Abstract
X-linked inhibitor of apoptosis protein (XIAP) is a potent inhibitor of caspases 3, 7 and 9, and mitochondrial Smac (second mitochondria-derived activator of caspase) release during apoptosis inhibits the activity of XIAP. In this study we show that cytosolic XIAP also feeds back to mitochondria to impair Smac release. We constructed a fluorescent XIAP-fusion protein by labelling NH2- and COOH-termini with Cerulean fluorescent protein (C-XIAP-C). Immunoprecipitation confirmed that C-XIAP-C retained the ability to interact with Smac and impaired extrinsically and intrinsically activated apoptosis in response to tumour necrosis factor-related apoptosis-inducing ligand/cycloheximide and staurosporine. In C-XIAP-C-expressing cells, cytochrome c release from mitochondria proceeded normally, whereas Smac release was significantly prolonged and incomplete. In addition, physiological expression of native XIAP prolonged or limited Smac release in HCT-116 colon cancer cells and primary mouse cortical neurons. The Smac-binding capacity of XIAP, but not caspase inhibition, was central for mitochondrial Smac retention, as evidenced in experiments using XIAP mutants that cannot bind to Smac or effector caspases. Similarly, the release of a Smac mutant that cannot bind to XIAP was not impaired by C-XIAP-C expression. Full Smac release could however be provoked by rapid cytosolic C-XIAP-C depletion upon digitonin-induced plasma membrane permeabilization. Our findings suggest that although mitochondria may already contain pores sufficient for cytochrome c release, elevated amounts of XIAP can selectively impair and limit the release of Smac.
Keywords: apoptosis, mitochondrial outer membrane permeabilization, XIAP, Smac, cytochrome c
A balance between cell proliferation and apoptotic cell death is of vital importance for all metazoans during both development and adulthood. Consequently, loss- or gain-of-function mutations in genes regulating apoptosis have been implicated in various proliferative and degenerative diseases. In most scenarios, apoptosis execution depends on proteases of the caspase family. During apoptosis, initiator caspase-9 and executioner caspases 3 and 7 are activated in response to cytochrome c (cyt-c) release from mitochondria into the cytosol. Activation of the executioner caspases proceeds in a rapid all-or-none manner,1 but can be inhibited if the main inhibitor of these caspases, X-linked inhibitor of apoptosis protein (XIAP), is present in concentrations above a critical threshold.2, 3 XIAP has been reported to prevent execution of apoptotic cell death in both physiological and pathophysiological scenarios. Upon cell differentiation, XIAP contributes to the high apoptosis resistance of various postmitotic cell types such as neurons, cardiomyocytes and myotubes.4, 5, 6 Similarly, XIAP expression was shown to increase with advancing tumour stages and is associated with a poor prognosis in prostate and renal cancers, as well as acute myeloid leukaemias.7, 8, 9 In contrast, XIAP deficiency in humans was associated with excessive lymphocyte apoptosis, which resulted in reduced numbers of natural killer T cells.10
XIAP comprises three baculoviral IAP repeat domains (BIR1–3), as well as a zinc-finger RING (really interesting new gene) domain, which exerts E3 ubiquitin ligase activity. The dimerization of caspase-9 monomers to yield the active caspase-9 conformation is inhibited by the BIR3 domain of XIAP through binding to the caspase-9 dimerization interface.11 Executioner caspases 3 and 7 instead are inhibited by a region preceding the BIR2 domain. This region occupies the substrate-binding groove of executioner caspases 3 and 7, but requires further interactions of the effector caspases with the BIR2 domain to stabilize this inhibition.12
The main antagonist of XIAP is the second mitochondria-derived activator of caspases (Smac). Dimeric Smac sterically and/or competitively occludes the caspase 3, 7 and 9 binding sites of XIAP and thereby can pave the way for efficient cell death execution.13, 14 Smac also binds to other IAPs, such as livin/ML-IAP, Bruce/Apollon, cIAPs 1 and 2,15, 16, 17 which interfere with caspase-mediated cell death and/or have additional roles in pro-survival and proliferation signalling. During apoptosis, Smac, similar to cyt-c, is released into the cytosol through pores formed by activated Bax or Bak proteins. Bax/Bak pores seem to be rather unspecific towards the proteins they release. Using single cell imaging, we and others have provided evidence that cyt-c, Smac and other soluble mitochondrial intermembrane space proteins are released simultaneously during apoptosis,18, 19 although Smac may be released with slightly slower kinetics.19 On the basis of the assumption that cytosolic proteins are able to enter the mitochondrial intermembrane space once Bax/Bak pores are formed, we investigated whether physiological or elevated XIAP levels can affect the kinetics or the extent of Smac release.
Results
Generation and molecular characterization of a fluorescent XIAP fusion protein
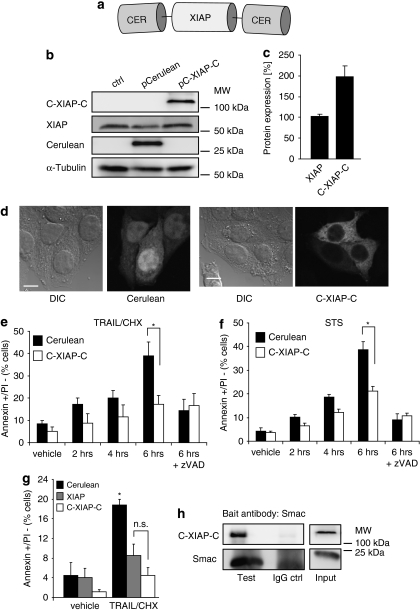
To investigate the influence of XIAP on apoptotic signalling, we generated a fluorescent XIAP fusion protein to unambiguously identify transfected cells within the analysed populations. For highly sensitive detection of XIAP expression, we fused XIAP at both NH2 and COOH termini with Cerulean fluorescent protein, a blue-shifted variant of green fluorescent protein (Figure 1a) that surpasses the more regularly used cyan fluorescent protein in fluorescence quantum efficiency and photostability.20 As XIAP can be cleaved by caspase-3, caspase-3-deficient MCF-7 breast cancer cells were initially used as a model system to obtain unperturbed information on the function of the full-length XIAP protein. Cerulean-XIAP-Cerulean (C-XIAP-C) was expressed at the expected molecular weight of approximately 109 kDa (Figure 1b) and expression levels of native XIAP were not affected by C-XIAP-C expression (Figure 1b). On average, the amounts of C-XIAP-C expressed resembled twice the amount of native XIAP (Figure 1c). Cells presenting low fluorescence intensities were therefore selected in subsequent imaging experiments to avoid excessive overexpression scenarios. C-XIAP-C was predominantly localized in the cytosol, and transfected MCF-7 cells morphologically did not differ noticeably from untransfected cells or control cells expressing Cerulean only (Figure 1d). C-XIAP-C efficiently blocked effector caspase-dependent apoptosis execution, as evidenced by reduced cell surface exposure of phosphatidylserine in response to tumour necrosis factor-related apoptosis-inducing ligand/cycloheximide (TRAIL/CHX) and broad-spectrum kinase inhibitor staurosporine (STS), drugs that target either the extrinsic or intrinsic apoptosis pathways, respectively (Figure 1e and f). Additional control experiments showed that the anti-apoptotic potential of C-XIAP-C was comparable to native XIAP (Figure 1g), and that C-XIAP-C could interact with its physiological binding partner and antagonist Smac (Figure 1h), as well as with Smac-yellow fluorescent protein (YFP) fusion proteins (Supplementary Figure 1) that were used later in this study. We therefore concluded that the C-XIAP-C probe reliably reflected the key features of native XIAP during apoptosis signalling.
Figure 1.
Characterization of a fluorescent XIAP fusion protein. (a) The generated XIAP fusion protein (C-XIAP-C). Full-length XIAP was fused at both NH2 and COOH termini with Cerulean fluorescent protein to allow for highly sensitive XIAP detection. (b) Parental MCF-7 cells as well as MCF-7 cells transiently transfected to express Cerulean or C-XIAP-C were probed with antibodies against Cerulean or XIAP. C-XIAP-C was detected at its expected molecular weight of 109 kD. α-Tubulin served as loading control. (c) Comparative quantification of XIAP and C-XIAP-C expression in MCF-7 cells. Densitometry was performed on digitally recorded 12-bit images of immunoblots probed with a XIAP antibody. Data are means+S.E.M. from n=3 detections. (d) Expression of C-XIAP-C does not affect cellular morphology. Cells were imaged 24 h after transfection. Cerulean fluorescence was found throughout the cell, whereas C-XIAP-C was predominantly found in the cytosol. Scale bar=10 μm. (e, f) C-XIAP-C inhibits apoptosis. MCF-7 cells transiently transfected to express Cerulean or C-XIAP-C were exposed to TRAIL/CHX (100 ng/ml plus 1 μg/ml) or STS (1 μM) for the indicated times and analysed by flow cytometry. Apoptosis was measured as the percentage of annexin V-FITC-positive/PI-negative cells. Addition of caspase inhibitor zVAD-fmk (50 μM) indicated that the apoptotic response to TRAIL/CHX or STS was caspase dependent. Data are shown as mean+S.D. from n=3 samples. *Significant differences (P<0.05; one-way ANOVA and subsequent Tukey's test). Experiments were repeated with similar results. (g) C-XIAP-C has an anti-apoptotic potency comparable to native XIAP. MCF-7 cells transiently transfected to express Cerulean, XIAP or C-XIAP-C were exposed to TRAIL/CHX (100 ng/ml plus 1 μg/ml) for 4 h and analysed by flow cytometry. Apoptosis was measured as the percentage of annexin V-FITC-positive/PI-negative cells. Data are shown as mean+S.D. from n=3 independent samples. *Significant induction of apoptosis (P<0.05); NS, not significant (one-way ANOVA and subsequent Tukey's test). Experiment was repeated with similar results. (h) C-XIAP-C interacts with Smac. XIAP-deficient HCT-116 cells transiently transfected to express C-XIAP-C were lysed and immunoprecipitation was performed in native cell extracts with a Smac bait antibody. After western blotting, C-XIAP-C was detected with an antibody directed against XIAP
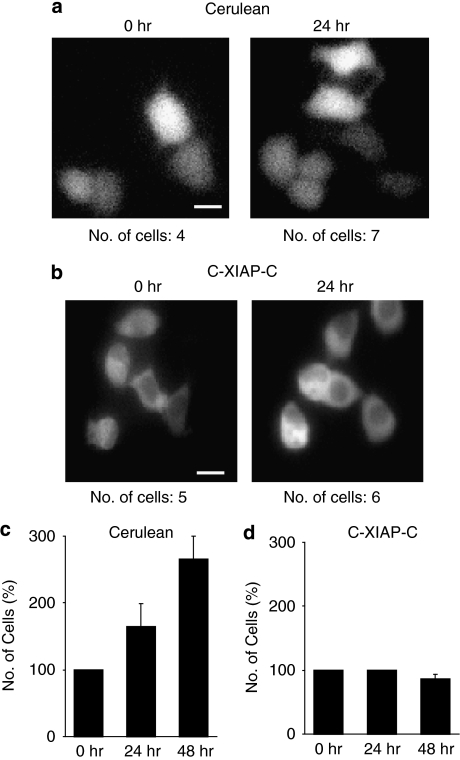
Attempts at generating proliferating MCF-7 cell clones that stably expressed C-XIAP-C or elevated amounts of native untagged XIAP failed. Indeed, when observing MCF-7 cells expressing C-XIAP-C for up to 2 days, we noticed that these cells ceased to proliferate, whereas control cells expressing Cerulean continued to divide (Figure 2a–d). These results suggested that elevated XIAP expression induced cell cycle arrest in MCF-7 breast cancer cells, a finding previously also reported for human endothelial cells.21 We therefore concluded that selection for clonal populations with stable ectopic XIAP expression is either impossible or could yield mutants with a further deregulated cell cycle. For a quantitative single-cell analysis, fluorescently labelled XIAP variants were therefore required to unambiguously identify cells with elevated XIAP expression after lipotransfection.
Figure 2.
Expression of C-XIAP-C impairs cell proliferation. (a, b) Cell proliferation was observed by fluorescence microscopy of Cerulean- or C-XIAP-C-expressing MCF-7 cells. Comparison of cell numbers after 24 h suggested that proliferation of C-XIAP-C-expressing cells was impaired. Scale bars=20 μm. (c, d) Identical fields of view of Cerulean- or C-XIAP-C-expressing cells were re-examined after 24 and 48 h and the number of fluorescent cells were recorded. Quantifications from n=3 experiments indicated that Cerulean-expressing MCF-7 cells continued to proliferate whereas expression of C-XIAP-C induced growth arrest. Error bars represent S.E.M.
Time-lapse fluorescence imaging indicates that C-XIAP-C impairs the mitochondrial release of Smac but not cyt-c
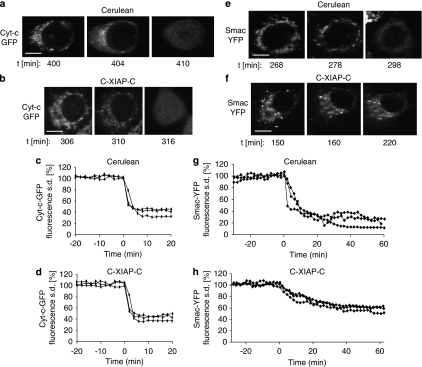
We next analysed the release of the pro-apoptotic mitochondrial intermembrane space proteins cyt-c and Smac in the presence of C-XIAP-C expression. As cells within a population do not commit to mitochondrial permeabilization synchronously, we resorted to single-cell time-lapse imaging to investigate the real-time kinetics of the release processes. To this end we used cyt-c and Smac fluorescent fusion proteins, which were previously shown to reliably show the behaviour of the native proteins.18, 19, 22 After apoptosis induction by TRAIL/CHX, cyt-c-green fluorescent protein (GFP) was released from mitochondria and distributed homogeneously throughout the cytosol in both Cerulean- and C-XIAP-C-expressing cells (Figure 3a and b). Plotting the release kinetics indicated that cyt-c-GFP release, once initiated, proceeded rapidly without differing noticeably between cells expressing Cerulean or C-XIAP-C (Figure 3c and d). Interestingly however, Smac-YFP release was impaired in cells expressing C-XIAP-C, whereas control cells expressing Cerulean released Smac-YFP fully (Figure 3e and f). Smac-YFP release from mitochondria of Cerulean-expressing cells progressed swiftly, whereas the release in the presence of C-XIAP-C expression was significantly prolonged and incomplete, as indicated by higher values in the cellular fluorescence S.D. after release (Figure 3g and h).
Figure 3.
Cyt-c-GFP and Smac-YFP release in the presence of C-XIAP-C expression. (a, b) A Cerulean-expressing or a C-XIAP-C-expressing MCF-7 cell releasing cyt-c-GFP in response to 100 ng/ml TRAIL plus 1 μg/ml CHX is shown. Upon release, cyt-c-GFP fluorescence redistributed from the mitochondria into the cytosol. Time indicates minutes after stimulus addition. Scale bar=10 μm. (c, d) Temporal profiles of cyt-c-GFP release in Cerulean-expressing (c) or C-XIAP-C-expressing (d) MCF-7 cells. The cyt-c-GFP release process was plotted as the change in cellular fluorescence S.D. and shown for three representative cells. Release onset was set to time zero. Cyt-c-GFP release proceeded rapidly. A total of n=10 Cerulean-expressing cells or n=19 C-XIAP-C-expressing cells from 4 or 6 experiments were analysed, respectively. (e) A Cerulean-expressing MCF-7 cell releasing Smac-YFP in response to 100 ng/ml TRAIL plus 1 μg/ml CHX is shown. Upon release, Smac-YFP fluorescence redistributed from the mitochondria to the cytosol. Time indicates minutes after stimulus addition. Scale bar=10 μm. (f) A C-XIAP-C-expressing MCF-7 cell releasing Smac-YFP in response to 100 ng/ml TRAIL plus 1 μg/ml CHX is shown. Smac-YFP fluorescence redistribution from the mitochondria to the cytosol seemed incomplete even after prolonged times. Time indicates minutes after stimulus addition. Scale bar=10 μm. (g, h) Temporal profiles of Smac-YFP release in Cerulean-expressing (g) or C-XIAP-C-expressing (h) MCF-7 cells. The Smac-YFP release process was plotted as the change in cellular fluorescence S.D. and shown for three representative cells. Release onset was set to time zero. Smac-YFP release in C-XIAP-C-expressing cells was slow and the loss in fluorescence S.D. was less pronounced when compared with Cerulean-expressing cells. A total of n=13 Cerulean-expressing cells or n=17 C-XIAP-C-expressing cells from 6 or 8 experiments were analysed, respectively
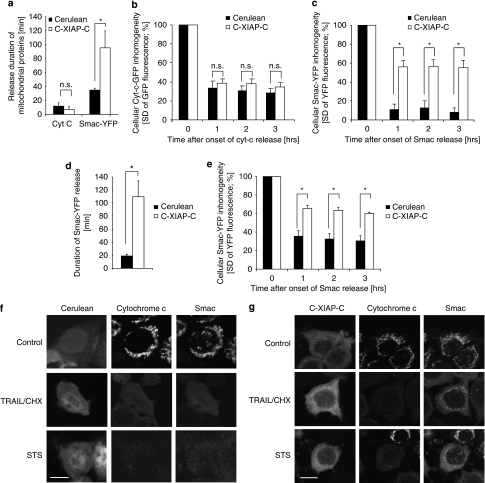
Pooling the data from all cells measured for statistical comparison further substantiated these observations. Although cyt-c-GFP release durations did not significantly differ between Cerulean- or C-XIAP-C-expressing cells, C-XIAP-C expression significantly extended Smac-YFP release up to a duration of approximately 90 min (Figure 4a). Comparing the cellular fluorescence homogeneities after release as a measure for the extent of cyt-c-GFP or Smac-YFP redistribution indicated that C-XIAP-C expression selectively prevented Smac-YFP from being fully released from mitochondria (Figure 4b and c). Similarly, we detected that Smac-YFP release was also prolonged and incomplete in C-XIAP-C-expressing MCF-7 cells during intrinsic apoptosis signalling induced by 1 μM STS (Figure 4d and e).
Figure 4.
Elevated XIAP expression results in significantly prolonged and submaximal release of Smac. (a) The duration of cyt-c-GFP or Smac-YFP release in response to 100 ng/ml TRAIL/1 μg/ml CHX was quantified for all MCF-7 cells measured and statistically compared. Cyt-c-GFP release durations were from 10 and 19 cells expressing Cerulean or C-XIAP-C, respectively. Smac-YFP release durations were from 13 and 17 cells expressing Cerulean or C-XIAP-C, respectively. Error bars represent S.E.M. *P<0.05; NS, not significant (Student's t-tests). (b) Comparison of amounts of cyt-c-GFP released in response to 100 ng/ml TRAIL/1 μg/ml CHX in cells expressing Cerulean or C-XIAP-C. Cellular fluorescence S.D. for 10 and 19 cells per group were compared at 1, 2 and 3 h after onset of cyt-c-GFP release. No significant difference could be detected between Cerulean- or C-XIAP-C-expressing MCF-7 cells. Error bars represent S.E.M. NS, not significant (Student's t-tests). (c) Comparison of amounts of Smac-YFP released in response to 100 ng/ml TRAIL/1 μg/ml CHX in cells expressing Cerulean or C-XIAP-C. Cellular fluorescence S.D. for 13 and 17 cells per group were compared at 1, 2 and 3 h after onset of Smac-YFP release. Significantly less Smac-YFP was released in MCF-7 cells expressing C-XIAP-C. Error bars represent S.E.M. *P<0.05, Student's t-tests. (d) As in (a), release durations of Smac-YFP were compared in MCF-7 cells exposed to 1 μM STS (n=8 or 11 cells per group). Smac release was significantly prolonged in cells expressing C-XIAP-C. Error bars represent S.E.M. *P<0.05, Student's t-test. (e) Comparison of amounts of Smac-YFP released in response to 1 μM STS in cells expressing Cerulean or C-XIAP-C. Cellular fluorescence S.D. for 8 and 11 cells per group were compared at 1, 2 and 3 h after onset of Smac-YFP release. Significantly less Smac-YFP was released in MCF-7 cells expressing C-XIAP-C. Error bars represent S.E.M. *P<0.05, Student's t-tests. (f, g) Immunofluorescence scans of native cyt-c and Smac in Cerulean-expressing (f) or C-XIAP-C-expressing (g) MCF-7 cells. Cells expressing Cerulean co-released both cyt-c and Smac (f). In C-XIAP-C-expressing cells, Smac was partially retained in the mitochondria, whereas cyt-c was apparently fully released in response to both TRAIL/CHX and STS (g). Scale bars=10 μm
We next performed immunofluorescence staining of TRAIL/CHX or STS-treated MCF-7 cells to investigate whether differential release patterns for cyt-c or Smac could phenotypically also be observed for the native proteins. Cells expressing Cerulean co-released both cyt-c and Smac (Figure 4f). In contrast, in C-XIAP-C-expressing cells we could indeed identify that Smac was partially retained in the mitochondria, whereas cyt-c was fully released in response to both TRAIL/CHX and STS (Figure 4g). When applying an edge detection algorithm to investigate cells for inhomogeneities in subcellular protein distributions, we found that in control cells XIAP was homogeneously distributed within the cytosol, whereas areas with slightly increased XIAP concentrations could be detected after TRAIL/CHX or STS treatment (Supplementary Figure 2A). These areas resembled edges detected for cells in which Smac release was incomplete (Supplementary Figure 2B), suggesting that after mitochondrial outer membrane permeabilization (MOMP), a fraction of XIAP might accumulate at or within mitochondria.
Physiological XIAP expression can impair Smac release
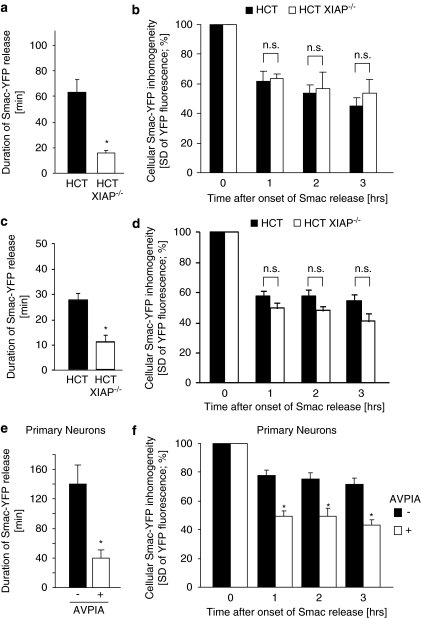
We previously identified that native XIAP expression in parental HCT-116 colon cancer cells causes a pronounced delay between MOMP and subsequent effector caspase activation.23 We therefore hypothesized that in these cells native XIAP might be sufficiently highly expressed to also influence Smac release. When comparing Smac-YFP release between parental and XIAP-deficient HCT-116 cells, we found that in the absence of XIAP, Smac-YFP release proceeded significantly faster during both extrinsic and intrinsic apoptosis (Figure 5a and c). The extent of Smac-YFP release was similar in all scenarios, indicating that the amounts of XIAP expressed in HCT-116 cells were not high enough to retain Smac efficiently inside mitochondria (Figure 5b and d). Physiological XIAP expression was described to exert a particularly prominent role in preventing apoptosis execution in differentiated neurons.5 Smac-YFP release in primary mouse cortical neurons indeed proceeded slowly and was incomplete, but could be accelerated significantly and brought to completion by brief pre-treatment of neuronal cultures with an IAP-antagonizing peptide (Figure 5e and f). These results therefore indicate that native XIAP can impair Smac release and that physiological IAP expression can limit Smac release in primary neurons.
Figure 5.
Physiological XIAP expression can impair Smac release. (a) The duration of Smac-YFP release in response to 100 ng/ml TRAIL/1 μg/ml CHX was quantified for parental and XIAP-deficient HCT-116 cells. Data are means from n=10 and 7 cells per group. Error bars represent S.E.M. *Significant acceleration of Smac release (P<0.05; Student's t-test). (b) Comparison of amounts of Smac-YFP released in response to 100 ng/ml TRAIL/1 μg/ml CHX. Cellular fluorescence S.D. for 10 and 7 cells per group were compared at 1, 2 and 3 h after onset of Smac release. Data are shown as means; error bars represent S.E.M. NS, not significant (Student's t-test). (c) The duration of Smac-YFP release in response to 1 μM STS was quantified for parental and XIAP-deficient HCT-116 cells. Data are means from n=8 and 5 cells per group. Error bars represent S.E.M. *Significant acceleration of Smac release (P<0.05; Student's t-test). (d) Comparison of amounts of Smac-YFP released in response to 1 μM STS. Cellular fluorescence S.D. for 8 and 5 cells per group were compared at 1, 2 and 3 h after onset of Smac release. Data are shown as means; error bars represent S.E.M. NS, not significant (Student's t-test). (e) The duration of Smac-YFP release in mouse cortical neurons in response to 300 nM STS was quantified either in the presence or absence of an IAP-antagonizing peptide. Data are means from n=10 and 12 cells per group. Error bars represent S.E.M. *Significant acceleration of Smac release (P<0.05; Student's t-test). (f) Comparison of amounts of Smac-YFP released in mouse cortical neurons. Cellular fluorescence S.D. for 10 and 12 cells per group were compared at 1, 2 and 3 h after onset of Smac release. Data are shown as means, error bars represent S.E.M. *Significant differences in Smac release (P<0.05; Student's t-test)
Impairment of Smac release during extrinsic or intrinsic apoptosis requires direct Smac–XIAP interaction
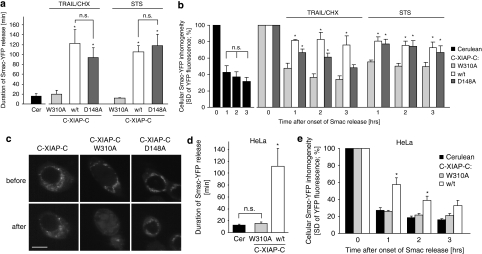
We next investigated what binding capacities of XIAP were crucial for the impaired Smac release that we observed in the presence of C-XIAP-C. To this end we expressed XIAP variants not interacting with either Smac or effector caspases in HCT-116 cells deficient of native XIAP. Consistent with our findings in MCF-7 cells, C-XIAP-C expression resulted in significantly prolonged and incomplete Smac release in response to both TRAIL/CHX and STS (Figure 6a–c). Using a C-XIAP-C W310A mutant to abrogate Smac binding13 restored rapid and complete Smac release as observed in control cells (Figure 6a–c). We also introduced a D148A mutation into C-XIAP-C to express a variant that can still bind to Smac but cannot efficiently inhibit effector caspases.12, 24 Presence of C-XIAP-C D148A resulted in prolonged Smac release, comparable to cells expressing wild-type C-XIAP-C (Figure 6a). In response to intrinsic apoptosis induced by STS, C-XIAP-C D148A was as potent as wild-type C-XIAP-C in preventing full Smac release (Figure 6b). In response to TRAIL/CHX, C-XIAP-C D148A was slightly less efficient in maintaining Smac inside mitochondria at late times (Figure 6b), probably because of caspase-8 directly activating caspase-3 and the latter cleaving XIAP between the BIR2 and 3 domains into fragments that can no longer bind efficiently to Smac.25 However, as C-XIAP-C D148A-expressing cells continued to proceed through apoptosis execution in response to TRAIL/CHX, we cannot exclude that morphological changes such as cellular shrinkage and blebbing also contributed to this reading. Expressing C-XIAP-C or the W310A mutant in HeLa cervical cancer at concentrations not fully inhibiting apoptosis execution similarly showed that the Smac-binding capacity of XIAP was critical to prolonging Smac release and to cause mitochondrial Smac retention (Figure 6d and e).
Figure 6.
Impairment of Smac release requires Smac–XIAP interaction but not effector caspase inhibition. (a) The duration of Smac-YFP release in response to TRAIL/CHX (100 ng/ml, 1 μg/ml) or STS (1 μM) was quantified for XIAP-deficient HCT-116 cells expressing Cerulean, C-XIAP-C or variants thereof bearing W310A or D148A point mutations. Data are means from n=4 control cells, or 8, 11 and 10 cells per group (TRAIL/CHX) or 6, 5 and 7 cells per group (STS). Error bars represent S.E.M. *Significantly prolonged Smac release (P<0.05; ANOVA and subsequent Tukey's test; NS, not significant). (b) Comparison of amounts of Smac-YFP released in response to TRAIL/CHX or STS. Cellular fluorescence S.D. for n=4 control cells, or 8, 11 and 10 cells per group (TRAIL/CHX) or 6, 5 and 7 cells per group (STS) were compared at 1, 2 and 3 h after onset of Smac release. Data are shown as means; error bars represent S.E.M. *Significant mitochondrial Smac retention (P<0.05; ANOVA and subsequent Tukey's test). (c) Representative images of Smac-YFP fluorescence in HCT-116 XIAP−/− cells expressing C-XIAP-C or its variants before and 60 min after onset of Smac-YFP release. Full Smac release was only observed in cells expressing C-XIAP-C W310A. Scale bar=10 μm. (d) The duration of Smac-YFP release in response to 100 ng/ml TRAIL/1 μg/ml CHX was quantified for HeLa cells expressing Cerulean, C-XIAP-C or C-XIAP-C W310A. Data are means from n=9, 7 and 7 cells per group. Error bars represent S.E.M. *Significantly prolonged Smac release (P<0.05; ANOVA and subsequent Tukey's test; NS, not significant). (e) Comparison of amounts of Smac-YFP released in response to 100 ng/ml TRAIL/1 μg/ml CHX. Cellular fluorescence S.D. for n=9, 7 and 7 cells per group were compared at 1, 2 and 3 h after onset of Smac release. Data are shown as means; error bars represent S.E.M. *Significant mitochondrial Smac retention (P<0.05; ANOVA and subsequent Tukey's test)
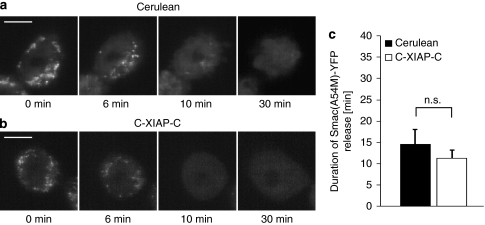
We also expressed a Smac mutant that cannot bind to XIAP (Smac(A54M)-YFP) in Smac-deficient HCT-116 cells, and investigated its release in the presence of Cerulean or C-XIAP-C expression. Irrespective of whether C-XIAP-C was present or not, we observed a complete and rapid release of Smac(A54M)-YFP in all cells investigated (Figure 7a–c). In addition, we compared the release kinetics of Smac-YFP to those of a spectrally compatible intermembrane space red fluorescent protein (RFP) that uses the same import mechanism as the immature Smac pro-form but cannot interact with XIAP (IMS-RP). Both proteins were co-released during TRAIL/CHX-induced apoptosis (Δt <2 min; not shown). The release duration of Smac-YFP was significantly increased at conditions of elevated XIAP expression, whereas the duration of IMS-RP release was not affected (Supplementary Figure 3). These findings showed that at elevated XIAP expression, mitochondrial intermembrane space proteins are still co-released but that binding of XIAP to Smac significantly modulates the kinetics and amounts of Smac release.
Figure 7.
Release of Smac(A54M)-YFP is not impaired by XIAP. (a, b) Representative Cerulean-expressing (a) or C-XIAP-C-expressing (b) HCT Smac−/− cells releasing Smac(A54M)-YFP in response to 100 ng/ml TRAIL+1 μg/ml CHX are shown. Upon release, Smac(A54M)-YFP fluorescence redistributed from the mitochondria into the cytosol. Onset of release was set to time zero. Release was complete in all cells investigated. Scale bar=10 μm. (c) The duration of Smac(A54M)-YFP release was quantified for all HCT Smac−/− cells measured. Data are means from n=4 Cerulean-expressing and n=6 C-XIAP-C-expressing cells. Error bars represent S.E.M. (NS, not significant; Student's t-test)
Mitochondrial retention of Smac by C-XIAP-C requires cellular integrity
Our above results indicated that XIAP may retain Smac inside mitochondria that already underwent MOMP. We therefore hypothesized that in cell populations induced to undergo apoptosis, individual cells could be identified that apparently had not yet released Smac-YFP in significant amounts, but might spontaneously do so upon reducing the cytosolic C-XIAP-C concentration. In contrast, in untreated cells, Smac-YFP would be expected to remain inside the mitochondrial compartment.
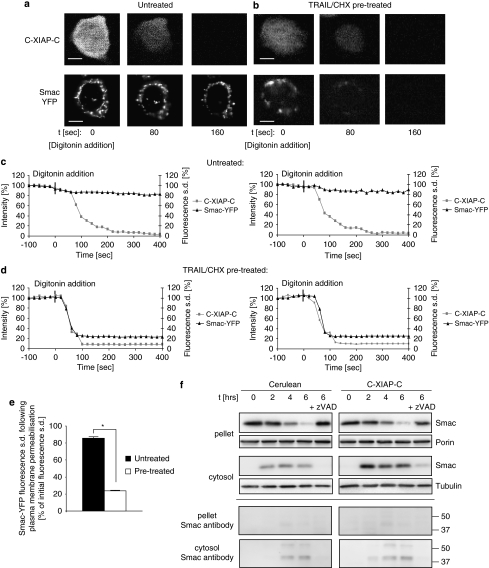
To rapidly reduce the C-XIAP-C concentration, we selectively permeabilized the plasma membrane of MCF-7 cells on stage of the microscope by addition of mild extracellular concentrations of digitonin (see Materials and Methods). In untreated cells, no mitochondrial Smac release was observed upon plasma membrane permeabilization (Figure 8a and c). In cells that were pre-treated with TRAIL/CHX for 4 h, the depletion of C-XIAP-C was instead accompanied by a simultaneous release of Smac from the mitochondria in all cells investigated (Figure 8b and d). These results were further substantiated after quantification and comparison of Smac release in the two scenarios (Figure 8e). These results therefore indicated that mitochondria in pre-treated cells had already undergone outer membrane permeabilization and that prolonged mitochondrial retention of Smac-YFP was dependent on sufficiently high levels of cytosolic XIAP.
Figure 8.
Plasma membrane permeabilization causes rapid cytosolic depletion of XIAP and re-establishes Smac release. (a) Representative images of the response of an untreated MCF-7 Smac-YFP cell to digitonin-based plasma membrane permeabilization. Upon permeabilization, C-XIAP-C fluorescence was lost from the cytosol. Mitochondrial Smac-YFP fluorescence did not noticeably change. Scale bar=10 μm. (b) Representative images of the response of a TRAIL/CHX pre-treated MCF-7 Smac-YFP cell to digitonin-based plasma membrane permeabilization. Upon permeabilization, C-XIAP-C fluorescence was lost from the cytosol. Mitochondrial Smac-YFP was lost in parallel. Scale bar=10 μm. (c, d) Temporal profiles of Smac-YFP and C-XIAP-C release in untreated (c) or TRAIL/CHX pre-treated (d) MCF-7 Smac-YFP cells. Smac-YFP release was plotted as the change in cellular fluorescence S.D., C-XIAP-C loss was plotted as the change in cellular fluorescence intensity. Two representative cells are shown each. Release onset was set to time zero. A total of n=4 untreated cells or n=8 pre-treated cells from three experiments each were analysed. (e) Comparison of amounts of Smac-YFP released after permeabilization of untreated or TRAIL/CHX pre-treated cells expressing C-XIAP-C. Cellular fluorescence S.D. for 4 and 8 cells per group were compared at 5 min after onset of C-XIAP-C loss. Error bars represent S.E.M. *P<0.05 (Student's t-test). (f) Mitochondrial Smac retention cannot be detected by biochemical fractionation. MCF-7 cells transfected to express Cerulean or C-XIAP-C were treated with TRAIL/CHX for the indicated times and subjected to digitonin-based biochemical fractionation. Smac release was detected by immunoblotting. Longer exposures were used to detect signals corresponding to poly-ubiquitinated Smac (calculated molecular weights of 44 and 52.5 kDa, respectively). Porin and α-tubulin served as loading controls. Experiment was repeated eight times with similar results
Digitonin-based plasma membrane permeabilization is an initial step for biochemical cell fractionation and is widely used to analyse changes in the subcellular localization of mitochondrial proteins during apoptosis. As our results above indicated, classical biochemical fractionation seems inappropriate to investigate the XIAP-dependent mitochondrial retention of Smac after MOMP. Indeed, immunoblots from mitochondria-containing pellet fractions showed identical patterns of mitochondrial Smac loss between Cerulean- or C-XIAP-C-expressing cells (Figure 8f, pellet fraction), confirming that the Smac-retaining function of XIAP is situated downstream of MOMP. It is noteworthy that the amounts of Smac that could be detected in cytosolic extracts subsequent to cell disruption were significantly higher in C-XIAP-C-expressing cells (Figure 8f, cytosolic fraction). This finding suggested that the additional mitochondrial Smac that is released upon cell disruption was inaccessible to the otherwise highly efficient cytosolic proteasomal degradation of Smac.26, 27 Correspondingly, the higher amounts of Smac detected were not accompanied by similar increases in signals that corresponded to the molecular weights of poly-ubiquitinated cytosolic Smac (Figure 8f).
Discussion
We identified that the presence of XIAP can prolong and limit the release of Smac from the mitochondria during extrinsically and intrinsically induced apoptosis. Our data indicate that this function of XIAP is primarily mediated by its Smac-binding capacity and is situated downstream of the permeabilization of the outer mitochondrial membrane.
As mitochondrial Bax/Bak pores are not selective towards the proteins passing through,18, 19 it is conceivable that apart from cyt-c and Smac leaving the mitochondria, XIAP may diffuse into Bax/Bak pores or gain access to the intermembrane space. Similarly, it was previously also shown that active effector caspases might enter the intermembrane space after pore formation.28 Mature Smac has a molecular weight of 21 kDa and dimerizes at physiological conditions.29, 30 Smac dimers migrate at an apparent molecular weight of approximately 100 kDa during native gel electrophoresis because of their non-globular, arch-shaped conformation.29, 30 XIAP, which has a molecular weight of 56 kDa, similarly dimerizes at elevated concentrations through its BIR1 domain.31 Stoichiometrically, native XIAP*Smac aggregates that may form at the outer mitochondrial membrane or within the intermembrane space therefore could constitute two and four molecules of XIAP and Smac, respectively. Such a protein complex may reach a size that no longer passes efficiently through Bax/Bak pores. Cyt-c instead, which is co-released with Smac, is a small (12 kDa) and predominantly monomeric protein that only transiently binds to and activates its cytosolic binding partner Apaf-1.
Although it has been shown that Bax, in cooperation with cardiolipin, can form pores sufficient to release megadalton dextran molecules from liposomes,32 our data indicate that inside living cells pore sizes may be limited. Limitations could potentially arise from the restricted availability of cardiolipin in the outer mitochondrial membrane and/or sterical hindrance through other intramembranous or membrane-associated proteins.33 In cells with deficiencies in pore forming Bcl-2 family members, the release of Smac has also been shown to be more readily impaired than cyt-c release,34 suggesting that scenarios may exist in which limitations in Bax/Bak amounts restrict efficient Smac release.
XIAP also exerts functions besides interacting with Smac and caspases 9, 3 and 7. For example, XIAP was shown to regulate bone morphogenic factor signalling, tumour necrosis factor signalling, transforming growth factor-β signalling as well as signalling through NOD-like receptors.31, 35, 36, 37 These functions are mediated by interactions through the BIR1 or BIR2 domains as well as by ubiquitination of binding partners through the XIAP RING domain, and can induce kinase signalling cascades and, at least when overexpressed, NF-κB activation. A role for XIAP in inducing NF-κB-mediated cell cycle arrest was previously described in human endothelial cells and attributed to NF-κB-dependent induction of inhibitors of cyclin-dependent kinases p21 and p27, as well as to a downregulation of cyclins A and D.21 Interestingly, we found that elevated XIAP expression also blocked cell cycle progression in MCF-7 breast cancer cells. In addition to caspase inhibition, elevated levels of XIAP therefore may induce a safeguard mechanism that prevents the proliferation of highly apoptosis-resistant and therefore potentially dangerous cells. This would suggest that tumours showing an overexpression of XIAP would very likely also present an overactivation or deregulation of cyclin-dependent kinase pathways, a feature that indeed is characteristic for many types of human cancers. We cannot fully exclude that induction of NF-κB signalling by XIAP contributed to the limited Smac release that we observed. However, as plasma membrane permeabilization abrogated the mitochondrial retention of Smac within seconds, we suggest that impaired Smac release is an immediate consequence of sufficiently high cytosolic XIAP concentrations rather than an indirect effect of alternative XIAP-dependent signalling. This is further supported by the Smac-binding capacity of XIAP being critical for limiting the release of Smac in our experiments.
It needs to be mentioned that if the entire fraction of mitochondrially retained Smac could still bind XIAP, which enters through pores, then mitochondrial Smac retention would not potentiate apoptosis inhibition. However, a pronounced mitochondrial accumulation of high amounts of XIAP could not be detected. Cells with incomplete Smac release only presented with a partial, if detectable, redistribution of XIAP to mitochondria. The lack of pronounced mitochondrial XIAP accumulation is consistent with a model in which a sufficiently high XIAP influx gradient counters Smac efflux and results in XIAP*Smac complexes at or within mitochondria that are too large to diffuse through Bax/Bak pores and might even block them for further Smac–XIAP interaction. Our biochemical data also indicated that a seemingly disproportionate amount of Smac was sealed off from cytosolic ubiquitination and degradation processes. The cytosolic caspase-inhibitory potential of XIAP thus could be significantly enhanced if the interaction of a small pool of XIAP and Smac is sufficient to prevent further Smac release. In this context, it is also interesting to note that the differentiation into postmitotic/non-proliferating cells, such as neurons, cardiomyocytes and myotube-like cells, is accompanied by XIAP obtaining a particularly dominant role in preventing apoptosis execution in these cells.4, 5, 6 Our findings in primary neurons indicated that mitochondrial retention of Smac could contribute to the potency of XIAP in such scenarios. Preventing Smac from entering the cytosol would also promote apoptosis prevention beyond immediate XIAP-mediated caspase inhibition. The binding of cytosolic Smac to cIAPs 1 and 2 induces rapid autoubiquitination and proteasomal degradation of Smac and cIAPs, resulting in an NF-κB-dependent secretion of TNF-α and subsequent autocrine apoptosis induction.26, 38, 39
Taken together, our data showed that the activity of XIAP includes a hitherto unknown function at the mitochondria: the selective mitochondrial retention of Smac subsequent to MOMP. This function may strengthen the anti-apoptotic potential of XIAP and seems of relevance for cancer cells and postmitotic cells that express high levels of XIAP.
Materials and Methods
Materials
Recombinant soluble human ‘killer' TRAIL and STS were from Alexis (San Diego, CA, USA). The broad-spectrum caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp(O-methyl)-fmk (zVAD) was purchased from Bachem (St. Helen's, UK). Embryo-tested mineral oil and CHX as well as all other chemicals came in analytical grade purity from Sigma-Aldrich (Tallaght, Dublin, Ireland).
Plasmids and molecular cloning
pSmac-YFP and pIMS-RP were described before.19, 40 To express C-XIAP-C, Cerulean was amplified from pmCerulean-C120 and ligated back into pmCerulean C1 using EcoRI and SalI restriction sites, yielding pmCerulean-2. The sequence of XIAP was amplified from plasmid pXIAP21 by polymerase chain reaction and the product was ligated into the EcoRI and XhoI sites of the pmCerulean2 plasmid, yielding pC-XIAP-C. Point mutations were introduced by site-directed mutagenesis using KOD Hot Start Polymerase (Novagen, Merck Biosciences, Nottingham, UK). Sense and antisense primers for the different mutations were as follows:
pC-XIAP-C D148A:
5′-GAACTGGGCAGGTTGTAGCTATATCAGACACCATATAC-3′ and 5′ GTATATGGTGTCTGATATAGCTACAACCTGCCCAGTTC 3′
pC-XIAP-C W310A, 5′-GGAGGGCTAACTGATGCGAAGCCCAGTGAAGAC-3′ and 5′-GTCTTCACTGGGCTTCGCATCAGTTAGCCCTCC-3′
p-Smac-YFP (A54M):
5′-GATTTGGCATGACACTGTGTATGGTTCCTATTGCTCAGAAATC-3′ and 5′-GATTTCTGAGCAATAGGAACCATACACAGTGTCATGCCAAATC-3′.
All coding sequences were verified by sequencing.
Peptide synthesis
A peptide comprised an NH2-terminal Smac-derived IAP antagonizing sequence (AVPIAQK), and a COOH-terminal HIV-TAT protein-transduction domain (GRKKRRQRRRPPQ) was prepared by standard solid-phase peptide synthesis. Single coupling cycles, using a 10-fold excess of fluorenylmethyloxycarbonyl chloride (Fmoc)-amino acid derivatives to resin-bound peptide were used, except for V2, R17 and P18, for which double coupling cycles were used. Assembly of the amino acid sequence, starting from a Rink Amide methoxybenzohydroxamic acid (MBHA) resin, was carried out on an automated peptide synthesizer (model 433A; Applied Biosystems, Warrington, UK). Peptides were deprotected and cleaved from the synthesis resin using a mixture of 80% trifluoroacetic acid, 5% water, 5% tri-isopropylsilane and 10% thioanisole at 20°C for 4 h. Peptides were precipitated and washed three times with 10 ml portions of diethyl ether, dried, dissolved in distilled water and lyophilized. Chromatographic analysis and purification were performed on a BioCAD SPRINT Perfusion Chromatography Workstation (PerSeptive Biosystems, Framingham, MN, USA) using Gemini columns (Phenomenex, Torrance, CA, USA). Purified peptides were finally characterized by MALDITOF mass spectrometry using an α-cyano-4-hydroxy-cinnamic acid matrix.
Cell culture and transfection
Human cancer cell lines were cultured in RPMI-1640 medium supplemented with penicillin (100 μg/ml), streptomycin (100 μg/ml) and 10% heat-inactivated fetal calf serum (Sigma-Aldrich) at 37°C and 5% CO2. For transfection, 0.5 μg of plasmid DNA and 1 μl Turbofect (Fermentas GmbH, St Leon Rot, Germany) were incubated in 100 μl of serum-free medium for 20 min at room temperature. The transfection mix was added dropwise to 1 ml culture media and left on the cells at 37°C overnight. MCF-7 and HeLa cells stably expressing Smac-YFP or cyt-c-GFP were described before.19 For the isolation of mouse cortical neurons, C57BL6 mouse neocortex was isolated from embryos at gestation day 16. The isolated tissue was transferred to dissection medium on ice (phosphate-buffered saline (PBS) with 0.25% glucose, 0.3% BSA) and incubated with trypsin-EDTA (0.25%) at 37°C for 15 min and neurons were dissociated by gentle pipetting after centrifugation (400 × g for 3 min). Neurons were re-suspended in fresh plating medium (MEM containing 5% fetal calf serum, 5% horse serum, 100 U/ml penicillin/streptomycin, 0.5 mM -glutamine and 0.6% -glucose) and plated in dishes. Plating medium was exchanged for embryonic neurobasal medium (Invitrogen, Paisley, UK) containing 100 U/ml of penicillin/streptomycin, 2% B27 (Invitrogen), 0.5 mM -glutamine and 600 nM cytosine arabinofuranoside (Sigma) the following day. One-third of the medium was exchanged for fresh medium every 2to 3 days. Neurons were transfected on days 4–7 with pSmac-YFP using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions. Ethical approval was provided by the Royal College of Surgeons in Ireland for all animal work carried out within this study.
Immunoprecipitation
For immunoprecipitation studies, cells were cultivated in a T75 culture flask in 12 ml of medium and grown to 80% confluency. Cells were transfected with pmCerulean-C1, pC-XIAP-C or pSmac-YFP as described above. Cells were trypsinized and collected at 1000 r.p.m. for 3 min and washed with PBS. The cell pellet was resuspended in NP-40 lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5 M EDTA and protease inhibitor cocktail 1 : 100) and homogenized with a needle and syringe. Samples were first incubated overnight at 4°C with 5 μg rabbit polyclonal Smac antibody (AF-789, R&D Systems, Minneapolis, MN, USA) or 5 μg anti-rabbit IgG antibody (control), then incubated with anti-rabbit peroxidase-conjugated secondary antibody (Jackson Laboratories, Bar Harbor, ME, USA) for 1 h at 4°C and finally incubated with protein A/G agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 2 h at 4°C. The protein–bead complex was collected by centrifugation, boiled and samples were subjected to western blotting.
Preparation of whole cell extracts, cell fractionation and western blotting
For whole cell extracts, cells were collected at 400 × g for 3 min and washed with PBS. To analyze the release of Smac, cells were fractioned into cytosolic extracts and pellets by selective plasma membrane permeabilization using permeablization buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 250 nM sucrose and 100 μg/ml digitonin) as described before.19 Cell pellets, cytosolic cell extracts or whole cell extracts were re-suspended in lysis buffer (62.5 mM Tris-HCl, pH 6.8, 10% (v/v) glycerin, 2% (w/v) SDS, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin A, 1 μg/ml leupeptin and 5 μg/ml aprotinin) and heated at 95°C for 20 min. Protein content was determined using Pierce Micro-BCA protein assay (Pierce, Northumberland, UK). An equal amount of protein (20 μg) was loaded onto SDS-polyacrylamide gels. Proteins were separated at 100 V for 2.5 h and then blotted to nitrocellulose membranes (Protean BA 83; 2 μm; Schleicher & Schuell, Whatman, Kent, UK) in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol (v/v) and 0.01% SDS) at 18 V for 60 min. The blots were blocked with 5% nonfat dry milk in TBST (15 mM Tris-HCl, pH 7.5, 200 mM NaCl and 0.1% Tween 20) at room temperature for 1 h. Membranes were incubated with the following antibodies: a mouse monoclonal XIAP antibody (610763, clone 48; BD Transduction Laboratories, San Jose, CA, USA); a rabbit polyclonal GFP antibody (PC 408; Calbiochem, Merck, Darmstadt, Germany); a mouse monoclonal α-tubulin antibody (Sigma); a rabbit polyclonal Smac antibody (AF-789, R&D Systems); or a mouse monoclonal porin antibody (Sigma). Membranes were washed with TBST three times for 5 min and incubated with anti-mouse or anti-rabbit peroxidase-conjugated secondary antibodies (Jackson Laboratories) for 1 h. Blots were washed and developed using the enhanced chemiluminescence detection reagent (Millipore, Cork, Ireland). Chemiluminescence was detected at 12-bit dynamic range using a Fuji LAS 4000 CCD system (Fujifilm UK Ltd., Bedfordshire, UK).
Flow cytometry
Flow cytometry was performed on a Partec Cyflow ML16 flow cytometer (Partec, Münster, Germany) equipped with a 488-nm argon ion laser, 532-nm diode laser and a 405-nm diode laser. In all, 10 000 gated events were acquired for each sample. For cell death analysis, cells were incubated in binding buffer (10 mM HEPES, 135 mM NaCl and 5 mM CaCl2) containing Annexin-V fluorescein isothiocyanate (FITC) conjugate (5 ml/ml) (BioVision, Mountain View, CA, USA) and propidium iodide (PI; 1 mg/ml) at 37°C for 15 min. Cells were re-suspended in ice-cold binding buffer and analysed. Cells were excited with the 405 nm laser to identify cells positively transfected with pCerulean, pCerulean/pXIAP or pC-XIAP-C. Cerulean fluorescence was collected through a 455 nm band-pass filter. FITC was exited with the 488 nm laser and fluorescence emission was collected through a 520-nm band-pass filter; PI was excited with the 488 nm laser and fluorescence emission was collected through a 620-nm long pass filter. Samples were analyzed using Flowmax software (Partec).
Live cell microscopy and digital imaging
Cells were cultivated on glass bottom dishes (Willco BV, Amsterdam, The Netherlands) in 200 μl of medium at least overnight to let them attach firmly. For MCF-7 cell proliferation analysis, 24 h after transfection, images were taken using a Nikon TE 300 equipped with a × 20, NA 0.45 phase contrast objective and a CCD camera (SPOT RT SE 6; Diagnostic Instruments, Sterling Heights, MI, USA) using the appropriate CFP filter sets (excitation 436±10 nm and emission 480±20 nm). To quantify cell proliferation, identical regions were re-examined every 24 h.
For time-lapse imaging of human cancer cells, the culture media was buffered with HEPES (10 mM, pH 7.4) and covered with mineral oil. Apoptosis was induced on stage with 1 μM STS or 100 ng/ml TRAIL plus 1 μg/ml CHX. For the imaging of mouse cortical neurons, the culture medium was exchanged for imaging buffer (120 mM NaCl, 3.5 mM KCl, 0.4 mM KH2PO4, 20 mM HEPES, 5 mM NaHCO3, 1.2 mM Na2SO4, 1.2 mM CaCl2, 1.2 mM MgCl2 and 15 mM glucose; pH 7.4). IAP-antagonizing peptide (AVPIAQKGRKKRRQRRRPPQ) was added at a final concentration of 100 μM 2 h before addition of 300 nM STS. Control experiments verified non-toxicity of peptide and excluded phototoxicity of the imaging procedure. Cells were monitored confocally using an LSM 5 live microscope (Carl Zeiss MicroImaging, Inc., Jena, Germany) equipped with a × 40 oil fluorescence objective and a temperature-controlled incubation chamber. The confocal laser scanning unit was equipped with a 405-nm argon laser (CFP excitation), a 489-nm argon laser (YFP/GFP excitation) and a 561-nm argon laser (RFP excitation). Images were analysed using Metamorph software (Molecular Devices, Coventry, UK). The release kinetics of cyt-c or Smac were shown as S.D. from the average pixel intensity of individual cells. Compartmentalized cyt-c-GFP or Smac-YFP contributes to a high S.D. and homogeneously distributed cyt-c-GFP or Smac-YFP was represented by a low S.D. Baselines before the release were scaled to 100%.
On stage plasma membrane permeabilization
MCF-7 Smac-YFP cells expressing C-XIAP-C were pre-treated with 100 ng/ml TRAIL/1 μg/ml CHX or vehicle for 4 h. C-XIAP-C and Smac-YFP images were then acquired confocally at 20 s temporal resolution. To permeabilize the plasma membrane, 100 μl of media was replaced with 100 μl of permeabilization buffer (20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 250 nM sucrose and 100 μg/ml digitonin). The confocal plane was chosen close to the glass coverslip surface to minimize mitochondrial movements upon plasma membrane permeabilization. Images were analysed using Metamorph software.
Immunofluorescence staining
Cells were cultivated on eight-well chamber slides (Ibidi, Integrated Biodiagnostics, Martinsried, (Munich), Germany) in 200 μl of medium. Cells were treated with 1 μM STS or 100 ng/ml TRAIL plus 1 μg/ml CHX. After 4 h of treatment, cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Cells were permeabilized for 3 min using 0.1% Triton X-100 in ice-cold PBS. Cells were washed in PBS and blocked in 5% horse serum, 0.3% Triton X-100 in PBS for 1 h at room temperature. Cells were once again washed in PBS and incubated in primary antibodies (a mouse monoclonal cyt-c antibody (55432, BD Biosciences) and a rabbit polyclonal Smac antibody (AF-789; R&D Systems)) at a dilution of 1 : 500 overnight at 4°C. Cells were washed in PBS and the secondary antibodies (FITC-conjugated anti-rabbit IgG, Rhodamine-conjugated anti-mouse IgG; Jackson Laboratories) were applied for 1 h at room temperature, at a dilution of 1 : 2000. Cells were washed once again in PBS and imaged at 12-bit dynamic range using an LSM 710 confocal microscope (Carl Zeiss). The laser scanning unit was equipped with a 405-nm argon laser (CFP excitation), a 488-nm argon laser (FITC excitation) and a 543-nm argon laser (Rhodamine excitation). For edge detection, raw images were processed using the ‘find edges' algorithm in ImageJ 1.42i (National Institute of Mental Health, MD, USA).
Statistics
Data are given as means±S.D. or S.E.M. For statistical comparison, Student's t-test or ANOVA and subsequent Tukey's test were used for normally distributed data. The P-values of <0.05 were considered to be statistically significant.
Acknowledgments
We are grateful to Drs. B Vogelstein, PK Sorger, JG Albeck, DW Piston and B Levkau for supplying cell lines and plasmids. We thank Dr. Heiko Düssmann for assistance with confocal imaging. This work was supported by grants from Science Foundation Ireland (07/RFP/BICF601), the Health Research Board Ireland (RP/2006/258), the National Biophotonics and Imaging Platform Ireland (Higher Education Authority PRTLI Cycle 4) and the European Union (FP 7 Health – APO-SYS). Agata Lichawska was supported by a Science Foundation Ireland Undergraduate Research Experience & Knowledge Award (SFI UREKA; 06/UR/B920).
Glossary
- BIR
baculoviral IAP repeat
- CHX
cycloheximide
- C-XIAP-C
Cerulean-XIAP-Cerulean
- Cyt-c
cytochrome c
- FITC
fluorescein isothiocyanate
- Fmoc
fluorenylmethyloxycarbonyl chloride
- GFP
green fluorescent protein
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- MBHA
methoxybenzohydroxamic acid
- MOMP
mitochondrial outer membrane permeabilization
- PBS
phosphate-buffered saline
- PI
propidium iodide
- RFP
red fluorescent protein
- RING
really interesting new gene
- Smac
second mitochondria-derived activator of caspase
- STS
staurosporine
- TRAIL
tumour necrosis factor-related apoptosis-inducing ligand
- XIAP
X-linked inhibitor of apoptosis protein
- YFP
yellow fluorescent protein
- zVAD
benzyloxycarbonyl-Val-Ala-Asp(O-methyl)-fmk
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Supplementary Material
References
- Rehm M, Dussmann H, Janicke RU, Tavare JM, Kogel D, Prehn JH. Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process. Role of caspase-3. J Biol Chem. 2002;277:24506–24514. doi: 10.1074/jbc.M110789200. [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, et al. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm M, Huber HJ, Dussmann H, Prehn JH. Systems analysis of effector caspase activation and its control by X-linked inhibitor of apoptosis protein. EMBO J. 2006;25:4338–4349. doi: 10.1038/sj.emboj.7601295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts MB, Vaughn AE, McDonough H, Patterson C, Deshmukh M. Reduced Apaf-1 levels in cardiomyocytes engage strict regulation of apoptosis by endogenous XIAP. J Cell Biol. 2005;171:925–930. doi: 10.1083/jcb.200504082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Singh S, Knezek M, Thompson CB, Deshmukh M. Critical function of endogenous XIAP in regulating caspase activation during sympathetic neuronal apoptosis. J Cell Biol. 2003;163:789–799. doi: 10.1083/jcb.200307130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MI, Huang YY, Deshmukh M. Skeletal muscle differentiation evokes endogenous XIAP to restrict the apoptotic pathway. PLoS One. 2009;4:e5097. doi: 10.1371/journal.pone.0005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson DB, Hongo F, Huerta-Yepez S, Mizutani Y, Miki T, Yu H, et al. Expression of X-linked inhibitor of apoptosis protein is a strong predictor of human prostate cancer recurrence. Clin Cancer Res. 2007;13:6056–6063. doi: 10.1158/1078-0432.CCR-07-0960. [DOI] [PubMed] [Google Scholar]
- Tamm I, Kornblau SM, Segall H, Krajewski S, Welsh K, Kitada S, et al. Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias. Clin Cancer Res. 2000;6:1796–1803. [PubMed] [Google Scholar]
- Yan Y, Mahotka C, Heikaus S, Shibata T, Wethkamp N, Liebmann J, et al. Disturbed balance of expression between XIAP and Smac/DIABLO during tumour progression in renal cell carcinomas. Br J Cancer. 2004;91:1349–1357. doi: 10.1038/sj.bjc.6602127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud S, Fondaneche MC, Lambert N, Pasquier B, Mateo V, Soulas P, et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, et al. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J. 2005;24:645–655. doi: 10.1038/sj.emboj.7600544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Sun C, Olejniczak ET, Meadows RP, Betz SF, Oost T, et al. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature. 2000;408:1004–1008. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, et al. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- Hao Y, Sekine K, Kawabata A, Nakamura H, Ishioka T, Ohata H, et al. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat Cell Biol. 2004;6:849–860. doi: 10.1038/ncb1159. [DOI] [PubMed] [Google Scholar]
- Bartke T, Pohl C, Pyrowolakis G, Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell. 2004;14:801–811. doi: 10.1016/j.molcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Vucic D, Deshayes K, Ackerly H, Pisabarro MT, Kadkhodayan S, Fairbrother WJ, et al. SMAC negatively regulates the anti-apoptotic activity of melanoma inhibitor of apoptosis (ML-IAP) J Biol Chem. 2002;277:12275–12279. doi: 10.1074/jbc.M112045200. [DOI] [PubMed] [Google Scholar]
- Munoz-Pinedo C, Guio-Carrion A, Goldstein JC, Fitzgerald P, Newmeyer DD, Green DR. Different mitochondrial intermembrane space proteins are released during apoptosis in a manner that is coordinately initiated but can vary in duration. Proc Natl Acad Sci USA. 2006;103:11573–11578. doi: 10.1073/pnas.0603007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm M, Dussmann H, Prehn JH. Real-time single cell analysis of Smac/DIABLO release during apoptosis. J Cell Biol. 2003;162:1031–1043. doi: 10.1083/jcb.200303123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Levkau B, Garton KJ, Ferri N, Kloke K, Nofer JR, Baba HA, et al. xIAP induces cell-cycle arrest and activates nuclear factor-kappaB: new survival pathways disabled by caspase-mediated cleavage during apoptosis of human endothelial cells. Circ Res. 2001;88:282–290. doi: 10.1161/01.res.88.3.282. [DOI] [PubMed] [Google Scholar]
- Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat Cell Biol. 2000;2:156–162. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- O'Connor CL, Anguissola S, Huber HJ, Dussmann H, Prehn JH, Rehm M. Intracellular signaling dynamics during apoptosis execution in the presence or absence of X-linked-inhibitor-of-apoptosis-protein. Biochim Biophys Acta. 2008;1783:1903–1913. doi: 10.1016/j.bbamcr.2008.05.025. [DOI] [PubMed] [Google Scholar]
- Huang Y, Park YC, Rich RL, Segal D, Myszka DG, Wu H. Structural basis of caspase inhibition by XIAP: differential roles of the linker versus the BIR domain. Cell. 2001;104:781–790. [PubMed] [Google Scholar]
- Deveraux QL, Leo E, Stennicke HR, Welsh K, Salvesen GS, Reed JC. Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J. 1999;18:5242–5251. doi: 10.1093/emboj/18.19.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Yang X. Cellular inhibitor of apoptosis 1 and 2 are ubiquitin ligases for the apoptosis inducer Smac/DIABLO. J Biol Chem. 2003;278:10055–10060. doi: 10.1074/jbc.M207197200. [DOI] [PubMed] [Google Scholar]
- MacFarlane M, Merrison W, Bratton SB, Cohen GM. Proteasome-mediated degradation of Smac during apoptosis: XIAP promotes Smac ubiquitination in vitro. J Biol Chem. 2002;277:36611–36616. doi: 10.1074/jbc.M200317200. [DOI] [PubMed] [Google Scholar]
- Ricci JE, Munoz-Pinedo C, Fitzgerald P, Bailly-Maitre B, Perkins GA, Yadava N, et al. Disruption of mitochondrial function during apoptosis is mediated by caspase cleavage of the p75 subunit of complex I of the electron transport chain. Cell. 2004;117:773–786. doi: 10.1016/j.cell.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Chai J, Du C, Wu JW, Kyin S, Wang X, Shi Y. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature. 2000;406:855–862. doi: 10.1038/35022514. [DOI] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Lu M, Lin SC, Huang Y, Kang YJ, Rich R, Lo YC, et al. XIAP induces NF-kappaB activation via the BIR1/TAB1 interaction and BIR1 dimerization. Mol Cell. 2007;26:689–702. doi: 10.1016/j.molcel.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- Distler AM, Kerner J, Hoppel CL. Proteomics of mitochondrial inner and outer membranes. Proteomics. 2008;8:4066–4082. doi: 10.1002/pmic.200800102. [DOI] [PubMed] [Google Scholar]
- Kandasamy K, Srinivasula SM, Alnemri ES, Thompson CB, Korsmeyer SJ, Bryant JL, et al. Involvement of proapoptotic molecules Bax and Bak in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced mitochondrial disruption and apoptosis: differential regulation of cytochrome c and Smac/DIABLO release. Cancer Res. 2003;63:1712–1721. [PubMed] [Google Scholar]
- Yamaguchi K, Nagai S, Ninomiya-Tsuji J, Nishita M, Tamai K, Irie K, et al. XIAP, a cellular member of the inhibitor of apoptosis protein family, links the receptors to TAB1-TAK1 in the BMP signaling pathway. EMBO J. 1999;18:179–187. doi: 10.1093/emboj/18.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkey Reffey S, Wurthner JU, Parks WT, Roberts AB, Duckett CS. X-linked inhibitor of apoptosis protein functions as a cofactor in transforming growth factor-beta signaling. J Biol Chem. 2001;276:26542–26549. doi: 10.1074/jbc.M100331200. [DOI] [PubMed] [Google Scholar]
- Krieg A, Correa RG, Garrison JB, Le Negrate G, Welsh K, Huang Z, et al. XIAP mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci USA. 2009;106:14524–14529. doi: 10.1073/pnas.0907131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.