Abstract
One proposed strategy to suppress the proliferation of imatinib-resistant cells in chronic myeloid leukemia (CML) is to inhibit key proteins downstream of Bcr-Abl. The PI3K/Akt pathway is activated by Bcr-Abl and is specifically required for the growth of CML cells. To identify targets of this pathway, we undertook a proteomic screen and identified several proteins that differentially bind 14-3-3, dependent on Bcr-Abl kinase activity. An siRNA screen of candidates selected by bioinformatics analysis reveals cold-shock domain protein A (CSDA), shown previously to regulate cell cycle progression in epithelial cells, to be a positive regulator of proliferation in a CML cell line. We show that Akt can phosphorylate the serine 134 residue of CSDA but, downstream of Bcr-Abl activity, this modification is mediated through the activation of MEK/p90 ribosomal S6 kinase (RSK) signaling. Inhibition of RSK, similarly to treatment with imatinib, blocked proliferation specifically in Bcr-Abl-positive leukemia cell lines, as well as cells from CML patients. Furthermore, these primary CML cells showed an increase in CSDA phosphorylation. Expression of a CSDA phospho-deficient mutant resulted in the decrease of Bcr-Abl-dependent transformation in Rat1 cells. Our results support a model whereby phosphorylation of CSDA downstream of Bcr-Abl enhances proliferation in CML cells to drive leukemogenesis.
Keywords: CML, Bcr-Abl, CSDA, phosphorylation, proteomics, proliferation, mass spectrometry
Chronic myeloid leukemia (CML) is a stem cell disease, in which neoplastic cells carry a translocated Philadelphia chromosome, in which a hybrid Bcr-ABL gene encodes a fusion oncoprotein, Bcr-Abl, with constitutive tyrosine kinase activity. Bcr-Abl induces multiple signaling pathways to transduce the oncogenic signal, which ultimately results in uncontrolled proliferation and neoplastic expansion.1 Bcr-Abl also reduces adhesion of CML cells to the extracellular matrix and stroma cells, allowing them to bypass the negative influence these interactions have on proliferation.2, 3 Furthermore, the oncoprotein is believed to inhibit the apoptotic response to mutagenic stimuli,4 providing a survival advantage for the leukemic clone, in addition to increasing the likelihood of genomic instability and, therefore, further oncogenic mutations.
Bcr-Abl-dependent pathways include PI3K, MAPK and JAK/STAT, which ultimately control transcription. Targeting the kinase activity of Bcr-Abl via competition with ATP for its binding to the kinase pocket is the basis of the therapeutic action of imatinib mesylate (IM), the preferred drug for front-line treatment of CML.5 However, persistence of residual disease or emergence of secondary resistance to IM is a major cause of concern, especially in the advanced phases of the disease.6, 7
One proposed strategy to suppress the proliferation of IM-resistant cells is to inhibit key proteins downstream of Bcr-Abl, such as Akt. Previous reports have shown that 14-3-3-affinity purification can be used to identify novel Akt substrates in cells in which Akt is activated through exposure to epidermal growth factor.8 Owing to the fact that the PI3K/Akt pathway plays a crucial role in the leukemogenesis of CML9, 10 and 14-3-3 binds to a number of well-characterized Akt targets in cancer signaling,11, 12 we postulated that such 14-3-3 binding would significantly contribute to Bcr-Abl-mediated leukemogenesis. We therefore utilized 14-3-3 affinity binding methodology to identify proteins that are regulated by Akt downstream of Bcr-Abl. Here we report that cold-shock domain protein A (CSDA) is a target of Bcr-Abl-induced phosphorylation, regulates proliferation and is critical for Bcr-Abl-induced transformation.
Results
Identification and comparative analysis of IM-sensitive 14-3-3 binding proteins in Bcr-Abl-positive CML cells
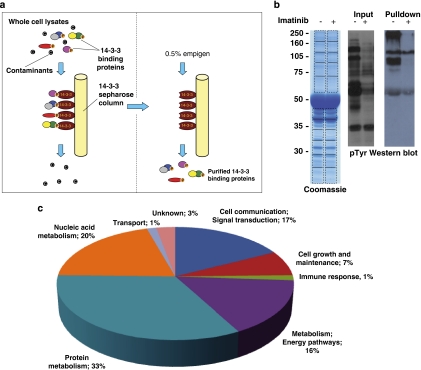
To identify Bcr-Abl-dependent 14-3-3 binding proteins, GST-14-3-3-affinity purification was utilized in combination with mass spectrometry to identify sufficient numbers of 14-3-3 bound proteins to detect differential binding after inhibition of Bcr-Abl kinase activity. The affinity purification was undertaken in whole-cell lysates from LAMA84 CML cells cultured in the presence or absence of IM as described in Materials and Methods (Figure 1a). Phosphotyrosine western blots of lysate inputs for the affinity purification displays a significant reduction in the level of tyrosine phosphorylation of multiple proteins after treatment with IM, confirming efficacy of the inhibitor in our assay (Figure 1b). Furthermore, enrichment of tyrosine-phosphorylated proteins was detected among the untreated 14-3-3 binding proteins in the pulldown (Figure 1b).
Figure 1.
Identification and analysis of Bcr-Abl-dependent 14-3-3-interacting proteins. (a) Schema of GST-14-3-3-affinity purification from IM-sensitive CML cells. Lysates from LAMA84 cells untreated or treated with 5 μM for 2 h were incubated with GST-14-3-3-sepharose; bound proteins were eluted and identified by mass spectrometry as described in Materials and Methods. (b) Aliquots of eluates from affinity purification were analyzed by Coomassie staining and western blot with phospho-tyrosine antibody. (c) Proteins that bound 14-3-3 only in the absence of IM were grouped by biological functions as described in the text
In total, 318 proteins were identified by mass spectrometry as being bound to 14-3-3 (Supplementary Table 1). In order to assess the robustness of the GST-14-3-3-affinity purification technique, we performed bioinformatic analysis of published interactions (www.hrpd.org) and confirmed that a number of proteins that bound to 14-3-3 in our affinity purification screen had previously been shown to bind 14-3-3 (Supplementary Table 2). The previous identification of these proteins as 14-3-3 binding proteins was achieved through a variety of techniques, including GST-affinity purification, Tandem-affinity purification and immunoprecipitation.13, 14, 15, 16, 17, 18, 19, 20, 21, 22 This reassured us that the eluates from the GST-14-3-3-affinity purifications were highly enriched for 14-3-3 binding proteins and that the proteins we identified were not artifactual results generated by the use of the GST-14-3-3-affinity purification technique.
Of the 318 proteins, 69 bound 14-3-3 only in the absence of IM treatment. We focused on these proteins since we were interested only in putative Akt targets that were regulated by Bcr-Abl activity in CML. In accord with other 14-3-3-affinity purification screens and the critical role of PI3K/Akt signaling in cancer cell biology,15, 17, 23 grouping of these proteins by biological function, as described by the Human Protein Reference Database (www.hrpd.org), enriched for protein metabolism (33%), nucleic acid metabolism (20%), signal transduction (17%), as well as energy and metabolism (16%) (Supplementary Table 3, Figure 1c).
siRNA screen for proliferation of candidate Bcr-Abl-dependent Akt targets
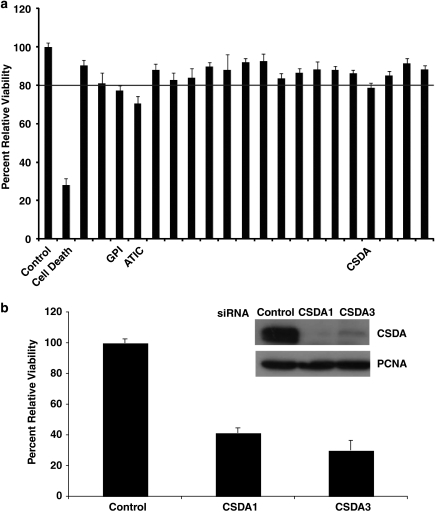
On the basis of phosphorylation-site motif analysis for 14-3-3 binding and Akt kinase motifs (http://scansite.mit.edu), as well as functional analysis, 20 of the 69 proteins that bound to 14-3-3 only in cells with active Bcr-Abl were selected for inclusion in a customized siRNA screen for proliferation (Supplementary Table 4). Two siRNA sequences were used for any gene that had pre-validated siRNAs, whereas three siRNA sequences were used for genes for which two pre-validated siRNAs were not available (Supplementary Table 5). The K562 cell line was used for the siRNA screen because of their relatively high transfection efficiency as well as their wide use in studying CML. Cells were transfected with the siRNA oligonucleotides and proliferation assessed by MTS 48 h post transfection as described in Materials and Methods. Proliferation was normalized to a non-targeting siRNA control and a ‘cell death' siRNA was included as a positive control for siRNA-mediated decrease of proliferation (Figure 2a).
Figure 2.
Targeted siRNA screen of selected candidate Akt effectors reveal CSDA to regulate proliferation in CML. (a) Twenty IM-sensitive 14-3-3 interactors were selected based on biological function and presence of putative Akt phosphorylation motif for inclusion in the siRNA screen. Two or three targeting siRNA sequences to each candidate were transfected in K562 cells for 48 h and averaged for readings of proliferation by MTS as described in Methods and Methods. The three candidates whose silencing decreased proliferation greater than 20% are indicated. (b) Transfection of CSDA siRNA targeting sequences 1 and 3 effectively silenced protein expression on K562 cells and greatly decreased proliferation after 72 h
As shown in Figure 2a, transfection of three of the 20 candidates included in the screen resulted in >20% decrease of proliferation: 5-aminoimidazol-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC, Entrez Gene ID 471), glucose phosphate isomerase (GPI, Entrez Gene ID 2821) and CSDA (Entrez Gene ID 8531). The latter has been shown to be a transcription factor, shuttling between tight junctions and nuclei, promoting cell proliferation.24, 25 We selected CSDA (also called ZONAB) for further analysis as besides enhancing cell proliferation, it has recently been implicated in breast cancer metastasis and shown to be upregulated in gastric cancer.26, 27 Its role in CML or other hematological malignancies, however, has not been in investigated. Two of the three siRNAs to CSDA could effectively silence endogenous protein expression in K562 cells as shown by western blot and transfection with these individual siRNAs for 72 h exhibited an even more profound effect on cell proliferation (Figure 2b). We further analyzed CSDA silenced K562 cells by Annexin V/propidium iodide (PI) staining as well as cell cycle analysis by PI and determined that CSDA expression regulates proliferation and not apoptosis (Supplementary Figure 1A) or cell cycle arrest (Supplementary Figure 1B).
CSDA is phosphorylated in a Bcr-Abl-dependent manner in CML cells and CML patient samples
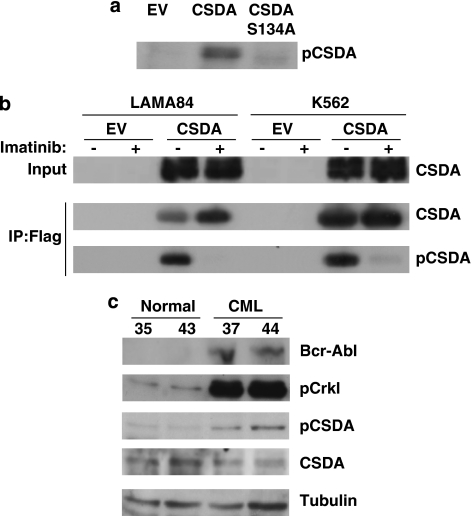
Examining the CSDA amino-acid sequence, we noted striking similarity with the recently identified Akt site on YB-1,28 a closely related cold-shock domain containing protein. We generated a phospho-deficient serine to alanine construct (CSDAS134A) targeting the putative Akt site and expressed it along with wild-type CSDA in 293 cells. Western blot analysis with a phospho-specific commercial antibody (pYB1) to the homologous site on YB-1 (serine 102) determined that this reagent could be used to detect specific phosphorylation on serine S134 of CSDA (Figure 3a).
Figure 3.
CSDA is phosphorylated at serine 134 in a Bcr-Abl-dependent manner in CML cell lines and CD34+-purified primary CML cells. (a) The 293 cells were transfected with empty vector, pCMV2B-CSDA or pCMV2B-CSDAS134A for 48 h and lysates analyzed by western blot with pYB-1 antibody to detect CSDA phosphorylation as described in the text. (b) LAMA84 and K562 cells were stably transfected with empty vector or pBABE_flagCSDA and treated or not (DMSO) with 1 μM IM for 2 h. Lysates were immunoprecipitated with FLAG M2 beads and inputs and eluates were analyzed by western blot for the expression of phosphorylated CSDA and of total CSDA by anti-ZONAB. (c) CD34+-purified cells from leukaphareses from normal individuals (sample ID 35, 43) or CML patients (sample ID 37, 44) were lysed and analyzed by western blot for the expression of Bcr-Abl by anti-c-Abl, phosphorylated CrkL, phosphorylated CSDA, CSDA and tubulin
We next developed FLAG-CSDA-expressing stable LAMA84 and K562 cell lines to assess Bcr-Abl-dependent phosphorylation of CSDA in CML. As shown in Figure 3b, immunoprecipitation of FLAG-CSDA followed by immunoblotting with pYB-1 of lysates from both stable cell lines treated with IM results in profound decrease in S134 phosphorylation of CSDA, revealing Bcr-Abl-dependent phosphorylation.
To determine whether Bcr-Abl-dependent CSDA phosphorylation is detectable in actual CML, we purified CD34+ cells from leukaphareses from CML patients or normal donors as described in Materials and methods. Western blot analysis (Figure 3c) revealed that lysates derived from CML cells (sample ID 37 and 44) show Bcr-Abl expression and a concomitant increase in phosphorylation of CrkL, a substrate for Bcr-Abl and a widely used readout for Bcr-Abl activity29, 30 relative to lysates from healthy individuals (sample ID 35 and 43). Tellingly, CML cells also exhibited increased phosphorylated CSDA signal relative to CSDA expression as compared with normal cells, indicating greater specific phosphorylation of CSDA at serine 134 in Bcr-Abl-positive CML.
Bcr-Abl-induced CSDA phosphorylation is independent of Akt, but dependent on MEK/RSK pathway
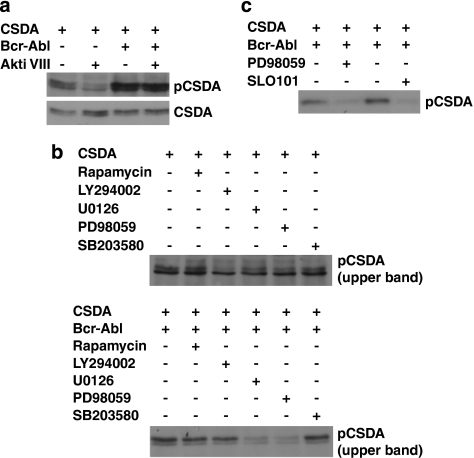
Bcr-Abl activates a number of downstream signaling pathways besides PI3K/Akt. To investigate whether Bcr-Abl-induced CSDA phosphorylation is dependent on Akt activity, we co-expressed CSDA with either empty vector or Bcr-Abl in 293 cells, and treated them with control vehicle or a specific Akt inhibitor (Akti VIII). Western blot analysis revealed that Akt inhibition is effective in decreasing the basal CSDA phosphorylation in vector control cells, but not in Bcr-Abl-transfected cells (Figure 4a).
Figure 4.
CSDA phosphorylation is mediated by Akt in the absence of Bcr-Abl, but by MEK/RSK signaling downstream of Bcr-Abl. (a) The 293 cells were co-transfected with pCMV2B-CSDA and empty vector or pCDNA3.1Bcr-Abl as indicated for 48 h. Cells were then serum-starved overnight, treated or not (DMSO control) with 10 μM Akti VIII for 2 h, and then serum was reintroduced (to 10%) for 30 min. Lysates were analyzed by western blot for CSDA and phosphorylated CSDA expression as described previously. (b) The 293 cells were co-transfected with pCMV2B-CSDA and empty vector (top panel) or pCDNA3.1Bcr-Abl (bottom panel), serum starved as in (a), and treated or not (DMSO control) with the inhibitors rapamycin (80 nM), LY294002 (50 μM), U0126 (20 μM), PD98059 (100 μM) or SB203580 (10 μM) for 2 h, after which 10% serum was added for 30 min. Lysates were analyzed by western blot for CSDA and phosphorylated CSDA expression as described previously. (c) The 293 cells were co-transfected with pCMV2B-CSDA and pCDNA3.1Bcr-Abl and treated with PD98059 (100 μM) or SL0101 (20 μM) and lysates analyzed for CSDA and phosphorylated CSDA expression as before
In order to determine which kinases are responsible for Bcr-Abl-induced CSDA phosphorylation, we co-transfected 293 cells with CSDA and empty vector or Bcr-Abl, and treated them with control vehicle, mTOR inhibitor (rapamycin), PI3K inhibitor (LY294002), active MEK inhibitor (U0126), MEK inhibitor (PD98059) or p38 MAPK inhibitor (SB203580). As shown in the top panel of Figure 4b, treatment with the PI3K inhibitor but not the other inhibitors reduced basal CSDA phosphorylation. Similar to our findings with Akt (Figure 4a), PI3K inhibition did not affect Bcr-Abl-induced CSDA phosphorylation (Figure 4b, bottom panel). Strikingly, both MEK inhibitors were ineffective in reducing basal phosphorylation (Figure 4b, top panel), but robustly reduced Bcr-Abl-induced CSDA phosphorylation.
Sequence analysis led us to expect that MEK does not directly phosphorylate CSDA at serine 134. However, the p90 ribosomal S6 kinase (RSK) was recently reported to phosphorylate the conserved site on YB-1 and RSK signaling downstream of MEK activity is well established.31, 32 Tellingly, treatment with an RSK inhibitor (SL0101) blocked Bcr-Abl-induced CSDA phosphorylation as effectively as MEK inhibition (Figure 4c). These data suggest that although Akt phosphorylates CSDA at serine 134 in non-CML cells, Bcr-Abl activity results in MEK-dependent RSK phosphorylation of CSDA at the same site.
RSK inhibition specifically blocks proliferation in Bcr-Abl-positive cell lines and primary CML cells
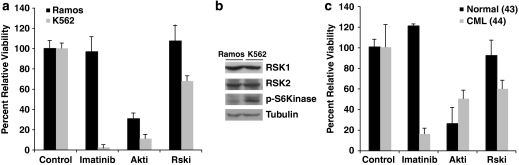
We investigated whether RSK activity is selectively critical in Bcr-Abl-positive cells by undertaking a proliferation assay comparing K562 and Ramos cell lines. As expected, treatment with IM selectively blocked proliferation in K562 CML cells while having negligible effect on the Bcr-Abl-negative, Ramos Burkitt's lymphoma line, whereas Akt inhibition decreased proliferation in both cell lines (Figure 5a). Strikingly, similar to IM, RSK inhibition reduced proliferation only in the K562 cells (Figure 5a). We assessed whether the difference in sensitivity to RSK inhibitor may be a function of differential S6 kinase activity in these cells. Indeed, although Ramos and K562 cells express similar levels of both RSK1 and RSK2, the K562 CML lines exhibit markedly increased S6 kinase activity as detected by an antibody specific to S6 kinase phosphorylated at Thr389 (Figure 5b).
Figure 5.
RSK inhibition selectively blocks proliferation in CML cell lines and primary CML cells. (a) Ramos or K562 cells were seeded and treated with DMSO control, IM (2 μM), Akt inhibitor (10 μM Akti VIII) or RSK inhibitor (20 μM SL0101) as described in Materials and methods. Proliferation was assessed by MTS after 72 h treatment. (b) Ramos or K562 cell lysates were analyzed by western blot for RSK1, RSK2, phospho-S6 kinase and tubulin as indicated. (c) CD34+-purified cells from leukapharesis from a normal individual (sample ID 43) or a CML patient (sample ID 44) were cultured for 16 h followed by treatment with DMSO control or indicated inhibitors as in (a). Proliferation was assessed by MTS after 72 h treatment. Three independent experiments were each carried out in triplicate and data presented are mean value±SD of one representative experiment
To determine whether specificity to RSK inhibition is also evidenced in primary CML cells, we compared the proliferation rate of normal and CML CD34+ progenitors treated with IM, Akt inhibitor or RSK inhibitor. Similar to what we observed in the cell lines (Figure 5a), IM-induced Bcr-Abl inhibition and RSK inhibition affected growth only of CML progenitor cells, whereas Akt inhibition abrogated proliferation in both CML and normal cells (Figure 5c). These data indicate that inhibition of RSK specifically reduces proliferation in Bcr-Abl-positive cells, both in cell lines and primary CML.
CSDA expression and S134 phosphorylation regulates Bcr-Abl-dependent transformation
We have shown that CSDA expression and RSK activity are both critical for proliferation in CML (Figures 2b and 5). We have also discovered that CSDA is phosphorylated at serine 134 downstream of Bcr-Abl in an RSK-dependent manner (Figure 4). To determine whether CSDA S134 phosphorylation is critical for Bcr-Abl-dependent transformation, we generated stable lines expressing empty vector or co-expressing Bcr-Abl and empty vector, CSDA or the CSDAS134A phospho-deficient mutant in Rat1 cells to employ in soft agar colony formation assays.33, 34
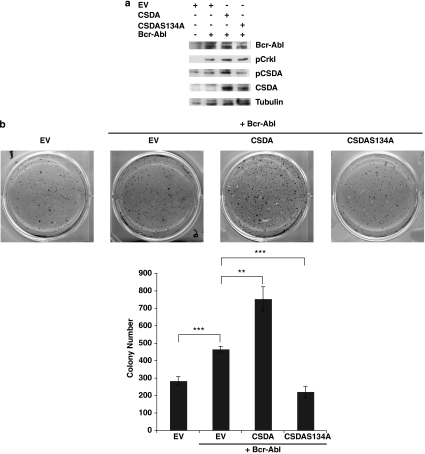
After selection, we validated the expression of Bcr-Abl and CSDA constructs (Figure 6a). Furthermore, using phospho-specific antibodies to CrkL and CSDA, we confirmed that expressed Bcr-Abl was active, and that expressed CSDA, but not CSDAS134A protein, exhibits increased phosphorylation at serine 134A with Bcr-Abl co-expression (Figure 6a). As shown in Figure 6b, cells stably expressing Bcr-Abl generated significantly greater number of colonies when cultured in soft agar than cells only expressing empty vector. Colony formation was further and significantly enhanced from cells co-expressing Bcr-Abl and CSDA, but not the phospho-deficient CSDAS134A construct compared with Bcr-Abl and empty vector (Figure 6b). In fact, co-expression of Bcr-Abl and CSDAS134A resulted in lower number of colonies than Bcr-Abl alone. These findings reveal that CSDA phosphorylation as well as expression is critical for Bcr-Abl-mediated malignant transformation.
Figure 6.
CSDA expression and phosphorylation at serine 134 regulates Bcr-Abl-dependent transformation. (a) Rat1 fibroblasts were stably co-transfected with relevant empty vectors (EV) or pCDNA3.1Bcr-Abl and empty vector or pBabe_flagCSDA or pBabe_flagCSDA as indicated, and lysates analyzed by western blot for the expression of Bcr-Abl, phosphorylated CrkL, phosphorylated CSDA, CSDA and tubulin as described previously. (b) Indicated Rat1 stable cell lines were cultured in soft agar and analyzed for colony formation as described in Materials and methods. Three independent experiments were each carried out in triplicate and data presented are mean value±SD of one representative experiment. EV versus EV+Bcr-Abl: ***t=10.75, P=0.0017; Bcr-Abl+EV versus Bcr-Abl+CSDA: **t=6.971, P=0.02; Bcr-Abl+EV versus Bcr-Abl+CSDAS134A: ***t=11.9, P=0.0003. Representative pictures are provided
Discussion
We showed that CSDA is specifically phosphorylated at serine 134 downstream of Bcr-Abl in CML cell lines and primary progenitor cells from CML patients (Figure 3). However, we unexpectedly determined that CSDAS134 phosphorylation is dependent on Akt in the absence of Bcr-Abl, but dependent on MEK/RSK downstream of Bcr-Abl (Figure 4). RSK targets such as S6 are key regulators of translation and S6 phosphorylation downstream of MEK activity was shown to be via RSK rather than mTOR to stimulate cap-dependant translation.35, 36 Although CSDA has been reported to function as a transcription factor, activating target genes such as PCNA and cyclin D1 that regulate proliferation and morphogenesis in epithelial cells,24, 37 we did not detect significant changes in specific luciferase activity of either PCNA- or cyclin D1-luciferase constructs, co-expressed with Bcr-Abl and either CSDA or CSDAS134A constructs (data not shown).
It may be that Bcr-Abl-MEK-RSK signaling induces CSDA to regulate translation rather than transcription to promote proliferation in CML. Interestingly, the closely related cold-shock domain-containing protein, YB-1, has been shown to regulate translation in breast cancer progression by repressing cap-dependent and promoting cap-independent mechanisms, with Akt phosphorylation at serine 102 resulting in decreased cap-complex binding of YB-1, and thus decreased translational repression in breast and other epithelial cells.28, 38, 39, 40 It will be interesting to determine if there are similar cap-dependent and -independent differences in RSK versus Akt phosphorylation of CSDA at serine 134 (homologous to serine 102 in YB-1) in regulating translation in CML- and Bcr-Abl-negative leukemias, and if blocking CSDAS134 phosphorylation can be exploited to specifically target CML.
We also showed that inhibition of RSK activity specifically blocks proliferation, similarly to CSDA silencing, in an established Bcr-Abl-positive cell line as well as in primary progenitor cells from CML patients (Figure 5). Furthermore, we showed that CSDA, but not the RSK phospho-site mutant, enhances Bcr-Abl-dependent transformation. In fact, co-expression of CSDAS134A blocks Bcr-Abl-dependent transformation. However, it is probable that other RSK phosphorylation targets, both known and undiscovered, are also critical for tumorigenesis, and future work will determine if particular substrates are relevant for specific cancers. Unsurprisingly, proteins involved in protein synthesis, processing and fate were enriched in our bioinformatics analysis of 14-3-3 binding proteins in our initial proteomic screen for Bcr-Abl effectors in CML (Figure 1c).
In this study, we selected CSDA for further investigation based on its previous known function in regulating cell cycle progression and upregulated expression in epithelial cancers,24, 26, 27 as well as its scoring highly in a secondary targeted siRNA screen for proliferation in K562 CML cells (Figures 2a). Gene ontology analysis of the Bcr-Abl-dependent 14-3-3 binders revealed enrichment for proteins involved in energy and metabolism (Supplementary Table 3, Figure 1c). Interestingly, the other two proteins that scored highly in the siRNA proliferation screen, ATIC, which catalyzes the last two steps of de novo purine biosynthesis, and GPI, which catalyzes the reversible isomerization of glucose-6-phosphate and fructose-6-phosphate, are both key metabolic enzymes linked to hematological malignancies41, 42 (Figure 2a). Therefore, metabolic regulation downstream of Bcr-Abl can also be targeted in CML, as it has been in therapy for other cancers.43, 44 The findings in this study indicate that CSDA phosphorylation and RSK signaling in general may offer an alternative to PI3K/Akt inhibition in targeting Bcr-Abl-dependent leukemogenesis.
Materials and Methods
Cell culture, transfection and treatment
LAMA84, K562 and Ramos cell lines were cultured in suspension in RPMI 1640 medium (Invitrogen, Paisley, UK), supplemented with 10% heat-inactivated fetal calf serum (Harlan Sera-Lab Ltd, Loughborough, UK), 5000 IU/ml penicillin, 5000 μm/ml streptomycin and 200 mM -glutamine. The 293T and Rat1 cell lines were cultured in Dulbecco's modified Eagle's media (Invitrogen) supplemented as above.
Transfections of cDNA in 293T and Rat1 cells were performed with Effectene (Qiagen, Crawley, UK) according to the manufacturer's instructions. Rat1 stable cell lines were generated with co-selection in G418 and puromycin (Invitrogen). Retroviral infections of LAMA84 and K562 were performed as described previously,45 and stable cell lines were generated with selection in puromycin. siRNA transfection in K562 and Ramos cells were performed with Interferin (Polyplus, Illkirch, France) according to the manufacturer's instructions.
IM was kindly provided by Dr. E Buchdunger (Novartis Pharma, Basel, Switzerland). Akt inhibtor VIII, LY294002, rapamycin, U0126, PD98059 and SB203580 were all purchased from Calbiochem (Nottingham, UK). Cells were treated with inhibitors at the concentrations detailed in the figure legends.
Patient sample preparation
Peripheral blood cells were obtained by leukapheresis from newly diagnosed patients with CML or from normal donors. These cells were portions of leukaphereses processed by the Stem Cell Laboratory, Hammersmith Hospital (London, UK), in excess of clinical requirements. Informed consent for the use of cells for research was obtained with approval from the Hammersmith and Queen Charlotte's and Chelsea Research Ethics Committee Institutional Review Board. CD34+ cells were labeled using MiniMACS technology according to the manufacturer's instructions (Miltenyi Biotec, Bisley, UK). Cells were resuspended in 350 μl of MACS buffer (phosphate-buffered saline (Gibco, Paisley, UK), 0.5% bovine serum albumin (PAA Laboratories, Pasching, AT) and 2 mM EDTA (Sigma, Gillingham, UK) per 108 cells with 100 μl FcR blocking reagent and 50 μl of microbeads conjugated to monoclonal anti-CD34 antibody (QBEND10) and incubated for 15 min at 4°C. The cells were then washed and resuspended in 2 ml of MACS buffer per 108 cells. Labeled cells were then passed through a pre-washed MiniMACS column mounted on a magnet. Following this, the column was washed four times with MACS buffer, removed from the magnet and the cells eluted with 2 ml MACS buffer. The purity of the CD34+ fraction was consistently above 96% as determined by flow cytometry (FACScalibur, Becton Dickinson, Oxford, UK) with anti-CD34 staining. Aliquots were immediately frozen for subsequent lysis for western blot analysis or cultured in leukemic cell growth media for proliferation assays.
Plasmids
pGEX6P1/14-3-3τ was generated by subcloning the open-reading frame of human 14-3-3τ into pGEX6P1 (Amersham Biosciences, Little Chalfont, UK). pCLAmpho and pcDNA3.1Bcr-Abl were described previously.46, 47 pCMV2B_CSDA was generated by subcloning the open-reading frame of human CSDA (Origene, Rockville, MD, USA) into pCMV_Tag2B (Stratagene, La Jolla, CA, USA). pBabe_flagCSDA was cloned by PCR-mediated addition of a 5′ BamHI restriction site and FLAG tag sequence (DYKDDDDK) and 3′ EcoRI restriction site. The CDS with 5′ FLAG tag was then ligated with BamHI- and EcoRI-digested pBabe. Construction of all vectors was confirmed by both restriction digestion and DNA sequencing CSDAS134A point mutant was constructed by site-directed mutagenesis kit (QuickChange XL, Stratagene) according to the manufacturer's instruction using the following primer synthesized by SigmaGenosys (Gillingham, UK): 5′-ACGGAAATATCTGCGCGCTGTAGGAGATGGAGAAA-3′. Point mutation was confirmed by DNA sequencing.
Western blotting and immunoprecipitation
Cells were lysed in ice-cold buffer containing 1% (v/v) Triton X-100, 0.05% (v/v) SDS,10 mM NaH2PO4, 150 mM NaCl, 5 mM EDTA, 10 mM NaF, 1 mM NaVO3, 5 nM molybdic acid, 100 nM okadaic acid, 1 mM DTT, 1 × Protease Inhibitor Cocktail (PIC) (Roche, Burgess Hill, UK), 1 × PhosSTOP phosphatase inhibitor cocktail (Roche) and resolved by SDS-PAGE transferred to PVDF membrane (Hybond-P, GE Healthcare, Little Chalfont, UK). Anti-phosphotyrosine antibody (4G10) was a kind gift from Professor B Druker (Oregon Health & Science University, Portland, OR, USA) and anti-CSDA (ZONAB) antibody was a kind gift from Professor Karl Matter (UCL, London, UK). Anti-FLAG (M2) antibody was purchased from Sigma, and anti-c-Abl (Ab3) was purchased from Calbiochem. Anti-PCNA (sc-25280), anti-tubulin (sc-53140), anti-RSK1 (sc-231), anti-RSK2 (sc-9986) and anti-YB1 (sc-101198) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-phospho-CrkL (3181), anti-phospho-p70S6 kinase (9206) and anti-phosphoYB-1 (2900) were purchased from Cell Signalling Technology (Hitchin, UK). All antibodies were used at 1:1000 dilution. Anti-FLAG immunoprecipitation was performed with M2 agarose beads (Sigma) according to the manufacturer's instructions.
GST-14-3-3-affinity purification
Recombinant GST-14-3-3 was produced from pGEX6P1/14-3-3τ transformed in BL21-DE3 bacteria (Stratagene) and purified by binding to glutathione–sepharose beads (GE Healthcare). LAMA84 cells (5 × 109) were left untreated or treated with 2 μM IM and lysed in buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, 1% (v/v) Triton X-100, 10 mM b-glycerophosphate, 50 mM NaF, 1 mM sodium orthovanadate, 5 mM sodium pyrophosphate, 100 nM okadaic acid, 0.27 M sucrose, 1 mM DTT and 1 × PIC. Lysates were clarified and incubated with GST-14-3-3 sepharose beads and 14-3-3 binding proteins eluted with 0.5% Empigen after stringent washing. Eluates were desalted in Zeba columns (Pierce, Cramlington, UK) and concentrated in YM-10 centrifugal concentrator (Microcon, Watford, UK) and resolved by 4–12% SDS-PAGE (NuPAGE, Invitrogen). Gels were either stained with Gelcode (Pierce) colloidal Coomassie for total protein expression or transferred for western blot analysis with anti-phosphotyrosine antibody (4G10).
Mass spectrometry
Entire sample lanes from Coomassie-stained gels were sectioned into equivalent-sized bands and digested with trypsin. Resultant peptides were subject to LC-MS/MS (Q-TOF, Waters, Elstree, UK) at the Protein Analysis Unit (WHRI, Barts and The London School of Medicine and Dentistry, London, UK; QMUL). Spectra were analyzed using MS/MS Ion Search feature of the MASCOT search engine (Matrix, www.matrixscience.com). Bioinformatic analyses were performed using the Scansite (www.scansite.mit.edu) and Human Protein Reference Database (HPRD, www.hprd.org) on-line software programs.
siRNA screen
Ninety-six-well plates with siRNA targeting sequences (annotated in Supplementary Table 5) to 20 selected candidates (Supplementary Table 4) were custom ordered from Qiagen. A total of 2 × 104 K562 cells were transfected with 0.5 μl Interferin and 10 nM final siRNA concentration according to the manufacturer's instructions.
Cell proliferation
Proliferation was assessed with MTS reagent (Promega, Madison, WI, USA) according to the manufacturer's instructions. Cells were analyzed at 48 h for siRNA screen and 72 h for individual CSDA siRNAs, post-transfection. For inhibitor treatments, 2 × 104 K562 or Ramos and 5 × 104 CD34+ progenitor primary cells were treated 24 and 16 h, respectively, after seeding with inhibitors at indicated concentrations and cultured for 72 h before MTS reading.
Apoptosis
Cells were analyzed for apoptosis by Annexin V/PI staining as before.48 Cells were analyzed at 72 h post-transfection with control and individual CSDA siRNAs.
Cell cycle
Cells were harvested 72 h post-transfection with control and individual CSDA siRNAs and fixed in 75% ice-cold ethanol at 4°C for 2 h. Then, cells were stained with PI (Molecular Probes, Eugene, OR, USA) and analyzed by flow cytometry.
Cell transformation
After selection of the transfected stable cell lines, 1 × 104 Rat1 cells were added to 1.5 ml of growth medium with 0.35% agar and layered onto 2 ml of 0.5% agar base in six-well plates. Cells were fed with 2 ml of medium every 3 days for 4 weeks, after which colonies were stained with MTT (0.5 mg/ml) for 30 min and counted. Colonies visible under a microscope were counted as positive for growth.
Statistics
We performed statistical analysis using ANOVA. Results were considered significant at P<0.05.
Acknowledgments
This work was supported by Cancer Research UK and Leuka. Author contributions: DS designed and performed experiments and helped write manuscript; PL, MY, GN and YKSM performed experiments; JVM co-conceived, designed and supervised initial project execution and helped write manuscript; and SB co-conceived, designed and supervised execution of project and wrote final manuscript.
Glossary
- CML
chronic myeloid leukemia
- IM
imatinib mesylate
- ATIC
5-aminoimidazol-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
- GPI
glucose phosphate isomerase
- CSDA
cold-shock domain protein A
- RSK
p90 ribosomal S6 kinase
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Supplementary Material
References
- Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- Gordon MY, Dowding CR, Riley GP, Goldman JM, Greaves MF. Altered adhesive interactions with marrow stroma of haematopoietic progenitor cells in chronic myeloid leukaemia. Nature. 1987;328:342–344. doi: 10.1038/328342a0. [DOI] [PubMed] [Google Scholar]
- Verfaillie CM, Hurley R, Lundell BI, Zhao C, Bhatia R. Integrin-mediated regulation of hematopoiesis: do BCR/ABL-induced defects in integrin function underlie the abnormal circulation and proliferation of CML progenitors. Acta Haematol. 1997;97:40–52. doi: 10.1159/000203658. [DOI] [PubMed] [Google Scholar]
- Bedi A, Zehnbauer BA, Barber JP, Sharkis SJ, Jones RJ. Inhibition of apoptosis by BCR-ABL in chronic myeloid leukemia. Blood. 1994;83:2038–2044. [PubMed] [Google Scholar]
- Goldman JM, Melo JV. BCR-ABL in chronic myelogenous leukemia—how does it work. Acta Haematol. 2008;119:212–217. doi: 10.1159/000140633. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- Sawyers CL, Hochhaus A, Feldman E, Goldman JM, Miller CB, Ottmann OG, et al. Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: results of a phase II study. Blood. 2002;99:3530–3539. doi: 10.1182/blood.v99.10.3530. [DOI] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi JK, et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorski T, Kanakaraj P, Nieborowska-Skorska M, Ratajczak MZ, Wen SC, Zon G, et al. Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood. 1995;86:726–736. [PubMed] [Google Scholar]
- Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. The 14-3-3 cancer connection. Nat Rev Cancer. 2003;3:931–943. doi: 10.1038/nrc1230. [DOI] [PubMed] [Google Scholar]
- Sano H, Liu SC, Lane WS, Piletz JE, Lienhard GE. Insulin receptor substrate 4 associates with the protein IRAS. J Biol Chem. 2002;277:19439–19447. doi: 10.1074/jbc.M111838200. [DOI] [PubMed] [Google Scholar]
- Aitken A, Baxter H, Dubois T, Clokie S, Mackie S, Mitchell K, et al. Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem Soc Trans. 2002;30:351–360. doi: 10.1042/bst0300351. [DOI] [PubMed] [Google Scholar]
- Benzinger A, Muster N, Koch HB, Yates JR, III, Hermeking H. Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer. Mol Cell Proteomics. 2005;4:785–795. doi: 10.1074/mcp.M500021-MCP200. [DOI] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, et al. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Nellist M, Goedbloed MA, de Winter C, Verhaaf B, Jankie A, Reuser AJ, et al. Identification and characterization of the interaction between tuberin and 14-3-3zeta. J Biol Chem. 2002;277:39417–39424. doi: 10.1074/jbc.M204802200. [DOI] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Peggie M, Wong BH, Morrice N, MacKintosh C. 14-3-3s regulate fructose-2,6-bisphosphate levels by binding to PKB-phosphorylated cardiac fructose-2,6-bisphosphate kinase/phosphatase. EMBO J. 2003;22:3514–3523. doi: 10.1093/emboj/cdg363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinczek B, Brajenovic M, Ebneth A, Drewes G. MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes. J Biol Chem. 2004;279:5915–5923. doi: 10.1074/jbc.M304528200. [DOI] [PubMed] [Google Scholar]
- Tzivion G, Luo ZJ, Avruch J. Calyculin A-induced vimentin phosphorylation sequesters 14-3-3 and displaces other 14-3-3 partners in vivo. J Biol Chem. 2000;275:29772–29778. doi: 10.1074/jbc.M001207200. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Yamaguchi T, Natsume T, Kufe D, Miki Y. JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat Cell Biol. 2005;7:278–285. doi: 10.1038/ncb1228. [DOI] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, et al. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379 (Part 2:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourisseau T, Georgiadis A, Tsapara A, Ali RR, Pestell R, Matter K, et al. Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol Cell Biol. 2006;26:2387–2398. doi: 10.1128/MCB.26.6.2387-2398.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda MS, Garrett MD, Matter K. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J Cell Biol. 2003;160:423–432. doi: 10.1083/jcb.200210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GR, Zheng Y, Che XM, Wang XY, Zhao JH, Wu KJ, et al. Upregulation of human DNA binding protein A (dbpA) in gastric cancer cells. Acta Pharmacol Sin. 2009;30:1436–1442. doi: 10.1038/aps.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NP, Yang H, Mattaini KR, Hunter KW. The metastasis efficiency modifier ribosomal RNA processing 1 homolog B (RRP1B) is a chromatin-associated factor. J Biol Chem. 2009;284:28660–28673. doi: 10.1074/jbc.M109.023457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland BW, Kucab J, Wu J, Lee C, Cheang MC, Yorida E, et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24:4281–4292. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348–6371. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- Sattler M, Salgia R, Okuda K, Uemura N, Durstin MA, Pisick E, et al. The proto-oncogene product p120CBL and the adaptor proteins CRKL and c-CRK link c-ABL, p190BCR/ABL and p210BCR/ABL to the phosphatidylinositol-3′ kinase pathway. Oncogene. 1996;12:839–846. [PubMed] [Google Scholar]
- Roux PP, Richards SA, Blenis J. Phosphorylation of p90 ribosomal S6 kinase (RSK) regulates extracellular signal-regulated kinase docking and RSK activity. Mol Cell Biol. 2003;23:4796–4804. doi: 10.1128/MCB.23.14.4796-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards SA, Dreisbach VC, Murphy LO, Blenis J. Characterization of regulatory events associated with membrane targeting of p90 ribosomal S6 kinase 1. Mol Cell Biol. 2001;21:7470–7480. doi: 10.1128/MCB.21.21.7470-7480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kin Y, Li G, Shibuya M, Maru Y. The Dbl homology domain of BCR is not a simple spacer in P210BCR-ABL of the Philadelphia chromosome. J Biol Chem. 2001;276:39462–39468. doi: 10.1074/jbc.M105484200. [DOI] [PubMed] [Google Scholar]
- Pendergast AM, Quilliam LA, Cripe LD, Bassing CH, Dai Z, Li N, et al. BCR-ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the GRB-2 adaptor protein. Cell. 1993;75:175–185. [PubMed] [Google Scholar]
- Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282:14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima WR, Parreira KS, Devuyst O, Caplanusi A, N′Kuli F, Marien B, et al. ZONAB promotes proliferation and represses differentiation of proximal tubule epithelial cells. J Am Soc Nephrol. 2010;21:478–488. doi: 10.1681/ASN.2009070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Vogt PK. Phosphorylation by Akt disables the anti-oncogenic activity of YB-1. Oncogene. 2008;27:1179–1182. doi: 10.1038/sj.onc.1210719. [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, et al. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol Cell Biol. 2006;26:277–292. doi: 10.1128/MCB.26.1.277-292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial–mesenchymal transition. Cancer Cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Ma Z, Cools J, Marynen P, Cui X, Siebert R, Gesk S, et al. Inv(2)(p23q35) in anaplastic large-cell lymphoma induces constitutive anaplastic lymphoma kinase (ALK) tyrosine kinase activation by fusion to ATIC, an enzyme involved in purine nucleotide biosynthesis. Blood. 2000;95:2144–2149. [PubMed] [Google Scholar]
- Kanno H, Fujii H, Hirono A, Ishida Y, Ohga S, Fukumoto Y, et al. Molecular analysis of glucose phosphate isomerase deficiency associated with hereditary hemolytic anemia. Blood. 1996;88:2321–2325. [PubMed] [Google Scholar]
- Scatena R, Bottoni P, Pontoglio A, Mastrototaro L, Giardina B. Glycolytic enzyme inhibitors in cancer treatment. Expert Opin Invest Drugs. 2008;17:1533–1545. doi: 10.1517/13543784.17.10.1533. [DOI] [PubMed] [Google Scholar]
- Sheng H, Niu B, Sun H. Metabolic targeting of cancers: from molecular mechanisms to therapeutic strategies. Curr Med Chem. 2009;16:1561–1587. doi: 10.2174/092986709788186255. [DOI] [PubMed] [Google Scholar]
- Yoshida C, Yoshida F, Sears DE, Hart SM, Ikebe D, Muto A, et al. Bcr-Abl signaling through the PI-3/S6 kinase pathway inhibits nuclear translocation of the transcription factor Bach2, which represses the antiapoptotic factor heme oxygenase-1. Blood. 2007;109:1211–1219. doi: 10.1182/blood-2005-12-040972. [DOI] [PubMed] [Google Scholar]
- Griswold IJ, MacPartlin M, Bumm T, Goss VL, O′Hare T, Lee KA, et al. Kinase domain mutants of Bcr-Abl exhibit altered transformation potency, kinase activity, and substrate utilization, irrespective of sensitivity to imatinib. Mol Cell Biol. 2006;26:6082–6093. doi: 10.1128/MCB.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson V, Gudmundsdottir K, Luong P, Leung KY, Knebel A, Basu S. JNK phosphorylates Yes-associated protein (YAP) to regulate apoptosis. Cell Death Dis. 2010;1:e29. doi: 10.1038/cddis.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.