Abstract
The sarcomeres form the molecular motor of the cardiomyocyte and consist of a complex multi-protein of thick and thin filaments which are anchored to the cytoskeleton. The thick filament, composed of myosin and associated proteins, and the thin filament composed of actin, tropomyosin and the troponins develop actinmyosin crossbridges which cycle in response to calcium resulting in sliding of the filaments and contraction. The thin filament in fixed to the cardiomyocyte cytoskeleton at the Z-disc, a complex of structural and regulatory proteins. A giant protein, titin, provides an external scaffold and regulates passive force in diastole. Both genetic disorders and acquired conditions may affect proteins of the sarcomere. Genetic disorders of the thick and thin filament proteins are the predominant cause of hypertrophic cardiomyopathy. These mutations lead to abnormal sarcomere function, often an enhanced sensitivity to calcium, and impaired relaxation. This may result in secondary changes in calcium cycling and amplification of hypertrophic signaling cascades. Correcting the abnormal function of the sarcomere as well as intervening in later stages of the pathophysiologic cascades may ameliorate disease. In dilated cardiomyopathy genetic abnormalities in the sarcomere, Z-disc, calcium regulatory and cytoskeletal proteins as well as the dystrophin complex may be causal for disease. In dilated cardiomyopathy, disturbances in post-translational modifications of the sarcomere my also play a prominent role. Experimental models indicate that altered phosphorylation of sarcomeric proteins may impair systolic and diastolic function as well as the response to heart rate and afterload. Thus correcting these post-translational changes are legitimate targets for future therapeutic strategies for dilated cardiomyopathy.
Keywords: hypertrophic cardiomyopathy, dilated cardiomyopathy, sarcomere
Introduction
The sarcomere is the molecular motor of the cardiomyocyte. It is composed of interdigitated thick and thin filaments which slide along each other in response to systolic levels of intracellular calcium leading to shortening of the cardiomyocyte and thus contraction of the heart. During diastole, calcium is actively removed from the cytosol via a calcium pump in the sarcoplasmic reticulum as well as through calcium extrusion across the cell membrane. Disarray of the sarcomere was recognized as a feature of hypertrophic cardiomyopathy many decades ago. In 1990, however, it was discovered that mutations in the beta myosin heavy chain caused hypertrophic cardiomyopathy, and subsequent findings of additional mutations in sarcomeric proteins defined familial hypertrophic cardiomyopathy as a “disease of the sarcomere” [1, 2]. Numerous studies have been performed to measure abnormal sacromeric function caused by these mutations and have attempted to link the cellular dysfunction to the complex phenotype of the disease which includes diastolic dysfunction, hypertrophy, obstruction to left ventricular outflow, risk of arrhythmia and sudden cardiac death and in some cases progression to cardiac dilation and heart failure.
Dysfunction of the sarcomere is not limited to hypertrophic cardiomyopathy. Studies of dilated cardiomyopathy have revealed mutations in both sarcomeric and related proteins leading to dilated cardiomyopathy (reviewed in [3, 4]). In addition, altered posttranslational modifications of the sarcomere are found in dilated cardiomyopathy and may contribute to cardiac dysfunction. In the following review we will discuss sarcomeric dysfunction in both hypertrophic and dilated cardiomyopathy and suggest how understanding the pathophysiology of dysfunction may guide future therapies.
Composition and function of the sarcomere
The myofilament consists of the thick and thin filaments, which form and cycle (?) crossbridges in response to changes in intracellular Ca2+ as recently reviewed by Hinken and Solaro[5]. The thick filament consists of myosin heavy chains (MHC), predominantly the β isoform in human heart, and associated light chains which in ventricle are the essential light chain (MLC1) and regulatory light chain (MLC2), as well as myosin binding protein C (MyBP-C). Titin is a large protein that serves as a molecular scaffold but also appears to play a regulatory role in control of passive force. The thin filament is comprised of cardiac actin, tropomyosin (Tm) and troponin (Tn). Troponin is a complex which consists of 3 subunits: cardiac troponin I (TnI), which inhibits actin myosin cross bridge formation at diastolic levels of calcium; cardiac troponin T (TnT) which fixes troponin to the thin filament by binding tropomyosin; and cardiac troponin C (TnC) which binds calcium. With release of calcium from the sarcoplasmic reticulum during systole, it binds to cardiac TnC. This results in a change of affinities of binding domains of TnI from actin-Tm to TnC as well as other alterations in thin filament protein interactions. This alteration in binding promotes the interaction of actin with myosin, through both steric changes in the Tm position and allosteric changes in the thin filament, and permits strong crossbrige formation and contraction. It is also important to point out that the myosin head contains an ATPase which hydrolyses adenosine triphosphate (ATP) with each cycle of the actin-myosin crossbridge.
There are numerous methods that may be used to assess sarcomeric function. The function of the sarcomere may be measured by isolating the sarcomeric proteins or myofilaments in vitro. These preparations maintain the actin-myosin ATPase function in response to calcium, permitting measurements of the relationship to this activity to calcium in vitro. It is also feasible to measure tension in single cardiomyocytes or small bundles of cardiomyocytes such as small trabeculae. When these preparations are intact, delicate transducers can be fixed to the end of the muscle to measure tension of the preparations with electrical pacing of the cell. Another methodology used in intact myocytes is to load calcium sensitive dyes into the cell to simultaneously measure calcium and tension developed in response to electrical pacing. Alternatively, these muscle preparations may be studied after chemical treatment to remove cell membranes, permitting direct steady state measurement of the response to calcium levels set by the investigator. Isolated sarcomeric proteins, either native or produced recombinantly, may also be used to study aspects of protein-protein interactions, calcium binding or ATPase activity, or to perform motility assays which measure the velocity of actin filament movement along myosin heads fixed to glass slides.
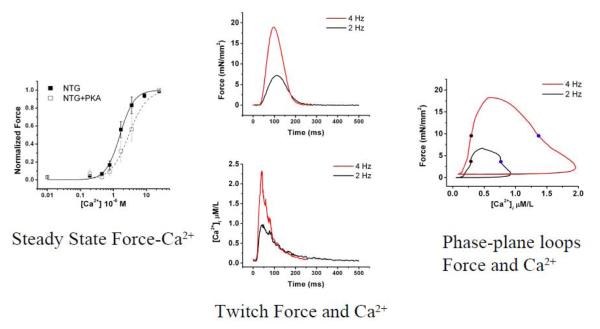
Figure 1 illustrates some different type of measurements such as steady state force-calcium relationships in panel to the left. In this muscle, the cell membrane has been removed and sequential measurements of force are made at varied calcium concentrations. In the middle panel of this figure, the measurements of force and calcium are measured in a muscle trabecula with intact membrane. Calcium-sensitive dye permits measurement of calcium during muscle twitch while force is simultaneously measured. As the frequency of electrical stimulation increases from 2 Hz to 4 Hz both peak calcium and peak force increases. The rightward panel provides simultaneous traces of force and calcium plotted orthogonally during a muscle twitch at two different pacing frequencies.
Figure 1. Measurements of sarcomeric function.
The leftward panel illustrates steady state force-calcium relationship in a non-transgenic (NTG) control mouse cardiac trabeculae which has been chemically skinned and exposed to varying calcium concentrations. The force-calcium relationship is shifted rightward (desensitized) after treatment with protein kinase A (PKA). The middle panel demonstrates calcium transient measurements (lower) and force measurements (upper) from intact cardiac trabeculae paced at 2 Hz and 4 Hz. The rightward panel illustrates simultaneous calcium-force measurements plotted orthogonally throughout a cardiac twitch in a phase-plane hysteresis loop. Figure panels reprinted from figures 1, 5 and 7 from the Journal of Molecular and Cellular Cardiology, Volume 48, Issue 5, G.A. Ramirez-Correa et al. entitled, “Calcium sensitivity, force frequency relationship and cardiac troponin I: Critical role of PKA and PKC phosphorylation sites”, 943-953, 2010 with permission from Elsevier.
Pathophysiologic mechanisms in hypertrophic cardiomyopathy
Following discovery of disease-causing mutations of sacromeric proteins, a great deal of work has been performed to study sarcomeric function in hypertrophic cardiomyopathy and to begin to understand the progression of this disorder, which is typically asymptomatic in the young. These studies have been undertaken in animal models, typically genetically modified mice or rats, and in few cases in rabbits, as well as by studying human tissue and performing in vitro studies on recombinantly produced proteins. A host of abnormalities in sarcomeric function have been documented in numerous studies. A comprehensive discussion of these studies in detail is beyond the scope of this review but has been covered in detail by others [3, 4, 6]. With β–MHC mutations, maximal force activation was found to be enhanced [7, 8], however in the study of human tissue explanted a heart transplantation from an individual homozygous for R403W mutant, enhanced hydrolysis of ATP by the actin-myosin ATPase was even more prominent than the enhanced motility assay suggesting that the energetic cost of contraction is increased [9]. Disturbances in ATP utilization may have secondary effects such as production of reactive oxygen species leading to further protein dysfunction. Increased energetic cost of contraction was also found with TnT mutations in murine models, though this was found to be variable in severity depending on the mutation. [10, 11].
In addition to altered maximal force or motility, increased calcium sensitivity of force or ATPase activity appears to be a common pathophysiology as reviewed by Morimoto [3]. This has been noted in a number of thick and thin filament protein mutations (See Table 1). This may result in increased passive (diastolic) tension of muscle and provides a rationale for diastolic dysfunction in hypertrophic cardiomyopathy (HCM). Functional studies of childhood onset restrictive cardiomyopathy mutations have revealed very profound functional abnormalities resulting in severe and high-risk disease [12-14]. It is important to point out that many of these assays are undertaken under steady state conditions, and more explicit findings may be present in dynamic studies such as illustrated in figure 1.
Table 1.
Examples of sarcomere dysfunction associated with hypertrophic cardiomyopathy mutations.
| Gene | Protein | Mutation | Sacromere dysfunction | Comments and References |
||
|---|---|---|---|---|---|---|
| Ca2+ sensitivity |
Maximal Force/ Passive force |
Sliding velocity/ATPase activity |
||||
| MYH7 | β-MHC | R403Q | ↔ to ↑ at sub-max Ca2+ |
↑ Max | ↑* | [8, 32, 33] Reviewed in [3, 6, 34], * Some earlier papers found decreased ATPase or sliding velocity see discussion in reviews. |
| MYBPC3 | MyBP- C |
truncation | ↑ | ↓ Max power |
[35] | |
| MYL2 | MLC- 2s/v |
E22K | ↑ | [36] | ||
| TNNT2 | cTnT | R92Q | ↑ | ↑ Passive |
[3, 10, 37- 40] |
|
| I79N | ↑ | ↑ Passive |
||||
| TNNI3 | cTnI | R145G | ↑ | [41] | ||
| TMP1 | α-TM | E180G | ↑ | [42] | ||
| D175N | ↑ | [43, 44] | ||||
| ACTC | 〈- cardiac actin |
E99K | ↓ Max | ↓ | [45] | |
Detailed studies of disease progression in animal models have pointed out a pathway of altered sarcomere function and diastolic dysfunction [15] as well as the development of altered calcium cycling which occurs prior to development of hypertrophy and histologic changes including fibrosis [16]. This pathway of early findings of diastolic dysfunction prior to onset of hypertrophy has been mirrored in human studies [17]. Altered calcium dynamics may lead to abnormal signaling cascades ultimately resulting in muscle hypertrophy and abnormal extracellular matrix. Indeed, the Seidman lab has demonstrated that early interruption of altered calcium dynamics in a murine model by treatment with the calcium channel blocker diltiazem avoided the progression of disease [18]. Modifying the contractile properties of the sarcomere in the presence of a disease-causing mutation also interrupts the pathway of abnormal calcium sensitivity and rescues Tm mutant mice from the HCM phenotype [19]. Finally, potential strategies based on experimental models have been recently discussed in a detailed review [20]. These types of studies offer hope for preventing disease progression in asymptomatic individuals with hypertrophic cardiomyopathy mutations. Indeed this is the basis of a current randomized clinical trial of diltiazem in genotype-positive individuals who have not yet developed hypertrophy: (http://clinicaltrials.gov/ct2/show/record/NCT00319982).
It is important to consider several caveats to the above discussion. The profile of functional defects and their progression may vary with the protein affected, the specific mutation within the protein or even the specific amino acid change within the same site of the protein. Certainly, other disease modifiers may influence disease presentation and course. Furthermore, children with severe early onset hypertrophic cardiomyopathy may have more than one sacromeric mutation either within the same gene or a different sarcomeric gene leading to more profound functional alterations.
The therapeutic implications of these studies will likely impact the future care of children with hypertrophic cardiomyopathy. Targeted therapies may become available to either primarily repair the function of the sarcomere, or to avoid secondary effects such as abnormal calcium cycling, hypertrophic signaling cascades, oxidative stress or signaling which mediates fibrosis development. This might be approached with genetically mediated therapies to repair the effects of the mutant protein or to turn off the mutant allele, or use of small molecules targeted to the myofilament. The availability of a clinical trial and the possibility of additional trials in the future provide additional impetus for pediatric cardiologists to consider referral of children at risk for HCM for genetic counseling and testing for familial HCM causing mutations when a proband has been identified in a family.
The sarcomere in dilated cardiomyopathy
Sarcomeric mutations are one cause of familial dilated cardiomyopathy (DCM) and genetic abnormalities in the sarcomere, Z-disc, calcium regulatory and cytoskeletal proteins as well as the dystrophin complex may also cause DCM [21-23]. Many of these proteins are known to have a key role in transmission of the force to result in cell shortening with the contractile cycle. Muscle studies of sarcomeric mutants causing DCM have in some cases revealed sacromeric dysfunction such as decreased maximal tension or decreased calcium sensitivity as reviewed by Morimoto [3] (See Table 2). However, many cases of DCM in childhood are of unclear etiology or occur as a result of inflammatory diseases such as myocarditis or toxic exposures such as anthracycline cardiomyopathy. Despite the etiology of disease, however, secondary effects on the sarcomere may occur due to altered post-translational modification of proteins. These changes included altered phosphorylation, oxidative injury and possibly novel modifications of proteins.
Table 2.
Examples of sarcomere dysfunction associated with dilated cardiomyopathy mutations.
| Gene | Protein | Mutation | Sacromere dysfunction | Comments and References |
||
|---|---|---|---|---|---|---|
| Ca2+ sensitivity |
Force Generation or Passive Force |
Sliding velocity/ATPase activity |
||||
| MYH7 | β-MHC | S532P | ↓ shortening (S.L.) |
↓ sliding velocity | [46, 47] | |
| TNNT2 | cTnT | K210 Del | ↓ Muscle and ATPase |
↔ maximal Force (Fmax) or cooperativity ↓ Fmax |
↓ sliding velocity ↓ ATPase activity ↓ n Hill (cooperativity) |
[48-51] |
| TNNI3 | cTnI | K36Q/ N185K |
↓ | ↓maximal ATPase activity ↓ n Hill (cooperativity) |
[52] | |
| TNNC1 | cTnC | G159D | ↓ ATPase ↑ human skinned myocytes |
↓ ATPase activity ↓ or ↔ In vitro motility ↓ n Hill (cooperativity) |
[48, 53] | |
| TPM1 | α-Tm | E40K | ↔ | ↓ ATPase activity ↓ n Hill (cooperativity) |
[48, 54] | |
| ACTC1 |
α- cardiac actin |
E361G | ↓ mouse model after dephos |
↔ In vitro motility | [55, 56] | |
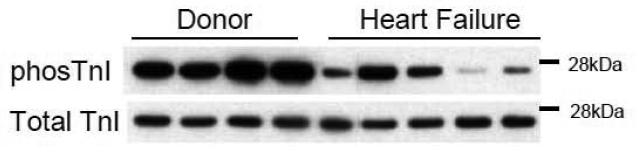
A prominent change in sarcomeric proteins which has been noted by several groups in both samples from failing human heart is decreased phosphorylation of two serines (Ser 23,24) on troponin I [24, 25] (see figure 2). These sites, which appear to be predominantly phosphorylated by protein kinase A (PKA), are in an amino terminal region that is unique to the cardiac isoform of TnI. Phosphorylation of these sites decreases the steady state force-calcium relationship, though it is thought to augment relaxation in diastole. When the heart is stimulated by β-adrenergic agonists, the prominent targets for phosphorylation include the sarcomere, particularly TnI and MyBP-C, and the protein phospholamban which regulates the calcium pump in sarcoplasmic reticulum. Therefore, β-adrenergic stimulation has both an impact on the sarcomere as well as accelerated calcium pumping and increased calcium release during systole. We sought to understand how TnI phosphorylation altered cardiac function by creating mutant transgenic mice in which a phospho-mimicking amino acid was introduced into these sites[26]. Other labs have performed similar studies [27]. We found that the transgenic mouse line with pseudophosphorylated TnI at the PKA sites had an enhanced force frequency response, as well as enhanced relaxation when exposed to an acute increase in afterload. Heart failure is also known to increase protein kinase C (PKC) activation in heart, so we also studied a murine line with mutations that mimicked diminished PKA phosphorylation in addition to other TnI mutations that mimicked enhanced PKC phosphorylation of TnI[28]. These mice had a decreased force frequency response and an exaggerated impairment of relaxation with imposition of afterload. A recent study has demonstrated how this decreased TnI phosphorylation at PKA sites might be attacked therapeutically. Lai et al. developed a model of post-infarct heart failure and were able to ameliorate the phenotype as well as restoring TnI Ser 23/24 phosphorylation by overexpressing adenylyl cyclase VI in mice [29]. This provides some proof of principle that remodeling of sarcomeres that are dysfunctional as a result of altered phosphorylation could be a legitimate therapy for heart failure.
Figure 2. Decreased phosphorylation of troponin I Ser 23, 24 in heart failure.
Immunoblot with antibodies recognizing phosphorylation at troponin I Ser 23, 24 sites, the target for protein kinase A (top panel) and antibody recognizing total troponin I (bottom panel). Samples are from tissue bank provided by Dr. Cris dos Remidios, Sydney, Australia. Human cardiac ventricular tissue was explanted from donors without heart disease in which tissue was not utilized at transplant (lanes 1-4), or from end stage heart failure patients (lanes 5-9). Phosphorylation at troponin I Ser 23,24 was significantly reduced in samples from end-stage heart failure.
Studies of human cardiac tissue from end stage explanted hearts have also confirmed sarcomeric dysfunction as recently reviewed by Hamdani et al. [30]. Both altered calcium sensitivity and altered kinetic properties with delayed rate constant of force development have been noted. Decreases in maximal force have been noted by some investigators [31]. These changes are likely secondary to a set of changes at specific phosphorylation sites and additional post-translational modifications, rather than changes restricted to TnI.
Summary
The sarcomere is the essential molecular motor of the cardiomyocyte. Sarcomeric dysfunction is the underlying primary cause of many genetically mediated HCM and DCM disorders. In additional, altered post-translational modifications of sarcomeric proteins appear to contribute to the phenotype of acquired DCM. Therapies directed toward ameliorating sarcomere dysfunction will likely contribute to the treatment of both DCM and HCM in the future.
Acknowledgements
This work was supported in part by NIH NHLBI Proteomics Centers Contract N01-HV-28180 and NIH RO1 HL63038. Genaro A. Ramirez-Correa thanks the Minority Mentoring Program of AHA for their support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Geisterfer-Lowrance AA, Kass S, Tanigawa G, et al. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62(5):999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 2.Thierfelder L, Watkins H, MacRae C, et al. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77(5):701–12. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 3.Morimoto S. Sarcomeric proteins and inherited cardiomyopathies. Cardiovasc Res. 2008;77(4):659–66. doi: 10.1093/cvr/cvm084. [DOI] [PubMed] [Google Scholar]
- 4.Xu Q, Dewey S, Nguyen S, Gomes AV. Malignant and benign mutations in familial cardiomyopathies: insights into mutations linked to complex cardiovascular phenotypes. J Mol Cell Cardiol. 2010;48(5):899–909. doi: 10.1016/j.yjmcc.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Hinken AC, Solaro RJ. A dominant role of cardiac molecular motors in the intrinsic regulation of ventricular ejection and relaxation. Physiology (Bethesda) 2007;22:73–80. doi: 10.1152/physiol.00043.2006. [DOI] [PubMed] [Google Scholar]
- 6.Tardiff JC. Sarcomeric proteins and familial hypertrophic cardiomyopathy: linking mutations in structural proteins to complex cardiovascular phenotypes. Heart Fail Rev. 2005;10(3):237–48. doi: 10.1007/s10741-005-5253-5. [DOI] [PubMed] [Google Scholar]
- 7.Debold EP, Schmitt JP, Patlak JB, et al. Hypertrophic and dilated cardiomyopathy mutations differentially affect the molecular force generation of mouse alpha-cardiac myosin in the laser trap assay. Am J Physiol Heart Circ Physiol. 2007;293(1):H284–91. doi: 10.1152/ajpheart.00128.2007. [DOI] [PubMed] [Google Scholar]
- 8.Tyska MJ, Hayes E, Giewat M, Seidman CE, Seidman JG, Warshaw DM. Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ Res. 2000;86(7):737–44. doi: 10.1161/01.res.86.7.737. [DOI] [PubMed] [Google Scholar]
- 9.Keller DI, Coirault C, Rau T, et al. Human homozygous R403W mutant cardiac myosin presents disproportionate enhancement of mechanical and enzymatic properties. J Mol Cell Cardiol. 2004;36(3):355–62. doi: 10.1016/j.yjmcc.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Chandra M, Tschirgi ML, Tardiff JC. Increase in tension-dependent ATP consumption induced by cardiac troponin T mutation. Am J Physiol Heart Circ Physiol. 2005;289(5):H2112–9. doi: 10.1152/ajpheart.00571.2005. [DOI] [PubMed] [Google Scholar]
- 11.He H, Javadpour MM, Latif F, Tardiff JC, Ingwall JS. R-92L and R-92W mutations in cardiac troponin T lead to distinct energetic phenotypes in intact mouse hearts. Biophys J. 2007;93(5):1834–44. doi: 10.1529/biophysj.107.107557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis J, Wen H, Edwards T, Metzger JM. Allele and species dependent contractile defects by restrictive and hypertrophic cardiomyopathy-linked troponin I mutants. J Mol Cell Cardiol. 2008;44(5):891–904. doi: 10.1016/j.yjmcc.2008.02.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes AV, Liang J, Potter JD. Mutations in human cardiac troponin I that are associated with restrictive cardiomyopathy affect basal ATPase activity and the calcium sensitivity of force development. J Biol Chem. 2005;280(35):30909–15. doi: 10.1074/jbc.M500287200. [DOI] [PubMed] [Google Scholar]
- 14.Yumoto F, Lu QW, Morimoto S, et al. Drastic Ca2+ sensitization of myofilament associated with a small structural change in troponin I in inherited restrictive cardiomyopathy. Biochem Biophys Res Commun. 2005;338(3):1519–26. doi: 10.1016/j.bbrc.2005.10.116. [DOI] [PubMed] [Google Scholar]
- 15.Georgakopoulos D, Christe ME, Giewat M, Seidman CM, Seidman JG, Kass DA. The pathogenesis of familial hypertrophic cardiomyopathy: early and evolving effects from an alpha-cardiac myosin heavy chain missense mutation. Nat Med. 1999;5(3):327–30. doi: 10.1038/6549. [DOI] [PubMed] [Google Scholar]
- 16.Fatkin D, McConnell BK, Mudd JO, et al. An abnormal Ca(2+) response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy. J Clin Invest. 2000;106(11):1351–9. doi: 10.1172/JCI11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho CY, Sweitzer NK, McDonough B, et al. Assessment of diastolic function with Doppler tissue imaging to predict genotype in preclinical hypertrophic cardiomyopathy. Circulation. 2002;105(25):2992–7. doi: 10.1161/01.cir.0000019070.70491.6d. [DOI] [PubMed] [Google Scholar]
- 18.Semsarian C, Ahmad I, Giewat M, et al. The L-type calcium channel inhibitor diltiazem prevents cardiomyopathy in a mouse model. J Clin Invest. 2002;109(8):1013–20. doi: 10.1172/JCI14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jagatheesan G, Rajan S, Petrashevskaya N, et al. Rescue of tropomyosin-induced familial hypertrophic cardiomyopathy mice by transgenesis. Am J Physiol Heart Circ Physiol. 2007;293(2):H949–58. doi: 10.1152/ajpheart.01341.2006. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Seidman JG, Seidman CE. Narrative review: harnessing molecular genetics for the diagnosis and management of hypertrophic cardiomyopathy. Ann Intern Med. 2010;152(8):513–20. doi: 10.1059/0003-4819-152-8-201004200-00008. W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fatkin D, Otway R, Richmond Z. Genetics of dilated cardiomyopathy. Heart Fail Clin. 2010;6(2):129–40. doi: 10.1016/j.hfc.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Jefferies JL, Towbin JA. Dilated cardiomyopathy. Lancet. 2010;375(9716):752–62. doi: 10.1016/S0140-6736(09)62023-7. [DOI] [PubMed] [Google Scholar]
- 23.Judge DP. Use of genetics in the clinical evaluation of cardiomyopathy. Jama. 2009;302(22):2471–6. doi: 10.1001/jama.2009.1787. [DOI] [PubMed] [Google Scholar]
- 24.Bodor GS, Oakeley AE, Allen PD, Crimmins DL, Ladenson JH, Anderson PA. Troponin I phosphorylation in the normal and failing adult human heart. Circulation. 1997;96(5):1495–500. doi: 10.1161/01.cir.96.5.1495. [DOI] [PubMed] [Google Scholar]
- 25.Zakhary DR, Moravec CS, Stewart RW, Bond M. Protein kinase A (PKA)-dependent troponin-I phosphorylation and PKA regulatory subunits are decreased in human dilated cardiomyopathy. Circulation. 1999;99(4):505–10. doi: 10.1161/01.cir.99.4.505. [DOI] [PubMed] [Google Scholar]
- 26.Takimoto E, Soergel DG, Janssen PM, Stull LB, Kass DA, Murphy AM. Frequency- and afterload-dependent cardiac modulation in vivo by troponin I with constitutively active protein kinase A phosphorylation sites. Circ Res. 2004;94(4):496–504. doi: 10.1161/01.RES.0000117307.57798.F5. [DOI] [PubMed] [Google Scholar]
- 27.Sakthivel S, Finley NL, Rosevear PR, et al. In vivo and in vitro analysis of cardiac troponin I phosphorylation. J Biol Chem. 2005;280(1):703–14. doi: 10.1074/jbc.M409513200. [DOI] [PubMed] [Google Scholar]
- 28.Bilchick KC, Duncan JG, Ravi R, et al. Heart failure-associated alterations in troponin I phosphorylation impair ventricular relaxation-afterload and force-frequency responses and systolic function. Am J Physiol Heart Circ Physiol. 2007;292(1):H318–25. doi: 10.1152/ajpheart.00283.2006. [DOI] [PubMed] [Google Scholar]
- 29.Lai NC, Tang T, Gao MH, et al. Activation of cardiac adenylyl cyclase expression increases function of the failing ischemic heart in mice. J Am Coll Cardiol. 2008;51(15):1490–7. doi: 10.1016/j.jacc.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamdani N, Kooij V, van Dijk S, et al. Sarcomeric dysfunction in heart failure. Cardiovasc Res. 2008;77(4):649–58. doi: 10.1093/cvr/cvm079. [DOI] [PubMed] [Google Scholar]
- 31.Noguchi T, Hunlich M, Camp PC, et al. Thin-filament-based modulation of contractile performance in human heart failure. Circulation. 2004;110(8):982–7. doi: 10.1161/01.CIR.0000139334.43109.F9. [DOI] [PubMed] [Google Scholar]
- 32.Gao WD, Perez NG, Seidman CE, Seidman JG, Marban E. Altered cardiac excitation-contraction coupling in mutant mice with familial hypertrophic cardiomyopathy. J Clin Invest. 1999;103(5):661–6. doi: 10.1172/JCI5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer BM, Fishbaugher DE, Schmitt JP, et al. Differential cross-bridge kinetics of FHC myosin mutations R403Q and R453C in heterozygous mouse myocardium. Am J Physiol Heart Circ Physiol. 2004;287(1):H91–9. doi: 10.1152/ajpheart.01015.2003. [DOI] [PubMed] [Google Scholar]
- 34.Lowey S. Functional consequences of mutations in the myosin heavy chain at sites implicated in familial hypertrophic cardiomyopathy. Trends Cardiovasc Med. 2002;12(8):348–54. doi: 10.1016/s1050-1738(02)00181-0. [DOI] [PubMed] [Google Scholar]
- 35.Yang Q, Sanbe A, Osinska H, Hewett TE, Klevitsky R, Robbins J. A mouse model of myosin binding protein C human familial hypertrophic cardiomyopathy. J Clin Invest. 1998;102(7):1292–300. doi: 10.1172/JCI3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szczesna-Cordary D, Guzman G, Zhao J, Hernandez O, Wei J, Diaz-Perez Z. The E22K mutation of myosin RLC that causes familial hypertrophic cardiomyopathy increases calcium sensitivity of force and ATPase in transgenic mice. J Cell Sci. 2005;118(Pt 16):3675–83. doi: 10.1242/jcs.02492. [DOI] [PubMed] [Google Scholar]
- 37.Chandra M, Rundell VL, Tardiff JC, Leinwand LA, De Tombe PP, Solaro RJ. Ca(2+) activation of myofilaments from transgenic mouse hearts expressing R92Q mutant cardiac troponin T. Am J Physiol Heart Circ Physiol. 2001;280(2):H705–13. doi: 10.1152/ajpheart.2001.280.2.H705. [DOI] [PubMed] [Google Scholar]
- 38.Miller T, Szczesna D, Housmans PR, et al. Abnormal contractile function in transgenic mice expressing a familial hypertrophic cardiomyopathy-linked troponin T (I79N) mutation. J Biol Chem. 2001;276(6):3743–55. doi: 10.1074/jbc.M006746200. [DOI] [PubMed] [Google Scholar]
- 39.Szczesna D, Zhang R, Zhao J, Jones M, Guzman G, Potter JD. Altered regulation of cardiac muscle contraction by troponin T mutations that cause familial hypertrophic cardiomyopathy. J Biol Chem. 2000;275(1):624–30. doi: 10.1074/jbc.275.1.624. [DOI] [PubMed] [Google Scholar]
- 40.Tardiff JC, Hewett TE, Palmer BM, et al. Cardiac troponin T mutations result in allele-specific phenotypes in a mouse model for hypertrophic cardiomyopathy. J Clin Invest. 1999;104(4):469–81. doi: 10.1172/JCI6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.James J, Zhang Y, Osinska H, et al. Transgenic modeling of a cardiac troponin I mutation linked to familial hypertrophic cardiomyopathy. Circ Res. 2000;87(9):805–11. doi: 10.1161/01.res.87.9.805. [DOI] [PubMed] [Google Scholar]
- 42.Prabhakar R, Boivin GP, Grupp IL, et al. A familial hypertrophic cardiomyopathy alpha-tropomyosin mutation causes severe cardiac hypertrophy and death in mice. J Mol Cell Cardiol. 2001;33(10):1815–28. doi: 10.1006/jmcc.2001.1445. [DOI] [PubMed] [Google Scholar]
- 43.Evans CC, Pena JR, Phillips RM, et al. Altered hemodynamics in transgenic mice harboring mutant tropomyosin linked to hypertrophic cardiomyopathy. Am J Physiol Heart Circ Physiol. 2000;279(5):H2414–23. doi: 10.1152/ajpheart.2000.279.5.H2414. [DOI] [PubMed] [Google Scholar]
- 44.Muthuchamy M, Pieples K, Rethinasamy P, et al. Mouse model of a familial hypertrophic cardiomyopathy mutation in alpha-tropomyosin manifests cardiac dysfunction. Circ Res. 1999;85(1):47–56. doi: 10.1161/01.res.85.1.47. [DOI] [PubMed] [Google Scholar]
- 45.Bookwalter CS, Trybus KM. Functional consequences of a mutation in an expressed human alpha-cardiac actin at a site implicated in familial hypertrophic cardiomyopathy. J Biol Chem. 2006;281(24):16777–84. doi: 10.1074/jbc.M512935200. [DOI] [PubMed] [Google Scholar]
- 46.Kamisago M, Sharma SD, DePalma SR, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343(23):1688–96. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 47.Schmitt JP, Debold EP, Ahmad F, et al. Cardiac myosin missense mutations cause dilated cardiomyopathy in mouse models and depress molecular motor function. Proc Natl Acad Sci U S A. 2006;103(39):14525–30. doi: 10.1073/pnas.0606383103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirza M, Marston S, Willott R, et al. Dilated cardiomyopathy mutations in three thin filament regulatory proteins result in a common functional phenotype. J Biol Chem. 2005;280(31):28498–506. doi: 10.1074/jbc.M412281200. [DOI] [PubMed] [Google Scholar]
- 49.Morimoto S, Lu QW, Harada K, et al. Ca(2+)-desensitizing effect of a deletion mutation Delta K210 in cardiac troponin T that causes familial dilated cardiomyopathy. Proc Natl Acad Sci U S A. 2002;99(2):913–8. doi: 10.1073/pnas.022628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson P, Mirza M, Knott A, et al. Alterations in thin filament regulation induced by a human cardiac troponin T mutant that causes dilated cardiomyopathy are distinct from those induced by troponin T mutants that cause hypertrophic cardiomyopathy. J Biol Chem. 2002;277(43):40710–6. doi: 10.1074/jbc.M203446200. [DOI] [PubMed] [Google Scholar]
- 51.Venkatraman G, Harada K, Gomes AV, Kerrick WG, Potter JD. Different functional properties of troponin T mutants that cause dilated cardiomyopathy. J Biol Chem. 2003;278(43):41670–6. doi: 10.1074/jbc.M302148200. [DOI] [PubMed] [Google Scholar]
- 52.Carballo S, Robinson P, Otway R, et al. Identification and functional characterization of cardiac troponin I as a novel disease gene in autosomal dominant dilated cardiomyopathy. Circ Res. 2009;105(4):375–82. doi: 10.1161/CIRCRESAHA.109.196055. [DOI] [PubMed] [Google Scholar]
- 53.Dyer EC, Jacques AM, Hoskins AC, et al. Functional analysis of a unique troponin c mutation, GLY159ASP, that causes familial dilated cardiomyopathy, studied in explanted heart muscle. Circ Heart Fail. 2009;2(5):456–64. doi: 10.1161/CIRCHEARTFAILURE.108.818237. [DOI] [PubMed] [Google Scholar]
- 54.Mirza M, Robinson P, Kremneva E, et al. The effect of mutations in alpha-tropomyosin (E40K and E54K) that cause familial dilated cardiomyopathy on the regulatory mechanism of cardiac muscle thin filaments. J Biol Chem. 2007;282(18):13487–97. doi: 10.1074/jbc.M701071200. [DOI] [PubMed] [Google Scholar]
- 55.Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280(5364):750–2. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 56.Song W, Dyer E, Stuckey D, et al. Investigation of a transgenic mouse model of familial dilated cardiomyopathy. J Mol Cell Cardiol. 2010;49(3):380–9. doi: 10.1016/j.yjmcc.2010.05.009. [DOI] [PubMed] [Google Scholar]