Abstract
Intracisternal A-type particle (IAP) elements are high copy number long terminal repeat (LTR) rodent retrotransposons. Some IAP elements can transpose, and are responsible for ~12% of spontaneous mouse mutations. Inbred mouse strains show variation in genomic IAP distribution, contributing to inter-strain genetic variability. Additionally IAP elements can influence the transcriptional regulation of neighbouring genes through their strong LTR promoter, effecting phenotypic variation. This genetic and phenotypic variability can translate into experimental variability between mouse strains. For example, it has been demonstrated that strain-specific genetic/epigenetic factors can interact to yield variable responses to drugs. Therefore, in experimental contexts it is essential to unequivocally identify mouse strains. Recently it was estimated that any two inbred strains share only ~40% of their IAP insertions. Of the remaining 60%, some insertions will be strain specific, fixed during inbreeding. These fixed insertions can be exploited as genetic markers to identify inbred strains, if they can be identified simply and efficiently. Here, we report the development of a PCR-based system allowing direct acquisition of strain-specific IAP insertions. In a pilot study, we identified 21 IAP loci, genotyped IAP insertions at 9 loci, and discovered two strain-specific insertions that could reliably identify these strains.
Keywords: IAP element, Retrotransposon, Inbred mouse strain, Genetic identification, Display PCR
Introduction
Mouse IAP (Intra-cisternal A-type particle) elements are high copy number (~1000 copies per haploid genome) long terminal repeat (LTR) retrotransposons. They are categorized as class II Endogenous RetroViruses (ERVs) [1] and are currently actively mobile in the mouse genome [2]. The copy number of IAP elements varies between inbred mouse strains [3] and IAP insertions are responsible for 10–12% of spontaneous mutations [2]. On average, there are ~700 full length and ~300 truncated IAP copies in a given mouse genome [1].
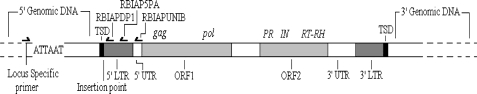
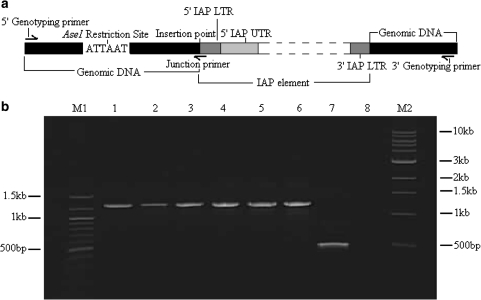
A full length IAP element comprises two open reading frames (ORFs) flanked by two identical LTRs, as illustrated in Fig. 1. The 5′ and 3′ LTRs are flanked by direct repeats of the insertion site, known as target site duplications (TSDs). The 5′ LTR houses a strong promoter that drives transcription of ORF1 (gag and pol) and ORF2 (PR, IN and RT-RH) [4]. The genomic size of IAP elements varies, with full length IAP elements being approximately 7.2 kilobases (kb) in length [5]. Shorter, inactive copies, usually derived from full length elements by internal deletions [6, 7] are highly variable in size, but all retain conserved 5′ UTR sequences adjacent to the 5′ LTR (see Fig. 1). Solitary (or solo) LTRs represent the shortest IAP sequences and are likely derived from inter-LTR recombination, as seen in murine leukaemia viruses [8]. As such they are thus evolutionarily older than “dual” LTR elements.
Fig. 1.
IAP diagram, showing primer placement. The figure shows a map of an IAP element depicting the typical features. ATTAAT is the AseI restriction site. TSDs (black) at the 5′ and 3′ end are target site duplications, succeeded and preceded by the 5′ and 3′ LTRs (long terminal repeats). The 5′ and 3′ UTRs (white) are preceded and followed by the 5′ and 3′ LTRs, respectively. ORF1 (open reading frame—grey) comprises of gag and pol genes and ORF2 houses PR, IN and RT-RH genes. Positions of RBIAPDP1, RBIAP5PA and RBIAPUNIB (IAP specific primers utilized in the IAP display system) are marked by directional arrows. Figure adapted from [4]
Recently, there have been calls to improve independent genetic validation of cell lines that are maintained in laboratories and stock centers [9]. The HeLa cell line has been known to contaminate other cell lines from various tissues and outgrow them [10]. As a result, cell line misidentification is an issue for researchers interpreting or replicating other groups’ results [11]. A genetic test for identifying HeLa cell line contamination, that utilises a HeLa-specific L1 retrotransposon insertion, was reported by Rahbari et al. [12] in 2009. As with human cell lines, verification of mouse strain identity could conceivably become a condition for publication: the development of equivalent systems for mice is timely.
The need for independent, genetic validation of mouse strain identity is imperative due to the mouse’s key role as a model system. It is well established that strain background has significant effects on the experimental responses. For example, Deutsch et al. [13] reported that epigenetic mechanisms and strain-specific genetic differences interact to influence the abilities of Flurazepam and MK801 to alleviate electrically induced seizures in mice. In 1997, Crawley et al. [14] performed a meta-analysis of responses of inbred strains to behavioral stress tests and drug treatments. They concluded that incompatible results between studies can result from differing laboratory conditions, but that strain-specific genetic differences are also significant. Independent validation of mouse strains identity would be of benefit for replication and might be required for publication, as required by some publishers for human cell lines [9].
As active transposons and insertional mutagens, IAP elements are of great interest to researchers studying the evolutionary dynamics of retrotransposons [15]. However, their recent activity in inbred mouse strains also enables these elements to be used as strain-specific genetic markers. IAP profiles vary greatly between inbred mouse strains [16], and individual insertions are fixed in a homozygous state by inbreeding. A method to rapidly identify insertions that are unique to particular strains (other than by whole genome sequencing) is a necessary first step in exploiting the potential of these insertions as markers.
The IAP display system described here is a PCR-mediated genome-wide transposon display system that is selective (by targeting the conserved 5′ UTR) for young, “dual” LTR IAP elements. It is a robust and reliable method that can be used to simultaneously access a large number of IAP insertions from all the classes. As a proof of principle, we applied this method to seven inbred strains, and characterised two insertions as strain specific, enabling PCR-based genetic identification of strains C3H and 129Sv. Thus, the system enables the acquisition of strain-specific insertions for genetic identification.
Methods and Materials
DNA Extraction
DNA for creating libraries was extracted from mouse spleen tissue, provided by our collaborators. Qiagen MaXtract kits (Qiagen, Crawley, UK) were used with a modified protocol, as follows. 1/4th of a mouse spleen was incubated overnight (14–16 h) at 55°C in 1 ml of lysis solution A (100 mM NaCl, 25 mM EDTA, 20 mM Tris-HCl pH 8.0) and 1 ml of solution B (1% SDS, 12.5 mM EDTA, 10 mM Tris-HCl pH 8.0) to which 30 μl of Proteinase K solution (25 mg/ml) was added in a 15 ml Qiagen MaXtract tube. DNA was extracted from the lysed digested tissue by standard phenol/chloroform extraction and ethanol precipitation procedures.
Mouse Strains
Seven inbred mouse strains were utilised, and were assigned numerical and shortened identifiers as follows (full strain identifier is bracketed): (1) 129SV [129S2/Sv Hsd2], (2) A/J [A/J cla Hsd], (3) BALB/C [Balb/C Ola Hsd], (4) CBA [CBA/Ca Ola Hsd], (5) C3H [C3H/HeN Hsd], (6) C57 [C57BL/6J Ola Hsd], (7) DBA [DBA/1 Ola Hsd]. All the mice were purchased from Harlan Laboratories, UK.
IAP Display
A schematic representation of the IAP display system is shown in Fig. 2. The method, derived from that of Badge et al. [17], comprises the following steps:
Fig. 2.
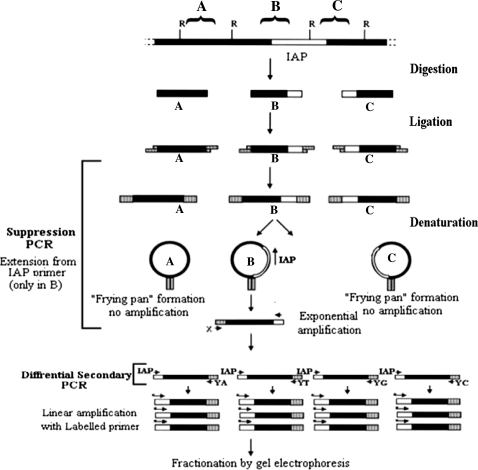
IAP display system schematic. The flow chart depicts the stages of the IAP display system. Digestion of genomic DNA and ligation to a GC-rich oligonucleotide linker create a library of fragments flanked by defined sequences (A–C) (top). The denatured single-stranded DNA can self-anneal within the common linker sequences to form a frying pan-shaped intermediate that is inefficiently amplified (A and C), whereas IAP-specific primer can anneal within the frying pan (B), leading to extension products containing a single linker, which subsequently can undergo exponential amplification (bottom). Black bars represent genomic DNA sequence, white bars represent IAP sequence, and speckled bars represent linker sequences. Restriction sites (R) that are compatible with the linker and the IAP specific (IAP) and linker-specific (X and Y-A/T/G/C) primers are shown. Figure adapted from [17]
Library Construction
600 ng of genomic DNA was digested to completion with 24 units of AseI (NEB, Hitchin, UK) in the manufacturer’s recommended buffer at 37°C, overnight A control reaction containing C57BL/6J DNA, but lacking AseI was also setup, hereafter referred to as the “enzyme absent” control. After digestion, reactions were heated to 65°C for 20 min and snap cooled on ice to inactivate the restriction enzyme. Prior to setting up the ligation reaction, linker oligonucleotides (Sigma–Aldrich, Dorset, UK) were freshly annealed by mixing equal volumes of 20 μM RBMSL2 and RBD3, heating to 65°C for 10 min, and then cooling to room temperature over 30 min. 100 ng of the digested DNA was ligated to a 40-fold molar excess of the annealed suppression linker (2.7 μl of 10 μM annealed linker for AseI libraries) with 4 Weiss units T4 DNA ligase (Promega, Southhampton, UK) in 1× Ligase Buffer (Invitrogen, Paisley, UK) overnight (~16 h) at 15°C, in a final volume of 20 μl. All the ligation reactions were setup in duplicate, including the “enzyme absent” control. Two additional control reactions containing all other components but lacking ligase (“ligase absent”) or annealed linker (“linker absent”) were also setup at this stage. After ligation the reaction was incubated at 70°C for 10 min to inactivate the ligase. Excess linkers and short DNA fragments (i.e., <100 bp) were removed with the Qiaquick PCR purification system (Qiagen, Crawley, UK), following the manufacturers protocol, but eluting the DNA in 30 μl 5 mM Tris-HCl pH 7.5. In our hands the purification is generally ~80% efficient, resulting in a purified library containing approximately 2.7 ng/μl of genomic DNA. Libraries are sensitive to freeze/thaw, and so were aliquoted and stored frozen at −80°C. All the oligonucleotide sequences are listed in Table 1.
Table 1.
Oligonucleotides utilised
| Name | Sequence (5′-3′) | Comments |
|---|---|---|
| RBIAP281A | CTGGCCAGCCTCAACTCAGA | 281 IAP genotyping primer |
| RBIAP281B | CCACCACAGTGGGAAAGCTG | 281 IAP genotyping primer |
| RRIAP674B | TTCATTCTCCATTGGATGGACT | 016 IAP genotyping primer |
| RR674C | CTCTCCTGCCAGATGAGCTTC | 016 IAP genotyping primer |
| ARIAPSV3GC | TGTGGGCGCGGCTCCCAACATTTAACAAATG | SV3G-IAP Junction genotyping primer |
| ARIAPSV3GA | CCAGCACCTTCCCATTAGGC | SV3G IAP genotyping primer |
| ARIAPSV3GB | CCGCCATCAGGAAACATCAA | SV3G IAP genotyping primer |
| RBIAP5PA2 | GCAGATTATTTGTTTACCACT | IAP 5′ LTR primer |
| RBIAPDP1 | GTCAGCGCCATCTTGTAACG | IAP display primer |
| RBIAPUNIB | GCAGTTCTGGTTCTGGAATG | IAP 5′ UTR primer |
| RRYATA | GGCGCGTAGCATAGAACGTAATA | Nested linker primer |
| RRYATT | GGCGCGTAGCATAGAACGTAATT | Nested linker primer |
| RRYATG | GCGCGTAGCATAGAACGTAATG | Nested linker primer |
| RRYATC | GCGCGTAGCATAGAACGTAATC | Nested linker primer |
| RBX4 | GTGGCGGCCAGTATTC | Linker primer |
| RBMSL2 | GTGGCGGCCAGTATTCGTAGGAGGGCGCGTAGCATAGAACG | Linker oligonucleotide |
| RBD3 | TACGTTCTATGCTAC | “Dummy” linker oligonucleotide |
The table lists the name, sequence and comments concerning the function of the oligonucleotide employed in the IAP display system and IAP insertion genotyping
Primary Suppression PCR
1 μl of ligated genomic DNA was amplified in 15 μl PCR reactions containing 1× PCR buffer (45 mM Tris-HCl pH 8.8, 11 mM NH4SO4, 0.9 mM MgCl2, 6.7 mM β-mercaptoethanol, 113 μg/ml BSA (Ambion, Huntingdon UK) 1 mM dNTPs),1.25 μM RBX4, 1.25 μM RBIAPUNIB and 0.05 units/μl Taq DNA polymerase (ABgene, Epsom, UK). Reactions were cycled in a MJ DNA tetrad PCT250 (MJ Research/Biorad, Hercules, CA) using the following conditions: 96°C-30 s; 30× [96°C-30 s; 64°C-30 s; 72°C-2 min]; 72°C-10 min. All the controls from the previous steps were carried forward and a “primary PCR DNA −ve control”, lacking input DNA, was added.
Secondary PCR
Primary suppression PCR reactions were diluted 1:10 in 5 mM Tris-HCl pH 7.5 and 1 μl diluted PCR reaction was added to 9 μl secondary PCR reactions containing 1× PCR buffer (45 mM Tris-HCl pH 8.8, 11 mM NH4SO4, 0.9 mM MgCl2, 6.7 mM β-mercaptoethanol, 113 μg/ml BSA (Ambion, Huntingdon UK), 1.1 mM dNTPs), 0.625 mM RRYATA/T/G/C, 0.625 mM RBIAP5PA2, 0.05 units/μl Taq DNA polymerase (ABgene, Epsom, UK) Reactions were cycled in a MJ DNA tetrad PCT250 (MJ Research/Biorad, Hercules, CA) using the following conditions: 96°C-30 s; 30× [96°C-30 s; 64°C-30 s; 72°C-2 min]; 72°C-10 min. All the controls from the previous steps were carried forward and a “secondary PCR DNA −ve control”, lacking input DNA, was added for each of the differential primer sets (A, T, G and C).
Display PCR
1 μl of secondary PCR reaction was added to 9 μl display PCR reactions containing 1× PCR buffer C (45 mM Tris-HCl pH 8.8, 11 mM NH4SO4, 0.9 mM MgCl2, 6.7 mM β-mercaptoethanol, 113 μg/ml BSA (Ambion, Huntingdon UK), 0.25 mM dNTPs,), 0.9–1.9 pmol of γ32P labelled RBIAPDP1 and 0.02 units/μl Taq DNA polymerase (ABgene, Epsom, UK). Reactions were cycled in a Perkin Elmer (Cambridge, UK) Cetus GeneAmp 9600 PCR machine using the following conditions: 96°C-2 min; 30× [96°C-30 s; 64°C-30 s; 72°C-30 s]; 72°C-10 min. All the controls from previous steps were carried forward in duplicate, and duplicate “display PCR DNA −ve controls” added. All the strains were represented four times in each of the differential primer sets (G, A, T and C) on the display gel. All the controls were chosen from one differential set and duplicated to accommodate all the samples on the display gel.
Fractionation and Recovery
The display PCR products were fractionated on 6% polyacrylamide, 50% urea, denaturing gels (Gel mix: 50 g Urea, 12 ml Long Ranger 50% concentrate (Lonza, Rockland, USA), 5 ml 20× Glycerol Tolerant Gel Buffer (USB, Cleveland, USA) and water to 100 ml). Dried gels were autoradiographed overnight. Gel fragments of interest were excised, using the autoradiograph for registration, and frozen overnight in 5 mM Tris-HCl pH7.5. 1 μl of the thawed mixture was re-amplified in a 9 μl PCR reaction containing 1× PCR buffer (45 mM Tris-HCl pH 8.8, 11 mM NH4SO4, 0.9 mM MgCl2, 6.7 mM β-mercaptoethanol, 113 μg/ml BSA, 1.1 mM dNTPs), 0.625 mM RRYATA/T/G/C, 0.625 mM RBIAP5PA2, 0.05 units/μl Taq DNA polymerase (ABgene, Epsom, UK). Reactions were cycled in a MJ DNA tetrad PCT250 (MJ Research/Biorad, Hercules, CA) using the following conditions: 96°C-30 s; 30× [96°C-30 s; 64°C-30s; 72°C-2 min]; 72°C-10 min. The amplified products were fractionated on 1% agarose gels (Seakem LE agarose, Cambrex, Wiesbaden, Germany). Excised PCR products were purified using the Qiagen Minelute system (Qiagen, Crawley, UK) following the manufacturer’s protocol, but eluting the DNA in 10 μl of 5 mM Tris-HCl pH 7.5. Purified PCR products were directly sequenced using ABI BigDye Ver.3 Ready Reactions by the PNACL core DNA sequencing service (University of Leicester).
PCR Genotyping
Novel IAP insertions were genotyped across seven inbred mouse strains for presence/absence polymorphism. Upon identification of the insertion point of novel insertion, flanking DNA sequence was downloaded from UCSC genome browser [18, 19] and Repeatmasked [20] prior to the design of PCR primers using Primer 3 [21]; where possible primers were placed such that the 5′ flanking primer (upstream of the 5′ IAP LTR) was located 5′ of the restriction site to which the library linker was ligated, to enable independent verification of the ligation point.
Results
IAP Display
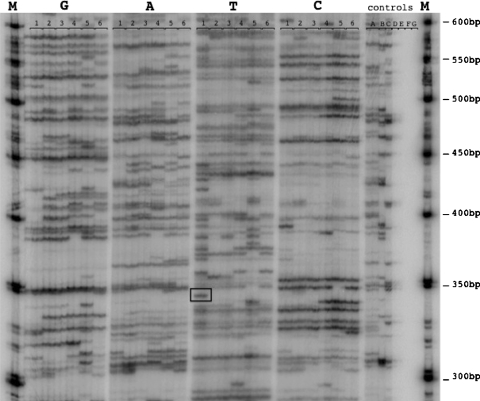
The IAP display system reported here exploits suppression PCR to selectively visualize genome-wide transposon distribution in the mouse. The genomic libraries were subjected to three successive PCRs, the first of which selects for full length IAP pro-viruses. Subsequent nested PCRs fractionate the linkered fragments by their restriction site flanking nucleotides (G, A, T or C) and the last PCR, with a radiolabelled primer, yields linear ssDNA extension products (shown schematically in Fig. 2). The radiolabelled products were fractionated on large format denaturing polyacrylamide gels, and visualised by autoradiography. A representative display gel is shown in Fig. 3. All the amplicons represent putative IAP insertions, but amplicons showing presence/absence variation or strain specificity (Fig. 3, black rectangle) were selected for further analysis. Selected amplicons were excised from the gel using the autoradiograph as a guide. Recovered amplicons were re-amplified, and subsequently characterised by sequencing.
Fig. 3.
IAP display gel. A representative IAP display gel showing differential display patterns relative to the secondary linker primer utilised (RRYATA/T/G/C). Six mouse strains (1 DBA, 2 C3H, 3 CBA, 4 A/J, 5 129Sv, 6 BALB/) were analysed using duplicate display reactions. Control Lanes: A duplicated “enzyme absent” control, B linker absent control, C ligase absent control, D primary PCR DNA −ve control, E secondary PCR −ve control, F and G display PCR DNA −ve controls. Lanes marked M contain radiolabelled 50 bp ladder molecular weight marker (NEB). A putative strain-specific IAP insertion is boxed
Sequences of amplicons were analysed using BLAT searches at the UCSC genome browser website, and the flanking genomic DNA sequences were used to design PCR-based genotyping assays for IAP insertions. All the acquired IAP insertions demonstrated homozygous presence/absence dimorphism as expected, since the strains are inbred. The polymorphic status (presence in multiple strains) or strain-specificity of IAP insertions was verified by genotyping assays.
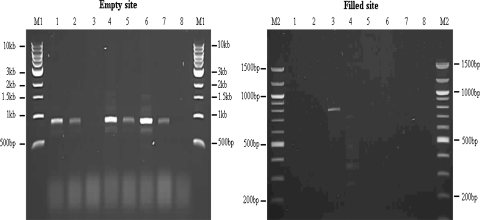
PCR genotyping assays were designed using two approaches. The first was a 2 reaction, 3 primer PCR approach. Separate reactions were carried out for the IAP absent (empty) site (5′ flanking genomic primer to 3′ flanking genomic primer) and the IAP present (filled) site (5′ flanking genomic primer to 5′ IAP LTR primer—RBIAPUNIB or a long range PCR using the 5′ and 3′ flanking genomic primers). A map showing the placement of primers is depicted in Fig. 4a. The second approach utilises a 1 reaction, 3 primer system where empty site primers (5′ flanking genomic primer to 3′ flanking genomic primer) were combined with an IAP LTR/genomic flank junction specific primer enabling simultaneous amplification of the differently sized amplicons, as illustrated in Fig. 6a.
Fig. 4.
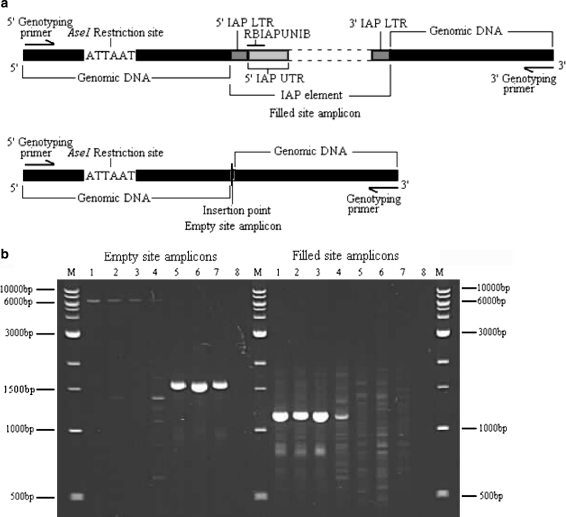
a Generic IAP map with positions of 3-primer PCR genotyping primers. Generic map of an IAP depicting the genotyping primers positions and the filled and empty site amplicons. The black boxes represent the genomic DNA flanking the IAP insertion. The brown boxes depict the 5′ and 3′ IAP LTRs, the dark grey box represents the 5′ IAP UTR. RBIAPUNIB is a 5′ IAP UTR-specific oligonucleotide. Directional arrows at either end represent the locus specific primers. b Locus 281 genotyping. Agarose gel showing the products amplified using primer combinations of RIAP281A/RBIAP281B (empty site) and RBIAP281A/RBIAPUNIB (filled site). Lanes Labelled 1–7 represent different mouse strains. 1 A/J, 2 Balb/C, 3 C3H, 4 C57, 5 CBA, 6 DBA, and 7 129Sv. Lanes labelled 8 represent the −ve control and lanes marked M contain 1 kb DNA ladder (NEB)
Fig. 6.
a Generic IAP map with positions of junction PCR genotyping primers. The black boxes represent the genomic DNA flanking the IAP insertion. The dark grey boxes depict the 5′ and 3′ IAP LTRs, the light grey box represents the 5′ IAP UTR. Junction primer bridges the flanking genomic DNA and IAP 5′ LTR DNA through the IAP insertion point. Directional arrows at either end represent the locus specific primers. b Locus SV3G genotyping. Agarose gel image showing fractionation of SV3G junction primer PCR amplicons. Lanes 1–7 represent different inbred strains: 1 A/J, 2 Balb/C, 3 CBA, 4 C3H, 5 C57, 6 DBA, 7 129Sv. Lane marked 8 is the negative hood control. Lanes marked M1 and M2 represent 100 bp and 1 kb molecular markers (NEB), respectively. Empty site amplicons, amplified by ARIAPSV3GA/ARIAPSV3GB, are approximately 1,200 bp in size and filled site amplicon, amplified by ARIAPSV3GA/ARIAPSV3GC, is approximately 500 bp in size
Polymorphic Insertions
One polymorphic insertion acquired from strain BALB/C (Insertion 281) was genotyped using the two reaction approach. Amplification across the full-length of the IAP element was carried out using primers RBIAP281A and RBIAP281B. Strains with an IAP insertion at locus 281 yielded a filled site product of ~6.2 kb and those without an insertion yielded an empty site product of ~1.6 kb (Fig. 4b, left panel). The filled site amplification reaction, using IAP-specific primer RBIAPUNIB with locus-specific primer RBIAP281A, yielded a product of ~1.2 kb for strains that were homozygous present for insertion 281, and no product for strains that were homozygous absent (Fig. 4b, right panel). Strains A/J, BALB/C, C3H were determined to be homozygous present for the insertion at locus 281 and strains CBA, DBA and 129Sv were shown to be homozygous absent. Strain C57 (Lane 4, right panel, Fig. 4b) gave consistent results with filled site amplification, strongly suggesting it is homozygous present for this insertion, but consistently showed an additional smaller amplicon in the empty site amplifications.
Strain-Specific Insertions
Insertion 674 was genotyped using the 2 primer approach and found to be strain specific. Insertion locus 674 was acquired from strain C3H and genotyped using locus-specific flanking genomic primers (RRIAP674C and RRIAP674B) and IAP-specific primer (RBIAPDP1). See Fig. 4a for the primer placement. Empty site reactions were carried out using primer RRIAP674C and RRIAP674B, generating the predicted ~900 bp amplicon. Filled site reactions (RRIAP674C and RBIAPDP1) yield an expected product size of ~680 bp. Only strain C3H was found to be homozygous for an IAP insertion at locus 674, as shown in Fig. 5b. As such, insertion 674 could be used for genetic identification of the C3H strain.
Fig. 5.
Locus 674 genotyping. Agarose gels show the fractionation of empty site and filled site amplification products. Lanes marked 1–7 represent the seven strains. Lane 1 A/J, lane 2 Balb/C, lane 3 C3H, lane 4 C57, lane 5 CBA, lane 6 DBA and lane 7 129Sv. Lane 8 represents the hood negative control. Lanes marked M1 indicate the 1 kb DNA ladder (NEB) and lanes marked M2 contain 100 bp ladder (NEB)
One Reaction Genotyping
To simplify the genotyping of IAP insertions, junction primers were designed such that they bridged the 5′ LTR of the IAP element and the 5′ flanking genomic DNA at the insertion point. Empty and filled site amplifications were carried out simultaneously in the same reaction using 3 primers (5′ and 3′ flanking genomic primers and the IAP junction primer). Strains lacking an insertion at the locus yielded only an empty site product, but strains with the insertion yielded a smaller product amplified between the 5′ flanking genomic DNA primer and the junction primer. A map depicting the positions of primers in these assays is shown in Fig. 6a. As illustrated in Fig. 6b this approach was utilised to genotype an insertion known as SV3g. This insertion was isolated from strain 129Sv and found to be strain specific (Fig. 6b). Strains lacking the insertion yielded a ~1.2 kb product, and strains with an insertion yield a product of ~500 bp. The smaller (filled site) fragment is only amplified from strain 129Sv (Fig. 6b).
Discussion
Here we demonstrate a reliable molecular genomic technique to locate IAP insertions that differ between individual mice, in this case from different inbred strains. In this context we explored the technique’s utility for the discovery of strain-specific genetic markers and verified, by independent PCR assays, that such IAP insertions could be used to discriminate between particular strains. Using the IAP display system, we isolated 21 IAP insertions of which nine were genotyped across seven inbred strains and found two strain-specific insertions.
The IAP display system described here is a suppression PCR-mediated global transposon display methodology. As IAP retrotransposons are active in the mouse genome (being responsible for 10–12% of spontaneous mutations [2]), we expect that the diversity of IAP display patterns between strains reflects that of the original population with the caveat that many heterozygous insertions will have been fixed and many will have been lost by strong drift in inbred populations. Novel insertions that occur after inbred strain establishment will likely be rapidly fixed in the homozygous state. As a result inbreeding can create stable, but diverse, IAP profiles for different inbred mouse strains. One advantage of our display system is that it facilitates the direct identification of genetic markers which can be used for a number of purposes. While strain-specific IAP insertions can be used to unequivocally identify individual mice or cell lines from particular strains, polymorphic insertions present in a number of strains can likewise be used for exclusion purposes (i.e., to demonstrate a mouse or cell line is NOT derived from a particular strain.) This can be particularly useful in animal husbandry within core animal housing facilities which maintain multiple inbred strains and laboratories which utilise cell lines from multiple strains. Genetic markers offer a simple way to verify written documentation and serve as a proxy if this is unavailable. There are other DNA variations such as microsatellite repeats and single nucleotide polymorphisms (SNPs) that could be used as genetic markers in a similar fashion. However, IAP elements have three properties that make them excellent for this application: they are intrinsically unlikely to revert precisely, resulting in identity by descent (as opposed to identity by state for microsatellites), their ancestral state is known (i.e., absence of the insertion) in contrast to SNPs, and straight forward PCR product presence/absence genotyping. The proposed IAP-based identification system can provide a genetic basis for strain identification. This can be used to locally validate the identity of newly acquired strains or newly generated cell lines. It can also be utilised for strain verification purposes where the provenance of the strain is in question. Using the IAP display system, we isolated 21 IAP insertions, of these nine were genotyped in seven inbred strains and we found two strain-specific insertions.
An alternative approach for identifying strain-specific IAP insertions involves comparing the genome sequences of inbred strains. Trace sequences of four inbred strains (129S1, 129X1, A/J, DBA) mapped to the genome assembly of the C57BL/6J strain (mm9) have been annotated with respect to insertion/deletion polymorphism and are available via MouseIndelDB: http://variation.osu.edu/mouse_indel/mm9/index.html [22]. However, the genomes of other widely used strains are yet to be sequenced and so the discovery of strain-specific insertions is dependent upon progress in this area. In addition, such bioinformatic approaches may be plagued by false positives (e.g., fixed insertions excluded during assembly but identified by trace analysis) and false negatives (traces containing genomic/IAP sequence junctions that cannot be mapped reliably to the assembly). As a result, it is highly probable that a proportion of strain-specific IAP insertions identified in silico may not be validated. Our method, by enabling direct selection of insertions with appropriate properties (presence/absence in particular strains or strain specificity) enables rapid marker system development with high validation rates. In this pilot study, of the nine loci genotyped, all were confirmed as showing presence/absence polymorphism.
Apart from being a potential genetic marker, IAP retrotransposons can also be utilized to conduct epigenetic studies. The effects of the IAP promoter on genes in its vicinity are well documented. A study by Perry et al. [23] identified five alleles of the agouti gene, with each allele characterised by an IAP insertion at varying distances upstream of the agouti locus. This suggests that the IAP 5′ LTR promoter’s exerts an epigenetic influence over transcriptional activity in its vicinity. Another study by Horie et al. [24] in 2007, established differential epigenetic suppression of IAP elements across the genome by measuring expression levels of chimeric IAP elements with GFP cassettes. Cytosine methylation plays a critical role in suppressing IAP retrotransposition [25]. High levels of DNA methyl transferase-1 (Dnmt-1) enzyme are maintained during embryogenesis, appearing to keep IAP retrotransposition in check [26]. The IAP display system presented here can be modified to study methylation at many IAP loci simultaneously because the 5′ LTR sequences amplified include CpG dinucleotides, whose methylation status can be queried using differential restriction enzyme digests (using the isoschizomers HpaII and MspI).
The IAP display system could also be used to identify de novo insertions via analysis of clonal cell lines or through pedigree analysis to potentially determine the transposition frequency of IAP elements. IAP insertions are effective mutagens and can cause gene inactivation or, occasionally, altered transcription of a gene [2]. Although IAP elements are implicated in approximately 10–12% of spontaneous mutations in the mouse genome [2], the majority of these are visible mutations and so suffer from a strong acquisition bias. An IAP display system could be used as an objective tool to monitor the rate of spontaneous mutations at the DNA level caused by the activity of IAP elements, informing on these elements’ biology and evolution.
Acknowledgments
We are indebted to Dr Ruth Barber for supplying mouse DNA. This research was funded by a Wellcome Trust Project Grant (075163/Z/04/Z) to R.M.B and Prof. Sir Alec Jeffreys, FRS.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Kuff EL, Lueders KK. The intracisternal A-particle gene family: Structure and functional aspects. Advances in Cancer Research. 1988;51:183–276. doi: 10.1016/S0065-230X(08)60223-7. [DOI] [PubMed] [Google Scholar]
- 2.Maksakova IA, Romanish MT, Gagnier L, et al. Retroviral elements and their hosts: Insertional mutagenesis in the mouse germ line. PLoS Genetics. 2006;2(1):e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen-Ong GL, Cole MD. Amplification of a specific set of intracisternal A-particle genes in a mouse plasmacytoma. Journal of Virology. 1984;49(1):171–177. doi: 10.1128/jvi.49.1.171-177.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivics Z, Izsvák Z. A whole lotta jumpin’ goin’ on: New transposon tools for vertebrate functional genomics. Trends in Genetics. 2005;21(1):8–11. doi: 10.1016/j.tig.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Dupressoir A, Heidmann T. Expression of intracisternal A-particle retrotransposons in primary tumors of oncogene-expressing transgenic mice. Oncogene. 1997;14(24):2951–2958. doi: 10.1038/sj.onc.1201148. [DOI] [PubMed] [Google Scholar]
- 6.Ishihara H, Tanaka I, Wan H, Nojima K, Yoshida K. Retrotransposition of limited deletion type of intracisternal A-particle elements in the myeloid leukemia Clls of C3H/He mice. Journal of Radiation Research. 2004;45(1):25–32. doi: 10.1269/jrr.45.25. [DOI] [PubMed] [Google Scholar]
- 7.Rakyan VK, Chong S, Champ ME, et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(5):2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel WN, Stoye JP, Taylor BA, Coffin JM. A linkage map of endogenous murine leukemia proviruses. Genetics. 1990;124(2):221–236. doi: 10.1093/genetics/124.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katsnelson A. Biologists tackle cells’ identity crisis. Nature. 2010;465(7298):537. doi: 10.1038/465537a. [DOI] [PubMed] [Google Scholar]
- 10.MacLeod RA, Dirks WG, Matsuo Y, et al. Widespread intraspecies cross-contamination of human tumor cell lines arising at source. International Journal of Cancer. 1999;83(4):555–563. doi: 10.1002/(SICI)1097-0215(19991112)83:4<555::AID-IJC19>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Lucey BP, Nelson-Rees WA, Hutchins GM. Henrietta Lacks, HeLa cells, and cell culture contamination. Archives of Pathology and Laboratory Medicine. 2009;133(9):1463–1467. doi: 10.5858/133.9.1463. [DOI] [PubMed] [Google Scholar]
- 12.Rahbari R, Sheahan T, Modes V, et al. A novel L1 retrotransposon marker for HeLa cell line identification. BioTechniques. 2009;46(4):277–284. doi: 10.2144/000113089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutsch SI, Mastropaolo J, Burket JA, Rosse RB. An epigenetic intervention interacts with genetic strain differences to modulate the stress-induced reduction of flurazepam’s antiseizure efficacy in the mouse. European Neuropsychopharmacology. 2009;19(6):398–401. doi: 10.1016/j.euroneuro.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Crawley JN, Belknap JK, Collins A, et al. Behavioral phenotypes of inbred mouse strains: Implications and recommendations for molecular studies. Psychopharmacology (Berl) 1997;132(2):107–124. doi: 10.1007/s002130050327. [DOI] [PubMed] [Google Scholar]
- 15.Ribet D, Harper F, Dupressoir A, et al. An infectious progenitor for the murine IAP retrotransposon: Emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Research. 2008;18(4):597–609. doi: 10.1101/gr.073486.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoye JP, Coffin JM. Polymorphism of murine endogenous proviruses revealed by using virus class-specific oligonucleotide probes. Journal of Virology. 1988;62(1):168–175. doi: 10.1128/jvi.62.1.168-175.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badge RM, Alisch RS, Moran JV. ATLAS: A system to selectively identify human-specific L1 insertions. American Journal of Human Genetics. 2003;72(4):823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Research. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.UCSC Genome Browser Home. http://genome.ucsc.edu/. Accessed 4 June 2010.
- 20.Smit, A.F.A., Hubley, R., Green, P. (1996). RepeatMasker Home Page. http://repeatmasker.org/. Accessed 4 June 2010.
- 21.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 22.Akagi K, Stephens RM, Li J, et al. MouseIndelDB: A database integrating genomic indel polymorphisms that distinguish mouse strains. Nucleic Acids Research. 2010;38(Database issue):D600–D606. doi: 10.1093/nar/gkp1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry WL, Copeland NG, Jenkins NA. The molecular basis for dominant yellow agouti coat color mutations. Bioessays. 1994;16(10):705–707. doi: 10.1002/bies.950161002. [DOI] [PubMed] [Google Scholar]
- 24.Horie K, Saito E, Keng VW, et al. Retrotransposons influence the mouse transcriptome: Implication for the divergence of genetic traits. Genetics. 2007;176(2):815–827. doi: 10.1534/genetics.107.071647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genetics. 1998;20(2):116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 26.Gaudet F, Rideout WM, Meissner A, et al. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Molecular and Cellular Biology. 2004;24(4):1640–1648. doi: 10.1128/MCB.24.4.1640-1648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]