Abstract
Accumulating evidence from epidemiological studies indicates that chronic inflammation and oxidative stress play critical roles in neoplastic development. The aim of this study was to investigate the anti-inflammatory, anti-oxidative stress activities, and differential regulation of Nrf2-mediated genes by tea Chrysanthemum zawadskii (CZ) and licorice Glycyrrhiza uralensis (LE) extracts. The anti-inflammatory and anti-oxidative stress activities of hexane/ethanol extracts of CZ and LE were investigated using in vitro and in vivo approaches, including quantitative real-time PCR (qPCR) and microarray. Additionally, the role of the transcriptional factor Nrf2 (nuclear erythroid-related factor 2) signaling pathways was examined. Our results show that CZ and LE extracts exhibited potent anti-inflammatory activities by suppressing the mRNA and protein expression levels of pro-inflammatory biomarkers IL-1β, IL-6, COX-2 and iNOS in LPS-stimulated murine RAW 264.7 macrophage cells. CZ and LE also significantly suppressed the NO production of LPS-stimulated RAW 264.7 cells. Additionally, CZ and LE suppressed the NF-κB luciferase activity in human HT-29 colon cancer cells. Both extracts also showed strong Nrf2-mediated antioxidant/Phase II detoxifying enzymes induction. CZ and LE induced NQO1, Nrf2, and UGT and antioxidant response element (ARE)-luciferase activity in human hepatoma HepG2 C8 cells. Using Nrf2 knockout [Nrf2 (−/−)] and Nrf2 wild-type (+/+) mice, LE and CZ showed Nrf2-dependent transactivation of Nrf2-mediated antioxidant and phase II detoxifying genes. In summary, CZ and LE possess strong inhibitory effects against NF-κB-mediated inflammatory as well as strong activation of the Nrf2-ARE-anti-oxidative stress signaling pathways, which would contribute to their overall health promoting pharmacological effects against diseases including cancer.
Key words: anti-inflammatory, anti-oxidative stress, chrysanthemum, licorice, Nrf2, phase II drug metabolizing/detoxifying enzymes
INTRODUCTION
Normal inflammation in general is a process involving interactions between pro-inflammatory and anti-inflammatory signaling pathways. Inflammation occurs when the tissue is injured or infected by external challenges (1). In this context, generally acute inflammation is self-limiting and recovered by itself (1). However, chronic inflammation could increase the risk of developing diseases such as cancer in the inflamed tissues (2).
In Eastern Asia, the use of plants including roots and fruits as herbal medicines is common. Licorice and tea chrysanthemum are two popular herbal medicines used to treat various inflammatory diseases. Licorice (Glycyrrhiza) species have been used in Europe as herbal medicines for centuries as well. Licorice root is used for the treatment of gastric or duodenal ulcers, hepatitis, sore throats, coughs, bronchitis, arthritis, allergies, and cardiovascular disease (1,3). There are numerous dietary supplements stomach ulcers, bronchitis, and sore throat, as well as infections caused by viruses, such as hepatitis but have not been approved by the Food and Drug Administration (FDA; National Center for Complementary and Alternative Medicines (NCCAM; http://nccam.nih.gov/health/licoriceroot/)). Licorice, including glycyrrhizin, isoliquiritigenin, liquiritigenin, licochalcone A and B, and β-glycyhrritinic acid (4–6), have been shown to possess anti-inflammatory activities and glycyrrhizin can inhibit reactive oxygen species (ROS). Licorice has also been shown to inhibit the expression of COX-2 and other pro-inflammatory proteins (3,7,8). There are currently (as of 09/29/2010) 12 on-going, completed or terminated clinical trials on “licorice or licorice-related” dietary supplement listed on Clinicaltrial.gov (http://clinicaltrials.gov). Chrysanthemum which includes five germacrane-type sesquiterpenes, kikkanols D, D monoacetate, E, F, and F monoacetate, were isolated from the ethyl acetate-soluble portion and two flavanone glycosides, (2S)- and (2R)-eriodictyol 7-O-beta-d-glucopyranosiduronic acids, and a phenylbutanoid glycoside, (2S, 3S)-1-phenyl-2,3-butanediol 3-O-beta-d-glucopyranoside, isolated from the flowers of Chrysanthemum, has been used to treat vertigo, hypertension, bacterial and viral infectious diseases (9–12). Botanical chrysanthemum tea or extract are available as dietary supplements and promoted as health enhancing products. Extract of chrysanthemum has been shown to possess strong anti-oxidative stress, anti-inflammatory effects and previous studies revealed that different extraction methods yielding different soluble fraction of extracts would possess different effects of anti-inflammation and immunomodulation (9,13). A completed clinical trial of chrysanthemum extract as dietary supplement to lower serum LDL cholesterol and raising HDL cholesterol is listed on Clinicaltrial.gov (http://clinicaltrials.gov).
Despite these diverse potential health beneficial and pharmacological effects of licorice and chrysanthemum, the molecular and the signaling mechanisms leading to these biological effects are still unclear. We theorized that the non-polar fractions of licorice and chrysanthemum, may possess anti-inflammatory and anti-oxidative stress properties that could be further developed for diseases prevention including cancer chemoprevention. In this study, we aim to investigate the transcription regulation of LE and CZ on the transcriptional factor nuclear erythroid-related factor 2 (Nrf2) signaling pathway that controls the expression of many anti-oxidative stress and phase II drug metabolizing (DM)/detoxifying enzymes, which are typically elicited by many chemopreventive compounds (14–16).
Nrf2 plays an important role to mediate phase II detoxifying/antioxidant enzymes expression. Under normal conditions, Nrf2 appears to be associated with actin-binding Keap 1 that forms Nrf2-Keap1 complex preventing Nrf2 from entering into the nuclear and promoting its proteasomal degradation. Typically, the half-life of Nrf2 in un-stimulated mammalian cells is 15–45 min. Upon treatments of the cells with oxidants such as H2O2, oxidative stress or cancer chemopreventive compounds, conformational changes occur due to oxidation of thiol-sensitive amino acids present in the Nrf2-Keap 1 complex and would drive the dissociation of Nrf2 from Keap 1, thereby allowing the translocation of Nrf2 into the nucleus, Nrf2 binds to the antioxidant response element (ARE) of ARE-target genes and leads to enhanced phase II detoxifying/antioxidant enzymes expression (17,18). Therefore, in the current study, we utilized the Nrf2-deficient mice (Nrf2−/−; KO) and Nrf2 wild-type (Nrf2+/+; WT) mice to examine whether the in vivo phase II DM/detoxifying/anti-oxidative/properties elicited by the extracts would be mediated by Nrf2.
MATERIALS AND METHODS
Plant Extracts
Whole plants of Chrysanthemum zawadskii (CZ) Herbich var. latilobum (Maxim.) Kitamura and licorice roots, derived from Glycyrrhiza uralensis (LE) Fisch. were purchased from a local drug store (Dea Guang Medical, Chunchon, South Korea) and identified by Emeritus Professor Hyung Jun Ji (Seoul National University, Seoul, Korea). Dried and ground C. zawadskii (5 kg) (CZ) and roots of G. uralensis (5 kg) (LE) were dip-extracted with hexane:ethanol (70 L) at a ratio 9:1 (v/v) at room temperature for 24 h. The slurry was then filtered through filter paper and the residue was re-extracted twice. The combined extracts were filtered, and the filtrates were concentrated under reduced pressure at 40°C to yield the hexane/ethanol extract of CZ (412 g, 0.82% yield) and LE (455 g, 0.91% yield).
Cell Culture and Treatment
The murine RAW 264.7 macrophage cells, a well-established model system for many inflammatory studies as well as the luciferase reporter assay of nuclear factor kappa-light chain enhancer of B cells (NFκB) stabilized in human colon cancer cells HT-29 (HT-29-N9) were used to investigate the anti-inflammatory effects of licorice and chrysanthemum extracts (19). Similarly, the Nrf2-mediated ARE luciferase assay stabilized in human hepatoma HepG2 cell (HepG2-C8) was used to investigate the potential of the extracts in activating the Nrf2/ARE signaling pathway (20). Mouse macrophage cell line RAW 246.7, was obtained from American Type Culture Collection. HepG2-C8 and HT-29-N9 cells were generated in our laboratory as described previously (21–24). All cells were cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen Corp., Carlsbad, CA, USA) supplemented with 10% (V/V) FBS (Lifeblood Medical, Inc), penicillin 100 U/ml, and streptomycin 100 μg/ml. Cells were maintained in a humidified incubator with 5% CO2 at 37°C. The cells were treated with LPS (1 μg/ml, Sigma, St. Louis, MO, USA) alone or pretreated 1 h with LE 25 μg/ml, CZ 25 μg/ml, or curcumin (CUR) 10 μg/ml (as positive control) dissolved in DMSO before they were challenged with LPS (19,23)
Nitrite Assay
The culture medium of the cells treated with different compounds was mixed with a Griess reagent in an equal volume of 0.1% (1 mg/ml) N-(1-naphthyl)ethylenediamine dihydrochloride in deionized water and 1.0% (10 mg/ml) sulfanilic acid in 5% phosphoric acid solution. The mixed sample was incubated at 37°C for 30 min. Absorbance at 548 nm was measured and concentrations were calculated using a sodium nitrite standard curve.
RNA Isolation and Reverse Transcription Polymerase Chain Reaction Analysis
The RAW 246.7 cells were cultured in six-well plates and were challenged by LPS 1 μg/ml with or without pretreatment with LE, CZ, or CUR for 8 h at the 37°C incubator. Total RNA was isolated by TRIZOL® according to the manufacturer’s protocol (Invitrogen Corp., Carlsbad, CA, USA). First-strand cDNA was synthesized from 5 μg of total RNA using SuperScript III First-strand Reverse Transcriptase (Invitrogen Corp. Carlsbad, CA, USA) and oligo dT primers according to the manufacturer’s instructions. After reverse transcription, the polymerase chain reaction (PCR) reactions were performed by using 1 μl of reverse transcription product, 1 μl of primer mixture (final concentration, 10 μmol/L), and 8 μl of Platinum® Taq DNA Polymerase kit (Invitrogen Corp. Carlsbad, CA, USA), and performed with initial denaturation at 94°C for 2 min, 25 cycles of amplification, and extension at 72°C for 10 min. PCR products were fractionated on 1.5% agarose gel. The primers used in this experiment are shown in Table I.
Table I.
Murine Primers for PCR
| Gene | Forward | Reverse |
|---|---|---|
| GAPDH | 5′-TGC TCG AGA TGT CAT GAA GG-3′ | 5′-TTG CGC TCA TCG TAG GCT TT-3′ |
| IL-1β | 5′-GAG TGT GGA TCC CAA GCA AT-3′ | 5′-CTC AGT GCA GGC TAT GAC CA-3′ |
| IL-6 | 5′-AGT TGC CTT CTT GGG ACT GA-3′ | 5′-GCC ACT CCT TCT GTG ACT CC-3′ |
| TNF-α | 5′-ACG GCA TGG ATC TCA AAG AC-3′ | 5′-GGT CAC TGT CCC AGC TT-3′ |
| iNOS | 5′-GTG GTG ACA AGC ACA TTT GG-3′ | 5′-GGC TGG ACT TTT CAC TCT GC-3′ |
| COX-2 | 5′-TCC TCC TGG AAC ATG GAC TC-3′ | 5′-TGA TGG TGG CTG TTT TGG TA-3′ |
| Nrf-2 | 5′-AGC AGG ACA TGG AGC AAG TT-3′ | 5′-TTC TTT TTC CAG CGA GGA GA-3′ |
| UGT1A1 | 5′-GTG GCC CAG TAC CTG ACT GT-3′ | 5′-CGA TGG TCT AGT TCC GGT GT-3′ |
| NQO-1 | 5′-CAG ATC CTG GAA GGA TGG AA-3′ | 5′-AAG TTA GTC CCT CGG CCA TT-3′ |
Western Blotting
The RAW 246.7 cells were challenged by LPS 1 μg/ml with or without pretreatment with LE, CZ, or CUR. After 24 h, the cells were washed with ice-cold phosphate buffer saline (PBS) (pH 7.4), and scraped into microcentrifuge tubes and pelleted. Cells were resuspended and lysed in RIPA buffer (Sigma, St. Louis, MO). 20 μg protein per lane was loaded onto 4–15% SDS-PAGE (Bio-Rad Laboratories, Hercules, CA). After separation by SDS-PAGE, the protein was transferred onto nitrocellulose membrane (Millipore Corp., Billerica, MA, USA), and then was blocked in 5% bovine serum albumin (BSA; Fisher Scientific, Fair Law, NJ, USA) in tris–buffer saline tween-20 (TBST) solution for 1 h. Membranes were probed by respective antibodies including β-actin, COX2, cPLA2, and iNOS (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C. Blots were washed with TBST solution 15 min for four times and incubated with respective secondary antibodies for 1 h. After washing 15 min for four times with TBST solution, the immunoreactive bands were determined by adding SuperSignal West Femto mix (1:1 mix of stable peroxide buffer and luminol/enhancer solution, Thermo Scientific, Rockford, IL) to detect immunoreactive bands which were then visualized and quantified by Bio-Rad ChemiDoc XRS system (Hercules, CA).
Enzyme-Linked Immunosorbent Assay
The RAW 264.7 cells were cultured in 96-well plate with 200 μl medium. IL-6 and IL-1β enzyme-linked immunosorbent assay (ELISA) assay kits were purchased from Invitrogen Corporation, Carlsbad, CA, USA The assays were performed according to the manufacturer’s instructions. For the ELISA assay, 50 μl of incubation buffer was first added to all the wells. After adding incubation buffer, 50 μl standard diluent buffer and 50 μl of standards, controls, or samples were added to each well in a stepwise fashion.
Luciferase Reporter Assay
The NF-κB- and ARE luciferase activities were measured using a luciferase reporter assay system according to the manufacturer’s instructions (Promega, Madison, WI, USA). Briefly, after treatments, the cells were washed with ice-cold PBS and harvested in reporter lysis buffer. After centrifugation, 10 μl of the supernatants were mixed with 50 μl of luciferase assay substrate and measured for luciferase activity by using a Sirius Luminometer (Berthold Detection Systems GmbH D-75173 Pforzheim, Germany). The luciferase activity was normalized against known protein concentrations and expressed as fold induction of luciferase activity over the control cells, which were treated with 0.1% DMSO. The protein level was determined by Bio-Rad protein assay according to the manufacturer’s instructions as we have described previously (22,25).
Quantitative Real-Time PCR Assays
The HepG2-C8 cells were cultured in six-well plates and were treated with respective extracts for 8 h at 37°C and the total RNA collected respectively. The primers for qPCR are listed in Table II (Integrated DNA Technologies, Coralville, IA, USA). The qPCR reactions were carried out using 1 μl cDNA product, 50 nM of each primer, and Power SYBR Green master mix (Applied Biosystems, Foster City, CA, USA) in 10 μl reactions. The reactions were performed using an ABI Prism 7900HT sequence detection system amplified specificity was verified by first-derivative melting curve analysis using the ABI software (SDS2.3, Applied Biosystems, Foster City, CA, USA). Relative quantification of each gene expression profile was calculated using a ΔΔCt method and presented as relative quantitative value (RQ value) = 2−ΔΔCt (RQ manager, Applied Biosystems, Foster City, CA, USA) (26,27).
Table II.
Human Primers for Quantitative Real-Time PCR
| Gene | Forward | Reverse |
|---|---|---|
| GAPDH | 5′-TCG ACA GTC AGC CGC ATC TTC TTT-3′ | 5′-ACC AAA TCC GTT GAC TCC GAC CTT-3′ |
| UGT1A1 | 5′-TAA GTG GCT ACC CCA AAA CG-3′ | 5′-TCT TGG ATT TGT GGG CTT TC-3′ |
| NQO-1 | 5′-CTG GAG TGT GCC CAA TGC TA-3′ | 5′-CAT GAA TGT CAT TCT CTG GCC A-3′ |
| Nrf-2 | 5′-TGC TTT ATA GCG TGC AAA CCT CGC-3′ | 5′-ATC CAT GTC CCT TGA CAG CAC AGA-3′ |
Animal and In Vivo Study
The second generation (F2) Nrf2 (−/−) mice (C57BL/SV129) and the C57BL/6J wild-type mice (The Jackson Laboratory, Bar Harbor, ME) were used for the in vivo study to investigate if the induction of phase II detoxifying/antioxidizing enzymes by the extracts was Nrf2-dependent (28). Five animals were used in each group of Nrf2 (−/−) mice (Nrf2 KO) and wild-type mice (Nrf2 WT). The mice were treated with vehicle (as a negative control; cremophor: tween 80: ethyl alcohol: deionized water = 2:1:1:6), LE 150 mg/kg, LE 300 mg/kg, CZ 150 mg/kg, and CZ 300 mg/kg by oral gavage in a final volume of 100–110 μl (13). After 12 h, livers were collected for the RNA extraction, and total RNA were used for PCR, qPCR, and microarray analyses. Housing and care of the animals were in accordance with the guidelines established by the University’s Animal Research Committee consistent with the NIH Guidelines for the Care and Use of Laboratory Animals (Table III).
Table III.
Confirmation of Genotype of the Animals
| Gene | Primers |
|---|---|
| 3′-primer | 5′-GGA ATG GAA AAT AGC TCC TGC C-3′ |
| 5′-primer | 5′-GCC TGA GAG CTG TAG GCC C-3′ |
| lacZ primer | 5′-GGG TTT TCC CAG TCA CGA C-3′ |
Microarray Gene Expression Analysis
Affymetrix MOE_430 microarrays (containing 45,101 probes) were used to probe the global gene expression profile of pooled RNA from Nrf2 WT or KO mice after oral administration of CZ and LE of 150 mg/kg. These microarrays were conducted as published previously (27). The “.CEL” files containing intensity values were created from the scanned image by using Microarray Suite 5 (Affymetrix). The .CEL files and the .CDF file (information on the location and identity of different probe cells) were then analyzed using the dChip analysis software to identify genes that were differentially expressed in Nrf2-associated pathways in the liver samples of both treated and untreated controls (29–31). Normalization against the median using default settings in dChip (median chip) was used and the expression values of each probe of all arrays (median intensities range 110–280) were calculated using default dChip model-based algorithms with perfect match only for fluorescence intensities. A transformed normalized data was generated and saved in an Excel file, which was later being imported into the Ingenuity Pathway Analysis Program (www.ingenuity.com, IPA 8.0) for further data characterization (29,32). Over 2,000 highly differentially expressed genes were filtered using a cut-off intensity at 1,750 and a subset of Nrf2-mediated oxidative stress response genes was performed using the canonical pathway analysis function. Comparative analysis was performed between treated groups for CZ and LE with their respective counterparts’ control.
Statistical Analyses
Values were presented as means ± standard error of the mean (SEM). Statistical analysis of the data was performed by Student’s t test. p values lower than 0.05 were considered significant. Nonparametric statistical Mann–Whitney U test of the qPCR results for the in vivo animal study was performed using SPSS software (version 17, USA) (33,34).
Results
Inhibition of mRNA and Protein Levels of Pro-Inflammatory Markers by LE and CZ Extracts
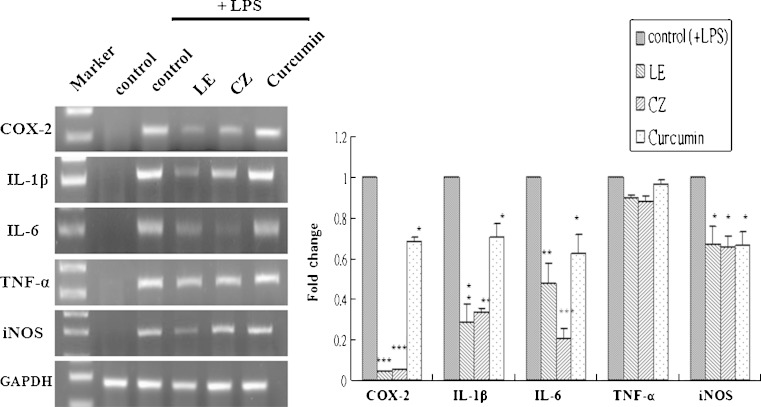
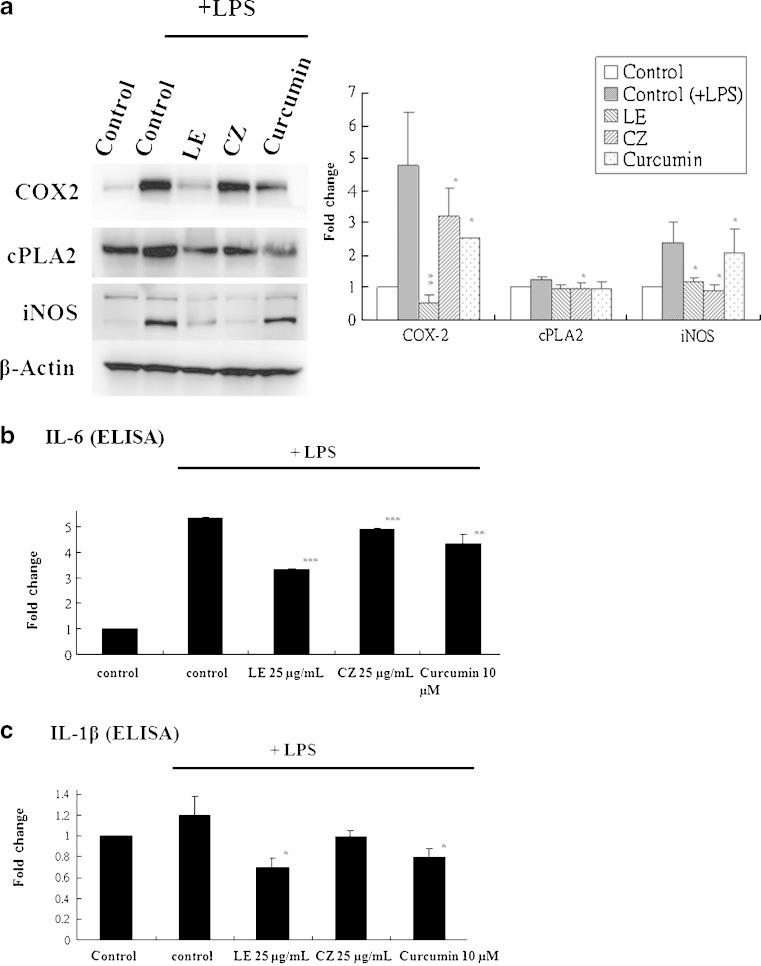
To demonstrate the anti-inflammatory effects of LE and CZ extracts, the mRNA expression levels of pro-inflammatory makers, COX-2, IL-1β, IL-6, and iNOS in LPS-stimulated RAW 246.7 cells were found to be strongly suppressed by LE and CZ (Fig. 1.) Similarly, the LE and CZ extracts also demonstrated strong inhibitory effect on the protein expression levels of COX-2, cPLA2 and iNOS (Fig. 2a).
Fig. 1.
Effect of LE and CZ extracts on the mRNA expression of pro-inflammatory biomarkers (mRNA) in LPS-stimulated RAW 246.7 cells. LE 25 μg/ml; CZ 25 μg/ml; Curcumin 10 μM. The gene bands were quantified by densitometry and normalized by GAPDH, ratio (*** compare with LPS-induced, p < 0.001; ** compare with LPS-induced, p < 0.01; * compare with LPS-induced, p < 0.05)
Fig. 2.
Effect of LE and CZ extracts on the protein expression of pro-inflammatory biomarkers (protein) in LPS-stimulated RAW 246.7 cells. LE 25 μg/ml; CZ 25 μg/ml; Curcumin 10 μM. a Pro-inflammatory proteins analyzed performed by western blotting, the protein bands were quantified by densitometry and normalized by β-actin, ratio. Three independent experiments were done and the data were represented as mean ± SEM. b Expression of IL-6 performed by ELISA (n = 3). c Expression of IL-1β performed by ELISA (*** compare with LPS-induced, p < 0.001; ** compare with LPS-induced, p < 0.01; * compare with LPS-induced, p < 0.05)
LE and CZ Extracts Inhibit IL-6 and IL-1β
To investigate the suppression effect of pro-inflammatory proteins levels by LE and CZ, the expression of IL-1β and IL-6 was analyzed by using enzyme-linked immunosorbent assay. LE significantly inhibited the expression level of LPS-induced IL-1β (p < 0.05) and IL-6 (p < 0.001), whereas, CZ significantly inhibited only the expression level of LPS-induced IL-6 (p < 0.001; Fig. 2b and c).
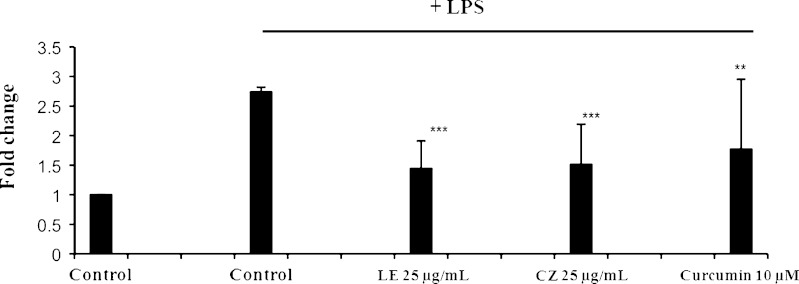
Inhibitory Effect of LE and CZ Extracts on LPS-Induced Nitrate Oxide Production
Nitric Oxide (NO) is a molecular mediator of many physiological processes, including vasodilation, inflammation, thrombosis, immunity and neurotransmission (35). The inhibitory effect of extracts on the production of nitric oxide, which could be due to inflammatory reaction, was determined by the Griess reaction to measure the level of nitrite, an indicator of NO synthesis. The extracts, CZ (25 μg/ml) and LE (25 μg/ml), significantly inhibited LPS-induced NO production (p < 0.001) by 44% and 48%, respectively (Fig. 3).
Fig. 3.
Effect of LE and CZ extracts on production of NO in LPS-stimulated RAW 246.7 cells (*** compare with LPS-induced, p < 0.001; ** compare with LPS-induced, p < 0.01)
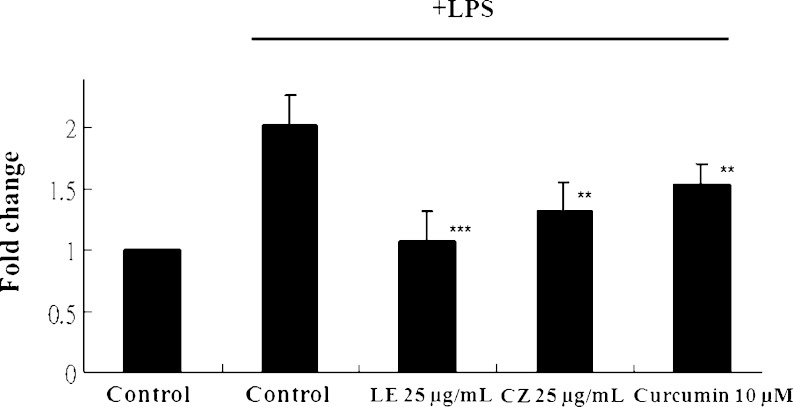
Effect of LE and CZ Extracts on NF-κB Luciferase Activity in HT-29-N9 Cells
NF-κB pathway can trigger the expression of pro-inflammatory proteins. To investigate the effect of inhibition of NF-κB by LE and CZ, HT-29-N9 cells were stimulated by LPS with or without pretreatment of LE and CZ. Both LE and CZ significantly suppressed the LPS-induced NF-κB luciferase activity (p < 0.05). LE had a slightly stronger inhibitory effect on NF-κB luciferase activity inhibition in comparison to CZ (p < 0.001; Fig. 4).
Fig. 4.
Effect of LE and CZ extracts on NF-κB luciferase activity in HT-29 cells (*** compare with LPS-induced, p < 0.001; **compare with LPS-induced, p < 0.01)
Effect of LE and CZ Extracts on ARE Luciferase Activity and Expression of Nrf2 and its Trans-Activated Target Genes in HepG2 C8 Cells
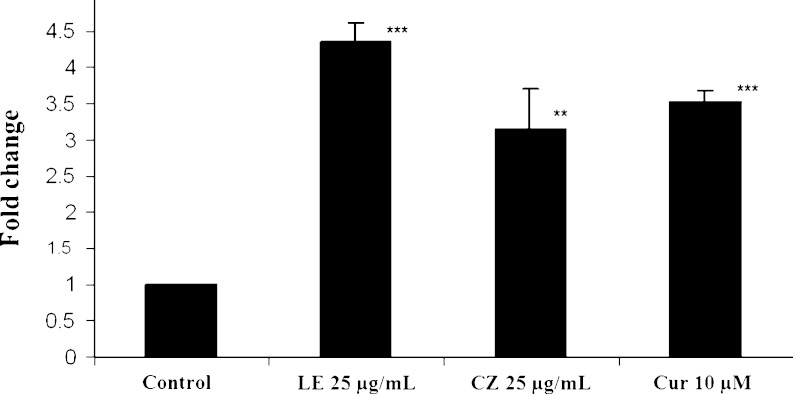
To investigate whether LE and CZ extracts could induce the Nrf2-mediated ARE luciferase activity as well as the transcriptional regulation of Nrf2 target genes, ARE luciferase assay and qPCR analysis were performed. LE and CZ extracts significantly induced the ARE luciferase activity in HepG2C8 cells, by four- (p < 0.001) and threefolds (p < 0.01), respectively, as compared to the control group (Fig. 5). Interestingly, LE and CZ significantly induced the endogenous Nrf2-target gene, NAD(P)H dehydrogenase (quinone) 1 (NQO-1), by 3.57 and 2.92 folds, respectively. The expression of phase II UDP-glucuronosyltransferase 1A1 (UGT1A1) was also induced by LE and CZ by 2.50 and 1.51 folds, respectively (Fig. 6).
Fig. 5.
Effect of LE and CZ extracts on ARE luciferase activity in HepG2 C8 cells (*** compare with control, p < 0.001; ** compare with control, p < 0.01)
Fig. 6.
Induction of effects of LE and CZ extracts on the mRNA expression of Nrf2 and Phase II genes in HepG2 C8 cell. LE 25 μg/ml; CZ 25 μg/ml; Curcumin 10 μM. Quantitative real-time PCR results were normalized by β-actin, ratios. Normalized RQ mRNA expression values for Nrf2, NQO1, and UGT1A1 were shown as bar charts (*** compare with control, p < 0.001; ** compare with control, p < 0.01; * compare with control, p < 0.05)
Effect of LE and CZ Extracts on the Expression of Nrf2 and its Trans-Activated Target Genes in the Liver of Nrf2 WT C57BL/6J Mice and Nrf2 Deficient (KO) Mice
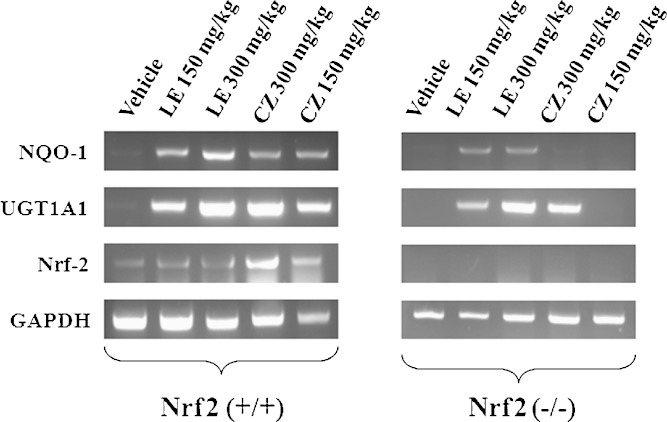
To investigate if the phase II detoxifying/antioxidant genes activation by the extracts was mediated through Nrf2 signaling pathway, the expression level of these genes in the liver of Nrf2 KO and WT mice treated with the extracts were measured using qPCR. Figure 7 shows that NQO-1 and UGT1A1 mRNA were substantially induced in the Nrf2 WT mice as compared to the Nrf2 KO. While LE extracts could induce the expression of NQO-1 and UGT1A1 genes in both Nrf2 KO and WT mice, the induction of NQO-1 by CZ extracts was only observed in the Nrf2 WT mice. In contrast, the induction of UGT1A1 gene in Nrf2 KO mice by CZ was only observed at the higher dose level. As expected, Nrf2 mRNA was only expressed in the Nrf2 WT mice and induced by CZ and LE, but not expressed nor induced by CZ and LE in the Nrf2 KO mice (Fig. 7).
Fig. 7.
Effect of LE and CZ extracts on the mRNA expression of Nrf2 and its trans-activated target genes such as phase II detoxifying/antioxidant genes in the liver of C57BL/6J mice (Nrf2 WT and Nrf2 KO)
Microarray Analysis of LE and CZ-Induced Nrf2-Dependent Genes in the Liver of Nrf2 WT Mice and Nrf2 KO Mice
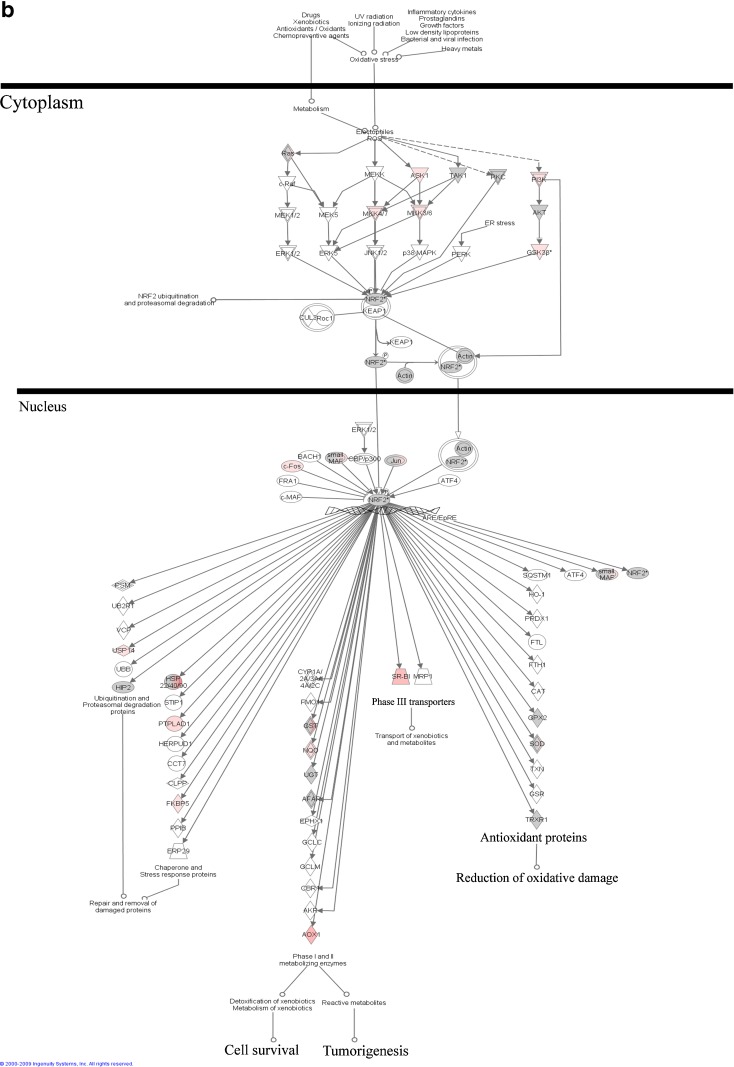
Using Affymetrix MOE_430 microarrays (containing 45,101 probes) the gene expression profiles of CZ and LE treated Nrf2 WT and KO mice were compared. In general, CZ treatment has a stronger effect on the expression of Nrf2-target genes as compared to LE (data not shown); therefore, we focused on reporting the CZ data. Genes that were induced by CZ only in the WT mice but not in the Nrf2 KO mice were considered as CZ-induced Nrf2-dependent genes. The canonical pathway of Nrf2-mediated oxidative stress response by CZ is presented in Fig. 8. Similar trends were also observed for LE treatment but to a lesser extent (data not shown). Among the Nrf2-dependent phase II detoxification and antioxidant genes that were found to be induced by CZ were aldehyde oxidase 1(AOX1), FK506 binding protein 5 (FKBP5), different isoforms of glutathione S-transferases, protein tyrosine phosphatase-like A domain containing 1 (PTPLAD1), ubiquitin specific peptidase 14 (USP14) (tRNA-guanine transglycosylase) and phase III transporter genes, solute carrier family 35 (UDP-galactose transporter), member A2 (SLC35A2), and scavenger receptor class B, member 1 (SCARB1).
Fig. 8.
a Nrf2 pathway induced by CZ treatment in Nrf2 (+/+) mice; b Nrf2 pathway induced by CZ treatment in Nrf2 (−/−) mice
Figure 9 shows that Nrf2 gene expression was induced by both LE and CZ treatments in the WT but not Nrf2 KO mice. AOX-1 was induced in WT mice to a higher level than in Nrf2 KO mice by both LE and CZ. GSTm3 and NQO-1 genes were induced in WT mice by both CZ and LE treatments.
Fig. 9.
Microarray assay for LE- and CZ-induced Nrf2-dependent genes in the liver of C57BL/6J mice and Nrf2 deficient mice. The figure shows the intensity of Nrf2, AOX1, GSTm3, and NQO1 genes expression in the six groups of mice, respectively
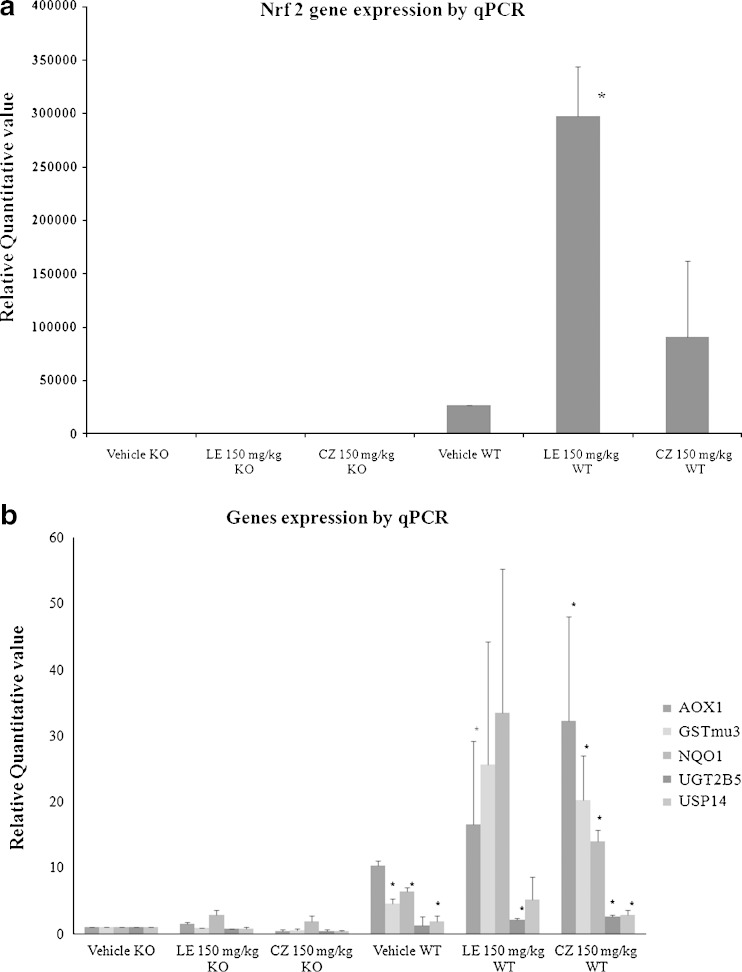
Validation of Microarray Data by Quantitative Real-Time PCR
To validate the results of the microarray, qPCR was performed to quantify the expression level of seven selected genes (Table IV). The values of each gene were normalized by the value of β-actin. Nonparametric Mann–Whitney U test was used to compare the differences between groups (Fig. 10). The qPCR data was in agreement with the trends observed in microarray such as Nrf2 (Fig. 10a), AOX1 (Fig. 10b), GSTm3 (Fig. 10b), and NQO-1 (Fig. 10b).
Table IV.
Murine Primers for Quantitative Real-Time PCR
| Gene | Forward | Reverse |
|---|---|---|
| β-actin | 5′-CGT TCA ATA CCC CAG CCA TG-3′ | 5′-GAC CCC GTC ACC AGA GTC C-3′ |
| AOX1 | 5′-TGC ACT GGT CTC TCA TGG TGG AAT-3′ | 5′-AGT CCA TTG AGA TCT GCC ACC ACA-3′ |
| FKBP5 | 5′-AGC GAG CAA CTG AGA AGA CTG GAT-3′ | 5′-ACC CGA GAA CAT CAT GGT CGA AGT-3′ |
| GSTm3 | 5′-ACT GTG GCT CCC GGT TCT CT-3′ | 5′-AAA TAA AGG CTG CAT GGG CTT-3′ |
| NQO1 | 5′-AGC CCA GAT ATT GTG GCC G-3′ | 5′-CCT TTC AGA ATG GCT GGC AC-3′ |
| Nrf2 | 5′-AGC AGG ACA TGG AGC AAG TT-3′ | 5′-TTC TTT TTC CAG CGA GGA GA-3′ |
| UGT2B5 | 5′-CAA TGG TGT CTA CGA GGC GAT-3′ | 5′-CTC CTT TGG CCA CCA TAT GG-3′ |
| USP14 | 5′-AAT GCA GAA GAC ATC TGG AGG CCA-3′ | 5′-TCC CAT ACA GCA TTT CCT GTG GGT-3′ |
Fig. 10.
Microarray verification of genes induced by LE- and CZ- induced Nrf2 dependent genes by qPCR results were normalized by β-actin, ratios. Normalized RQ values of each mouse was compared with Nrf2 KO mouse with vehicle treatment for a Nrf2, and b AOX1, FKBP5, GSTm3, NQO1, UGT2B5, and USP14 were shown, respectively. Genes were normalized to β-actin expression (* compare with Vehicle KO, p < 0.05)
DISCUSSION
Licorice and chrysanthemum have been used traditionally for many years as preventive and/or therapeutic agents for inflammatory related diseases in Eastern Asia and Europe (3,8,9). However, the precise cellular and molecular mechanisms remain unclear. Therefore, we investigated the non-polar fractions of LE and CZ on the expression of anti-inflammatory and anti-oxidative stress genes as well as Nrf2-mediated signaling pathways using in vitro (cell culture) and in vivo (Nrf2 KO and WT mice) approaches.
In agreement with previously published studies (1,7,8), our data show that the LE and CZ extracts possess strong anti-inflammatory properties (Figs. 1, 2, and 3). LE and CZ inhibited the NF-κB luciferase activity (Fig. 4), which implies that LE and CZ can attenuate the NF-κB signaling pathway. It has been reported that NF-κB pathway can trigger the expression of iNOS, COX-2, IL-6 via IKK and p38 (1,23). Therefore, the inhibition of NF-κB pathway by LE and CZ could potentially mediate the suppression effects of LE and CZ on the pro-inflammatory markers such as COX-2, IL-6, iNOS, and IL-1β (Figs. 1 and 2). In addition, our results also demonstrate that NO production was suppressed by LE and CZ (Fig. 3). The trend of NO suppression correlated well with iNOS mRNA and protein expression levels since NO is downstream of iNOS, and this result suggests that LE and CZ extracts suppressed the transcription and protein expression of iNOS.
The crosstalk between the Nrf2 and NF-κB mediated inflammatory signaling pathways is thought to be a potential mechanism (25,28). We have previously shown that the Nrf2 signaling pathway plays an important role in the down-regulation and defense of acute inflammation as well as induction of detoxifying and anti- oxidation (24,25). NF-κB which could mediate inflammatory signaling pathway is a redox-sensitive transcription factor regulated by intracellular redox status (36). Therefore, the potential of LE and CZ extracts in the induction of Nrf2 signaling pathway was investigated as we have performed previously (14,16,21,22,37–40). Our results showed that ARE was significantly induced by LE (p < 0.001) and CZ extracts (p < 0.01; Fig. 5). Our result on ARE luciferase assay were in agreement with previous findings that licorice and chrysanthemum possess strong antioxidant and free radical inhibitory effects (3,41). Furthermore, LE and CZ extracts were found to induce the mRNA transcription of Nrf2 and its downstream target genes such as NQO1 and UGT1A1 (Fig. 6). This data suggest that LE and CZ can transcriptionally activate Nrf2 and induced phase II detoxifying and antioxidant genes.
To investigate if the effect of LE and CZ is Nrf2-dependent, WT and Nrf2 KO mice were gavaged with LE and CZ. NQO-1 was not induced in the Nrf2 KO mice but in the WT mice treated with CZ. Similarly, CZ-induced UGT1A1 in the WT but not Nrf2 KO mice (Fig. 7). NQO-1 was induced more in the WT than in the Nrf2 KO mice when treated with both CZ and LE. Our data also demonstrate that the induction of Nrf2 target genes such as AOX1, GSTm3, UGT1A1, UGT2B5, and NQO-1 by LE and CZ extracts are Nrf2 dependent (Figs. 7, 8, 9, and 10b). The microarray data also revealed that these detoxifying and antioxidant genes are highly Nrf2-dependent. Although AOX1 gene was also expressed in Nrf2 KO mice in microarray, the expression intensities were lower than the Nrf2 WT mice. These genes were induced by LE and CZ extracts in the liver of WT but not Nrf2 KO mice (Fig. 8a and b). As compared to the in vitro cell culture study (Fig. 5), the in vivo data show a much higher degree of induction of Nrf2-mediated genes by CZ and LE. The reasons for these discrepancies are not clear, but could possibly be due to the presence of the active metabolites of CZ and LE in vivo (3,9), such as β-glycyhrritinic acid, licochalcone, and isoliquiritigenin from licorice or luteolin, luteolin-7-glucoside and acacetin-7-rhamnoglucoside from chrysanthemum extracts (13). Such active metabolites, together with the parent compounds, could collectively induce greater expression of the Nrf2-mediated genes especially in the WT mice in vivo.
CONCLUSION
In summary, our study shows that Nrf2 signaling pathway plays a very important role not only in the up-regulation of anti-oxidative and detoxifying enzymes but also down-regulation of inflammatory pathways. Our current findings show that the LE and chrysanthemum CZ extracts exert strong anti-inflammatory, anti-oxidative stress and detoxification properties. It is believed that chronic inflammation is related to 20% of human cancers. Thus, the anti-inflammatory and anti-oxidative stress abilities of LE and CZ can be potentially utilized for cancer chemoprevention in humans.
Acknowledgments
The authors thanks Mr. Curtis Krier at the Cancer Institute of New Jersey (CINJ) Core Expression Array Facility for his assistance with the microarray analyses. This work was supported in part by: Institutional Funds, NIH R01-CA094828 of A.N.K. and a grant (Code # 20070301034039) from Biogreen 21 Program, Rural Development Administration, Republic of Korea.
References
- 1.Kim JY, Park SJ, Yun KJ, Cho YW, Park HJ, Lee KT. Isoliquiritigenin isolated from the roots of Glycyrrhiza uralensis inhibits LPS-induced iNOS and COX-2 expression via the attenuation of NF-kappaB in RAW 264.7 macrophages. Eur J Pharmacol. 2008;584(1):175–84. doi: 10.1016/j.ejphar.2008.01.032. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. At the crossroads of inflammation and cancer. Cell. 2004;118(6):671–4. doi: 10.1016/j.cell.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Asl MN, Hosseinzadeh H. Review of pharmacological effects of Glycyrrhiza sp. and its bioactive compounds. Phytother Res. 2008;22(6):709–24. doi: 10.1002/ptr.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim SN, Kim MH, Min YK, Kim SH. Licochalcone A inhibits the formation and bone resorptive activity of osteoclasts. Cell Biol Int. 2008;32(9):1064–72. doi: 10.1016/j.cellbi.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 5.Lee CK, Son SH, Park KK, Park JH, Lim SS, Kim SH, et al. Licochalcone A inhibits the growth of colon carcinoma and attenuates cisplatin-induced toxicity without a loss of chemotherapeutic efficacy in mice. Basic Clin Pharmacol Toxicol. 2008;103(1):48–54. doi: 10.1111/j.1742-7843.2008.00238.x. [DOI] [PubMed] [Google Scholar]
- 6.Fu Y, Hsieh TC, Guo J, Kunicki J, Lee MY, Darzynkiewicz Z, et al. Licochalcone-A, a novel flavonoid isolated from licorice root (Glycyrrhiza glabra), causes G2 and late-G1 arrests in androgen-independent PC-3 prostate cancer cells. Biochem Biophys Res Commun. 2004;322(1):263–70. doi: 10.1016/j.bbrc.2004.07.094. [DOI] [PubMed] [Google Scholar]
- 7.Kim YW, Zhao RJ, Park SJ, Lee JR, Cho IJ, Yang CH, et al. Anti-inflammatory effects of liquiritigenin as a consequence of the inhibition of NF-kappaB-dependent iNOS and proinflammatory cytokines production. Br J Pharmacol. 2008;154(1):165–73. doi: 10.1038/bjp.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takei M, Kobayashi M, Herndon DN, Pollard RB, Suzuki F. Glycyrrhizin inhibits the manifestations of anti-inflammatory responses that appear in association with systemic inflammatory response syndrome (SIRS)-like reactions. Cytokine. 2006;35(5–6):295–301. doi: 10.1016/j.cyto.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shunying Z, Yang Y, Huaidong Y, Yue Y, Guolin Z. Chemical composition and antimicrobial activity of the essential oils of Chrysanthemum indicum. J Ethnopharmacol. 2005;96(1–2):151–8. doi: 10.1016/j.jep.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 10.Bor JY, Chen HY, Yen GC. Evaluation of antioxidant activity and inhibitory effect on nitric oxide production of some common vegetables. J Agric Food Chem. 2006;54(5):1680–6. doi: 10.1021/jf0527448. [DOI] [PubMed] [Google Scholar]
- 11.Matsuda H, Morikawa T, Toguchida I, Harima S, Yoshikawa M. Medicinal flowers. VI. Absolute stereostructures of two new flavanone glycosides and a phenylbutanoid glycoside from the flowers of Chrysanthemum indicum L.: their inhibitory activities for rat lens aldose reductase. Chem Pharm Bull Tokyo. 2002;50(7):972–5. doi: 10.1248/cpb.50.972. [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa M, Morikawa T, Toguchida I, Harima S, Matsuda H. Medicinal flowers. II. Inhibitors of nitric oxide production and absolute stereostructures of five new germacrane-type sesquiterpenes, kikkanols D, D monoacetate, E, F, and F monoacetate from the flowers of Chrysanthemum indicum L. Chem Pharm Bull Tokyo. 2000;48(5):651–6. doi: 10.1248/cpb.48.651. [DOI] [PubMed] [Google Scholar]
- 13.Cheng W, Li J, You T, Hu C. Anti-inflammatory and immunomodulatory activities of the extracts from the inflorescence of Chrysanthemum indicum Linne. J Ethnopharmacol. 2005;101(1–3):334–7. doi: 10.1016/j.jep.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 14.Jeong WS, Jun M, Kong AN. Nrf2: a potential molecular target for cancer chemoprevention by natural compounds. Antioxid Redox Signal. 2006;8(1–2):99–106. doi: 10.1089/ars.2006.8.99. [DOI] [PubMed] [Google Scholar]
- 15.Khor TO, Yu S, Kong AN. Dietary cancer chemopreventive agents—targeting inflammation and Nrf2 signaling pathway. Planta Med. 2008;74(13):1540–7. doi: 10.1055/s-0028-1088303. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Khor TO, Xu C, Shen G, Jeong WS, Yu S, et al. Activation of Nrf2-antioxidant signaling attenuates NFkappaB-inflammatory response and elicits apoptosis. Biochem Pharmacol. 2008;76(11):1485–9. doi: 10.1016/j.bcp.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal. 2010. [DOI] [PMC free article] [PubMed]
- 18.Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal. 2010. [DOI] [PubMed]
- 19.Jeong WS, Kim IW, Hu R, Kong AN. Modulatory properties of various natural chemopreventive agents on the activation of NF-kappaB signaling pathway. Pharm Res. 2004;21(4):661–70. doi: 10.1023/B:PHAM.0000022413.43212.cf. [DOI] [PubMed] [Google Scholar]
- 20.Yu R, Mandlekar S, Lei W, Fahl WE, Tan TH, Kong AN. p38 mitogen-activated protein kinase negatively regulates the induction of phase II drug-metabolizing enzymes that detoxify carcinogens. J Biol Chem. 2000;275(4):2322–7. doi: 10.1074/jbc.275.4.2322. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Yu R, Owuor ED, Kong AN. Activation of antioxidant-response element (ARE), mitogen-activated protein kinases (MAPKs) and caspases by major green tea polyphenol components during cell survival and death. Arch Pharm Res. 2000;23(6):605–12. doi: 10.1007/BF02975249. [DOI] [PubMed] [Google Scholar]
- 22.Prawan A, Keum YS, Khor TO, Yu S, Nair S, Li W, et al. Structural influence of isothiocyanates on the antioxidant response element (ARE)-mediated heme oxygenase-1 (HO-1) expression. Pharm Res. 2008;25(4):836–44. doi: 10.1007/s11095-007-9370-9. [DOI] [PubMed] [Google Scholar]
- 23.Prawan A, Saw CL, Khor TO, Keum YS, Yu S, Hu L, et al. Anti-NF-kappaB and anti-inflammatory activities of synthetic isothiocyanates: effect of chemical structures and cellular signaling. Chem Biol Interact. 2009;179(2–3):202–11. doi: 10.1016/j.cbi.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong WS, Keum YS, Chen C, Jain MR, Shen G, Kim JH, et al. Differential expression and stability of endogenous nuclear factor E2-related factor 2 (Nrf2) by natural chemopreventive compounds in HepG2 human hepatoma cells. J Biochem Mol Biol. 2005;38(2):167–76. doi: 10.5483/bmbrep.2005.38.2.167. [DOI] [PubMed] [Google Scholar]
- 25.Khor TO, Huang MT, Kwon KH, Chan JY, Reddy BS, Kong AN. Nrf2-deficient mice have an increased susceptibility to dextran sulfate sodium-induced colitis. Cancer Res. 2006;66(24):11580–4. doi: 10.1158/0008-5472.CAN-06-3562. [DOI] [PubMed] [Google Scholar]
- 26.Nair S, Hebbar V, Shen G, Gopalakrishnan A, Khor TO, Yu S, et al. Synergistic effects of a combination of dietary factors sulforaphane and (−) epigallocatechin-3-gallate in HT-29 AP-1 human colon carcinoma cells. Pharm Res. 2008;25(2):387–99. doi: 10.1007/s11095-007-9364-7. [DOI] [PubMed] [Google Scholar]
- 27.Barve A, Khor TO, Nair S, Lin W, Yu S, Jain MR, et al. Pharmacogenomic profile of soy isoflavone concentrate in the prostate of Nrf2 deficient and wild-type mice. J Pharm Sci. 2008;97(10):4528–45. doi: 10.1002/jps.21311. [DOI] [PubMed] [Google Scholar]
- 28.Lin W, Wu RT, Wu T, Khor TO, Wang H, Kong AN. Sulforaphane suppressed LPS-induced inflammation in mouse peritoneal macrophages through Nrf2 dependent pathway. Biochem Pharmacol. 2008;76(8):967–73. doi: 10.1016/j.bcp.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair S, Barve A, Khor TO, Shen GX, Lin W, Chan JY, et al. Regulation of Nrf2- and AP-1-mediated gene expression by epigallocatechin-3-gallate and sulforaphane in prostate of Nrf2-knockout or C57BL/6J mice and PC-3 AP-1 human prostate cancer cells. Acta Pharmacol Sin. 2010;31(9):1223–40. doi: 10.1038/aps.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98(1):31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2(8):RESEARCH0032. [DOI] [PMC free article] [PubMed]
- 32.Prawan A, Buranrat B, Kukongviriyapan U, Sripa B, Kukongviriyapan V. Inflammatory cytokines suppress NAD(P)H:quinone oxidoreductase-1 and induce oxidative stress in cholangiocarcinoma cells. J Cancer Res Clin Oncol. 2009;135(4):515–22. doi: 10.1007/s00432-008-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saw CL, Huang Y, Kong AN. Synergistic anti-inflammatory effects of low doses of curcumin in combination with polyunsaturated fatty acids: docosahexaenoic acid or eicosapentaenoic acid. Biochem Pharmacol. 79(3):421–30. [DOI] [PubMed]
- 34.Saw CL, Olivo M, Chin WW, Soo KC, Heng PW. Transport of hypericin across chick chorioallantoic membrane and photodynamic therapy vasculature assessment. Biol Pharm Bull. 2005;28(6):1054–60. doi: 10.1248/bpb.28.1054. [DOI] [PubMed] [Google Scholar]
- 35.Cirino G, Distrutti E, Wallace JL. Nitric oxide and inflammation. Inflamm Allergy Drug Targets. 2006;5(2):115–9. doi: 10.2174/187152806776383143. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira-Marques V, Marinho HS, Cyrne L, Antunes F. Role of hydrogen peroxide in NF-kappaB activation: from inducer to modulator. Antioxid Redox Signal. 2009;11(9):2223–43. doi: 10.1089/ars.2009.2601. [DOI] [PubMed] [Google Scholar]
- 37.Khor TO, Keum YS, Lin W, Kim JH, Hu R, Shen G, et al. Combined inhibitory effects of curcumin and phenethyl isothiocyanate on the growth of human PC-3 prostate xenografts in immunodeficient mice. Cancer Res. 2006;66(2):613–21. doi: 10.1158/0008-5472.CAN-05-2708. [DOI] [PubMed] [Google Scholar]
- 38.Shen G, Khor TO, Hu R, Yu S, Nair S, Ho CT, et al. Chemoprevention of familial adenomatous polyposis by natural dietary compounds sulforaphane and dibenzoylmethane alone and in combination in ApcMin/+ mouse. Cancer Res. 2007;67(20):9937–44. doi: 10.1158/0008-5472.CAN-07-1112. [DOI] [PubMed] [Google Scholar]
- 39.Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48(2):91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin W, Shen G, Yuan X, Jain MR, Yu S, Zhang A, et al. Regulation of Nrf2 transactivation domain activity by p160 RAC3/SRC3 and other nuclear co-regulators. J Biochem Mol Biol. 2006;39(3):304–10. doi: 10.5483/bmbrep.2006.39.3.304. [DOI] [PubMed] [Google Scholar]
- 41.Wang T, Jiang H, Ji Y, Xu J. Anti-oxidation effect of water extract of Flos chrysanthemi on heart and brain in vivo and in vitro. Zhong Yao Cai. 2001;24(2):122–4. [PubMed] [Google Scholar]