groups in response to exercise training, changes in the “adiponectin system” explained almost a quarter of the variance of the change in ISI. The modeling approach used in the present study enabled the elucidation of complex relationships that were not immediately obvious and showed that the adipokine correlations were both over and above, and dependent on, fat mass. This also highlighted relationships between the adipokine “systems” and ISI that differed by subject group at baseline and showed that the effects of subject group on ISI response to exercise may be mediated through changes to leptin variables.
Baseline circulating leptin values were similar to those reported in the literature for similar subject groups [22]. Although there were no group differences at baseline in circulating leptin concentration or expression of the leptin gene or its receptors, subject group was an important element of the leptin “system” modeling of ISI. The final model, which explains almost half of the variance in ISI, indicates that fat mass is more important in determining ISI in the offspring than the control group and that increased plasma leptin is correlated with reduced ISI in the control group, but increased ISI in the offspring group. One interpretation of this is that, after accounting for other variables, the role of plasma leptin, or the context in which it works in the 2 groups, is different. The notion of differing roles for leptin in different contexts is consistent with the ideas of Magni et al [23] who showed that free and bound leptin correlates with different aspects of biology: fat mass and resting energy expenditure, respectively. The influence of plasma leptin on ISI was also determined by the expression level of LEPRshort mRNA, as was the influence of leptin mRNA. The function of these short transcripts is unknown, although they are thought to play a role in clearance of leptin from the circulation [24], which would alter the availability of plasma leptin, and in transport of leptin across the blood-brain barrier [25], which would alter the site of action of leptin. Increased expression of LEPRlong mRNA correlated with increased ISI regardless of group as would have been predicted because the full-length leptin receptor is required for leptin signaling [26]. Interestingly, expression of LEPRlong only increased significantly in response to training in the offspring group—an effect that would increase adipose tissue sensitivity to a given level of plasma leptin.
Modeling of the change in ISI against the change in the leptin “system” explained 47.7% of the variance in the change in ISI but did not identify subject group as an important element. This is likely due to the difference between the groups in the change in plasma leptin and the change in LEPRlong mRNA, such that the “group information” is contained in the Δ values of these variables. The final model shows that the influence of fat mass cannot be interpreted without appreciating the concurrent change in plasma leptin. Both on average reduce, as does leptin mRNA, all resulting in an increase in ISI. Similarly, the effect of the change in plasma leptin cannot be interpreted without appreciating the change in the expression of LEPRlong mRNA, which is required for plasma leptin to elicit a signal [26]. On average, plasma leptin reduced with exercise and LEPRlong increased. The model predicts that this would result in an increase in ISI, perhaps through the predicted resulting increase in leptin signaling.
Baseline circulating adiponectin values were similar to those reported in the literature for similar subject groups [22]. In cross-sectional studies, high circulating adiponectin concentrations are associated with high insulin sensitivity [27], a finding also evident in the present study. However, the present data show a reduction in plasma adiponectin in response to endurance exercise training alongside a parallel increase in insulin sensitivity. The literature on the effects of exercise training on plasma adiponectin concentration is inconclusive, with increases [14], decreases [16], and no change [17] reported, although often this includes different types of exercise or durations of training. Interestingly, we observed no significant effect of exercise training on adipose tissue expression of the adiponectin gene or its receptors. Similar results were obtained after correcting for fat mass (data not shown); therefore, the results cannot be explained by a reduction in fat mass with exercise. However, the adipocytes were obtained from a single fat depot; and it is possible that the lower circulating adiponectin concentrations post–exercise training could reflect a reduction in adiponectin production by another depot, such as visceral fat, or by a level of posttranscriptional or posttranslational control. To understand how the expression of ADIPOQ relates to adiponectin production, it would have been necessary to measure adiponectin protein in adipose tissue biopsies by Western blotting.
The model for adiponectin explains almost half the variance in ISI and predicts that increased fat mass is correlated with reduced ISI, whereas, after correction for fat mass, increased plasma adiponectin is an independent predictor of, and correlated with, increased ISI. The influences of ADIPOQ, ADIPOR1, and ADIPOR2 mRNAs are dependent on subject group. Modeling of the change in ISI against the change in the adiponectin “system” explained 22.8% of the variance in Δ ISI and identified subject group as an important element likely because there were no significant group by exercise interaction effects for the adiponectin “system” that could match the significant interaction effect in ISI. The model predicts that reducing plasma adiponectin will reduce the control group's ISI, but increase the offspring group's ISI, although an understanding of the change in fat mass must also be factored in to this. Recent evidence suggests that the ratio of high–molecular-weight adiponectin to total adiponectin is more closely related to insulin resistance than simply the total adiponectin, as measured in the present study [28]. Furthermore, Schober et al [29] have shown that different forms of adiponectin can have opposing effects; high–molecular-weight adiponectin enhanced interleukin-6 release from isolated monocytes, whereas low–molecular-weight adiponectin reduced interleukin-6 release. Thus, these apparently contradictory effects of adiponectin on ISI may be the result of altered ratios of the forms of adiponectin in the 2 groups. The model also indicates an interaction between ADIPOR1 and ADIPOR2 mRNAs and suggests that a balance between the 2 receptors is important because they determine the pathway of adiponectin signaling.
Modeling of all variables at baseline explained 72.4% of the variance in ISI, showing that the effects of the 2 “systems” are at least partially independent. Importantly, it highlighted several apparent points of interaction between the “systems” that warrant further investigation: plasma adiponectin interacts statistically with LEPRlong; ADIPOR1 mRNA interacts with all the leptin variables, making it a crucial interaction point; and ADIPOR2 mRNA interacts with leptin mRNA. This is consistent with information in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, showing an interaction between the 2 pathways in the activation of adenosine monophosphate–activated protein kinase [3,30], and gives information about the critical components to understand.
This work shows that, although both groups showed beneficial changes with exercise, in fact improving cardiorespiratory fitness to a similar extent [13], at the molecular level, exercise had differing effects in those with and without a family history of diabetes. Changes in the leptin “system” were more strongly related to exercise-induced changes in insulin sensitivity than changes in the adiponectin “system,” although the statistical models revealed points of communication between the 2 “systems,” particularly at ADIPOR1. These findings increase our understanding about how exercise influences the expression of important metabolic genes in subcutaneous adipose tissue and may help explain, at least in part, why women with a family history of diabetes increase insulin sensitivity to a greater extent than their peers with no diabetes family history in response to exercise training.
Acknowledgment
This research was supported by a project grant from the British Heart Foundation (PG/03/145).
Footnotes
Institutional approval: The trial was approved by the Research Ethics Committee of the North Glasgow University Hospitals NHS Trust and was registered with ClinicalTrials.gov (Trial identifier: NCT00268541).
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.metabol.2009.12.026.
Appendix A.
.
References
- 1.Zierath J.R., Krook A., Wallberg-Henriksson H. Insulin action and insulin resistance in human skeletal muscle. Diabetologia. 2000;43:821–835. doi: 10.1007/s001250051457. [DOI] [PubMed] [Google Scholar]
- 2.Ronti T., Lupattelli G., Mannarino E. The endocrine function of adipose tissue: an update. Clin Endocrinol (Oxf) 2006;64:355–365. doi: 10.1111/j.1365-2265.2006.02474.x. [DOI] [PubMed] [Google Scholar]
- 3.Kadowaki T., Yamauchi T., Kubota N., Hara K., Ueki K., Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jansson P.A., Enerback S., Eriksson J., Elam M., Smith U. Insulin resistance and increased plasma leptin levels in healthy non-obese NIDDM relatives. Diabetes. 1997;46:1395. [Google Scholar]
- 5.Kobberling J. The predictive value of diagnostic measures. Deut Med Wochenschr. 1982;107:591–595. doi: 10.1055/s-2008-1069984. [DOI] [PubMed] [Google Scholar]
- 6.Ohlson L.O., Larsson B., Bjorntorp P., Eriksson H., Svardsudd K., Welin L. Risk-factors for type-2 (non–insulin-dependent) diabetes-mellitus—13-1/2 years of follow-up of the participants in a study of Swedish men born in 1913. Diabetologia. 1988;31:798–805. doi: 10.1007/BF00277480. [DOI] [PubMed] [Google Scholar]
- 7.Kriketos A.D., Greenfield J.R., Peake P.W., Furler S.M., Denyer G.S., Charlesworth J.A. Inflammation, insulin resistance, and adiposity—a study of first-degree relatives of type 2 diabetic subjects. Diabetes Care. 2004;27:2033–2040. doi: 10.2337/diacare.27.8.2033. [DOI] [PubMed] [Google Scholar]
- 8.Nyholm B., Nielsen M.F., Kristensen K., Nielsen S., Ostergard T., Pedersen S.B. Evidence of increased visceral obesity and reduced physical fitness in healthy insulin-resistant first-degree relatives of type 2 diabetic patients. Eur J Endocrinol. 2004;150:207–214. doi: 10.1530/eje.0.1500207. [DOI] [PubMed] [Google Scholar]
- 9.Perseghin G., Ghosh S., Gerow K., Shulman G.I. Metabolic defects in lean nondiabetic offspring of NIDDM parents—a cross-sectional study. Diabetes. 1997;46:1001–1009. doi: 10.2337/diab.46.6.1001. [DOI] [PubMed] [Google Scholar]
- 10.Kowalska F., Strazkowski M., Nikolajuk A., Krukowska A., Kinalska I., Gorska M. Plasma adiponectin concentration and tumor necrosis factor–alpha system activity in lean non-diabetic offspring of type 2 diabetic subjects. Eur J Endocrinol. 2006;154:319–324. doi: 10.1530/eje.1.02084. [DOI] [PubMed] [Google Scholar]
- 11.Lattuada G., Costantino F., Caumo A., Scifo P., Ragogna F., De Cobelli F. Reduced whole-body lipid oxidation is associated with insulin resistance, but not with intramyocellular lipid content in offspring of type 2 diabetic patients. Diabetologia. 2005;48:741–747. doi: 10.1007/s00125-005-1686-6. [DOI] [PubMed] [Google Scholar]
- 12.Nyholm B., Fisker S., Lund S., Moller N., Schmitz O. Increased circulating leptin concentrations in insulin-resistant first-degree relatives of patients with non–insulin-dependent diabetes mellitus: relationship to body composition and insulin sensitivity but not to family history of non-insulin-dependent diabetes mellitus. Eur J Endocrinol. 1997;136:173–179. doi: 10.1530/eje.0.1360173. [DOI] [PubMed] [Google Scholar]
- 13.Barwell N.D., Malkova D., Moran C.N., Cleland S.J., Packard C.J., Zammit V.A. Exercise training has greater effects on insulin sensitivity in daughters of patients with type 2 diabetes than in women with no family history of diabetes. Diabetologia. 2008;51:1912–1919. doi: 10.1007/s00125-008-1097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bluher M., Bullen J.W., Lee J.H., Kralisch S., Fasshauer M., Kloting N. Circulating adiponectin and expression of adiponectin receptors in human skeletal muscle: associations with metabolic parameters and insulin resistance and regulation by physical training. J Clin Endocr Metab. 2006;91:2310–2316. doi: 10.1210/jc.2005-2556. [DOI] [PubMed] [Google Scholar]
- 15.Fatouros I.G., Tournis S., Leontsini D., Jamurtas A.Z., Sxina M., Thomakos P. Leptin and adiponectin responses in overweight inactive elderly following resistance training and detraining are intensity related. J Clin Endocrinol Metab. 2005;90:5970–5977. doi: 10.1210/jc.2005-0261. [DOI] [PubMed] [Google Scholar]
- 16.Jurimae J., Purge P., Jurimae T. Adiponectin is altered after maximal exercise in highly trained male rowers. Eur J Appl Physiol. 2005;93:502–505. doi: 10.1007/s00421-004-1238-7. [DOI] [PubMed] [Google Scholar]
- 17.Polak J., Klimcakova E., Moro C., Viguerie N., Berlan M., Hejnova J. Effect of aerobic training on plasma levels and subcutaneous abdominal adipose tissue gene expression of adiponectin, leptin, interleukin 6, and tumor necrosis factor alpha in obese women. Metabolism. 2006;55:1375–1381. doi: 10.1016/j.metabol.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Matsuda M., DeFronzo R.A. Insulin sensitivity indices obtained from oral glucose tolerance testing—comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22:1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 19.Ramakers C., Ruijter J.M., Deprez R.H.L., Moorman A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 20.Andersen C.L., Jensen J.L., Orntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 21.R-Development-Core-Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, Vienna, Austria; 2008. p. R Development Core Team.
- 22.Jurimae J., Jurimae T., Ring-Dimitriou S., LeMura L.M., Arciero P.J., von Duvillard S.P. Plasma adiponectin and insulin sensitivity in overweight and normal-weight middle-aged premenopausal women. Metabolism. 2009;58:638–643. doi: 10.1016/j.metabol.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Magni P., Liuzzi A., Ruscica M., Dozio E., Ferrario S., Bussi I. Free and bound plasma leptin in normal weight and obese men and women: relationship with body composition, resting energy expenditure, insulin-sensitivity, lipid profile and macronutrient preference. Clin Endocrinol. 2005;62:189–196. doi: 10.1111/j.1365-2265.2005.02195.x. [DOI] [PubMed] [Google Scholar]
- 24.Cumin F., Baum H.P., Levens N. Leptin is cleared from the circulation primarily by the kidney. Int J Obesity. 1996;20:1120–1126. [PubMed] [Google Scholar]
- 25.Golden P.L., Maccagnan T.J., Pardridge W.M. Human blood-brain barrier leptin receptor - Binding and endocytosis in isolated human brain microvessels. J Clin Invest. 1997;99:14–18. doi: 10.1172/JCI119125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arora A., Arora S. Leptin and its metabolic interactions - an update. Diabetes Obes Metab. 2008 doi: 10.1111/j.1463-1326.2008.00852.x. [Review], 18th February 2008;epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 27.Saltevo J., Laakso M., Jokelainen J., Keinanen-Kiukaanniemi S., Kumpusalo E., Vanhala M. Levels of adiponectin, C-reactive protein and interleukin-1 receptor antagonist are associated with insulin sensitivity: a population-based study. Diabetes Metab Res Rev. 2008;24:378–383. doi: 10.1002/dmrr.831. [DOI] [PubMed] [Google Scholar]
- 28.Hara K., Horikoshi M., Yamauchi T., Yago H., Miyazaki O., Ebinuma H. Measurement of the high-molecular weight form of adiponectin in plasma is useful for the prediction of insulin resistance and metabolic syndrome. Diabetes Care. 2006;29:1357–1362. doi: 10.2337/dc05-1801. [DOI] [PubMed] [Google Scholar]
- 29.Schober F., Neumeler M., Weigert J., Wurm S., Wanninger J., Schaffler A. Low molecular weight adiponectin negatively correlates with the waist circumference and monocytic IL-6 release. Biochem Bioph Res Co. 2007;361:968–973. doi: 10.1016/j.bbrc.2007.07.106. [DOI] [PubMed] [Google Scholar]
- 30.Kahn B.B., Alquier T., Carling D., Hardie D.G. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
.