SUMMARY
Occult hepatitis B virus (O-HBV) infection is characterized by the presence of HBV DNA without detectable hepatitis B surface antigen (HBV DNA+/HBsAg−) in the serum. Although O-HBV is more prevalent during HBV/HIV co-infection, analysis of HBV mutations in co-infected patients is limited. In this preliminary study, HBV PreSurface (PreS) and surface (S) regions were amplified from 33 HIV-positive patient serum samples − 27 chronic HBV (C-HBV) and six O-HBV infections. HBV genotype was determined by phylogenetic analysis, while quasispecies diversity was quantified for the PreS, S and overlapping polymerase regions. C-HBV infections harboured genotypes A, D and G, compared to A, E, G and one mixed A/G infection for O-HBV. Interestingly, nonsynonymous-synonymous mutation values indicated positive immune selection in three regions for O-HBV vs one for CHBV. Sequence analysis further identified new O-HBV mutations, in addition to several previously reported mutations within the HBsAg antigenic determinant. Several of these O-HBV mutations likely contribute to the lack of detectable HBsAg in O-HBV infection by interfering with detection in serologic assays, altering antigen secretion and/or decreasing replicative fitness.
Keywords: diversity, HBV polymerase, HBV/HIV co-infection, hepatitis B surface antigen, occult hepatitis B virus
INTRODUCTION
There are 350 million chronic carriers of the hepatitis B virus (C-HBV) worldwide. C-HBV infection is characterized by detectable hepatitis B surface antigen (HBsAg) in the serum [1]. Occult HBV infection (O-HBV), in contrast, is defined as low level HBV replication without detectable circulating HBsAg [2]. Antibodies against HBV core protein (anti-HBc) had been considered the sole serological marker of O-HBV infection [3]; however, serologically negative individuals have been described with HBV DNA as the only detectable marker of infection [4].
Occult HBV infection is transmissible through blood transfusion from human to human [5] and human to chimpanzee [6]. In addition, C-HBV infection developed after a liver transplant from an O-HBV-infected donor [7]. Retrospective studies have also identified O-HBV infection in 16–68% of tumours in patients with hepatocellular carcinoma (HCC) [8,9]. Similarly, O-HBV infection was associated with development of cirrhosis and HCC [10]. Nonetheless, the clinical consequences of prolonged O-HBV infection remain unclear.
HIV co-infection is common because of shared blood-borne transmission routes. While advances in antiretroviral therapy (ART) have prolonged AIDS-free survival in HBV/ HIV co-infected patients, liver disease has emerged as a leading cause of morbidity and mortality [11]. In HIV-positive cohorts, the prevalence of O-HBV infection is highly variable: Nunez et al. [12] did not identify any patients with detectable HBV DNA among 85 HIV+/HBsAg−/anti-HBc+ injection drug users (IDUs); while Hofer et al. [13] detected serum HBV DNA in 51 of 57 patients (89.5%) who were solely anti-HBc+. O-HBV infections have also been identified in hepatitis C virus (HCV)-positive IDUs [14] and liver biopsies from patients with normal liver biochemistry and without prior liver disease [15].
It is unclear why HBsAg is undetectable during O-HBV infection, although several hypotheses exist. One possibility is that HBsAg is not produced or is expressed at levels below detectable limits of current diagnostic assays. Alternatively, HBsAg could be produced but not secreted from infected hepatocytes. Importantly, altered expression of HBsAg likely results from HBV mutation(s) [16]. Mutations in either the surface open reading frame (ORF), containing the PreS and surface (S) regions, or their corresponding spacer and reverse transcriptase (RT) regions of the overlapping polymerase ORF could directly affect HBsAg production. To date, a limited number of studies have investigated O-HBV mutations, in relatively few individuals [17–24]. Here, we describe a detailed evaluation of HBV genomic sequences from chronic (HBsAg+) and occult (HBsAg−) HBV-infected individuals in the same cohort and assess the presence of HBsAg mutations associated with O-HBV infection.
MATERIALS AND METHODS
Patient population
This prospective HIV-positive cohort was previously described in an analysis of O-HBV infection [4]. HBV DNA levels were determined by real-time PCR [lower limit of detection (LLD) = 67 copies/mL or 100 IU/mL] and HBV serologic markers, HBsAg, anti-HBc and anti-HBs, were evaluated by ELISA (LLD = 0.5 ng/mL, 2 NCU/mL and 10 mIU/mL, respectively) (Biochain, Hayward, CA, USA). HCV serostatus was determined by ELISA, measuring antibodies against HCV. As samples were collected between 1989 and 2004, HIV RNA levels were not available for all individuals. ALT levels above 92 U/L (two times the upper limit of local normal levels [25]) were considered elevated. Here, 23 C-HBV and six O-HBV serum samples were utilized, along with four HIV-positive, C-HBV samples from our serum repository.
PCR amplification
Hepatitis B virus DNA extracted from 200 to 400 μL of serum was used in separate nested PCRs for the PreS (549 bp) and S (339 bp) regions using primers [21,26] and amplification conditions listed in Supplemental Table S1. DNA amplification was performed using the GeneAmp PCR reagent kit with AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA, USA) according to manufacturer's instructions. Five microlitres of HBV DNA initially served as template, while 1 μL of first round product served as template for the second round. A previously amplified HBV-positive patient serum sample served as a positive control, while HBV DNA-negative serum samples and a reaction without template served as negative controls.
Cloning and sequencing
PCR products were electrophoresed, purified and cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA). Ten clones per region per individual were sequenced bidirectionally. All sequences have been submitted to GenBank under accession numbers EU769235–EU769292.
Phylogenetic analysis
Sequence alignments were created for the PreS, S and their corresponding polymerase spacer [Pol(PS)] and RT [Pol(S)] regions, respectively, using the neighbour-joining method in CLUSTAL X [27]. Published references [28] and additional GenBank sequences were chosen at random to achieve five per genotype (Supplemental Table S2). For phylogenetic trees, the statistical robustness and reliability of the branching order were assessed using bootstrap analysis with 100 replicates. Calculations for genetic distance and non-synonymous–synonymous (dN–dS) mutation values [29] were performed using mega 3.1 [30]. Shannon entropy (Sn = − Σ (pi ln pi/ ln N) was calculated, where pi was the frequency of each distinct nucleotide sequence and N was the total number of sequences analysed.
Mutational analysis
GenBank was searched for complete HBV genomes. One thousand nine hundred and ninety-three sequences were inspected, and all non-HBV, primate, recombinant and O-HBV genomes were excluded. The remaining full-length HBV sequences for genotypes A (n = 143), E (n = 58) and G (n = 23) were included in the mutational analysis.
GenBank references, as well as C-HBV sequences generated in this study, were compared to O-HBV sequences to identify distinct amino acid mutations in each genomic region that characterize O-HBV infections. Analyses were performed in a genotype-matched manner because of distinct biological and clinical differences among HBV genotypes [31]. The PreS and S regions analysed encode for all of PreS1 and PreS2, as well as amino acids 49–148 of S, and overlap with the entire polymerase spacer region, as well as amino acids 1–6 and 58–156 of the polymerase RT region (all numbered according to [32]). Mutations were considered to be associated with O-HBV infection if they were (i) identified in ≥1 O-HBV clonal sequence and (ii) not identified in any genotype-matched C-HBV sequences from this study or GenBank references analysed. Identified mutations were then compared to published mutations associated with O-HBV infection [17–24]. Mutation prevalence was also determined in all GenBank and C-HBV sequences after stratification by HBV genotype. To further identify signature amino acids, O-HBV sequences were compared to genotype-matched C-HBV sequences from this study using viral epidemiology signature pattern analysis [33] at a threshold of 0.5.
Statistical analysis
Wilcoxon rank-sum tests were performed on patient demographics, and P-values <0.05 were considered significant. For quasispecies parameters (genetic distance, entropy and dN–dS), values of each dependent variable were rank-ordered, and two-factor analysis of variance (ANOVA) tests were performed. Infection status was a between-subjects factor with two levels (occult or chronic), while Region was a within-subjects factor with four levels [PreS, S, Pol(S), Pol(PS)]. For each outcome, 12 planned pairwise comparisons of interest were defined and utilized a t-type procedure where the mean difference in ranks for the conditions being compared was divided by the appropriate ANOVA error term (between-region comparisons using the within-subject error term and between-infection comparisons using the between-subject error term). A critical threshold for significance of P = 0.0042 (P = 0.05/12) was used to control for the overall Type I error rate at alpha = 0.05.
RESULTS
HBV region amplification
PreS was amplified and cloned from 19 of 27 (70.4%) C-HBV and 4 of 12 (33.3%) O-HBV infections. S was amplified and cloned from 25 of 27 (92.6%) C-HBV and 6 of 12 (50%) O-HBV infections. There were no statistically significant differences in patient demographics between C- and O-HBV-infected patients (Table 1), although median HBV DNA level was higher in C-HBV infections − 9.0 × 106 IU/mL – than in O-HBV infections − 1.4 × 104 IU/mL (P < 0.05), as reported previously [4].
Table 1.
Patient demographics. Patient demographic and clinical data were included for the 27 chronic carriers of the hepatitis B virus infections and six occult hepatitis B virus infections that could be amplified by nested PCR
| HBV infection n = 33 | Chronic n = 27 | Occult n = 6 |
|---|---|---|
| Age* | 35.8 years (21.2–58) | 35.4 years (26.7–42.2) |
| Race | ||
| African-American | 14 (51.9%) | 3 (50.0%) |
| Caucasian | 12 (44.4%) | 2 (33.3%) |
| Other | 1 (3.7%) | 1 (16.7%) |
| Gender | ||
| Male | 27 (100%) | 5 (83.3%) |
| Female | 0 | 1 (16.7%) |
| ALT*,† | 56.0 U/L | 27.0 U/L |
| n = 31 | (17–140) | (23–87) |
| HCV serostatus† | 0 | 1 (25%) |
| n = 21 | ||
| HIV† | ||
| Detectable | 21/23 (91%) | 5/5 (100%) |
| Viral load* | 6.0 × 104 copies/mL (7.2 × 102–2.0 × 105) | 1.7 × 105 copies/mL (1.0 × 104–3.7 × 105) |
| ART | ||
| On therapy | 6 (22.2%) | 1 (16.7%) |
| With HBV activity | 7‡ (100%) | 0 |
| CD4+ count*,† | 330.5 cells/mL (7–665) | 141.0 cells/mL (6–559) |
| HBV DNA* | 9.0 × 106 IU/mL (2.1 × 102–7.6 × 108) | 1.4 × 104 IU/mL (1.1 × 103–7.6 × 105) |
Local normal ALT levels were 5–46 U/L [25]; ALT levels are considered elevated when >92 U/L (two times the upper limit of normal). HCV serostatus was evaluated using anti-HCV ELISA. ART, antiretroviral therapy.
Median values are given with the range in parentheses.
Information was not available for all individuals (the number of patients with available information is listed under the heading); therefore, the median ALT and CD4+ count and percent detectable HCV and HIV are given for the individuals tested.
One additional individual is on HBV mono-therapy but not ART.
Identification of HBV genotypes
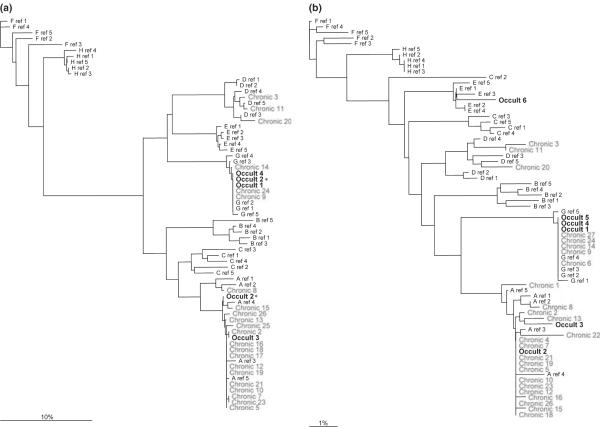
Consensus nucleotide sequences were generated and aligned with reference sequences. In the PreS region, genotypes A (n = 15), D (n = 1) and G (n = 2) were identified in C-HBV subjects; genotypes A (n = 1) and G (n = 2) were identified in O-HBV subjects (Fig. 1a). Additionally, one mixed infection with genotypes A and G was identified (Occult 2). In S, genotypes A (n = 17), D (n = 3) and G (n = 5) were identified in C-HBV subjects, and genotypes A (n = 2), E (n =1) and G (n = 3) were identified in O-HBV subjects, in agreement with PreS (Fig. 1b), although only genotype A was present for the mixed O-HBV infection.
Fig. 1.
Phylogenetic trees for the (a) PreSurface (PreS) and (b) S regions of HBV. Consensus nucleotide sequences were generated from 10 clones per individual. One mixed infection was identified (Occult 2) in the PreS region containing both genotypes A and G (*).
Evaluation of quasispecies diversity
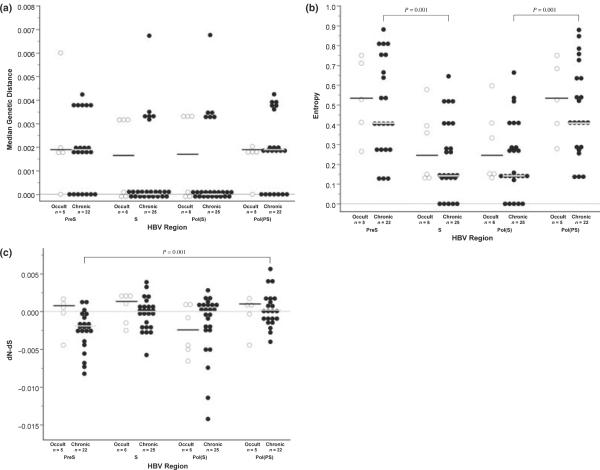
Clonal alignments were performed to assess quasispecies diversity. In PreS, all patients showed significant quasispecies diversity (≥2 distinct viral variants) (Supplemental Fig. S1). In S, while all patients with O-HBV showed significant quasispecies diversity, 5 of 27 C-HBV infections did not (Supplemental Fig. S2). No significant differences were found between C- and O-HBV infections with respect to genetic distance (Fig. 2a), entropy (Fig. 2b) or dN–dS (Fig. 2c), although dN–dS values were >0 – indicating positive selection – in three regions for O-HBV infections – PreS, S and Pol(PS) – compared to only Pol(PS) in C-HBV infections.
Fig. 2.
Dot plots for (a) median genetic distance, (b) Shannon entropy and (c) nonsynonymous–synonymous calculations. Although sequences were only available for four Occult hepatitis B virus (O-HBV)-infected patients for the PreSurface and Pol(PS) regions, n = 5 because of the mixed infection with genotypes A and G. (◯) represent values for O-HBV infections, while (•) represent values for chronic carriers of the hepatitis B virus (C-HBV) infections. Horizontal bars, black for O-HBV infections and grey for C-HBV infections, represent the median values for each group and dashed lines represent zero.
Among C-HBV patients, dN–dS values were significantly higher in Pol(PS) compared to PreS (P = 0.001). A signifi-cant increase in entropy was also noted in PreS and Pol(PS) compared to S and Pol(S), respectively (both, P = 0.001) (Fig. 2b).
Identification of occult hepatitis B virus sequence mutations
Genotype-matched sequence analysis was performed, identifying several novel mutations associated with O-HBV infection: five genotype A and 17 genotype G mutations in PreS, with three genotype A, one genotype E and six genotype G mutations, in addition to several previously published mutations in S (Table 2). Several S mutations identified reside within the antigenic determinant region of HBsAg (amino acids 100–165), suggesting that they may impact HBsAg detection.
Table 2.
PreSurface (PreS) and surface (S) mutations. S open reading frame mutations identified in Occult hepatitis B virus-infected individuals
| Surface region | HBV genotype | Newly identified | Previously identified | Reference | |
|---|---|---|---|---|---|
| PreS1 | A | N98Y | |||
| S109P | |||||
| G | M1I | D41N | |||
| G2R | P88L | ||||
| L3I | A89T | ||||
| S4P | P93L | ||||
| W5R | R102G | ||||
| S16P | T105I | ||||
| F23S | P106S | ||||
| D26Y | |||||
| PreS2 | A | S28G | |||
| H41Q | |||||
| D51G | |||||
| G | L20P | ||||
| T37A | |||||
| Surface | A | Y72H | |||
| I82T | G145A † | [21] | |||
| A128T† | |||||
| E | Y100F† | F85C | [24] | ||
| Y100S† | [24] | ||||
| G145R† | [18,22] | ||||
| G | S55P | ||||
| P62L | |||||
| F80S | |||||
| I86V | |||||
| L95W | |||||
| S136P† | |||||
Mutations also identified in the signature sequence analysis are indicated in bold.
Mutations that reside within the antigenic determinant loops of hepatitis B surface antigen (AA 100–165).
Additional mutations associated with O-HBV were identified in the corresponding polymerase ORF (Table 3). In the spacer region, four genotype A and 13 genotype G mutations were identified, with three mutations for both genotypes A and G and two for genotype E in the RT region.
Table 3.
Polymerase mutations. P open reading frame mutations identified in occult hepatitis B virus-infected individuals
| Polymerase region | HBV genotype | Newly identified | |
|---|---|---|---|
| Spacer | A | Q101L | Q150R |
| L112P | H164N | ||
| G | G5Q | R44* | |
| A6D | R44K | ||
| F7S | Q48R | ||
| L8* | S102P | ||
| G10S | E105G | ||
| F19S | S109F | ||
| R29L | |||
| RT | A | I103T | |
| C136Y | |||
| W153C | |||
| E | L66F | ||
| R153Q | |||
| G | V63A | ||
| T70A | |||
| H94R | |||
Mutations also identified in the signature sequence analysis are indicated in bold. RT, reverse transcriptase.
Indicates a stop codon.
Prevalence of mutations associated with occult hepatitis B virus infection across genotypes
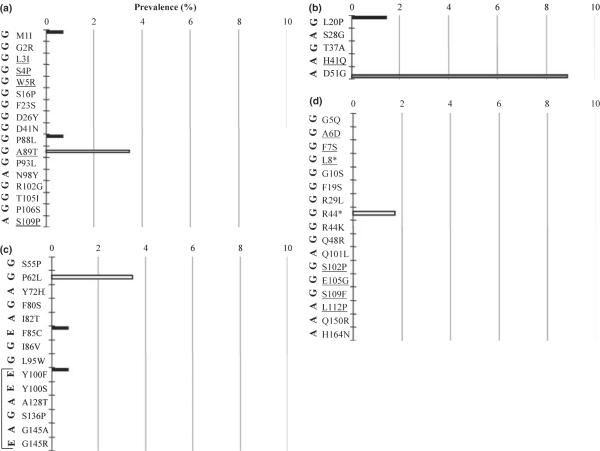
Occult hepatitis B virus mutation prevalence was also determined amongst distinct genotypes. Genotype A, E and G GenBank sequences and C-HBV sequences from this study were searched for all identified O-HBV mutations. Several OHBV mutations were identified at low frequencies in C-HBV sequences of different genotypes for the PreS1 (Fig. 3a), PreS2 (Fig. 3b), S (Fig. 3c) and polymerase spacer (Fig. 3d) regions. No RT mutations were identified in any other sequences.
Fig. 3.
Prevalence of (a) PreS1, (b) PreS2, (c) S and (d) polymerase spacer [Pol(PS)] mutations identified in Occult hepatitis B virus infections in genotype A, E and G reference sequences. None of the identified polymerase reverse transcriptase mutations [Pol(S)] were found in any of the additional reference sequences. (*) Indicates a stop codon. Black bars indicate prevalence in genotype A sequences, white bars indicate prevalence in genotype E sequences, and grey bars indicate prevalence in genotype G sequences. The original genotype in which each mutation was identified is indicated on the left in bold. Underlined mutations could not be analysed in all genotypes because of genotype-specific sequence differences. Bracket in (c) indicates mutations present in the antigenic determinant of hepatitis B surface antigen (amino acids 100–165).
Signature sequence analysis
Signature sequence analysis (SSA) was used to identify mutations more commonly found in O-HBV compared to C-HBV. SSA identified two PreS1 mutations in Occult 2 (genotype A), along with three corresponding polymerase spacer mutations, previously excluded because of low frequency in C-HBV sequences. In Occult 4 (genotype G), 50% of clones harboured a deletion of the first three N-terminal amino acids of PreS1. Another 40% of clones contained five mutations – M1I, G2R, L3I, S4P and W5R - below the 50% threshold for SSA. In the corresponding polymerase spacer, the first seven N-terminal amino acids were deleted in 50% of clones, and five mutations – G5Q, A6D, F7S, L8stop and G10S - were present in another 40%.
In S, SSA identified three mutations in Occult 3 (genotype A) – M103I, K122R and G145A (bold, Table 2), present in all clones, although M103I and K122R were found at low levels in C-HBV sequences. In the corresponding polymerase RT region, two mutations – V112I and W153C – were identified in all clones, but only W153C was absent from chronic genotype A sequences (bold, Table 3). This mutation coincides with the G145A mutation in the S region; while other mutations have been reported at RT position 153 [22,34], none were a tryptophan-to-cysteine mutation.
DISCUSSION
Potential virological differences between C-HBV and O-HBV have not been well defined. It is clear that mutations in the S and Pol ORFs have the potential to (i) alter protein secretion from hepatocytes, (ii) alter protein structure, thereby inhibiting antibody binding in commercial HBsAg detection assays and/or (iii) decrease the overall replication efficacy of the virus. In this preliminary study, we examined genotype distribution and quasispecies diversity within HIV-positive patients previously identified with C-HBV or OHBV infection [4]. Similar to previous reports, genotype A was most common [35–37]; however, genotype G (24%) also exists within this cohort. While genotype G mono-infection has been reported previously [38], it is most commonly found in mixed infections with genotype A [37,39], of which we identified one in an O-HBV-infected individual. To our knowledge, only one mixed infection has been previously reported in the setting of O-HBV infection [20]. These data suggest that genotype G mono-infection may be more common in the United States than previously reported and further support the existence of mixed O-HBV infections.
Although HBV has a lower mutation rate than RNA viruses, it is higher than most DNA viruses. To date, few studies have examined intrapatient diversity of HBV [40,41], and none have included patients with O-HBV infection. Sequencing 10 clones has previously been demonstrated to effectively evaluate quasispecies diversity in HCV [42]; therefore, this number should also be sufficient for HBV given its lower mutation rate. Although our sample size is relatively small, dN-dS values were greater than zero for three of four genomic regions analysed. This indicates that positive immune selection pressures are acting against these regions in O-HBV infection, potentially resulting in mutations that may adversely affect the production and/or detection of HBsAg. No significant differences in genetic distance, Shannon entropy or dN–dS were observed between C-HBV and O-HBV infections. When comparing regions within C-HBV infections, the significantly lower dN–dS for PreS vs Pol(PS) (P = 0.001) indicates increased synonymous mutation within PreS and highlights its intrinsic ability to tolerate mutations [43], although the spacer region is also highly tolerant of mutations/deletions. Significantly, higher entropies were also observed in PreS and Pol(PS) compared to S and Pol(S), respectively (P = 0.001), in C-HBV infections, again suggesting that functional constraints are less stringent for the PreS and spacer regions.
It is important to emphasize that mutations associated with O-HBV are frequently identified without a robust comparison with C-HBV infections in the same cohort, making it difficult to exclude natural polymorphisms and/or genotype-specific differences. To more effectively characterize virological differences between C- vs O-HBV, we performed genotype-matched sequence analysis of the PreS, S and polymerase spacer and RT regions to identify O-HBV mutations absent in C-HBV. Multiple mutations were identified, including several not previously described, although many were only found in one variant. New O-HBV mutations were particularly common for genotypes E and G, as previous investigations have focused primarily on the most common HBV genotypes (A–D). Because of the paucity of sequence data for these genotypes, a portion of these mutations could represent naturally polymorphic sites.
It is known that mutations within the S ORF affect the antigenicity [16,22] and detection of HBsAg [24,44,45]. Three O-HBV-infected individuals harboured mutations with direct importance to virus replication. Two sets of mutations in or near the antigenic determinant of HBsAg were of interest: Occult 3 (genotype A) – M103I, K122R and G145A – and Occult 6 (genotype E) – F85C, Y100S and G145R. While K122R is considered a polymorphism defining the d/y sub-serotypes, its effects in combination with the other two are unknown. The positions of these mutations – either alone or in combination – could alter the secondary structure of HBsAg and impair its detection. In particular, G145R resulted in undetectable HBsAg levels using three of four commercial assays [45]. RT mutations at position 153 – W153C and R153Q – result from the same nucleotide substitution as G145A and G145R in the S ORF. While G145A and G154R have been previously reported [18,21,22], these specific RT153 mutations have not been described. Although neither patient was receiving ART at the time, W153Q has been indicated in lamivudine resistance and was found to decrease replicative fitness in vitro along with additional mutations [34], although single RT mutations are also capable of decreasing HBV replication [46].
Most viral sequences for Occult 4 contained a defective or absent large HBsAg start codon. Although the M1I start codon mutation was also identified in 0.7% of genotype A GenBank reference sequences, these contain another potential start codon at position 12, where genotype G does not. Therefore, genotype G infections may not be able to overcome the consequences of this mutation. Defects in LHBsAg can lead to elimination of PreS1 synthesis or accumulation of large HBsAg (LHBsAg) within the endoplasmic reticulum of hepatocytes. Such mutations have been shown to arrest virion and subviral particle secretion [47,48], and could result in a lack of detectable HBsAg in the serum. Collectively, these data suggest that mutations at distinct amino acid positions in HBsAg or deletions at the N-terminus of the large HBsAg could play a role in persistence of O-HBV infection. Future studies are necessary to characterize additional genomic regions, and a functional analysis is ongoing to evaluate the effects of mutations identified here on HBsAg expression, retention and detection in vitro.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Bristol-Myers Squibb and by a K24 award from NIDDK (DK070528) to KES. Preliminary data were presented at the 15th Conference on Retroviruses and Opportunistic Infections in Boston, MA, February 3–6, 2008.
Abbreviations
- ART
antiretroviral therapy
- C-HBV
chronic carriers of the hepatitis B virus
- dN–dS
nonsynonymous–synonymous
- HBc
HBV core protein
- HBsAg
hepatitis B surface antigen
- HCC
hepatocellular carcinoma
- IDU
injection drug user
- LLD
lower limit of detection
- O-HBV
occult hepatitis B virus
- ORF
open reading frame
- PreS
PreSurface
- RT
reverse transcriptase
- SSA
signature sequence analysis
- S
surface
Footnotes
SUPPORTING INFORMATION Additional Supporting Information may be found in the online version of this article
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
REFERENCES
- 1.Lok ASF, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45(2):507–539. doi: 10.1002/hep.21513. [DOI] [PubMed] [Google Scholar]
- 2.Bodsworth NJ, Cooper DA, Donovan B. The influence of human immunodeficiency virus type 1 infection on the development of the hepatitis B virus carrier state. J Infect Dis. 1991;163(5):1138–1140. doi: 10.1093/infdis/163.5.1138. [DOI] [PubMed] [Google Scholar]
- 3.Jilg W, Sieger E, Zachoval R, Schatzl H. Individuals with antibodies against hepatitis B core antigen as the only serological marker for hepatitis B infection: high percentage of carriers of hepatitis B and C virus. J Hepatol. 1995;23(1):14–20. doi: 10.1016/0168-8278(95)80305-x. [DOI] [PubMed] [Google Scholar]
- 4.Shire NJ, Rouster SD, Stanford SD, et al. The prevalence and significance of occult hepatitis B virus in a prospective cohort of HIV-infected patients. J Acquir Immune Defic Syndr. 2007;44(3):309–314. doi: 10.1097/QAI.0b013e31802e29a9. [DOI] [PubMed] [Google Scholar]
- 5.Baginski I, Chemin I, Hantz O, et al. Transmission of serologically silent hepatitis B virus along with hepatitis C virus in two cases of posttransfusion hepatitis. Transfusion. 1992;32(3):215–220. doi: 10.1046/j.1537-2995.1992.32392213803.x. [DOI] [PubMed] [Google Scholar]
- 6.Thiers V, Nakajima E, Kremsdorf D, et al. Transmission of hepatitis B from hepatitis-B-seronegative subjects. Lancet. 1988;2(8623):1273–1276. doi: 10.1016/s0140-6736(88)92891-7. [DOI] [PubMed] [Google Scholar]
- 7.Campe H, Hillebrand GF, Mairhofer H, Nitschko H, Jager G. Undetected chronic hepatitis B virus infection of a vaccinated dialysis patient after liver transplantation. Nephrol Dial Transplant. 2005;20(7):1492–1494. doi: 10.1093/ndt/gfh823. [DOI] [PubMed] [Google Scholar]
- 8.Kannangai R, Molmenti E, Arrazola L, et al. Occult hepatitis B viral DNA in liver carcinomas from a region with a low prevalence of chronic hepatitis B infection. J Viral Hepat. 2004;11(4):297–301. doi: 10.1111/j.1365-2893.2004.00502.x. [DOI] [PubMed] [Google Scholar]
- 9.Tamori A, Nishiguchi S, Kubo S, et al. Sequencing of human-viral DNA junctions in hepatocellular carcinoma from patients with HCV and occult HBV infection. J Med Virol. 2003;69(4):475–481. doi: 10.1002/jmv.10334. [DOI] [PubMed] [Google Scholar]
- 10.Squadrito G, Pollicino T, Cacciola I, et al. Occult hepatitis B virus infection is associated with the development of hepatocellular carcinoma in chronic hepatitis C patients. Cancer. 2006;106(6):1326–1330. doi: 10.1002/cncr.21702. [DOI] [PubMed] [Google Scholar]
- 11.Thio CL, Seaberg EC, Skolasky JR, et al. HIV-1, hepatitis B virus, and risk of liver-related mortality in the Multicenter Cohort Study (MACS) Lancet. 2002;360(9349):1921–1926. doi: 10.1016/s0140-6736(02)11913-1. [DOI] [PubMed] [Google Scholar]
- 12.Nunez M, Rios P, Perez-Olmeda M, Soriano V. Lack of `occult' hepatitis B virus infection in HIV-infected patients. AIDS. 2002;16(15):2099–2101. doi: 10.1097/00002030-200210180-00024. [DOI] [PubMed] [Google Scholar]
- 13.Hofer M, Joller-Jemelka HI, Grob PJ, Luthy R, Opravil M. Frequent chronic hepatitis B virus infection in HIV-infected patients positive for antibody to hepatitis B core antigen only. Swiss HIV Cohort Study. Eur J Clin Microbiol Infect Dis. 1998;17(1):6–13. doi: 10.1007/BF01584356. [DOI] [PubMed] [Google Scholar]
- 14.Torbenson M, Kannangai R, Astemborski J, Strathdee SA, Vlahov D, Thomas DL. High prevalence of occult hepatitis B in Baltimore injection drug users. Hepatology. 2004;39(1):51–57. doi: 10.1002/hep.20025. [DOI] [PubMed] [Google Scholar]
- 15.Raimondo G, Navarra G, Mondello S, et al. Occult hepatitis B virus in liver tissue of individuals without hepatic disease. J Hepatol. 2008;48(5):743–746. doi: 10.1016/j.jhep.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 16.Hu K-Q. Occult hepatitis B virus infection and its clinical implications. J Viral Hepat. 2002;9(4):243–257. doi: 10.1046/j.1365-2893.2002.00344.x. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhuri V, Tayal R, Nayak B, Acharya SK, Panda SK. Occult hepatitis B virus infection in chronic liver disease: full-length genome and analysis of mutant surface promoter. Gastroenterology. 2004;127(5):1356–1371. doi: 10.1053/j.gastro.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Datta S, Banerjee A, Chandra PK, Chowdhury A, Chakravarty R. Genotype, phylogenetic analysis, and transmission pattern of occult hepatitis B virus (HBV) infection in families of asymptomatic HBsAg carriers. J Med Virol. 2006;78(1):53–59. doi: 10.1002/jmv.20503. [DOI] [PubMed] [Google Scholar]
- 19.Hass M, Hannoun C, Kalinina T, Sommer G, Manegold C, Günther S. Functional analysis of hepatitis B virus reactivating in hepatitis B surface antigen-negative individuals. Hepatology. 2005;42(1):93–103. doi: 10.1002/hep.20748. [DOI] [PubMed] [Google Scholar]
- 20.Jeantet D, Chemin I, Mandrand B, Zoulim F, Trepo C, Kay A. Characterization of two hepatitis B virus populations isolated from a hepatitis B surface antigen-negative patient. Hepatology. 2002;35(5):1215–1224. doi: 10.1053/jhep.2002.32710. [DOI] [PubMed] [Google Scholar]
- 21.Kao J-H, Chen P-J, Lai M-Y, Chen D-S. Sequence analysis of pre-S/surface and pre-core/core promoter genes of hepatitis B virus in chronic hepatitis C patients with occult HBV infection. J Med Virol. 2002;68(2):216–220. doi: 10.1002/jmv.10188. [DOI] [PubMed] [Google Scholar]
- 22.Schories M, Peters T, Rasenack J. Isolation, characterization and biological significance of hepatitis B virus mutants from serum of a patient with immunologically negative HBV infection. J Hepatol. 2000;33(5):799–811. doi: 10.1016/s0168-8278(00)80313-x. [DOI] [PubMed] [Google Scholar]
- 23.Wagner AA, Denis F, Weinbreck P, et al. Serological pattern `anti-hepatitis B core alone' in HIV or hepatitis C virus-infected patients is not fully explained by hepatitis B surface antigen mutants. AIDS. 2004;18(3):569–571. doi: 10.1097/00002030-200402200-00028. [DOI] [PubMed] [Google Scholar]
- 24.Weinberger KM, Bauer T, Bohm S, Jilg W. High genetic variability of the group-specific a-determinant of hepatitis B virus surface antigen (HBsAg) and the corresponding fragment of the viral polymerase in chronic virus carriers lacking detectable HBsAg in serum. J Gen Virol. 2000;81(5):1165–1174. doi: 10.1099/0022-1317-81-5-1165. [DOI] [PubMed] [Google Scholar]
- 25.Shire NJ, Rouster SD, Rajicic N, Sherman KE. Occult hepatitis B in HIV-infected patients. J Acquir Immune Defic Syndr. 2004;36(3):869–875. doi: 10.1097/00126334-200407010-00015. [DOI] [PubMed] [Google Scholar]
- 26.Gerken G, Kremsdorf D, Capel F, et al. Hepatitis B defective virus with rearrangements in the PreS gene during chronic HBV infection. Virol. 1991;183(2):555–565. doi: 10.1016/0042-6822(91)90984-j. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kidd-Ljunggren K, Miyakawa Y, Kidd AH. Genetic variability in hepatitis B viruses. J Gen Virol. 2002;83(6):1267–1280. doi: 10.1099/0022-1317-83-6-1267. [DOI] [PubMed] [Google Scholar]
- 29.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 30.Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 31.Fung SK, Lok ASF. Hepatitis B virus genotypes: do they play a role in the outcome of HBV infection? Hepatology. 2004;40(4):790–792. doi: 10.1002/hep.1840400407. [DOI] [PubMed] [Google Scholar]
- 32.Stuyver LJ, Locarnini SA, Lok A, et al. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology. 2001;33(3):751–757. doi: 10.1053/jhep.2001.22166. [DOI] [PubMed] [Google Scholar]
- 33.Korber B, Meyers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8(9):1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 34.Bock CT, Tillmann HL, Torresi J, et al. Selection of hepatitis B virus polymerase mutants with enhanced replication by lamivudine treatment after liver transplantation. Gastroenterology. 2002;122(2):264–273. doi: 10.1053/gast.2002.31015. [DOI] [PubMed] [Google Scholar]
- 35.Chu C-J, Keeffe EB, Han S-H, et al. Hepatitis B virus genotypes in the United States: results of a nationwide study. Gastroenterology. 2003;125(2):444–451. doi: 10.1016/s0016-5085(03)00895-3. [DOI] [PubMed] [Google Scholar]
- 36.Moriya T, Kuramoto IK, Yoshizawa H, Holland PV. Distribution of hepatitis B virus genotypes among American blood donors determined with a PreS2 epitope enzyme-linked immunosorbent assay kit. J Clin Microbiol. 2002;40(3):877–880. doi: 10.1128/JCM.40.3.877-880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuyver L, De Gendt S, Van Geyt C, et al. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81(1):67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 38.Chudy M, Schmidt M, Czudai V, et al. Hepatitis B virus genotype G monoinfection and its transmission by blood components. Hepatology. 2006;44(1):99–107. doi: 10.1002/hep.21220. [DOI] [PubMed] [Google Scholar]
- 39.Kato H, Orito E, Gish RG, et al. Hepatitis B e antigen in sera from individuals infected with hepatitis B virus of genotype G. Hepatology. 2002;35(4):922–929. doi: 10.1053/jhep.2002.32096. [DOI] [PubMed] [Google Scholar]
- 40.Amini-Bavil-Olyaee S, Alavian S-M, Adeli A, et al. Hepatitis B virus genotyping, core promoter, and precore/core mutations among Afghan patients infected with hepatitis B: a preliminary report. J Med Virol. 2006;78(3):358–364. doi: 10.1002/jmv.20547. [DOI] [PubMed] [Google Scholar]
- 41.Osiowy C, Giles E, Tanaka Y, Mizokami M, Minuk GY. Molecular evolution of hepatitis B virus over 25 years. J Virol. 2006;80(21):10307–10314. doi: 10.1128/JVI.00996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres-Puente M, Bracho MA, Jimenez N, Garcia-Robles I, Moya A, Gonzalez-Candelas F. Sampling and repeatability in the evaluation of hepatitis C virus genetic variability. J Gen Virol. 2003;84(9):2343–2350. doi: 10.1099/vir.0.19273-0. [DOI] [PubMed] [Google Scholar]
- 43.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64(2):613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeantet D, Chemin I, Mandrand B, et al. Cloning and expression of surface antigens from occult chronic hepatitis B virus infections and their recognition by commercial detection assays. J Med Virol. 2004;73(4):508–515. doi: 10.1002/jmv.20119. [DOI] [PubMed] [Google Scholar]
- 45.Moerman B, Moons V, Sommer H, Schmitt Y, Stetter M. Evaluation of sensitivity for wild type and mutant forms of hepatitis B surface antigen by four commercial HBsAg assays. Clin Lab. 2004;4:159–162. [PubMed] [Google Scholar]
- 46.Lin X, Yuan Z-H, Wu L, Ding J-P, Wen Y-M. A single amino acid in the reverse transcriptase domain of hepatitis B virus affects virus replication efficiency. J Virol. 2001;75(23):11827–11833. doi: 10.1128/JVI.75.23.11827-11833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruss V, Ganem D. Mutational analysis of hepatitis B surface antigen particle assembly and secretion. J Virol. 1991;65(7):3813–3820. doi: 10.1128/jvi.65.7.3813-3820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Z, Jensen G, Yen TS. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J Virol. 1997;71(10):7387–7392. doi: 10.1128/jvi.71.10.7387-7392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.