Abstract
The limited availability of genomic tools and data for nonmodel species impedes computational and systems biology approaches in nonmodel organisms. Here we describe the development, functional annotation, and utilization of genomic tools for the avian wildlife species Northern bobwhite (Colinus virginianus) to determine the molecular impacts of exposure to 2,6-dinitrotoluene (2,6-DNT), a field contaminant of military concern. Massively parallel pyrosequencing of a normalized multitissue library of Northern bobwhite cDNAs yielded 71,384 unique transcripts that were annotated with gene ontology (GO), pathway information, and protein domain analysis. Comparative genome analyses with model organisms revealed functional homologies in 8,825 unique Northern bobwhite genes that are orthologous to 48% of Gallus gallus protein-coding genes. Pathway analysis and GO enrichment of genes differentially expressed in livers of birds exposed for 60 days (d) to 10 and 60 mg/kg/d 2,6-DNT revealed several impacts validated by RT-qPCR including: prostaglandin pathway-mediated inflammation, increased expression of a heme synthesis pathway in response to anemia, and a shift in energy metabolism toward protein catabolism via inhibition of control points for glucose and lipid metabolic pathways, PCK1 and PPARGC1, respectively. This research effort provides the first comprehensive annotated gene library for Northern bobwhite. Transcript expression analysis provided insights into the metabolic perturbations underlying several observed toxicological phenotypes in a 2,6-DNT exposure case study. Furthermore, the systemic impact of dinitrotoluenes on liver function appears conserved across species as PPAR signaling is similarly affected in fathead minnow liver tissue after exposure to 2,4-DNT.
Keywords: toxicogenomics, next generation sequencing, gene orthologs, energy metabolism, dinitrotoluenes
the utility of genomic tools for advancing virtually all disciplines within the life sciences has been broadly documented. Although genomic tools development for ecologically relevant nonmodel species has lagged relative to model species, advancements in sequencing technology, bioinformatics processing, and gene-expression platforms have led to an increasing number of nonmodel species having deep-coverage, well-annotated transcriptomes from which high-quality genomic tools have been produced (19, 22, 45, 64, 67, 68). These tools will prove indispensable as the focus of biological research and regulatory decision frameworks continue to shift toward systems-biology approaches (29, 38, 56).
The advent of ultra-high throughput DNA sequencing techniques has greatly expanded sequencing depth and coverage (44) providing the ability to achieve robust de novo transcriptome characterization for virtually any species at relatively low cost (45, 67). In concert, in silico methods for high-throughput sequence assembly and annotation including expressed sequence tag (EST) characterization, Gene Ontology (GO) classification and assignment to functional pathways have risen to the challenge of maximizing biological information connected to the transcriptome (30, 66, 68). A number of commercial sources (e.g., Agilent Technologies, Nimblegen, Affymetrix) provide on-demand synthesis of high-density oligonucleotide arrays directly from users' annotated sequence databases yielding low-cost tools for investigation of highly integrated responses to various stimuli. Such expression-profiling tools are the engine for expanding the “universal language” with which to describe cellular responses (39).
Avian wildlife represents a diverse group of species that are critical to ecosystem function, yet few genomic tools are available to study them. Domestic chicken (Gallus gallus) represents the most robustly described avian species (11) and microarray tools developed for chicken have been shown to have diagnostic utility in avian cross-species investigations (13). However, the unique attributes of avian species extend beyond the repertoire of chicken to complex vocalizations and vocal learning which has led to a zebra finch (Taeniopygia guttata) genome sequencing project (http://www.genome.gov/11510730). The Northern bobwhite (Colinus virginianus) is important for numerous ecological (50, 52), methodological (34), and environmental management reasons (58), making it an excellent experimental avian wildlife model. The limited genomic description of Northern bobwhite consists mainly of 1,717 ESTs derived from a brain-tissue cDNA library (27) highlighting the need to greatly expand this information base.
In this work, we utilized pyrosequencing of a Northern bobwhite normalized cDNA library assembled from multiple tissues of both males and females followed by in silico annotation to robustly characterize the transcriptome. In addition to identifying 71,384 unique transcripts, comparison with the chicken genome provided 8,930 nonredundant transcript matches indicating that library provides ∼48% coverage of all chicken (G. gallus) protein-coding genes. Oligonucleotide microarrays were developed from the transcripts to assess the transcriptional responses of liver from Northern bobwhite exposed to the munitions compound 2,6-dinitrotoluene (2,6-DNT). 2,6-DNT is found as a soil and water contaminant on military facilities (49), posing potential risks to ground foraging birds including the Northern bobwhite. The Northern bobwhite microarray was used to characterize the systemic impacts of 2,6-DNT exposure and provide hypothetical mechanisms underlying the toxicological impacts of the environmental contaminant for use in ecological risk assessments.
MATERIALS AND METHODS
Tissue samples used to construct the normalized cDNA library for Northern bobwhite were collected in exposure studies conducted at the U.S. Army Center for Health Promotion and Preventative Medicine in Edgewood, MD. Exposure assays included two individual 14 day (d) exposures to the munitions constituent hexahydro-1,3,5-trinitro-1,3,5-triazine (35, 53), a 14d exposure to 2,6-DNT (35), and a 60d exposure to 2,6-DNT (52). In total, the RNA pool used to construct the Northern bobwhite cDNA library represents 179 tissue samples taken from 56 individual birds. Tissue types represented include brain, liver, testes, duodenum, colon, and feather pulp. All protocols were conducted consistent with good laboratory practices and were approved by the Institutional Animal Care and Use Committee at the U.S. Army Center for Health Promotion and Preventative Medicine.
Tissue Fixation and RNA Extraction
Immediately following euthanasia by CO2 asphyxia, tissue samples were fixed in RNA Later (Ambion, Austin, TX) following the manufacturer's recommendations. RNA extraction was conducted using RNeasy Mini RNA extraction kits (Qiagen, Valencia, CA). RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany) with RNA 6000 Nano LabChips RNA. Only samples with a 28s/18s ratio ≥1.4 and RNA integrity number ≥6 were used for downstream applications. The majority of RNA samples greatly exceeded these minimum requirements. The RNA compilation included 800 ng of total RNA from each of the 179 RNA samples, which was purified for poly(A) RNA using a NucleoTrap mRNA purification kit (Macherey-Nagel).
cDNA Library Construction and Normalization
The SMART PCR cDNA Synthesis Kit (Clonetech Laboratories Mountain View, CA) was utilized to reverse-transcribe 0.5 μg of the poly(A) RNA into full-length cDNAs. The cDNA library was normalized prior to sequencing to capture both high and low abundance transcripts using the Trimmer cDNA Normalization Kit (Evrogen JSC, Moscow, Russia).
cDNA Sequencing and Processing
The normalized cDNA library was sequenced by 454 Life Sciences (Branford, CT) using massively parallel pyrosequencing on a GS-FLX sequencer using a protocol to resolve 200 bp reads. Raw EST sequences and quality information are available at the Short Read Archive (SRA) division (http://www.ncbi.nlm.nih.gov/Traces/sra/) of GenBank under accession number SRA009460.5. Sequences were trimmed for flanking SMART Adapters using Codon Code Aligner (Codon Code Dedham, MA) with the sequences shorter than 50 bp discarded. Poly (A/T) tails were removed from raw EST sequences with Codon Code Aligner, resulting in 467,708 high-quality sequences out of a total 478,142 reads. Sequence assembly was performed using CAP3 (32) with the minimum percent identity (the minimum percentage of identical bases in the aligned region) ≥80% and minimum alignment score (overlap match plus counting mismatches) ≥ 30 bp. Contiguous sequences (contigs) and singlets <200 bp were removed. The resulting 71,384 sequences (35,904 contigs and 35,480 singletons) were considered putative unique genes.
Sequence Analysis, Open Reading Frame Prediction, and Domain Analysis
The homologs for the unique genes were interrogated with nonredundant database from the National Center for Biotechnology Information (NCBI) with Message Passing Interface (MPI) Blast (15) using high performance computing (http://cluster.vislab.usm.edu) on a 44-node cluster. Comparative genome analyses were performed using Refseq ID for protein, downloaded in February 2008 from http://www.ncbi.nlm.nih.gov. We derived the open reading frame (ORF) and frame direction of unique genes with the help of ESTScan (33, 48). Ortholog detection was performed with an in-house perl script to generate best basic local alignment tool (BLAST) E value and bit score pairs for chicken sequences (downloaded from NCBI, February 2008) and Northern bobwhite unique transcript dataset and further mapping between these two datasets performed in the database package MySQL (http://www.mysql.com). BLASTx orientations were preferentially used over frame directions from ESTScan in determining correct orientations for microarray probe design. For Interproscan domain analysis, the frame direction (and translated sequence) from ESTScan was used. Interproscan was used to search PROSITE, PRINTS, Pfam, ProDom, SMART, and Panther protein signatures for unique genes with significant blast hits (51).
GO and Kyoto Encyclopedia of Genes and Genomes Pathway Annotation
For the functional annotation of the Northern bobwhite orthologs, we performed the GO annotation against G. gallus annotation (downloaded from Ref. 20a, GOA Chicken 39.0, released December 15th 2008) using Web Gene Ontology Annotation Plot (WEGO) (72). The annotation derived was used to define second level GO function categories within each primary GO level (molecular function, cellular component, and biological process). The chicken and quail datasets were compared to find statistically significant relationships by Pearson χ2 test performed by WEGO on 2 × 2 matrices. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway analysis was conducted using KEGG Orthology-Based Annotation System (KOBAS) web server (71) investigating raw sequences that had a significant blast hit. Sequences with significant KEGG Orthology (KO) were compared against G. gallus as a reference organism.
Northern Bobwhite Oligonucleotide Microarray
We created a 15,000-probe microarray using a 8 × 15K custom oligonucleotide microarray platform (Agilent Technologies, Santa Clara, CA). The array was developed from a database consisting of sequences identifying putative transcript identities from BLASTx matches (E ≤ 10−5) to Northern bobwhite as well as from matches to closely related avian species including G. gallus, Numida meleagris, Coturnix japonica, Meleagris gallopavo, Callipepla gambelii, and Gallus varius to prioritize unique transcripts for inclusion on the array. A total of 8,454 nonredundant sequences with positive-frame orientations were identified and incorporated into the microarray design as described in the Supplemental Text.1 For the remaining positions available on the microarray, a total of 3,272 unique Refseq IDs from the remaining homologous sequences (nonavian species) in order of lowest E-value where E ≤ 10−5 were incorporated. Complementary sequences were created for each due to lack of confidence in frame direction yielding the 6,546 probe sequences needed to complete the microarray probe set (see below for anti-sense strand exclusion methods). The 15,000 target sequences were uploaded to eArray (Agilent Technologies) where 60-mer oligonucleotide probes were developed to represent each putative transcript on the microarray.
Microarray Experimental Design, Hybridizations, and Data Extraction
Male birds were selected for the investigation due to higher sensitivity to 2,6-DNT exposure relative to females (35). A completely randomized design was utilized to investigate differential gene expression in liver tissues among control birds and those exposed to 10 and 60 mg/kg/d doses of 2,6-DNT with each condition including four biological replicates. The Agilent One-Color Microarray Hybridization protocol (Agilent Technologies) was utilized for microarray hybridizations following manufacturer's recommendations. We utilized 1 μg of total RNA for all hybridizations. An Axon GenePix 4000B Microarray Scanner (MDS Analytical Technologies, Toronto, Canada) was used to scan microarrays at 5 μm resolution. Data were extracted from microarray images using Agilent Feature Extraction software (Agilent Technologies). Analysis of internal control spikes indicated that signal data was within the linear range of detection.
Microarray Data Analysis
Background-subtracted adjusted median signal intensities were normalized on a per-chip basis (54) with R (53a) that transforms the signal intensity by dividing signal intensity for all the genes with the mean intensity in each array. Microarray analysis was performed using a HDArray library of functions available at http://www.r-project.org/, which utilizes a Bayesian probabilistic framework-based t-test to tests for differences in gene expression (9). The normalized data were imported into HDArray using Bioconductor (9a) to measure the confidence value associated with fold change for each gene (P < 0.01, unless stated otherwise). Results of Bayesian analysis were compared against a parametric, nonpaired t-test. The t-test was conducted using GeneSpring version 7.2 (Agilent Technologies) where data were first normalized with per-chip median scaling and cross-gene error model. The t-test (P < 0.05) assumed nonequal variance and incorporated a >1.8 fold-change requirement investigating the 9,711 probes that had present flags for all four replicate samples for all conditions.
The 3,272 sense-antisense probe pairs printed on the microarray were utilized to QC the microarray analysis for cross hybridization. One probe in each probe pair represents a nonsense sequence, and our expectation was that no target should have specific binding to it. When differentially expressed genes (DEGs) were examined, 4 and 19 probe pairs were coexpressed in liver (10 and 60 mg, respectively). These nonspecific DEGs were removed from our DEG list. The number of nonspecific DEGs were limited relative to the overall number of properly functioning probe pairs, which provided confidence in the representation of unique probes on the microarray, hybridization quality, and microarray analysis.
Reverse-Transcription Quantitative Polymerase Chain Reaction
The accuracy of the microarray results was assessed by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) utilizing 35 primer sets (Supplemental Table S1) representing 34 unique genes of interest regarding the mechanistic toxicity of 2,6-DNT and one regulatory control (Northern bobwhite, 18S). Each condition included four biological replicates, identical to those used in the microarray experiment. See Supplemental Text for molecular methods. Applied Biosystems SDS 2.2 Software was utilized to resolve RT-qPCR data and the ΔΔCT (Applied Biosystems, Foster City, CA) method used to quantify results. RT-qPCR data for primer sets that did not generate a single dissociation peak were excluded from analysis. The 18S regulatory control was unaffected by 2,6-DNT treatment (data not shown) and was used as the internal normalizer for relative quantification. The 95% confidence interval was calculated around the mean relative expression for each 2,6-DNT dose. Confidence intervals that did not include unity were considered differentially expressed relative to controls.
Northern Bobwhite Database Construction and Web Interface
We developed a semiautomated pipeline for data extraction and parsing with PERL, BioPerl1.6. We incorporated the annotated unique transcripts to construct the Northern Bobwhite database with the relational database management system MySQL (5.0.45) and Apache Tomcat Web server (2.2.10), which was used as container for HTML and PHP scripts. Users can access information such as UniGene sequence, number and name of ESTs contained in contig, predicted coding region, NCBI information like BLAST E-value, gene name, Uniprot accession, PDB, and EMBL ID at http://www.quailgenomics.info/searchmetadata.php.
Public Availability of Gene Expression and Toxicology Data
Microarray and toxicological data are available at the National Institute of Environmental Health Sciences (NIEHS), Chemical Effects in Biological Systems (CEBS) database: http://cebs.niehs.nih.gov under the investigation title “Bobwhite Quail 2,6-DNT,” CEBS accession number: 011-00001-0001-000-4. Microarray data and description have been submitted to the Gene Expression Omnibus archive and can be accessed at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE18940.
RESULTS AND DISCUSSION
cDNA Sequencing, Sequence Assembly, and EST Processing
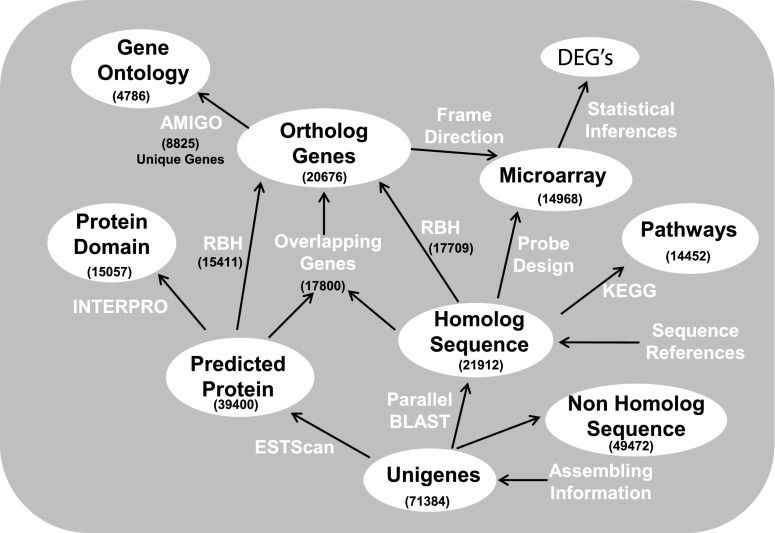
Sequencing of the normalized cDNA library yielded 114.9 megabases of sequence in 478,142 reads with an average read length of 240 bp and ∼95% of the total sequence read lengths distributed between 200 and 300 bp. We compared the unique gene transcripts derived from two unique sequence assembly approaches, CAP3 (32) and Newbler (454 Life Sciences) to determine the optimal assembly method for our sequence dataset. The results of the two sequence assembly algorithms were compared by searching each set against the NCBI nonredundant protein databases. The CAP3 assembly generated a greater number of significant matches than the Newbler assembly (21,912 vs. 16,175) at BLASTx, E ≤ 10−5 (Table 1) as well as a greater number of Refseq accession protein IDs (11,667 vs. 8,766). We also compared the gene and protein products in common among the two assemblies and found that 97% (6,158/6,349) of genes identified by Newbler were also present in the CAP3 assembly (Supplemental Table S2). The CAP3 assembly provided an additional 2,164 distinct genes compared with 191 identified by Newbler. Finally, in cross-species protein database comparisons, 80% (7,007/8,784) of Newbler protein hits were similar to CAP3 (Supplemental Table S3). The CAP3 assembly resulted in a higher number of significant matches, a greater number of distinct genes, and incorporated most of the protein database matches identified using Newbler. Given these optimal assembly results, the CAP3-assembled sequences were utilized for all the downstream analyses and annotations of ESTs summarized in Fig. 1.
Table 1.
Overview of BLASTx matches derived from contig assemblies conducted using CAP3 assembly (used for all downstream annotations and analysis) and compared to a Newbler assembly
| CAP3 Contig |
CAP3 Singleton |
Total CAP3 Assembly |
Total Newbler Assembly |
|||||
|---|---|---|---|---|---|---|---|---|
| Homology | n | % | n | % | n | % | n | % |
| 0≤ E ≤10−100 | 1,145 | 3 | 0 | 0 | 1,145 | 2 | 698 | 2 |
| 10−100< E ≤ 10−50 | 3,280 | 9 | 2 | 0 | 3,282 | 5 | 2,020 | 4 |
| 10−50 < E ≤ 10−20 | 6,641 | 18 | 4,883 | 14 | 11,524 | 16 | 7,896 | 17 |
| 10−20 < E ≤ 10−5 | 3,015 | 8 | 2,946 | 8 | 5,961 | 8 | 5,560 | 12 |
| Total significant match (E ≤ 10−5) | 14,081 | 39 | 7,831 | 22 | 21,912 | 31 | 16,174 | 35 |
| No hit or E > 10−5 | 21,823 | 61 | 27,649 | 78 | 49,472 | 69 | 30,254 | 65 |
| Total | 35,904 | 100 | 35,480 | 100 | 71,384 | 100 | 46,428 | 100 |
The results depict the distribution of expressed sequence tag (EST) data searched against nr protein database by BLASTx.
Fig. 1.
Flowchart describing the work process of the ortholog detection and annotation. DEG, differentially expressed gene; BLAST, basic local alignment tool; RBH, reciprocal BLAST hit; KEGG, Kyoto Encyclopedia of Genes and Genomes.
A total of 82,064 unique sequences were identified comprising 37,685 contigs and 44,379 singlets after vector cleaning, adapter and polyA tract removal, and CAP3 sequence assembly. Nearly 64.4% and 19.8% of the contigs were composed of greater than two and nine ESTs, respectively (Supplemental Fig. S1). Contigs and singlets having length >200 bp, totaling 71,384 unigenes, were selected for further analysis and annotation. The average length of the assembled contigs was 500 bp (min = 202 bp, max = 6,524 bp, and SD = 403). The number of transcripts having significant matches to known or postulated protein sequences, where BLAST (7) expectation scores (E) were ≤10−5, was 39.2% of contigs (14,081 out of 35,904) and 22% (7,831 out of 35,480) of singlets (Table 1).
We recognized that the 71,384 unique putative transcripts identified were an overrepresentation of the expected overall Northern bobwhite transcriptome that likely resulted from nonoverlapping contigs and singletons leading to splits in transcript sequence coverage. Furthermore, many of the nonhomologous sequences might actually represent noncoding RNA, artifacts of sequencing and assembling errors or represent protein coding genes that still remain unknown. To examine the transcripts for the presence of noncoding RNAs, the unique transcripts were compared with integrated noncoding RNA sequences from the fly, human, and other eukaryote genomes (61). A total of 2,296 unique transcripts (BLASTn E ≤ 10−5) showed similarity with noncoding RNA in eukaryotic genome databases as described in the Supplemental Text.
Comparative Sequence Analysis
We examined the degree to which genes present in Northern bobwhite are conserved across various species by comparison of the unique transcripts to genomes of several model species. Unique transcripts for Northern bobwhite were similar to 18,968 (26.57%), 16,579 (23.23%), 16,144 (22.62%), 6,100 (8.55%), 4,551 (6.38%), and 1,906 (2.67%) genes of chicken, human, mouse (Mus musculus), Drosophila melanogaster, Caenorhabditis elegans, and yeast (Saccharomyces cerevisiae), respectively at E ≤ 10−10 (Table 2). Approximately 850 genes (1.19%) were similar to all six model organisms as well as chicken and 15,498 genes (21.71%) in mammalian model organisms (human and mouse), at E < 10−10 (Table 3). Not surprisingly, many genes with functions in central metabolism and protein synthesis such as eukaryotic translation initiation factor 1, glyceraldehyde-3-phosphate dehydrogenase, ribosomal protein, methylenetetrahydrofolate dehydrogenase, actin, lysyl/glycyl/threonyl tRNA synthetase, proteasome 26S ATPase, UDP-glucose pyrophosphorylase, heat shock protein, and fumarate hydratase were found to be conserved across all organisms at E < 10−100. Overall, Northern bobwhite had the greatest protein coding gene homology to chicken when compared against genomes of model organisms (Table 2); therefore, we selected the chicken genome to further investigate phylogeny.
Table 2.
Genomic comparison of Northern bobwhite (Colinus virginianus) transcriptome against nr database for model organisms
| Organisms |
Human |
Mouse |
Fly |
C. elegans |
Yeast |
Chicken |
All Organisms |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Homology | n | % | n | % | n | % | n | % | n | % | n | % | n | % |
| E ≤ 10−100 | 735 | 1.03 | 714 | 1.00 | 155 | 0.22 | 101 | 0.14 | 46 | 0.06 | 1,090 | 1.53 | 1,145 | 1.60 |
| E ≤ 10−50 | 3,160 | 4.43 | 3,050 | 4.27 | 791 | 1.11 | 549 | 0.77 | 226 | 0.32 | 4,229 | 6.39 | 4,561 | 6.39 |
| E ≤ 10−20 | 11,938 | 16.72 | 11,506 | 16.12 | 3,260 | 4.57 | 2,299 | 3.22 | 927 | 1.30 | 15,166 | 22.90 | 16,347 | 22.90 |
| E ≤ 10−10 | 16,579 | 23.23 | 16,144 | 22.62 | 6,100 | 8.55 | 4,551 | 6.38 | 1,906 | 2.67 | 18,968 | 26.57 | 20,297 | 28.43 |
| E ≤ 10−5 | 18,386 | 25.76 | 17,964 | 25.17 | 7,912 | 11.08 | 6,180 | 8.66 | 2,770 | 3.88 | 20,734 | 29.04 | 21,912 | 30.70 |
| No match | 52,998 | 74.24 | 53,420 | 74.83 | 63,472 | 88.92 | 65,204 | 91.34 | 68,614 | 96.12 | 50,650 | 70.95 | 49,472 | 69.31 |
Table 3.
Similar genes among model organisms represented in Northern bobwhite (C. virginianus) transcriptome
| E ≤10−100 |
E ≤10−50 |
E ≤10−20 |
E ≤10−10 |
|||||
|---|---|---|---|---|---|---|---|---|
| Organism | n | % | n | % | n | % | n | % |
| Human + mouse | 697 | 0.98 | 2899 | 4.06 | 10958 | 15.35 | 15498 | 21.71 |
| Human + fly + mouse | 152 | 0.21 | 757 | 1.06 | 3087 | 4.32 | 5749 | 8.05 |
| Human + fly + mouse + chicken | 150 | 0.21 | 741 | 1.04 | 3021 | 4.23 | 5627 | 7.88 |
| Human + fly + mouse + C. elegans | 98 | 0.14 | 503 | 0.70 | 2016 | 2.82 | 3959 | 5.55 |
| Human + fly + mouse + C. elegans + yeast | 40 | 0.06 | 170 | 0.24 | 559 | 0.78 | 949 | 1.33 |
| Human + fly + mouse + C. elegans + yeast + chicken | 40 | 0.06 | 168 | 0.23 | 550 | 0.77 | 929 | 1.30 |
| Human + fly + C. elegans + yeast + mouse + A. thaliana | 35 | 0.05 | 151 | 0.21 | 501 | 0.70 | 866 | 1.21 |
| Human + fly + C. elegans + yeast + A. thaliana + mouse + chicken | 35 | 0.05 | 151 | 0.21 | 494 | 0.69 | 850 | 1.19 |
| Fly + C. elegans | 99 | 0.14 | 504 | 0.71 | 2029 | 2.84 | 3991 | 5.59 |
| Fly + C. elegans + yeast | 40 | 0.06 | 170 | 0.24 | 563 | 0.79 | 953 | 1.34 |
| Fly + C. elegans + yeast + A. thaliana | 35 | 0.05 | 151 | 0.21 | 505 | 0.71 | 870 | 1.22 |
| C. elegans + yeast | 40 | 0.06 | 170 | 0.24 | 568 | 0.80 | 969 | 1.36 |
| C. elegans + yeast + A. thaliana | 35 | 0.05 | 151 | 0.21 | 506 | 0.71 | 879 | 1.23 |
| Yeast + A. thaliana | 37 | 0.05 | 159 | 0.22 | 533 | 0.75 | 928 | 1.30 |
Assessment of Ortholog Detection
Gene ortholog detection is extensively utilized to mine the increasing amount of sequence data generated by complete or partial genome projects, which provides increasing accuracy in predicting ortholog and paralog relationships (12, 20). Prediction of protein-coding regions was established using ESTScan, which uses a hidden Markov model to identify coding regions, even if the quality is low and contains frame shifts, a common sequencing error (33). ORFs were found for 39,400 (22,432 contigs and 16,968 singlets) of the 71,384 unique transcripts with 20,660 located in the 5′ end and 18,740 in the 3′ end of the transcripts (see section “Protein domain prediction” below). The average read length for the ORFs was 352.3 bp (min = 60, max = 4,500; SD = 232.2). A total of 18,647 proteins from translated ORFs (47.3% of total) were found to have significant BLASTp matches at E ≤ 10−5 and 18,499 (99.2%) of these mapped to the putative homologs. These predicted proteins were analyzed using a high-throughput automated annotation pipeline, yielding annotation for an additional 2.3% of proteins not recognized by BLASTp (see section “High-Throughput Automated Annotation”). The remaining predicted proteins (50.4%) detected by the ESTScan model may represent false positives or homologs that might be detected by less stringent blast cutoff values.
Phylogenetic trees provide accurate, however computationally intensive, detection of orthology for putative homologs. A less computationally demanding method is reciprocal BLAST hit (RBH) analysis, an alternative method frequently used as a shortcut for ortholog detection (46). The rationale is that two genes in different genomes are considered orthologous if they match as best hits when each full genome is queried against the other (10). We performed RBH to assess ortholog detection among Northern bobwhite and chicken (G. gallus, an intraorder phylogenetic relative of Northern bobwhite) across both the putative homologs and predicted ORFs that might lead to orthologous genes (Fig. 1). The blast hits (E ≤ 10−5) from chicken-bobwhite and bobwhite-chicken were sorted on minimum E value (and maximum bit score if more than one hit with same E value existed). We found 17,709 orthologs for the putative homologs and 15,441 orthologs for the predicted proteins from translated ORFs found by ESTScan. Out of orthologs predicted by ESTScan, 14,724 (95.3%) were also present in orthologs from the putative homologs, and the remaining in each dataset either were unique or had different ortholog matches. We observed that 17,800 putative homolog-ORF pairs (where matching protein identifiers were sorted on minimum E value for BLAST hits) were complementary in 85% of comparisons to the RBH pairs. To maximize the ortholog count for the Northern bobwhite, we conjoined ortholog sets derived from each method, resulting in 20,676 nonredundant orthologous unique transcripts that represent 8,825 unique gene products (Fig. 1). The number of orthologs corresponds to ∼48% of the G. gallus transcriptome, suggesting that we have achieved coverage of approximately half of the Northern bobwhite transcriptome. The summation of these orthologs provides a broad representation of predicted molecular functions inherent in the Northern bobwhite transcriptome that we utilized to establish functional annotations as we describe in the following section.
Functional Annotation
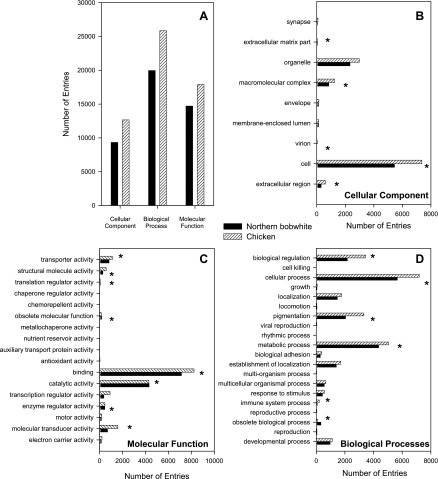
Annotation provides functional context to genes/orthologs. To functionally annotate the 8,825 Northern bobwhite orthologs, we investigated and inferred putative GO (8) annotations, finding matching annotations for 4,786 (54.2%) genes. The distribution of GO annotation within the first-level molecular function, biological process, and cellular component is shown in Fig. 2A. The number of second-level GO subcategories within each first-level GO category is represented for Northern bobwhite and chicken in Fig. 2, B–D. Statistically significant matches were observed among species for a variety of second-level GO categories including the top four most abundant categories for both for biological processes (“cellular process,” “metabolic process,” “biological regulation,” and “pigmentation”) and molecular functions (“binding,” “catalytic activity,” “molecular transducer activity,” and “transporter activity”). These results indicate similar enrichment in these ontology categories among Northern bobwhite and chicken. Annotations for these transcripts were inferred from the electronic annotation evidence level; however, with manual intervention, these orthologs are candidates that can be updated to Inferred from Sequence Orthology evidence level (http://www.geneontology.org/GO.evidence.shtml). The GO categories of these orthologs can be browsed through the GO browser implemented from amigo (28) that is available at Ref. 50a.
Fig. 2.
Gene ontology (GO) comparison between Northern bobwhite (Colinus virginianus) and domestic chicken (Gallus gallus). A: 1st-level GO information. B, C, and D: GO at the 2nd level for cellular component, biological process, and molecular function, respectively. A Pearson χ2 test was used to determine statistically significant matches (P value < 0.05) among species for GO categories (*statistically significant categories).
High-Throughput Automated Annotation
We utilized automated annotation of the Northern bobwhite transcript dataset so that we could efficiently identify protein domains and incorporate proteins within functional molecular pathways (43). We applied different approaches for annotation, pathway assignment, and protein domain prediction to develop insight into the Northern bobwhite genome, which has limited information in public repositories.
Pathway assignment.
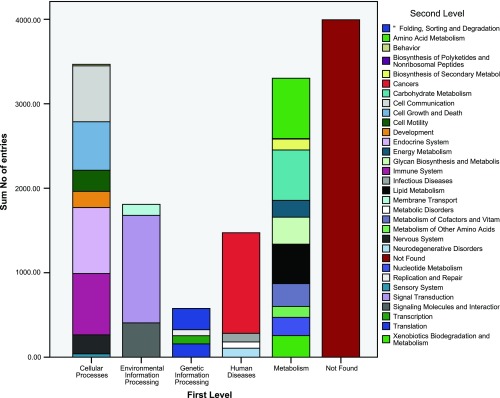
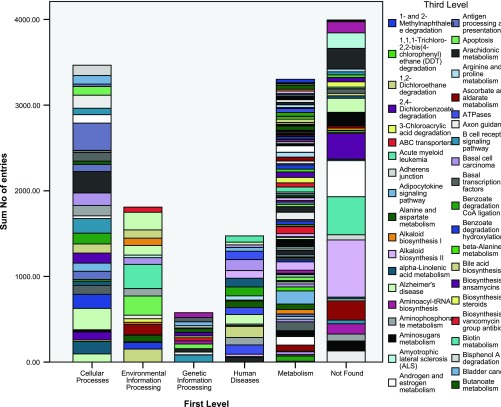
Pathway information provides an integrated representation of biochemical reactions and functional interactions compared with the gene-centric GO analysis (63, 69). We collected pathway information from the KEGG database using KO to annotate unique gene sequences (37). KO is a classification of ortholog and paralog groups based on highly confident sequence similarity scores and is directed acyclic graph with hierarchy layout in four tiers (43). We assigned metabolic pathways based on sequence similarity (E-value ≤ 10−5) to sequences with known KEGG pathways. Pathway identification of these unique transcripts were performed by statistical enrichment with Fisher exact test and Benjamini and Hochberg false discover rate correction against the G. gallus pathways, resulting in 14,452 KO entries. In the second level of the KO (Fig. 3), endocrine and immune system in cellular processes, signal transduction in environmental information processing, translation in genetic information processing, cancer in human diseases, amino acid and carbohydrate metabolism in metabolism were found to be most abundant. Among the most frequently resolved identities at the third level of organization (Fig. 4) were receptors and channels, protein kinases, MAPK signaling pathway, calcium signaling pathway, and neuroactive ligand-receptor interaction.
Fig. 3.
Distribution of KEGG orthology (KO) for Northern bobwhite (C. virginianus). Represented is the 2nd level of hierarchical organization.
Fig. 4.
Distribution of KO for Northern bobwhite (C. virginianus). Represented is the 3rd level of hierarchical organization.
Protein domain prediction.
Interproscan is used to scan for protein signature (protein families, domains, and functional sites) from the databases PROSITE, PRINTS, Pfam, and ProDom (73). The output also allows for the visual inspection of the protein signature that was performed for unique transcripts and is freely available (see section “Northern Bobwhite Database Construction and Web Interface”). We compared the 20,954 putative coding regions predicted by ESTSCAN for 21,912 unigenes (E ≤ 10−5) and found that 7,240 (33% of significant BLAST hits) were not assigned into any domain and family. The remaining 14,672 (66.95%) grouped into 2,453 domains and protein families. The most common domains were protein kinase (245), WD-40 repeats (122), ankyrin (96), zinc finger (91), and RNA recognition motif (84).
The 18,446 predicted coding regions for the remaining 49,475 nonsignificant unigenes (E > 10−5) queried resulted in 385 (.78%) unique matches in Interpro database. The most common domains were TonB box N-terminal (52), phosphopantetheine attachment site (18), and PS00014 (16) involved in posttranslation modification and β-tubulin autoregulation binding site (12).
Microarray Analysis: Effects of 2,6-DNT Exposure on Transcript Expression
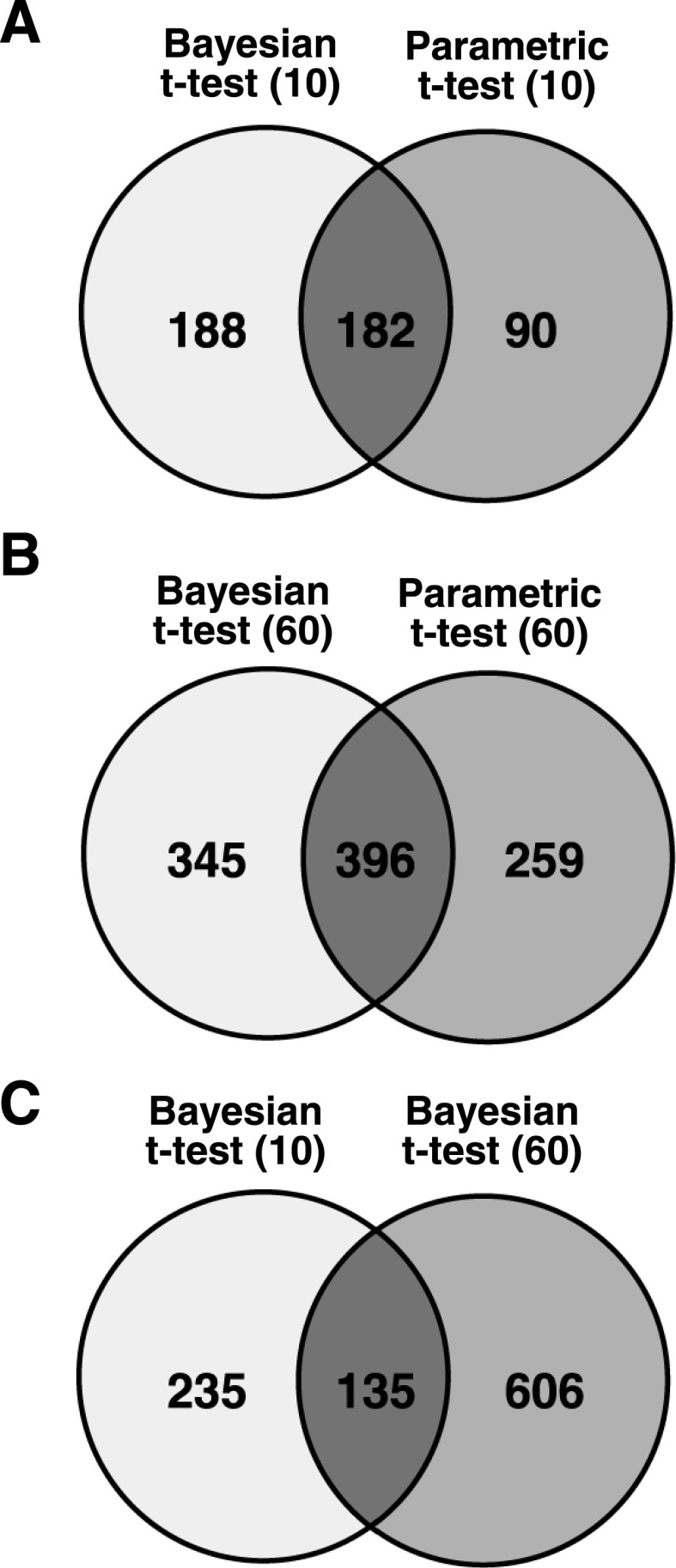
Several toxicological impacts have been observed in Northern bobwhite dosed with 2,6-DNT (52). The 10 and 60 mg/kg/d doses of 2,6-DNT were selected to represent the minimum dose at which significant impacts on toxicological endpoints were observed and a highly affective dose that elicited numerous significant impacts on toxicological endpoints approaching the threshold of mortality, respectively (52), hereafter referred to as L10 and L60. We compared the microarray results from Bayesian t-test with the results of parametric t-test and found a substantial overlap of DEG among the two analyses, including 182 and 396 common genes relative to controls for L10 and L60, respectively (Fig. 5, A and B). We proceeded by utilizing the results of Bayesian t-test for the purposes of investigating 2,6-DNT effects.
Fig. 5.
Results of microarray analyses identifying significant differential expression of transcripts relative to controls in response to a 60 day (d) exposure to 2,6-dinitrotoluene (2,6-DNT). A and B: comparison of the results of a parametric t-test (P value < 0.05 and fold change = 1.8) and Bayesian t-test (P value < 0.01) investigating differential expression in liver tissues at the 10 and 60 mg/kg/d doses, respectively. C: comparison of differentially expressed transcripts among 10 and 60 mg/kg/d 2,6-DNT doses.
The number of DEG relative to unexposed controls (0 mg/kg/d) at P < 0.01 nearly doubled with the increase in 2,6-DNT dose from 372 to 750 in the L10 and L60 treatments, respectively, (Supplemental Table S4) with 135 DEG in common among doses (Fig. 5C). KEGG pathway and GO associations were mined from the Northern bobwhite annotated library for all differentially expressed genes (Supplemental Tables S5–S7). Further enrichment of GO associations to establish biologically meaningful contexts connected to expression changes were performed with the help of the database for annotation, visualization, and integrated discovery (16, 31) and GenMAPP (14, 57).
Validation of Microarray Analysis
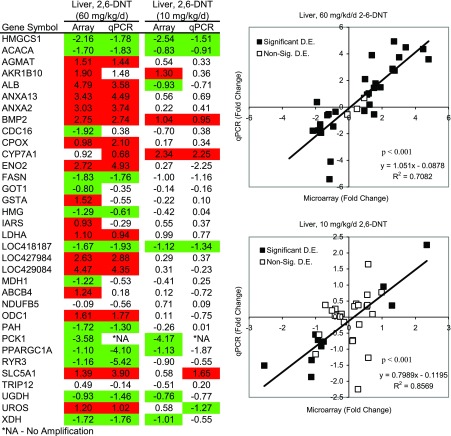
The correspondence among RT-qPCR and microarray results was generally good at 73.5% and 76.5% for the L10 and L60 treatments, respectively (Fig. 6). The correlation of fold-change results among the expression assays was highly significant (P < 0.001) and approximated a 1:1 relationship as evidenced by regression equations y = 1.05x − 0.09, R2 = 0.71 for L60 and y = 0.80x − 0.12, R2 = 0.86 for L10 (Fig. 6). These results indicate that the microarray analysis was generally accurate for liver tissue, lending confidence to the targets that were specifically assayed with RT-qPCR and to the greater microarray results set used to develop the global interpretation of 2,6-DNT toxicity in liver tissue.
Fig. 6.
Comparison of RT-qPCR and microarray results. Values represent fold change (Log2) in transcript copy number relative to controls. Red and green highlighted cells represent statistically significant increases and decreases in copy number, respectively. Regression analyses represent correlations in fold change among microarray and RT-qPCR results. Regression equations and R2 values provide correlations for significant differentially expressed transcripts. Gene names and primer sequences are provided in Supplemental Table S1.
Functional Impacts of 2,6-DNT Exposure
Exposure to 2,6-DNT caused a variety of toxicological impacts including gross-level effects and physiological effects as evidenced by alterations in blood chemistry in Northern bobwhite (52). On the basis of observable physiological effects, the lowest level at which 2,6-DNT adversely effected both male and female quail was 40 mg/kg/d, according to hematological measures, while the only statistically significant change observed at 10 mg/kg/d was reduced blood-glucose levels in males. The majority of impacts occurred in a dose-response manner, where the 10 mg/kg/d dose was affected in the same relative direction as the 60 mg/kg/d dose, however, not significantly. Here, we found significant changes in several physiological pathways at 10 and 60 mg/kg/d that were both dose responsive and dose dependent, corresponding with the observed toxicological phenotypes. Furthermore, genomic results detected 2,6-DNT impacts on gene expression (Fig. 6 and Supplemental Table S4) below the threshold at which adverse effects were manifested. We investigated pathways, GO terms and individual gene targets significantly impacted in 2,6-DNT treatments to determine potential mechanisms underlying observed gross-level effects and effects on blood chemistry and, further, explored genomics-directed observations to provide a systemic understanding of the general pharmacology of 2,6-DNT in Northern bobwhite (Table 4).
Table 4.
GO and KEGG pathway entries that best described the toxicological phenotypes observed in Northern bobwhite exposed to 2,6-DNT for 60d were used to gain toxicogenomic insights into the mechanisms underlying the toxicological effects
| Gross-level Effects |
| Edema in gastrointestinal tract |
| GenMapp: prostaglandin synthesis and regulation (L60: 4 up, 1 down) |
| Liver lesions in 11 of 12 males and 10 of 12 females in the 60 mg/kg/d treatments. Oval cell/biliary hyperplasia |
| GO:0006950: response to stress (L10: 5 up, 5 down) |
| GO:0009611: response to wounding (L60: 2 up, 5 down) |
| GO:0042060: wound healing (L60: 2 up, 3 down) |
| Brown pigmentation accumulation in the Kupffer cells (liver) in all birds @ 60 mg/kg/d and dark brown granular pigmentation accumulation in spleen |
| KEGG:gga00860: porphyrin and chlorophyll metabolism (L60: 2 up) |
| GO:0007596: blood coagulation (L60: 2up, 3 down) |
| GO:0007599: hemostasis (L60: 2 up, 3 down) |
| Diarrhea & weight loss as well as death in 3 and 4 birds in the 40 and 60 mg/kg/d treatments respectively |
| KEGG:gga00860: porphyrin and chlorophyll metabolism (L60: 2 up) |
| Blood Chemistry Results |
| Reduction in plasma glucose levels |
| KEGG:gga00010: glycolysis/gluconeogenesis (L60: 2 up, 5 down) |
| KEGG:gga00020: citrate cycle (TCA cycle) (L60: 1 up, 5 down; L10: 1 down. 1 common gene and response among doses) |
| KEGG:gga00030: pentose phosphate (L60: 1 down) |
| KEGG:gga00040: pentose and glucuronate interconversions (L60: 1 up, 1 down; L10: 1 up, 1 down). 2 common genes and responses among treatments) |
| KEGG:gga00051: fructose and mannose metabolism (L60: 1 up, 1 down; L10: 1 up, 1 down. 1 common gene and response among treatments) |
| KEGG:gga00052: galactose metabolism (L60: 1 up, 1 down; L10: 1 up. 1 common gene and response among treatments) |
| KEGG:gga00190: oxidative phosphorylation (L60: 3 up, 2 down, L10: 1 down; 1 common gene and response among treatments). |
| KEGG:gga00500: starch and sucrose metabolism (L60: 1 up, 5 down; L10: 1 down. 1 common gene and response among treatments) |
| KEGG:gga00620: pyruvate metabolism (L60: 2 up, 4 down; L10, 1 up, 2 down. 3 common gene responses and gene responses among treatments) |
| GO:0006091: generation of precursor metabolites and energy (L60: 7 up, 15 down) |
| GO:0016491: oxidoreductase activity (L60: 14 up, 25 down; L10: 9 up, 9 down) |
| Reduction in RBCs and hemoglobin concentrations |
| KEGG:gga00860: porphyrin and chlorophyll metabolism (L60: 2 up) |
| Reduction in albumin (>50%), globulin, and total protein |
| Albumin (ALB, NP_990592) (L60: up; L10 down) |
| KEGG:gga04120: ubiquitin-mediated proteolysis (L60: 4 down; L10: 3 up, 4 down) |
| GO:0006519: amino acid and derivative metabolic process (L60: 4 up, 8 down) |
| GO:0006520: amino acid metabolic process (L60: 4 up, 7 down) |
| GO:0009063: amino acid catabolic process (L60: 1 up, 3 down) |
| GO:0009308: amine metabolic process (L60: 8 up, 5 down) |
| GO:0009310: amine catabolic process (L60: 1 up, 3 down) |
| GO:0016504: protease activator activity (L10: 1 up, 1 down) |
| Reduction in aspartate amino transferase concetrations |
| Aspartate aminotransferase (also known as glutamic-oxaloacetic transaminase, GOT) (L60: 1 down) |
| Increase in uric acid |
| Aspartate aminotransferase (also known as glutamic-oxaloacetic transaminase, GOT) (L60: 1 down) |
| KEGG:gga00860: porphyrin and chlorophyll metabolism (L60: 2 up) |
| KEGG:gga00220: urea cycle and metabolism of amino groups (L60: 2 up, 1 down) |
| KEGG:gga00910: nitrogen metabolism (L 60: 2 down; L10, 1 down) |
| GO:0044270: nitrogen compound catabolic process (L60: 1 up, 3 down) |
| Reduction in Na+ and K+ ion concentration |
| GO:0030001: metal ion transport (L60: 11 up, 3 down) |
| GO:0006812: cation transport (L60: 13 up, 3 down) |
| GO:0015672: monovalent inorganic cation transport (L60: 12 up, 0 down) |
| Genomics-Directed Observations |
| 2,6-DNT metabolism |
| KEGG:gga00480: glutathione metabolism (L60: 3 up) |
| KEGG:gga00624: 1- and 2-methylnaphthalene degradation (L60: 1 up; L10; 1 up. 1 common gene and response among doses) |
| KEGG:gga00626: naphthalene and anthracene degradation (L10: 1 up). |
| KEGG:gga00980: metabolism of xenobiotics by cytochrome P450 (L60: 1 up) |
| GO:0006725: aromatic compound metabolic process (L60: 0 up, 6 down) |
| GO:0006807: nitrogen compound metabolic process (L60: 5 up, 9 down) |
| GO:0016775: phosphotransferase activity, nitrogenous group as acceptor (L60: 3 up, 1 down) |
| GO:0016840: carbon-nitrogen lyase activity (L60: 3 down) |
| Impacts on lipid metabolism |
| KEGG:gga00061: fatty acid biosynthesis (L60: 2 down; L10: 1 down. 1 common gene and response among doses) |
| KEGG:gga00071: fatty acid metabolism (L60: 2 down; L10: 1 down. 1 common gene and response among doses) |
| KEGG:gga00564: glycerophospholipid metabolism (L60: 3 up, 1 down; L10: 2 up. 2 common genes and responses among doses) |
| KEGG:gga00565: ether lipid metabolism (L60: 1 up, 2 down) |
| KEGG:gga00590: arachidonic acid metabolism (L60: 1 down) |
| KEGG:gga01040: polyunsaturated fatty acid biosynthesis (L60: 1 up, 2 down; L10: 1 up) |
Toxicogenomic assay investigated effects in liver (L) tissues of animals exposed to 2,6-dinitrotoluene (2,6-DNT) at 10 and 60 mg/kg/day (d) relative to tissue specific controls. Reference to “up” and “down” in the table refers to either increased or decreased transcript expression relative to controls. Detailed information regarding differentially expressed genes, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and Gene Ontology (GO) can be found in Supplemental Tables S4–S7.
Gross-level effects.
The high dose of 60 mg/kg/d, 2,6-DNT caused edema in the gastrointestinal (GI) tract of most birds with corresponding impacts on genes within the “prostaglandin synthesis and regulation” pathway (Table 4), which is key to the inflammatory response in mammalian models. Annexins (ANXA) are critical components of the inflammatory pathway (Supplemental Fig. S2), which are recognized to inhibit prostaglandin production in mammalian models (26). Anxa1,-2,-5, -13 were overexpressed (Fig. 6) at P value < 0.01 (Anxa7 and -8 were overexpressed at P value < 0.05) in the L60 treatment. Additionally, expression of the G protein-coupled receptor, endothelin A receptor (EDNRA), which regulates endothelin-1-mediated induction of prostaglandin E2 (40, 59), was reduced. Although prostaglandins are proinflammatory in most organs, they are recognized to have anti-inflammatory effects on GI mucosa (47). Increased expression of annexins as well as decreased expression of EDNRA can each contribute to reduced prostaglandin production, which is consistent with increased inflammation observed in the GI tract and liver of Northern bobwhite.
2,6-DNT exposure resulted in liver lesions and oval cell/biliary hyperplasia, in 11 of 12 males and 10 of 12 females in the L60 treatment group, ultimately contributing to four deaths (52). Oval cell hyperplasia is believed to occur in mammalian species as a part of a hepatic-repair process when liver injury exceeds the proliferation capacity of normal hepatocytes, and the observed biliary hyperplasia indicates that the primary site of injury involved the bile duct epithelium (25). Investigation of GO terms including “response to wounding,” “wound healing,” and “hemostasis” indicated differential expression of a number of genes (Supplemental Table S7) including greatly increased expression (4.35-fold, log2, Fig. 6) of tissue factor (LOC429084) as well as increased expression of interleukin 10 receptor beta (IL10RB) in the L60 group. Increased expression of clotting factors such as (LOC429084) is consistent with initiation of clotting at the site of lesion injury (24), while the increased expression of interleukin 10 receptor, beta (IL10RB), is an indicator of enhanced immune response against wound infection.
Brown pigmentation was observed to accumulate in quail liver Kupffer cells in all high-dose (L60) birds accompanied by dark brown granular pigmentation in spleen, diarrhea, and weight loss in the majority of birds. 2,6-DNT and related chemicals such as 2,4,6-trinitrotoluene are known to cause anemia in mammalian species via erythrolysis, which may in turn affect the primary blood conditioning organs including liver, kidney, and spleen (21, 62). Our results indicated that the gene coproporphyrinogen oxidase (CPOX) and a gene similar to uroporhyrinogen-III synthase (UROS, LOC426223), components of the porphyrin and chlorophyll metabolism pathway and an instrumental facilitator of heme biosynthesis, were overexpressed in the L60 treatment (Fig. 6 and Table 4). Increased expression of CPOX and UROS genes (Supplemental Fig. S3), if uncoupled from heme synthesis, is consistent with accumulation of porphyrin byproducts (uroporphyrinogen III, protoporphyrinogen IX) causing porphyria (18). Porphyrin byproducts are indicators of impeded heme cycling and can also result in symptoms similar to those observed in 2,6-DNT exposed quail including diarrhea, loss of sensation, low red blood cell (RBC) count, and abnormal liver function (22a, 44a). While some gross-level effects, such as weight loss arise from complex impacts on physiology, evidence from transcriptomics provides additional insight into systemic-level responses of the organism. Our gene expression results provide plausible mechanisms underlying several gross-level effects of 2,6-DNT.
Effects on blood chemistry.
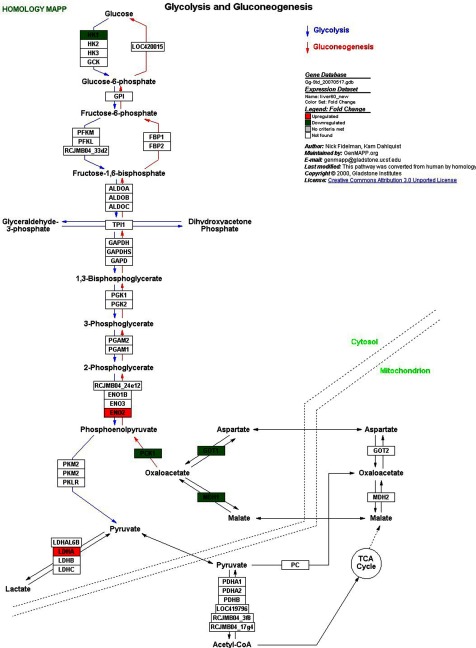
Changes in blood chemistry parameters were some of the most sensitive indicators of 2,6-DNT exposure with impacts occurring at doses as low as 10 mg/kg/day. A significant decrease was seen in plasma-glucose levels in L10 and L60 birds in absence of 2,6-DNT-related changes in feed consumption (52). Consistent with reduced glucose levels and weight loss observed in Northern bobwhite, expression of genes represented in nine pathways and two GO categories related to carbohydrate and energy metabolism were affected (Table 4) with the majority of genes having decreased expression in 2,6-DNT exposures (Supplemental Tables S5 and S7). Investigation of the glycolysis and gluconeogenesis pathway (Fig. 7) for the L60 treatment indicated impacts on genes involved in establishing equilibrium between the glycolysis and gluconeogenesis processes. Phosphoenolpyruvate carboxykinase 1 (PCK1), recognized as a major regulatory point for gluconeogenesis (23), had −2.7-fold log2 and −3.6-fold log2 decreased expression in L10 and L60 birds, respectively (Fig. 6). The concerted decreases in expression of (PCK1), aspartate aminotransferase (GOT1), and malate dehydrogenase 1 (MDH1) are consistent with a shift in equilibrium away from gluconeogenesis, therefore limiting glucose synthesis. In contrast, enolase 2 (ENO2), which utilizes the substrate formed in the PCK1-catalyzed reaction (phosphoenolpyruvate), was upregulated presumably in response to low substrate concentrations and in response to decreased plasma-glucose levels.
Fig. 7.
Effects of 2,6-DNT exposure on the glycolysis and gluconeogenesis pathway in liver tissue of Northern bobwhite dosed with 60 mg/kg/d, 2,6-DNT in a 60d exposure. Significant impacts on transcript expression relative to controls are represented by red (increased expression) and green (decreased expression) highlights.
Significant reductions in RBC concentrations, plasma albumin levels (>50% reduction), aspartate aminotransferase concentrations, total proteins, globulin concentration, and Na+ and K+ ion concentrations were also seen in L60 birds, whereas uric acid concentrations nearly doubled in L60 birds (52). As described above, the KEGG pathway “porphyrin and chlorophyll metabolism” involved in the “heme biosynthesis pathway” (Supplemental Fig. S3) was affected in the L60 treatments including over-expression of CPOX and UROS (Fig. 6, Supplemental Table S5). Increased expression of these genes is consistent with a compensatory effort to return hemoglobin and RBCs to normal levels.
A significant increase in albumin transcript copy number (ALB, 3.58-fold, log2, Fig. 6) is consistent with a response to compensate for marked reductions in plasma-albumin levels observed by Quinn et al. (52). Reductions in plasma-globulin and total proteins correspond with decreased expression of the majority of genes associated with the “ubiquitin-mediated proteolysis” pathway (Supplemental Table S5), as well as the GO categories related to amine and amino acid catabolism (Table 4, Supplemental Tables S6 and S7), indicating a potential inhibition of protein cycling initiated by 2,6-DNT exposure.
Reduced blood concentrations of aspartate aminotransferase, also known as glutamic-oxaloacetic transaminase (GOT1), at 60 mg/kg/day 2,6-DNT, corresponded with reduced GOT1 transcript expression in L60 (Fig. 6). In addition to the role of GOT1 in glycolysis and gluconeogenesis described above, it and number of additional genes are components of the metabolic pathways “nitrogen cycle,” “urea cycle and metabolism of amino acid groups,” and “porphyrin and chlorophyll metabolism,” as well as members of the GO category “nitrogen compound catabolic process,” which were impacted in the L60 treatment (Table 4, Supplemental Table S5 and S6). Additionally, genes including agmatine ureohydrolase (AGMAT) and ornithine decarboxylase 1 (ODC1), which are involved in the “urea cycle and metabolism of amino acid groups” pathway (Supplemental Table S5), had increased expression in L60 birds (Fig. 6). These overall impacts, and most specifically the increased expression of components of the “urea cycle and metabolism of amino acid groups” pathway, are consistent with elevated blood-urea concentrations observed in birds exposed to high doses of 2,6-DNT (52).
Finally, observed reductions in Na+ and K+ ion concentrations were accompanied by a predominant increase in transcript expression within the GO categories: “metal ion transport,” “cation transport,” and “monovalent inorganic cation transport” (Table 4 and Supplemental Table S7). Cation transport is fundamental to a variety of physiological processes and finding the ultimate cause of depletion is a difficult task. A potential result of depletion is manifested in the increased expression of the Na+/glucose cotransporter 1 (SLC5A1, Fig. 6) a probable response to increase absorption of glucose from dietary sources (65) to supplement already marginalized glucose supply to cells.
Genomics-directed observations.
In addition to having toxicological phenotypes guide our investigation of mechanisms of action, we utilized genomic results to identify impacts beyond the results provided in the toxicological bioassays to improve assessment of 2,6-DNT pharmacology and provide connectivity among systemic effects. Regarding 2,6-DNT pharmacology, pathways involved in phase I and phase II xenobiotic metabolism (“metabolism of xenobiotics by cytochrome P450” and “glutathione metabolism”, respectively) had increased expression in the L60 treatment (Table 4 and Supplemental Table S5). Specifically, glutathione-S transferase (GSTA) and cytochrome P450 (CYP7A1) had increased expression within these pathways (Fig. 6). TNT and related compounds (including 2,6-DNT) can induce these mechanisms (17, 36, 55), which may ultimately influence toxicity (60). Additionally, the numerous results indicating that pathways involved in nitrogen and urea metabolism (described above) are being impacted by 2,6-DNT suggest the potential for denitrification of 2,6-DNT and processing of resultant nitrogenous metabolites.
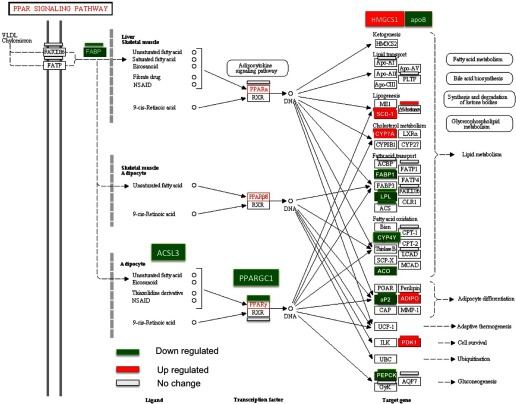
An additional observation provides a systemic link to the impacts on energy metabolism discussed above, in this case, involving perturbations caused by 2,6-DNT on transcript expression of genes and pathways involved in lipid metabolism. An isomer of 2,6-DNT, 2,4-dinitrotoluene (2,4-DNT), has been observed to impact lipid metabolism in the fish model, fathead minnow (Pimephales promelas), resulting in decreased expression of a number of genes that regulate fatty acid synthesis and causing phospholipid accumulation in the liver (70). Similarly, 2,6-DNT caused decreased expression of several genes involved in lipid metabolism in Northern bobwhite liver (L60) including fatty acid synthase (FASN), a gene similar to long-chain acyl-CoA synthetase 3 (LOC424810), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), a gene similar to delta7-sterol reductase (Loc422982), a gene similar to acyl-CoA synthetase long-chain family member (ACSL1), acetyl-CoA carboxylase alpha (ACACA), stearoyl CoA desaturase (SCD) apolipoprotein B (APOB), and acyl-CoA dehydrogenase family 11 (ACAD11) (Supplemental Table S4). In the fathead minnow model, impacts of 2,4-DNT appeared to be modulated via the peroxisome proliferative activated receptor α (PPARA1) and peroxisome proliferative activated receptor γ (PPARG1) pathways. No significant effect was observed on PPARA1 in Northern bobwhite liver; however, PPARG was downregulated (P = 0.03, −1.0 fold log2 in L10) in addition to peroxisome proliferative activated receptor γ coactivator 1 (PPARGC1A, −1.1-fold log2 in L10, −1.1-fold, log2 in L60). These impacts on components of the lipid metabolism pathway are an additional indicator of 2,6-DNT-induced perturbation of energy metabolism in Northern bobwhite, which ultimately may have forced the stress leading to many of the observed toxicological impacts (Fig. 8).
Fig. 8.
Effects of 2,6-DNT exposure on the peroxisome proliferative activated receptor (PPAR) pathway in liver tissue of Northern bobwhite dosed with 60 mg/kg/d, 2,6-DNT in a 60d exposure. Significant impacts on transcript expression relative to controls are represented by red (increased expression) and green (decreased expression) highlights.
Impacts on Energy Metabolism is Central to 2,6-DNT Toxicity
Consumption of 2,6-DNT impacted expression of genes that facilitate energy metabolism including both gluconeogenesis (Fig. 7) and lipid metabolism (Fig. 8). Gluconeogenesis is the process by which noncarbohydrate, carbon substrates are converted to glucose, the paramount short-term energy storage molecule. Marked reductions in PCK1 expression, in addition to reduced expression of GOT1 and MDH1, indicate impaired potential for glucose generation (23), which corresponds with reduced glucose levels in blood in the absence of feeding rate changes in Northern bobwhite. From the perspective of energy metabolism, lipids represent medium- to long-term storage molecules. Expression of PPARG1 and PPARGC1A, genes that represent key control points for initiation of fatty acid metabolism (Fig. 8) and mitochondrial biogenesis (41), was reduced, as was the case for a variety of additional genes involved in lipid metabolic pathways. Reduced expression of these genes is indicative of impairment of lipid catabolism, which can ultimately reduce availability of lipid as a substrate for production of cellular energy in times of energetic debt. A study investigating PPARA1 knockout mice indicated increased concentrations of long-chain fatty acids in plasma in addition to impairments in citric acid cycle flux, enhanced urea cycle activity, and increased amino acid catabolism (42). If PPAR impacts are conserved in bird species, these results indicate that reduced blood-protein levels and increased blood-urea levels may have resulted from enhanced catabolism of amino acids. Furthermore, these results suggest that 2,6-DNT-induced impairment of glucose and lipid metabolism in Northern bobwhite may have caused reduced availability of carbohydrate- and lipid-based substrates for energy generation, elevating amino acid-based biomolecules as a predominant substrate for this process. This would additionally contribute to a principle gross-level effect observed in 2,6-DNT exposure, weight loss/wasting.
Conclusions
In this paper, we have presented the entire life cycle of development of an annotated microarray from sequencing, annotation, and application to understanding sublethal impacts of 2,6-DNT dosing in Northern bobwhite. We performed comparative genome analyses with model organisms and GO annotation for 8,825 unique orthologous Northern bobwhite genes. Furthermore, we annotated this species with high-throughput annotation and transitioned this knowledge to develop a custom microarray for Northern bobwhite. Microarray analysis of liver tissue from Northern bobwhite exposed to 2,6-DNT indicated a variety of molecular impacts resulting from 2,6-DNT exposures that were consistent with overt toxicological symptoms. Key molecular-to-phenotype impacts included: prostaglandin pathway-mediated inflammation, increased expression of heme synthesis pathway in response to anemia, a shift in energy metabolism toward protein catabolism via inhibition of control points for glucose and lipid metabolic pathways, PCK1 and PPARGC1, respectively, as well as a variety of additional insights. The conservation of genomic responses among species including parallel impacts of a 2,6-DNT isomer on gene expression observed in fathead minnow (70) indicates the potential for commonality of mechanisms of action across distantly related phylogenetic relatives. Overall, our observations illustrate not only the utility of the genomic infrastructure developed for Northern bobwhite for assessment of systemic perturbations, but also the potential for generalization of genomic responses among phyla.
GRANTS
This work was supported by the U.S. Army Environmental Quality and Installations Basic Research Program and Toxicogenomics Focus Area. We also gratefully acknowledge the support of the Mississippi Functional Genomics Network (National Center for Research Resources Grant P20 RR-016476) and Engineer Research and Development Center Grant W912HZ-08-C-0032.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Glover George for assisting in establishing MPI-BLAST and use of http://cluster.vislab.usm.edu and Dr. Mehdi Pirooznia for providing the application BatchBlastExtractor available at http://mcbc.usm.edu.
Permission was granted by the Chief of Engineers to publish this information.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 7. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol 215: 403–410, 1995. [DOI] [PubMed] [Google Scholar]
- 8. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519, 2001. [DOI] [PubMed] [Google Scholar]
- 9a. Bioconductor. http://www.bioconductor.org/ 2009.
- 10. Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan YP. Predicting function: From genes to genomes and back. J Mol Biol 283: 707–725, 1998. [DOI] [PubMed] [Google Scholar]
- 11. Carre W, Wang X, Porter TE, Nys Y, Tang JBE, Morgan R, Burnside J, Aggrey SE, Simon J, Cogburn LA. Chicken functional genomics resource: sequencing and annotation of 35,407 ESTs from single and multiple tissue cDNA libraries and CAP3 assembly of a chicken gene index. Physiol Genomics 25: 514–524, 2006. [DOI] [PubMed] [Google Scholar]
- 12. Conte MG, Gaillard S, Droc G, Perin C. Phylogenomics of plant genomes: a methodology for genome-wide searches for orthologs in plants. BMC Genomics 9: 183, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crowley TM, Haring VR, Burggraaf S, Moore RJ. Application of chicken microarrays for gene expression analysis in other avian species. BMC Genomics 10: S3, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet 31: 19–20, 2002. [DOI] [PubMed] [Google Scholar]
- 15. Darling A, Carey L, Feng W. The design, implementation, and evaluation of mpiBLAST. 4th International Conference on Linux Clusters: the HPC Revolution 2003, in conjunction with ClusterWorld Conference & Expo San Jose, CA, 2003. [Google Scholar]
- 16. Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol 4: P3, 2003. [PubMed] [Google Scholar]
- 17. Ekman DR, Lorenz WW, Przybyla AE, Wolfe NL, Dean JFD. SAGE analysis of transcriptome responses in arabidopsis roots exposed to 2,4,6-trinitrotoluene1. Plant Physiol 133: 1397–1406, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elder GH. Genetic defects in the porphyrias: types and significance. Clinics Dermatol 16: 225–233, 1998. [DOI] [PubMed] [Google Scholar]
- 19. Ellegren H. Sequencing goes 454 and takes large-scale genomics into the wild. Mol Ecol 17: 1629–1631, 2008. [DOI] [PubMed] [Google Scholar]
- 20. Engelhardt BE, Jordan MI, Muratore KE, Brenner SE. Protein molecular function prediction by Bayesian phylogenomics. PLOS Comput Biol 1: e45, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a. European Bioinformatics Institute. Chicken GOA. http://www.ebi.ac.uk/ 2008.
- 21. Ferguson JW, McCain WC. Toxicological Study No. 6955-31-97-05-02, 14-day feeding study of hexahydro-1,3,5, trinitro-1,3,5-triazine (RDX) in the white-footed mouse, Peromyscus leucopus. U.S. Army Center for Health Promotion and Preventive Medicine (USACHPPM), 1999. [Google Scholar]
- 22. Garcia-Reyero N, Griffitt RJ, Liu L, Kroll KJ, Farmerie WG, Barber DS, Denslow ND. Construction of a robust microarray from a non-model species largemouth bass, Micropterus salmoides (Lacepede), using pyrosequencing technology. J Fish Biol 72: 2354–2376, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a. Genetics Home Reference. Porphyria. http://ghr.nlm.nih.gov/condition=porphyria 2009.
- 23. Gómez-Valadés AG, Méndez-Lucas A, Vidal-Alabró A, Blasco FX, Chillon M, Bartrons R, Bermúdez J, Perales JC. Pck1 Gene silencing in the liver improves glycemia control, insulin sensitivity, and dyslipidemia in db/db mice. Diabetes 57: 2199–2210, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gouault-Helimann M, Josso F. Initiation in vivo of blood coagulation. The role of white blood cells and tissue factor. Nouv Presse Med 8: 3249–3253, 1979. [PubMed] [Google Scholar]
- 25. Greaves P. Liver and pancreas. In: Histopathology of Preclinical Toxicity Studies: Interpretation and Relevance. New York: Elsevier Science, 2007. p. 457–528. [Google Scholar]
- 26. Green PG, Strausbaugh HG, Levine JD. Annexin I is a local mediator in neural-endocrine feedback control of inflammation. J Neurophysiol 80: 3120–3126, 1998. [DOI] [PubMed] [Google Scholar]
- 27. Gust KA, Pirooznia M, Quinn MJ, Jr, Johnson MS, Escalon L, Indest KJ, Guan X, Clarke J, Deng Y, Gong P, Perkins EJ. Neurotoxicogenomic investigations to assess mechanisms of action of the munitions constituents RDX and 2,6-DNT in Northern bobwhite (Colinus virginianus). Toxicol Sci 110: 168–180, 2009. [DOI] [PubMed] [Google Scholar]
- 28. Harris MA, Clark JI, Ireland A, Lomax J, Ashburner M, Collins R, Eilbeck K, Lewis S, Mungall C, Richter J, Rubin GM, Shu SQ, Blake JA, Bult CJ, Diehl AD, Dolan ME, Drabkin HJ, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Binkley G, Cherry JM, Christie KR, Costanzo MC, Dong Q, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Hong EL, Lane C, Miyasato S, Nash R, Sethuraman A, Skrzypek M, Theesfeld CL, Weng SA, Botstein D, Dolinski K, Oughtred R, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Mulder N, Chisholm R, Fey P, Gaudet P, Kibbe W, Pilcher K, Bastiani CA, Kishore R, Schwarz EM, Sternberg P, Van Auken K, Gwinn M, Hannick L, Wortman J, Aslett M, Berriman M, Wood V, Bromberg S, Foote C, Jacob H, Pasko D, Petri V, Reilly D, Seiler K, Shimoyama M, Smith J, Twigger S, Jaiswal P, Seigfried T, Collmer C, Howe D, Westerfield M. The Gene Ontology (GO) project in 2006. Nucl Acids Res 34: D322–D326, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heckmann L, Sibly RM, Connon R, Hooper HL, Hutchinson TH, Maund SJ, Hill CJ, Bouetard A, Callaghan A. Systems biology meets stress ecology: linking molecular and organismal stress responses in Daphnia magna. Genome Biol 9: R40, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu W, Yan Q, Shen DK, Liu F, Zhu ZD, Song HD, Xu XR, Wang ZJ, Rong YP, Zeng LC, Wu J, Zhang X, Wang JJ, Xu XN, Wang SY, Fu G, Zhang XL, Wang ZQ, Brindley PJ, McManus DP, Xue CL, Feng Z, Chen Z, Han ZG. Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nat Genet 35: 139–147, 2003. [DOI] [PubMed] [Google Scholar]
- 31. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols 4: 44–57, 2009. [DOI] [PubMed] [Google Scholar]
- 32. Huang XQ, Madan A. CAP3: a DNA sequence assembly program. Genome Res 9: 868–877, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequence. Proc Int Conf Intell Syst Mol Biol 138–147, 1999. [PubMed] [Google Scholar]
- 34. Johnson MS, Michie MW, Bazar MA, Gogal RM. Influence of oral 2,4-dinitrotoluene exposure to the Northern bobwhite (Colinus virginianus). Int J Toxicol 24: 265–274, 2005. [DOI] [PubMed] [Google Scholar]
- 35. Johnson MS, Quinn MJ, Bazar MA, Gust KA, Escalon BL, Perkins EJ. Subacute toxicity of oral 2,6-dinitrotoluene and 1,3,5-trinitro-1,3,5-triazine (RDX) exposure to the Northern bobwhite (Colinus virginianus). Environ Toxicol Chem 26: 1481–1487, 2007. [DOI] [PubMed] [Google Scholar]
- 36. Johnson MS, Vodela JK, Reddy G, Holladay SD. Fate and the biochemical effects of 2,4,6-trinitrotoluene exposure to tiger salamanders (Ambystoma tigrinum). Ecotoxicol Environ Saf 46: 186–191, 2000. [DOI] [PubMed] [Google Scholar]
- 37. Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucl Acids Res 34: D354–D357, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kavlock RJ, Ankley G, Blancato J, Breen M, Conolly R, Dix D, Houck K, Hubal E, Judson R, Rabinowitz J, Richard A, Setzer RW, Shah I, Villeneuve D, Weber E. Computational toxicology-a state of the science mini review. Toxicol Sci 103: 14–27, 2008. [DOI] [PubMed] [Google Scholar]
- 39. Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet J, Subramanian A, Ross KN, Reich N, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313: 1929–1935, 2006. [DOI] [PubMed] [Google Scholar]
- 40. Leis HJ, Zach D, Huber E, Windischhofer W. Prostaglandin endoperoxide synthase-2 contributes to the endothelin/sarafotoxin-induced prostaglandin E2 synthesis in mouse osteoblastic cells MC3T3–E1): evidence for a protein tyrosine kinase-signaling pathway and involvement of protein kinase C. Endocrinology 139: 1268–1277, 1998. [DOI] [PubMed] [Google Scholar]
- 41. Liang H, Ward WF. PGC-1alpha: a key regulator of energy metabolism. Adv Physiol Educ 30: 145–151, 2006. [DOI] [PubMed] [Google Scholar]
- 42. Makowski L, Noland RC, Koves TR, Xing W, Ilkayeva OR, Muehlbauer MJ, Stevens RD, Muoio DM. Metabolic profiling of PPAR-/- mice reveals defects in carnitine and amino acid homeostasis that are partially reversed by oral carnitine supplementation. FASEB J 23: 586–604, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mao XZ, Cai T, Olyarchuk JG, Wei LP. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21: 3787–3793, 2005. [DOI] [PubMed] [Google Scholar]
- 44. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a. Mayo Clinic. Porphyria. http://www.mayoclinic.com/health/ 2009.
- 45. Meyer E, Aglyamova GV, Wang S, Buchanan-Carter J, Abrego D, Colbourne JK, Willis BL, Matz MV. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics 10: 219, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moreno-Hagelsieb GLK. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24: 319–324, 2008. [DOI] [PubMed] [Google Scholar]
- 47. Morteau O. Cytokinase, cyclooxygenases and oral tolerance. In: Oral Tolerance: the Response of the Intestinal Mucosa to Dietary Antigens. New York: Springer, 2004, p. 70–71. [Google Scholar]
- 48. Nagaraj SH, Deshpande N, Gasser RB, Ranganathan S. ESTExplorer: an expressed sequence tag (EST) assembly and annotation platform. Nucl Acids Res 35: W143–W147, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pennington JC, Jenkins TF, Ampleman G, Thiboutot S, Brannon JM, Hewitt AD, Lewis J, Brochu S, Diaz E, Walsh MR, Walsh ME, Taylor S, Lynch JC, Clausen J, Ranney TA, Ramsey CA, Hayes CA, Grant CL, Collins CM, Bigl SR, Yost S, Dontsova K. Distribution and fate of energetics on DoD test and training ranges: final report. Vicksburg, MS, 2006. [Google Scholar]
- 50. Perea MT, Prados EA, Villajos AN, Prados JLA, Baudin JMG. Influence of avian reproduction ecotoxicological endpoints in the assessment of plant protection products. J Environ Sci Health B 44: 106–112, 2009. [DOI] [PubMed] [Google Scholar]
- 50a. Quail Genomics. http://quailgenomics.info 2009.
- 51. Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucl Acids Res 33: W116–W120, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Quinn MJJr, Bazar MA, McFarland CA, Perkins EJ, Gust KA, Gogal RM, Johnson MS. Effects of subchronic exposure to 2,6-dinitrotoluene in the Northern bobwhite (Colinus virginianus). Environ Toxicol Chem 26: 2202–2207, 2007. [DOI] [PubMed] [Google Scholar]
- 53. Quinn MJ, Jr, Bazar MA, McFarland CA, Perkins EJ, Gust KA, Johnson MS. Sublethal effects of subacute exposure to RDX (1,3,5-trinitro-1,3,5-triazine) in the Northern bobwhite (Colinus virginianus). Environ Toxicol Chem 28: 1266–1270, 2009. [DOI] [PubMed] [Google Scholar]
- 53a. R Project. http://cran.r-project.org/ 2009.
- 54. Rawat A, Deng YP. Novel implementation of conditional co-regulation by graph theory to derive co-expressed genes from microarray data. BMC Bioinformatics 9: S7, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reddy G, Chandra SAM, Lish JW, Qualls JWJ. Toxicity of 2,4,6-trinitrotoiuene (TNT) in hispid cotton rats (Sigmodon hispidus): hematological, biochemical, and pathological effects. Int J Toxicol 19: 169–177, 2000. [Google Scholar]
- 56. Robbens J, van der Ven K, Maras M, Blust R, De Coen W. Ecotoxicological risk assessment using DNA chips and cellular reporters. Trends Biotechnol 25: 460–466, 2007. [DOI] [PubMed] [Google Scholar]
- 57. Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics 8: 217, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sauer JR, Link WA, Nichols JD, Royle JA. Using the North American Breeding Bird Survey as a tool for conservation: a critique of BART et al. (2004). J Wildlife Manage 69: 1321–1326, 2005. [Google Scholar]
- 59. Shames DS, Minna JD, Gazdar AF. DNA methylation in health, disease, and cancer. Curr Mol Med 7: 85–102, 2007. [DOI] [PubMed] [Google Scholar]
- 60. Sims JG, Steevens JA. The role of metabolism in the toxicity of 2,4,6-trinitrotoluene and its degradation products to the aquatic amphipod Hyalella azteca. Ecotoxicol Environ Saf 70: 38–46, 2008. [DOI] [PubMed] [Google Scholar]
- 61. Szymanski M, Erdmann VA, Barciszewski J. Noncoding regulatory RNAs database. Nucl Acids Res 31: 429–431, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Talmage SC, Opresko DM, Maxwell CJ, Welsh CJE, Cretella FM, Reno PH, Daniel FB. Nitroaromatic munition compounds: environmental effects and screening values. Rev Environ Contam Toxicol 161: 1–156, 1999. [DOI] [PubMed] [Google Scholar]
- 63. Thomas PD, Mi HY, Lewis S. Ontology annotation: mapping genomic regions to biological function. Curr Opin Chem Biol 11: 4–11, 2007. [DOI] [PubMed] [Google Scholar]
- 64. Timmermans MJ, de Boer ME, Nota B, de Boer TE, Marien J, Klein-Lankhorst RM, van Straalen NM, Roelofs D. Collembase: a repository for springtail genomics and soil quality assessment. BMC Genomics 8: 341, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Turk E, Martin MG, Wright EM. Structure of the human Na+/glucose cotransporter GenSeG LTI. J Biol Chem 269: 15204–15209, 1994. [PubMed] [Google Scholar]
- 66. Udall JA, Swanson JM, Haller K, Rapp RA, Sparks ME, Hatfield J, Yu YS, Wu YR, Dowd C, Arpat AB, Sickler BA, Wilkins TA, Guo JY, Chen XY, Scheffler J, Taliercio E, Turley R, McFadden H, Payton P, Klueva N, Allen R, Zhang DS, Haigler C, Wilkerson C, Suo JF, Schulze SR, Pierce ML, Essenberg M, Kim H, Llewellyn DJ, Dennis ES, Kudrna D, Wing R, Paterson AH, Soderlund C, Wendel JF. A global assembly of cotton ESTs. Genome Res 16: 441–450, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol 17: 1636–1647, 2008. [DOI] [PubMed] [Google Scholar]
- 68. Wang J, Jemielity S, Uva P, Wurm Y, Graff J, Keller L. An annotated cDNA library and microarray for large-scale gene-expression studies in the ant Solenopsis invicta. Genome Biol 8: R9, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Werner T. Bioinformatics applications for pathway analysis of microarray data. Curr Opin Biotechnol 19: 50–54, 2008. [DOI] [PubMed] [Google Scholar]
- 70. Wintz H, Yoo LJ, Loguinov A, Wu Y, Steevens JA, Holland RD, Beger RD, Perkins EJ, Hughes O, Vulpe CD. Gene expression profiles in fathead minnow exposed to 2,4-DNT: correlation with toxicity in mammals. Toxicol Sci 94: 71–82, 2006. [DOI] [PubMed] [Google Scholar]
- 71. Wu JM, Mao XZ, Cai T, Luo JC, Wei LP. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucl Acids Res 34: W720–W724, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L, Wang J. WEGO: a web tool for plotting GO annotations. Nucl Acids Res 34: W293–W297, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zdobnov EM, Apweiler R. InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848, 2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.