Abstract
Bim is known to be critical in killing of melanoma cells by inhibition of the RAF/MEK/ERK pathway. However, the potential role of the most potent apoptosis-inducing isoform of Bim, BimS, remains largely unappreciated. Here, we show that inhibition of the mutant B-RAFV600E triggers preferential splicing to produce BimS, which is particularly important in induction of apoptosis in B-RAFV600E melanoma cells. Although the specific B-RAFV600E inhibitor PLX4720 upregulates all three major isoforms of Bim, BimEL, BimL, and BimS, at the protein and mRNA levels in B-RAFV600E melanoma cells, the increase in the ratios of BimS mRNA to BimEL and BimL mRNA indicates that it favours BimS splicing. Consistently, enforced expression of B-RAFV600E in wild-type B-RAF melanoma cells and melanocytes inhibits BimS expression. The splicing factor SRp55 appears necessary for the increase in BimS splicing, as SRp55 is upregulated, and its inhibition by small interfering RNA blocks induction of BimS and apoptosis induced by PLX4720. The PLX4720-induced, SRp55-mediated increase in BimS splicing is also mirrored in freshly isolated B-RAFV600E melanoma cells. These results identify a key mechanism for induction of apoptosis by PLX4720, and are instructive for sensitizing melanoma cells to B-RAFV600E inhibitors.
Keywords: BimS, B-RAFV600E, B-RAF inhibitors, apoptosis, melanoma
Results from clinical studies with small molecule inhibitors of the mutant B-RAFV600E have been very encouraging, and promise to provide a much needed breakthrough in the treatment of melanoma by targeting B-RAFV600E.1, 2 The latter is found in ∼50% of melanomas, leading to constitutive activation of the RAF/MEK/ERK pathway that is important for melanoma cell growth and survival, and is involved in resistance to many therapeutic approaches.3, 4 However, a number of questions have already been raised from these studies, such as the durability of responses and why some melanomas with mutant B-RAF have not shown major responses.1, 2
It is well established that blockade of the RAF/MEK/ERK pathway inhibits melanoma cell growth.5 In addition, a more desirable outcome, induction of apoptosis, has also been shown in varying in vitro systems, in particular, in B-RAFV600E melanoma cells.6, 7, 8, 9, 10 Apoptosis of such cells was clearly demonstrated in an ex vivo model after administration of the B-RAF inhibitor, PLX4720, that is selective for the mutant B-RAFV600E6. Consistently, regression of metastatic mutant B-RAF melanomas is a frequent sign of the response to administration of PLX4032, a close analogue to PLX4720,1, 2 suggesting that induction of apoptosis may be a major biological consequence of inhibition of mutant B-RAF.
Several mechanisms have been reported to contribute to apoptosis induced by inhibition of the RAF/MEK/ERK pathway. These include dephosphorylation of Bad, translocation of Bmf, upregulation of BimEL, and downregulation of Mcl-1.7, 8, 9, 10, 11 Among them, upregulation of BimEL via inhibition of its phosphorylation and subsequent proteasomal degradation may be the best documented7, 8 and is of particular interest, in that Bim, unlike other more selective Bcl-2 homology 3 (BH3)-only proteins such as Bad and Bmf, can bind with high affinity to and inhibit all prosurvival Bcl-2 family proteins.12 In addition, Bim can directly bind to and activate Bax.12 It is of note that besides posttranslational changes, inhibition of the RAF/MEK/ERK pathway has also been shown to cause upregulation of Bim mRNA.13
There are three major isoforms of Bim, BimEL, BimL, and BimS, that are generated by alternative splicing.14 Although BimS is encoded by exons 2, 5, and 6, BimL is encoded by exons 2, 4, 5, and 6, and BimEL by exons 2, 3, 4, 5, and 6. Both BimL and BimEL contain a binding site for dynein light chain 1,14, 15 hence, their proapoptotic activity is controlled by sequestration to the cytoskeleton-associated dynein motor complex.15 Because exon 3 encodes an ERK1/2-docking domain and ERK1/2 phosphorylation sites, BimEL is subject to phosphorylation by the MEK/ERK pathway that targets it for proteasomal degradation and also prevents its binding to Bax.16 BimS is not subject to any known posttranslational regulation and is the most potent apoptosis inducer among the three isofoms.13, 16, 17
Alternative splicing is a tightly regulated process that generates multiple functional variants from individual genes, thus enhancing protein diversity.18 Alternative splicing patterns are frequently altered in cancer cells, resulting in aberrant expression of mRNA and protein variants that have been proposed to have unique properties to confer biological characteristics of the cells.19, 20, 21, 22 The splicing process is catalyzed by the spliceosome that is composed of cis-acting elements, such as splicing enhancers and silencers, and trans-acting factors, including the serine/arginine-rich (SR) and heterogeneous ribonucleoprotein particle (hnRNP) protein families. SR proteins are characterized by one or two RNA recognition motifs at the N-terminal and have an important part in splice-site selection through association with splicing enhancers and silencers.18 Changes in the expression of a number of SR proteins have been found in various types of cancer cells.23, 24, 25, 26
To better understand the mechanism(s) by which inhibition of B-RAFV600E induces apoptosis of melanoma cells, we have examined completely the apoptotic response of B-RAFV600E melanoma cells to the B-RAFV600E inhibitor PLX4720. We show in this report that preferential splicing to produce BimS has an important role in induction of apoptosis by PLX4720 in B-RAFV600E melanoma cells. Moreover, we demonstrate that the increase in BimS splicing is mediated by the SR protein, SRp55.
Results
The B-RAFV600E inhibitor PLX4720 induces apoptosis in B-RAFV600E melanoma cells
Our initial studies confirmed that the small molecule compound PLX4720 is specific for inhibition of B-RAFV600E. This was shown by its inhibitory effect on ERK activation in B-RAFV600E melanoma cell lines, but not in those carrying the wild-type B-RAF even when it was used at 10 μM (Figure 1a and Supplementary Figure 1A). The inhibitory effect of PLX4720 at 10 μM on activation of ERK was sustained till 72 h after treatment (Figure 1a). Examination of the effect of PLX4720 on cell growth similarly demonstrated that it inhibited proliferation of B-RAFV600E melanoma cells, but had only minimal effects on growth of those harboring the wild-type B-RAF (Figure 1b and Supplementary Figures 1B).
Figure 1.
PLX4720 inhibits proliferation and induces apoptosis in mutant B-RAF melanoma cells. (a) Whole cell lysates from Mel-RM (wild-type B-RAF) and Mel-RMu (B-RAFV600E) cells treated with PLX4720 at indicated concentrations for 72 h (upper panel) or at 10 μM for indicated periods (lower panel), were subjected to western blot analysis of phosphorylated ERK1/2 and ERK1/2. (b) Mel-RM and Mel-RMu cells were treated with PLX4720 at indicated concentrations for 72 h. Cell viability (left panel) and apoptosis (right panel) were quantitated by the MTS assay and propidium iodide (PI) method, respectively. The data shown are the mean±S.E. of three individual experiments. (c) Mel-RM and Mel-RMu cells were treated with PLX4720 at 10 μM for indicated periods. Cell viability (left panel) and apoptosis (right panel) were quantitated by the MTS assay and PI method, respectively. The data shown are the mean±S.E. of three individual experiments. (d) A summary of the effect of PLX4720 on cell survival in a panel of mutant and wild-type B-RAF melanoma cell lines. Cells treated with PLX4720 at 10 μM for 72 h were subjected to MTS assays. The data shown are the mean±S.E. of three individual experiments. (e) Left panel: B-RAFV600E Mel-RMu and Mel-CV cells were transfected with the control or B-RAF siRNA. After 24 h, whole cell lysates were subjected to western blot analysis of B-RAF, phosphorylated ERK1/2, and ERK1/2. Western blot analysis of A-RAF and C-RAF was included as controls to show the specificity of the B-RAF siRNA. Right panel: Mel-RMu and Mel-CV cells were transfected with the control or B-RAF siRNA. After 24 h, cells were treated with PLX4720 (10 μM) for a further 72 h. Apoptotic cells were measured by the PI method. The data shown are either representative (left panel), or the mean±S.E. (right panel), of three individual experiments
We examined whether induction of apoptosis was involved in PLX4720-mediated inhibition of cell growth in Mel-RMu (B-RAFV600E) and Mel-RM (wild-type B-RAF) cells. At concentrations of up to 1 μM, PLX4720 did not induce significant apoptosis by 72 h (Figure 1b). At 3 μM, it induced apoptosis in ∼30% of Mel-RMu cells, but apoptosis in Mel-RM cells remained marginal. When it was used at 10 μM, >65% of Mel-RMu cells and ∼20% of Mel-RM cells underwent apoptosis, which corresponded well with the efficiency of inhibition of cell viability in both cell lines (Figure 1b). Predominant induction of apoptosis by PLX4720 at 10 μM was confirmed by treating the cells with the compound at the same concentration for varying time periods (Figure 1c). This was also shown by activation of caspase-3 and -7, and cleavage of the caspase-3 substrate PARP (Supplementary Figure 2).
Studies in a panel of melanoma cell lines indicated that B-RAFV600E lines were, in general, significantly more sensitive to PLX4720 than those harboring the wild-type B-RAF (P<0.01; two-tailed student's t-test) (Figure 1d). Similar to PLX4720, Small interfering RNA (siRNA) knockdown of B-RAF induced apoptosis in two B-RAFV600E melanoma cell lines, indicating that induction of apoptosis by PLX4720 is due to inhibition of B-RAFV600E (Figure 1e).
PLX4720 preferentially enhances splicing of BimS
Overexpression of Bcl-2 inhibited induction of apoptosis by PLX4720 in B-RAFV600E melanoma cells, indicating that the mitochondrial apoptotic pathway is essential in PLX4720-induced apoptosis (Figure 2a). In support of this, treatment with PLX4720 resulted in activation of Bax, and mitochondrial release of cytochrome c and apoptosis-inducing factor (AIF) (Supplementary Figure 3). These results suggest that activation of one or more BH3-only proteins of the Bcl-2 family is important in initiating PLX4720-mediated apoptotic signaling.27 As shown in Figure 2b, PLX4720 caused upregulation of the Bim isoforms, BimEL, BimL, and BimS, in B-RAFV600E Mel-RMu cells, but not in wild-type B-RAF Mel-RM cells. In particular, the increase in BimS was most prominent and sustained. The changes in BimEL expression was associated with reduction in the levels of an extra band, with reduced electrophoretic motility that corresponds to phosphorylated BimEL.13 Of note, PLX4720 also induced a novel protein product with an apparent molecular weight between BimL and BimS at 36 h after treatment (Figure 2b). In contrast to regulation of Bim, PLX4720 did not cause any significant changes in other Bcl-2 family proteins analyzed, except for downregulation of the anti-apoptotic proteins Mcl-1 and Bcl-2 at relatively late stages (36 h after treatment) in Mel-RMu cells (Figure 2b). Regulation of Bim by PLX4720 was confirmed in another three B-RAF-mutant melanoma cell lines (Supplementary Figure 4).
Figure 2.
PLX4720 upregulates Bim. (a) Upper panel: overexpression of Bcl-2 in Mel-RMu and Mel-CV cells stably transfected with cDNA encoding Bcl-2. Whole cell lysates were subjected to western blot analysis of Bcl-2 and GAPDH (as a loading control). Lower panel: Mel-RMu and Mel-CV cells overexpressing Bcl-2 were treated with PLX4720 (10 μM) for 72 h before apoptosis was quantitated by the propidium iodide (PI) method. The data shown are either representative (upper panel) or mean±S.E. (lower panel) of three individual experiments. (b) Whole cell lysates from Mel-RM and Mel-RMu cells treated with PLX4720 (10 μM) for indicated time periods were subjected to western blot analysis of Bim, Bid, PUMA, Noxa, Bax, Bak, Mcl-1, Bcl-2, and GAPDH (as a loading control). The data shown are representative of three individual western blot analyses. (c) Left panel: total RNA from Mel-RM and Mel-RMu cells treated with PLX4720 (10 μM) for indicated time periods was isolated and subjected to real-time PCR analysis for Bim mRNA expression. The relative abundance of mRNA expression before treatment was arbitrarily designated as 1. Right panel: total RNA from Mel-RM and Mel-RMu cells treated with PLX4720 (10 μM) for 16 h with or without pretreatment with actinomycin D (Act-D) (3 μg/ml) for 1 h were subjected to real-time PCR analysis. The relative abundance of mRNA expression before treatment was arbitrarily designated as 1. The data shown are the mean±S.E. of three individual experiments
The marked increase in BimS induced by PLX4720 was intriguing because, unlike BimEL and BimL, this isoform is not regulated by any known posttranslational mechanisms.13, 15 We reasoned that upregulation of BimS is a consequence of enhanced Bim transcription and a subsequent increase in splicing to produce BimS. To test this, we first quantitated the Bim mRNA expression before and after exposure to PLX4720. As shown in Figure 2c, PLX4720 triggered a rapid and sustained increase in Bim mRNA in Mel-RMu cells, which could be efficiently inhibited by pretreatment with actinomycin D (Figure 2c), suggesting that this was due to a transcriptional increase, rather than a change in the mRNA stability.
We next monitored the levels of the three major Bim mRNA species in Mel-RMu cells before and after exposure to PLX4720 in qPCR analysis. Figure 3a shows that, although they were all increased by treatment with PLX4720, the ratio of BimS mRNA to BimEL mRNA in Mel-RMu cells after treatment for 16 h was four times higher than that before treatment (Figure 3b). Similarly, the ratio of BimS mRNA to BimL mRNA was also increased, although to a lesser extent (Figure 3b). The increase in BimS mRNA relative to BimEL and BimL mRNA was confirmed in an additional three B-RAF-mutant melanoma cell lines (Supplementary Figure 5). Collectively, these results suggest that PLX4720 may cause preferential splicing to produce BimS.
Figure 3.
PLX4720 preferentially increases splicing to produce BimS. (a) Total RNA from Mel-RMu cells treated with PLX4720 (10 μM) for 16 h was subjected to real-time PCR analysis for BimEL, BimL, and BimS mRNA expression. The levels of the expression of individual species before treatment were arbitrarily designated as 1. The data shown are the mean±S.E. of three individual experiments. (b) Total RNA from Mel-RMu cells treated with PLX4720 (10 μM) for 16 h was subjected to BimEL, BimL, and BimS mRNA expression as in a. Left panel: the ratios between the levels of BimS mRNA and BimEL mRNA before and after treatment, respectively, were calculated as (ΔΔCt of BimS/ΔΔCt of BimEL). Right panel: the ratios between the levels of BimS mRNA and BimL mRNA before and after treatment, respectively, were calculated as (ΔΔCt of BimS/ΔΔCt of BimL). The data shown are the mean±S.E. of three individual experiments. (c) Upper panel: total RNA from Mel-RM and Mel-RMu cells treated with SBHA (10 μg/ml) for 16 h were subjected to real-time PCR analysis for Bim mRNA expression. The relative abundance of mRNA expression before treatment was arbitrarily designated as 1. Lower panel: whole cell lysates from cells treated as above were subjected to western blot analysis of Bim and GAPDH (as a loading control). The strong band denoted by the arrowhead is nonspecific, and was associated with the particular batch of the antibody against Bim used in the experiment. The data shown are either representative (lower panel) or the mean±S.E. (upper panel) of three individual experiments. (d) Total RNA from Mel-RM and Mel-RMu cells treated with SBHA (10 μg/ml) for 16 h were subjected to real-time PCR analysis for BimEL, BimL, and BimS mRNA expression. The ratios of BimS mRNA to BimEL and BimL mRNA before and after treatment, respectively, were calculated as in b. The data shown are the mean±S.E. of three individual experiments
To confirm that the increase in BimS splicing is specific to inhibition of B-RAFV600E by PLX4720, we treated Mel-RMu and Mel-RM cells with the histone deacetylase inhibitor suberic bishydroxamate (SBHA), which is known to upregulate Bim at the transcriptional level.28 Figure 3c shows that, as reported before, the Bim mRNA and protein levels were upregulated by SBHA in melanoma cells, regardless of their B-RAF mutational status. Although this increase was reflected at the levels of the three Bim mRNA species, the ratios of the BimS mRNA to the BimEL and BimL mRNA in both cell lines before and after treatment remained unaltered (Figure 3d).
Enforced expression of B-RAFV600E inhibits Bims expression in melanocytes and melanoma cells
We transfected cDNA encoding B-RAFV600E into wild-type B-RAF Mel-RM and Me1007 cells. Enforced expression of B-RAFV600E resulted in increases in activation of ERK (Figure 4a). Because the three Bim protein variants were all constitutively expressed at low levels in both cell lines, it was not feasible to judge whether enforced expression of B-RAFV600E resulted in downregulation of the proteins. To overcome this limitation, we treated the cells transfected with B-RAFV600E with SBHA, and monitored changes in the three mRNA species. Figure 4b shows that enforced expression of B-RAFV600E blocked increases in BimS mRNA induced by SBHA. Inhibition of SBHA-mediated induction of BimS by enforced expression of B-RAFV600E was also mirrored at the protein level (Figure 4c).
Figure 4.
Enforced expression of B-RAFV600E inhibited BimS expression in wild-type B-RAF melanoma cells and melanocytes. (a) Mel-RM and Me1007 cells were stably transfected with cDNA encoding B-RAF carrying the V600E mutation. Whole cell lysates were subjected to western blot analysis of B-RAF, pERK1/2, and ERK1/2. The data shown are representative of three individual western blot analyses. (b) Mel-RM and Me1007 cells were stably transfected with cDNA encoding B-RAF carrying the V600E mutation. Cells were treated with SBHA (10 μg/ml) for a further 16 h. Total RNA was isolated and subjected to real-time PCR analysis for BimEL, BimL, and BimS mRNA expression. The levels of the expression of individual species before treatment were arbitrarily designated as 1. The data shown are the mean±S.E. of three individual experiments. (c) Whole cell lysates from Mel-RM and Me1007 cells treated as in b were subjected to western blot analysis of Bim and GAPDH (as a loading control). The arrowhead points to nonspecific bands. The data shown are representative of three individual western blot analyses. (d) Melanocytes were transiently transfected with cDNA encoding B-RAF carrying the V600E mutation. After 24 h, whole cell lysates were subjected to western blot analysis of B-RAF, pERK1/2, ERK1/2, Bim, and Bcl-2. The arrowhead points to nonspecific bands. The data shown are representative of three individual western blot analyses
The effect of B-RAFV600E on the expression of the three Bim isoforms was also examined in a cultured melanocyte line that was transfected with cDNA encoding B-RAFV600E. It was notable that the BimEL, BimL, and BimS proteins were all constitutively expressed at detectable, although moderate, levels in cultured melanocytes (Figure 4d). Survival of melanocytes in the presence of these Bim isoforms is conceivable owing to sequestration of BimEL and BimL in the cytoskeleton,13, 14 and neutralization of BimS by anti-apoptotic Bcl-2 family proteins (Figure 4d).12 Consistent with its inhibitory effect on the expression of BimS in melanoma cells, enforced expression of B-RAFV600E caused a decrease in this isoform in melanocytes (Figure 4d). Intriguingly, there was an increase in the levels of BimL in melanocytes transfected with B-RAFV600E, suggesting that the effect of B-RAFV600E on regulation of Bim expression may be more complex than just impinging on BimEL and BimS, and may vary between different types of cells.
BimS has a dominant role in apoptosis of B-RAFV600E melanoma cells induced by PLX4720
To examine the relative importance of BimEL and BimS in PLX4720-induced apoptosis, we transfected siRNA specific for Bim in general, BimEL, and BimS into Mel-RMu cells (Figures 5a–c). Although siRNA knockdown of BimEL inhibited PLX4720-induced apoptosis by 30%, inhibition of BimS by siRNA blocked apoptosis induction by PLX4720 by 56% (Figure 5d). These results indicate that both BimEL and BimS are involved in induction of apoptosis of B-RAFV600E melanoma cells by PLX4720, but BimS has a greater part than BimEL. Although we did not specifically measure the role of BimL because of technical limitations in designing siRNA that specifically silences BimL, it is conceivable that this isoform also contributes to PLX4720-induced apoptosis. More potent inhibition of PLX4720-induced apoptosis by knockdown of BimS was also demonstrated in another two B-RAFV600E melanoma cell lines (Supplementary Figure 6).
Figure 5.
BimS has a critical role in PLX4720-induced apoptosis of mutant B-RAF melanoma cells. (a) Left panel: Mel-RMu and Mel-CV cells (B-RAFV600E) were transfected with the control or Bim siRNA. After 24 h, cells were treated with PLX4720 for 16 h. Total RNA was isolated and subjected to real-time PCR analysis for Bim mRNA expression. The relative abundance of mRNA expression in cells transfected with the control siRNA before treatment was arbitrarily designated as 1. Right panel: Mel-RMu and Mel-CV cells were transfected with the control or Bim siRNA. After 24 h, cells were treated with PLX4720 for a further 72 h. Apoptosis was quantitated by the propidium iodide (PI) method. The data shown are the mean±S.E. of three individual experiments. (b) Mel-RMu cells were transfected with the control, BimEL, or BimS siRNA. After 24 h, cells were treated with PLX4720 (10 μM) for 16 h. Total RNA was isolated and subjected to real-time PCR analysis for BimEL (left panel) and BimS (right panel) mRNA expression. The levels of the expression of individual species in cells transfected with the control siRNA without treatment were arbitrarily designated as 1. The data shown are the mean±S.E. of three individual experiments. (c) Whole cell lysates from Mel-RMu cells treated as in b were subjected to western blot analysis of Bim and GAPDH (as a loading control). The arrowhead points to nonspecific bands. The data shown are representative of three individual western blot analyses. (d) Mel-RMu cells with BimEL or BimS knocked down as in b were treated with PLX 4720 at 10 μM for 72 h. Apoptosis was quantitated by the PI method. The data shown are the mean±S.E. of three individual experiments. (e) Left panel: Mel-RMu and Mel-CV cells were transfected with pCMV6-AC-GFP or pCMV6-AC-BimS-GFP. After 24 h, whole cell lysates were subjected to western blot analysis of BimS-GFP using an antibody against GFP. Western blot analysis of GAPDH was then performed as a loading control. Right panel: Mel-RMu and Mel-CV cells were transfected with pCMV6-AC-GFP or pCMV6-AC-BimS-GFP. After 24 h, mitochondrial fractions were subjected to western blot analysis of BimS-GFP using an antibody against GFP. Western blot analysis of COX IV was then performed as a loading control. (f) Left panel: Representative flow cytometric histograms of measurement of apoptosis using PE-conjugated Annexin-V in Mel-RMu and Mel-CV cells transfected with pCMV6-AC-GFP or pCMV6-AC-BimS-GFP. PE-positive cells were quantitated in gated GFP-positive cell populations. The numbers represent percentages of positive cells. Right panel: Mel-RMu and Mel-CV cells were transfected with pCMV6-AC-GFP or pCMV6-AC-BimS-GFP for indicated time periods. Apoptotic cells were quantitated with PE-conjugated Annexin-V in gated GFP-positive cell populations. The data shown are representative of two experiments. (g) Mitochondrial fractions from Mel-RMu and Mel-CV cells treated with PLX4720 (10 μM) for indicated time periods were subjected to western blot analysis of Bim and COX IV (as a control). The data shown are representative of three individual western blot analyses
To further consolidate the role of BimS in induction of apoptosis of mutant B-RAF melanoma cells, the GFP-tagged open reading frame of human BimS cDNA cloned into the pCMV6-AC vector (pCMV6-AC-BimS-GFP) was transiently transfected into Mel-RMu and Mel-CV cells (Figure 5e). Figure 5f shows that overexpression of BimS induced apoptosis of the cells that could be detected as early as 16 h after transfection. By 48 h, ∼50% of the cells in both cell lines had committed to apoptosis. It is of note that BimS-GFP is readily detected in mitochondrial fractions at 24 h (Figure 5e), consistent with previous reports that BimS-induced apoptosis requires its mitochondrial localization.17 As shown in Figure 5g, exposure to PLX4720 similarly resulted in marked relocation of endogenous BimS onto mitochondria in Mel-RMu and Mel-CV cells.
The SR protein SRp55 is involved in increased splicing of BimS triggered by PLX4720 in B-RAFV600E melanoma cells
The gene encoding the SR protein SRp55, splicing factor arginine/serine-rich 6 (SFRS6), has been shown to be upregulated in B-RAFV600E melanoma cells.29 We therefore studied whether SRp55 is involved in regulation of alternative splicing of Bim in B-RAFV600E melanoma cells by inhibition of B-RAFV600E. Surprisingly, the levels of the SRp55 protein appeared similar between B-RAFV600E melanoma cell lines and those in the wild-type B-RAF (Figure 6a). However, in response to treatment with PLX4720, the levels were increased in B-RAFV600E Mel-RMu cells, but not in wild-type Mel-RM cells (Figure 6a). Similarly, treatment with PLX4720 resulted in a marked increase (fivefold) in the expression levels of the SFRS6 mRNA in Mel-RMu but not in Mel-RM cells (Figure 6b).
Figure 6.
SRp55 has a role in upregulation of BimS by PLX4720. (a) Left panel: whole cell lysates from a panel of mutant and wild-type B-RAF melanoma cell lines were subjected to western blot analysis of SRp55 and GAPDH (as a loading control). Right panel: whole cell lysates from Mel-RM and Mel-RMu cells treated with PLX4720 (10 μM) for indicated time periods were subjected to western blot analysis of SRp55 and GAPDH (as a loading control). The data shown are representative of three individual western blot analyses. (b) Total RNA from Mel-RM and Mel-RMu cells treated with PLX4720 (10 μM) for 16 h was isolated and subjected to real-time PCR analysis for SFRS6. The relative abundance of mRNA expression before treatment was arbitrarily designated as 1. The data shown are the mean±S.E. of three individual experiments. (c) Mutant B-RAF Mel-RMu and Mel-CV cells were transfected with the control, SFRS6, and SFRS12 siRNAs. After 24 h, total RNA was isolated and subjected to real-time PCR analysis for SFRS6 and SFRS12 mRNA expression. The relative abundance of mRNA expression in cells transfected with the control siRNA was arbitrarily designated as 1. The data shown are the mean±S.E. of three individual experiments. (d) Whole cell lysates from cells treated as in c were subjected to western blot analysis of SRp55, SRrp86, and GAPDH (as a loading control). The data shown are representative of three individual western blot analyses. (e) Mel-RMu and Mel-CV cells were transfected with the control, SFRS6, and SFRS12 siRNAs. After 24 h, cells were treated with PLX4720 (10 μM) for a further 16 h. Total RNA was isolated and subjected to real-time PCR analysis for BimS mRNA expression. The relative abundance of mRNA expression in cells transfected with the control siRNA without treatment with PLX4720 was arbitrarily designated as 1, which was not shown. The data shown are the mean±S.E. of three individual experiments. (f) Mel-RMu and Mel-CV cells were transfected with the control, SFRS6, and SFRS12 siRNA. After 24 h, total RNA was isolated and subjected to real-time PCR analysis for BimEL, BimL, and BimS mRNA expression. Left panel: the ratios between the levels of BimS mRNA and BimEL mRNA before and after treatment, respectively, were calculated as (ΔΔCt of BimS/ΔΔCt of BimEL). Right panel: the ratios between the levels of BimS mRNA and BimL mRNA before and after treatment, respectively, were calculated as (ΔΔCt of BimS/ΔΔCt of BimL). The data shown are the mean±S.E. of three individual experiments. (g) Mel-RMu and Mel-CV cells were transfected with the control and SFRS6 siRNA, respectively. After 24 h later, whole cell lysates were subjected to western blot analysis of Bim and GAPDH (as a loading control). The arrowhead points to nonspecific bands generated by the antibody against Bim. The data shown are representative of three individual western blot analyses. (h) Mel-RMu and Mel-CV cells were transfected with the control, SFRS6, and SFRS12 siRNA. After 24 h, cells were treated with PLX4720 (10 μM) for a further 72 h. Apoptosis was quantitated by the propidium iodide (PI) method. The data shown are the mean±S.E. of three individual experiments. (i) Left panel: Mel-Rmu and Mel-CV cells were transfected with with pCMV6-AC-GFP or pCMV6-AC-SFRS6-GFP. After 48 h, cells were harvested and apoptosis was measured with PE-conjugated Annexin-V in gated GFP-positive cell populations. The data shown are representative of two experiments. Right panel: Mel-Rmu and Mel-CV cells were transfected with with pCMV6-AC-GFP or pCMV6-AC-SFRS6-GFP. After 24 h, whole cell lysates were subjected to western blot analysis of SRp55-GFP and BimS. The arrowhead points to nonspecific bands. Western blot analysis of GAPDH was then performed as a loading control. The data shown are representative of three individual western blot analyses
We next transfected a siRNA pool for SFRS6 into Mel-RMu and Mel-CV cells. Transfection of a siRNA pool for splicing factor arginine/serine-rich 12 (SFRS12) was included as a control (Figures 6c and d). Inhibition of SRp55 but not of SRrp86, blocked the increase in the BimS mRNA and protein, and the increases in the ratios of the BimS mRNA to the BimEL and BimL mRNAs was induced by PLX4720 (Figures 6e–g). This was associated with attenuation of PLX4720-induced killing in both Mel-RMu and Mel-CV cells (Figure 6h). Figure 6i shows that overexpression of SRp55 resulted in moderate levels of apoptosis in Mel-RMu and Mel-CV cells in the absence of any further treatment. This was associated with an increase in BimS to varying degrees in Mel-RMu and Mel-CV cells (Figure 6i).
PLX4720 preferentially increases BimS and induces apoptosis in fresh melanoma isolates carrying B-RAFV600E
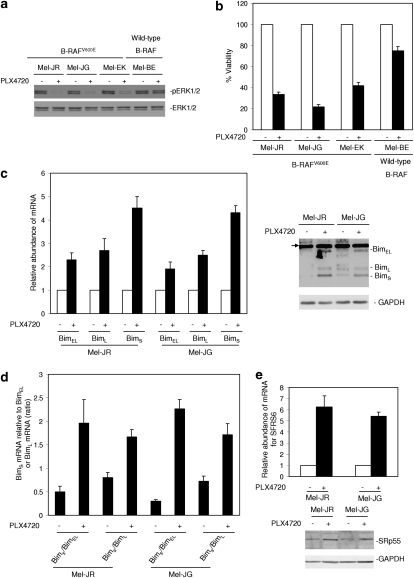
As shown in Figure 7a, PLX4720 inhibited activation of ERK1/2 in two fresh isolates with B-RAFV600E but not in the one with wild-type B-RAF. Consistently, PLX4720 markedly reduced the viability of the B-RAFV600E cells but not of wild-type B-RAF cells (Figure 7b). Two B-RAFV600E fresh isolates were used for further studies. PLX4720 upregulated the three Bim mRNA species in both fresh isolates (Figure 7c). It also upregulated all three protein variants, except for BimL, in Mel-JG cells (Figure 7c). Notably, there were also discrepancies in the levels of other mRNA species and corresponding protein variants after treatment with PLX4720. This suggests that mechanisms other than those mediated by inhibition of B-RAFV600E may be involved in regulation of the expression of Bim protein variants. Nevertheless, similar to results with melanoma cell lines, PLX4720 triggered increases in the ratios of the BimS mRNA to the BimEL and BimL mRNAs in both fresh isolates (Figure 8a). This was associated with an increase in the levels of the SRp55 protein and the SFRS6 mRNA (Figure 8b). Inhibition of BimS by siRNA partially restored viability of the cells, whereas inhibition of SRp55 by siRNA blocked the increase in BimS and similarly inhibited killing induced by PLX4720 (Figure 8c).
Figure 7.
PLX4720 inhibits activation of ERK1/2, reduces cell viability, upregulates BimS, increases the ratios of BimS mRNA to BimEL and BimL mRNA, and upregulates SRp55 in mutant B-RAF fresh melanoma isolates. (a) Whole cell lysates from fresh melanoma isolates harboring mutant B-RAF (Mel-JR, Mel-JG, and Mel-EK) or wild-type B-RAF (Mel-BE) treated with PLX4720 (10 μM) for 16 h were subjected to western blot analysis of pERK1/2 and ERK1/2. The data shown are representative of three individual western blot analyses. (b) Fresh melanoma isolates were treated with PLX4720 (10 μM) for 72 h before cell viability was quantitated by MTS assays. The data shown are the mean±S.E. of three individual experiments. (c) Left panel: freshly isolated Mel-JR and Mel-JG cells were treated with PLX4720 (10 μM) for 16 h. Total RNA was isolated and subjected to real-time PCR analysis for BimEL, BimL, and BimS mRNA expression. The levels of the expression of individual species before treatment were arbitrarily designated as 1. Right panel: whole cell lysates from freshly isolated Mel-JR and Mel-JG cells treated with PLX4720 (10 μM) for 16 h were subjected to western blot analysis of Bim and GAPDH (as a loading control). The arrowhead points to nonspecific bands. The data shown are either representative (right panel) or the mean±S.E. (left panel) of three individual experiments. (d) Freshly isolated Mel-JR and Mel-JG cells were treated with PLX4720 (10 μM) for 16 h. Total RNA was isolated and subjected to real-time PCR analysis for BimEL, BimL, and BimS mRNA expression. The ratios between the levels of BimS mRNA and BimEL mRNA before and after treatment, respectively, were calculated as (ΔΔCt of BimS/ΔΔCt of BimEL), and the ratios between the levels of BimS mRNA and BimL mRNA before and after treatment, respectively, were calculated as (ΔΔCt of BimS/ΔΔCt of BimL). The data shown are the mean±S.E. of three individual experiments. (e) Upper panel: total RNA from Mel-JR and Mel-JG cells treated with PLX4720 as in a was subjected to real-time PCR analysis for SFRS6 mRNA expression. The relative abundance of SFRS6 mRNA in cells before treatment was arbitrarily designated as 1. Lower panel: whole cell lysates from Mel-JR and Mel-JG cells treated as given above were subjected to western blot analysis of SRp55 and GAPDH (as a loading control). The data shown are either representative (lower panel) or the mean±S.E. (upper panel) of three individual experiments
Figure 8.
Inhibition of BimS by siRNA reverses the reduction in cell viability induced by PLX4720, whereas inhibition of SRp55 blocks induction of BimS and induction of apoptosis by PLX4720 in mutant B-RAF fresh melanoma isolates. (a) Left panel: Mel-JR and Mel-JG cells were transfected with the control and BimS siRNA, respectively. After 24 h, whole cell lysates were subjected to western blot analysis of Bim and GAPDH (as a loading control). The arrowhead points to nonspecific bands. Right panel: Mel-JR and Mel-JG cells were transfected with the control and BimS siRNA, respectively. After 24 h, cells were treated with PLX4720 (10 μM) for a further 72 h. Apoptosis was measured by the propidium iodide (PI) method. The data shown are either representative (left panel) or the mean±S.E. (right panel) of three individual experiments. (b) Mel-JR and Mel-JG cells were transfected with the control and SFRS6 siRNA, respectively. After 24 h, cells were treated with PLX4720 (10 μM) for a further 16 h. Whole cell lysates were subjected to western blot analysis of SRp55, Bim, and GAPDH (as a loading control). The arrowhead points to nonspecific bands. The data shown are representative of three individual western blot analyses. (c) Total RNA from Mel-JR and Mel-JG cells treated as in b was subjected to real-time PCR analysis for BimS mRNA expression. The relative abundance of BimS mRNA in cells transfected with the control siRNA without treatment with PLX4720 was designated as 1, which was not shown. The data shown are the mean±S.E. of three individual experiments. (d) Mel-JR and Mel-JG cells were transfected with the control and SFRS6 siRNA, respectively. After 24 h, cells were treated with PLX4720 (10 μM) for a further 72 h. Apoptosis was quantitated by the PI method. The data shown are either the mean±S.E. (right panel of a, c, and d) or representative (left panel of a and b) of three individual experiments. The data shown are the mean±S.E. of three individual experiments
Discussion
The above results extend the role of Bim in apoptosis induced by inhibition of B-RAFV600E beyond upregulation of BimEL by showing that PLX4720 triggers preferential splicing to produce BimS, which has a greater part in induction of apoptosis than BimEL.7, 8 In addition, the results demonstrate that the increase in splicing of BimS is due to a mechanism that is regulated by the splicing factor SRp55, which is increased in B-RAFV600E melanoma cells by PLX4720.
In support of previous observations,30 BimEL was upregulated by PLX4720 in B-RAF-mutant melanoma cells, which was associated with a reduction in the levels of an extra band with reduced electrophoretic motility that corresponds to phosphorylated BimEL.13 This was consistent with inhibition of activation of ERK1/2 by PLX4720,5 in that ERK1/2 can phosphorylate BimEL, thereby causing its ubiquitination and degradation by the proteasome system.13 Inhibition of this pathway has been suggested to account for a major part of the accumulation of BimEL.13, 30 Strikingly, BimL, and in particular, BimS, were also increased by PLX4720 in B-RAFV600E melanoma cells. Nevertheless, the kinetics and sustainability of upregulation of the three Bim isoforms varied from one another, suggesting that the mechanisms responsible for upregulation of the individual proteins may be different.
The marked increase in BimS induced by PLX4720 is of particular interest, in that BimS is rarely detectable at the protein level in cells.13, 16 This is presumably associated with its stronger potency in induction of apoptosis than other isoforms, as BimS is not subject to any posttranslational regulation and is instantly activated once it is expressed.13 When overexpressed, BimS can rapidly translocate onto the mitochondrial outer membrane where it recruits and activates Bax independently of inhibition of anti-apoptotic Bcl-2 family proteins.17 Further studies by real-time revealed that the BimS transcript was preferentially induced by PLX4720 in B-RAFV600E melanoma cells. Moreover, the preferential induction of splicing to produce BimS seemed to be specific for PLX4720, as treatment of B-RAFV600E melanoma cells with the histone deacetylase inhibitor SBHA, which is known to increase transcription of Bim, did not cause selective upregulation of the BimS transcript, although the levels of all three mRNA species were increased.28 The preferential splicing to produce BimS is of functional significance, in that specific inhibition of BimS resulted in a greater degree of inhibition of PLX4720-induced killing than selective inhibition of BimEL.
The preferential induction of splicing of BimS suggests that the mutant B-RAFV600E may regulate Bim alternative splicing in melanoma cells, and in particular, may suppress splicing to produce BimS. This was supported by the finding that enforced expression of B-RAFV600E in wild-type B-RAF melanoma cells blocked upregulation of the BimS transcript by SBHA, but had no effect on upregulation of the BimEL and BimL mRNA. Furthermore, enforced expression of B-RAFV600E in melanocytes resulted in decreases in the BimS mRNA and protein, but intriguingly, the levels of the BimL mRNA and protein were increased by enforced expression of B-RAFV600E These results suggest that regulation of Bim splicing by mutant B-RAF is more complex than inhibition of splicing of BimS, and may vary between different cell types. It is of interest that, in contrast to melanoma cells, melanocytes expressed readily detectable levels of BimS together with BimEL and BimL. This may indicate that BimS expression is lost during melanoma development as a consequence of mutations in B-RAF. Bim has been shown to be decreased with melanoma progression.31
Altered splicing patterns, in particular, changes in splicing patterns of apoptosis-related genes, are frequently found in various cancers.20, 21, 23 Although the mechanisms underlying this remain unclear, it is well established that alternative splicing is tightly regulated by splicing factors, including the SR and hnRNP protein families.25, 27, 32 There is a growing body of evidence showing that some protein kinases such as Akt and ERK1/2 that have important roles in cancer development also have roles in regulation of activity of SR proteins, probably by modulating their phosphorylation status.33, 34, 35 In this study, we found that one of the SR proteins, SRp55, was associated with preferential splicing to produce BimS after inhibition of mutant B-RAF by PLX4720. This was demonstrated by the findings that PLX4720 upregulated SRp55, and that inhibition of SRp55 with siRNA blocked upregulation of the BimS transcript and reversed the increases in ratios of the BimS mRNA to the BimEL and BimL mRNA induced by PLX4720. Consistently, knockdown of SRp55 partially inhibited apoptosis induced by PLX4720 in mutant B-RAF melanoma cells. Therefore, the mutant B-RAFV600E appears to regulate the expression of SRp55 that in turn has a role in regulating alternative splicing of Bim, in particular, in promoting splicing to produce BimS.
The finding that killing of mutant B-RAF fresh melanoma isolates by PLX4720 was similarly associated with upregulation of Bim, in particular, BimS, is of particular importance, in that this may reflect more closely the reaction of melanoma cells in vivo to treatment with B-RAF inhibitors. Together, the results reported in this study identify induction of BimS as a key mechanism for induction of apoptosis by PLX4720 in melanoma cells carrying B-RAFV600E. We speculate that this may be critical for long-term clinical responses to the inhibitor. Plasma concentrations of PLX4032 of around 60 μM were apparently not associated with significant adverse effects in phase I clinical trials with PLX4032,1 suggesting that the concentrations used in this study are achievable clinically.
Materials and Methods
Cell lines
Human melanoma cell lines Mel-RM, Me1007, Mel-RMu, MM200, Mel-CV, and Sk-Mel-110 have been described previously.36, 37 They were cultured in DMEM containing 5% FCS (Commonwealth Serum Laboratories, Melbourne, Vic, Australia). Melanocytes were kindly provided by Dr P Parsons (Queensland Institute of Medical Research, Qld, Australia) and cultured in medium supplied by Clonetics (Edward Kellar, Vic, Australia).
Fresh melanoma isolates
Isolation of melanoma cells from fresh surgical specimens was carried out as described previously.38 Protocols were approved by the Human Research Ethics Committee of Hunter New England Health, Australia.
Antibodies, recombinant proteins, and other reagents
PLX4720 was provided from Plexxikon Inc (Berkeley, CA, USA). It was dissolved in DMSO and made up in stock solutions of 4 mM. Actinomycin D and SBHA were purchased from Sigma-Aldrich (Castle Hill, NSW, Australia). SBHA was dissolved in distilled water and made up in a stock solution of 10 mg/ml. The mouse MAbs against pERK, Bcl-2, Mcl-1, Bad, and AIF, and the rabbit polyclonal antibodies (Abs) against B-RAF were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The MAb against Noxa and the polyclonal Ab against Bim were purchased from Imgenex (San Diego, CA, USA). The rabbit polyclonal Abs against PUMA, ERK, COX IV, A-RAF, C-RAF, and Bid were from Cell Signalling Technology (Beverly, MA, USA). The rabbit polyclonal Abs against SFRS6, SFRS12, and β-actin were from Sigma-Aldrich. The mouse MAbs against cytochrome c, PARP were from Pharmingen (Bioclone, Marrickville, NSW, Australia). The rabbit polyclonal anti-Bax against amino acids 1 through to 20 was purchased from Upstate Biotechnology (Lake Placid, NY, USA). The mouse MAb against Bak (Ab-1) was purchased from Calbiochem (La Jolla, CA, USA). The mouse MAb against caspase-7 and the rabbit polyclonal Ab against caspase-3 were from Stressgen (Victoria, BC, Canada). The cell-permeable general caspase inhibitor Z-Val-Ala-Asp(OMe)-CH2F (z-VAD-fmk) was purchased from Calbiochem. Isotype control Abs used were the ID4.5 (mouse IgG2a) MAb against Salmonella typhi supplied by Dr L Ashman (Institute for Medical and Veterinary Science, Adelaide, Australia), the 107.3 mouse IgG1 MAb purchased from PharMingen (San Diego, CA, USA), and rabbit IgG from Sigma-Aldrich .
Cell viability assays (MTS assays)
The cytotoxic effect of PLX4720 on melanoma cells was determined using VisionBlue Fluorescence Cell Viability Assay Kit (Biovision Inc., Mountain View, CA, USA) as described previously.38 Briefly, cells were seeded at 5000 cells per well onto flat-bottomed 96-well culture plates and allowed to grow for 24 h followed by the desired treatment. Cells were then labeled with the VisionBlue reagent and detected by Synergy two multi-detection microplate reader (BioTek, Winooski, VT, USA) according to the manufacturer's instructions.
Apoptosis
Quantitation of apoptotic cells by measurement of sub-G1 DNA content using the propidium iodide (Sigma-Aldrich) method or by Annexin-V staining was carried out as described elsewhere.36, 37
Western blot analysis
Western blot analysis was carried out as described previously.36, 37 Labeled bands were detected by Immun-Star HRP Chemiluminescent Kit, and images were captured and the intensity of the bands was quantitated with the Bio-Rad VersaDoc image system (Bio-Rad, Regents Park, NSW, Australia).
Preparation of mitochondrial and cytosolic fractions
The mitochondrial and cytosolic fractions were prepared by using Qproteome Mitochondrial Isolation Kit (Qiagen, Doncaster, Vic, Australia) according to the manufacturer's instruction. In brief, after trypsinization, cells were washed with PBS and followed by 0.9% sodium chloride solution. Cell pellets were resuspended in ice-cold lysis buffer and incubated for 10 min at 4°C on an end-over-end shaker. After incubation, cell lysates were spun at 1000 × g for 10 min at 4°C and the supernatant for cytosolic fraction was collected. Cell pellets were resuspended in disruption buffer, purification buffer, and followed by storage buffer with spins in between. The mitochondrial pellets were finally resuspended in lysis buffer for western blot analysis.
Plasmid vector and transfection
Stable Mel-RMu and Mel-CV transfectants of Bcl-2 were established by electroporation of the pEF-puro vector carrying human Bcl-2 cDNA provided by Dr David Vaux (Walter and Eliza Hall Institute, Melbourne, Vic, Australia) and described elsewhere.39 The pCDH-CMV-MCS-EF1-Puro (CD510B-1) vector carrying B-RAFV600E cDNA was kindly provided by Professor Richard Marais (The Institute of Cancer Research, UK). The pCMV6-AC-BimS-GFP vector and the pCMV6-AC-SFRS6-GFP vector were purchased from OriGene (Rockville, MD, USA). Melanoma cells were seeded at 1 × 105 cells per well in 24-well plate, 24 h before transfection. Cells were transfected with 1 μg plasmid as well as the empty vector (Sigma-Aldrich) in Opti-MEM medium (Invitrogen, Carlsbad, CA, USA) with Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer's protocol. At 6 h after transfection, the cells were switched into antibiotic-free medium containing 5% FCS for a further 24 h. Cells were then passaged at 1:10 ratio into fresh medium for further 24 h, followed by G418 or puromycin (Sigma-Aldrich) selection.
Real-time PCR
Real-time PCR was performed using the ABI Fast 7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA). For Bim mRNA detection, 25 μl mixture was used for reaction, which contains 5 μl cDNA sample (0.5–1 μg/μl), 300 nM forward primers for Bim 5′-TGCAGACATTTTGCTTGTTCAA-3′ and β-actin 5′-GGCACCCAGCACAATGAAG-3′, 300 nM reverse primers for Bim 5′-GAACCGCTGGCTGCATAATAAT-3′ and β-actin 5′-GCCGATCCACACGGAGTACT-3′, 200 nM probes for Bim 6FAM-CCAACAAGACCCAGCACCGCG-TAMRA and β-actin 6FAM-TCAAGATCATTGCTCCTCCTGAGCGC-TAMRA, and 9 mM MgCI2. To specifically detect individual Bim isoforms, forward primers were designed to span the junctions of exons 3 and 4, exons 2 and 4, and exons 4 and 5, which are unique to BimEL, BimL, and BimS, respectively. BimEL forward primer is 5′-GTGGGTATTTCTCTTTTGACACAGAC-3′, BimL forward primer is 5′-TACAGACAGAGCCACAAGACAG-3′, and common reverse primer for both BimEL and BimL is 5′-GTTCAGCCTGCCTCATGGAAG-3′ BimS forward primer is 5′-TGACCGAGAAGGTAGACAAT-3′, BimS reverse primer is 5′-GCCATACAAATCTAAGCCAGT-3′. Real-time PCR for three Bim isoforms was done by Fast SYBR Green Master Mix (Applied Biosystems). For SFRS6 and SFRS12, assay-on-demand for SFRS6 (assay ID: Hs00740177_g1), SFRS12 (assay ID: Hs00377948_m1), and GAPDH (assay ID: Hs99999905_m1) were used according to manufacturer's protocol (Applied Biosystems). Analysis of cDNA for β-actin or GAPDH was included as a control. The threshold cycle value (Ct) was normalized against β-actin or GAPDH cycle numbers. The relative abundance of mRNA expression of a control sample was arbitrarily designated as 1, and the values of the relative abundance of mRNA of other samples were calculated accordingly.
Small interfering RNA
The siRNA constructs for Bim, B-RAF, SFRS6, and SFRS12 were obtained as the siGENOME SMARTpool reagents (Dharmacon, Lafayette, CO, USA), the siGENOME SMARTpool Bim (M-004383-01-0010), the siGENOME SMARTpool B-RAF (M-003460-03-0010), the siGENOME SMARTpool SFRS6 (M-016067-01-0010), and the siGENOME SMARTpool SFRS12 (M-016865-01-0010). The nontargeting siRNA control, SiConTRolNon-targeting SiRNA pool (D-001206-13-20) was also obtained from Dharmacon.
To specifically knockdown BimEL and BimS, siRNAs were designed to target exons 3 and 5, which are unique to BimEL and BimS, respectively. The oligonucleotides used were BimEL sense 5′-CUGCUGUCUCGAUCCUCCAdTdT-3′, BimEL antisense 5′-UGGAGGAUCGAGACAGCAGdTdT-3′ BimS sense 5′-CAUAUGGUCAUUGGUGAUUdTdT-3′, BimS antisense 5′-AAUCACCAAUGACCAUAUGdTdT-3′ control sense 5′-GGCUGUAACUUACGUGUACUUdTdT-3′, control antisense, 5′-AAGUACACGUAAGUUACAGCCdTdT-3′. Transfection of siRNA pools was carried out as described previously.39
Acknowledgments
This work was supported by research grants from the NSW State Cancer Council, the Melanoma and Skin Cancer Research Institute Sydney, the Hunter Melanoma Foundation, NSW, and the National Health and Medical Research Council (NHMRC), Australia. XD Zhang, RF Thorne, and H Rizos are Cancer Institute NSW Fellows. We thank Dr. David Vaux (Walter and Eliza Hall Institute, Melbourne, Vic, Australia) for the pEF-puro vector carrying human Bcl-2 cDNA and Professor Richard Marais (The Institute of Cancer Research, UK) for the pCDH-CMV-MCS-EF1-Puro (CD510B-1) vector carrying B-RAFV600E cDNA. We also greatly thank Dr. Gideon Bollag for supply of PLX4720.
Glossary
- BH3
Bcl-2 homology 3
- SR protein
serine/arginine-rich protein
- SBHA
suberic bishydroxamate
- SFRS6
splicing factor arginine/serine-rich 6
- SFRS12
splicing factor serine/arginine-rich 12
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Supplementary Material
References
- Flaherty KT, Puzanov I, Sosman J, Kim K, Ribas A, McArthur A, et al. Phase I study of PLX4032: Proof of concept for V600E BRAF mutation as a therapeutic target in human cancer J Clin Oncol 200927Abstract 9000. [Google Scholar]
- Chapman P, Puzanov I, Sosman J, Kim K, Ribas A, McArthur A, et al. Early efficacy signal demonstrated in advanced melanoma in a phase I trial of the oncogenic BRAF-selective inhibitor PLX4032. Eur J Cancer Suppl. 2009;7:5. [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Platz A, Egyhazi S, Ringborg U, Hansson J. Human cutaneous melanoma; a review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site. Mol Oncol. 2008;1:395–405. doi: 10.1016/j.molonc.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellbrock C, Rana S, Paterson H, Pickersgill H, Brummelkamp T, Marais R. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS One. 2008;3:e2734. doi: 10.1371/journal.pone.0002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J Lee JT, Wang W, Zhang J, Cho H, Mamo S, Bremer R, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg MS, Jansen ES, Cook M, Harris C, Strasser A, Scott CL. Treatment of B-RAF mutant human tumor cells with a MEK inhibitor requires Bim and is enhanced by a BH3 mimetic. J Clin Invest. 2008;118:3651–3659. doi: 10.1172/JCI35437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YF, Jiang CC, Kiejda KA, Gillespie S, Zhang XD, Hersey P. Induction of apoptosis in human melanoma cells by inhibition of MEK is caspase independent, and is mediated by the Bcl-2 family members PUMA, Bim and Mcl-1. Clin Cancer Res. 2007;13:4934–4942. doi: 10.1158/1078-0432.CCR-07-0665. [DOI] [PubMed] [Google Scholar]
- Panka DJ, Atkins MB, Mier JW. Targeting the mitogen-activated protein kinase pathway in the treatment of malignant melanoma. Clin Cancer Res. 2006;12:2371s–2375s. doi: 10.1158/1078-0432.CCR-05-2539. [DOI] [PubMed] [Google Scholar]
- VanBrocklin MW, Verhaegen M, Soengas MS, Holmen SL. Mitogen-activated protein kinase inhibition induces translocation of Bmf to promote apoptosis in melanoma. Cancer Res. 2009;69:1985–1994. doi: 10.1158/0008-5472.CAN-08-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann KM, VanBrocklin MW, Staffend NA, Kitchen SM, Koo HM. Mitogen-activated protein kinase pathway-dependent tumor-specific survival signaling in melanoma cells through inactivation of the proapoptotic protein bad. Cancer Res. 2003;63:8330–8337. [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ. 2005;12:1008–1104. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell. 1999;3:287–296. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Zhang LC, Huang DC, Webb GC, Bottema CD, Shore P, et al. Gene structure alternative splicing, and chromosomal localization of pro-apoptotic Bcl-2 relative Bim. Mamm Genome. 2001;12:163–168. doi: 10.1007/s003350010242. [DOI] [PubMed] [Google Scholar]
- Weber A, Paschen SA, Heger K, Wilfling F, Frankenberg T, Bauerschmitt H, et al. BimS-induced apoptosis requires mitochondrial localization but not interaction with anti-apoptotic Bcl-2 proteins. J Cell Biol. 2007;177:625–636. doi: 10.1083/jcb.200610148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajares MJ, Ezponda T, Catena R, Calvo A, Pio R, Montuenga LM. Alternative splicing: an emerging topic in molecular and clinical oncology. Lancet Oncol. 2007;8:349–357. doi: 10.1016/S1470-2045(07)70104-3. [DOI] [PubMed] [Google Scholar]
- Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Revil T, Shkreta L, Chabot B. Pre-mRNA alternative splicing in cancer: functional impact, molecular mechanisms and therapeutic perspectives. Bull Cancer. 2006;93:909–919. [PubMed] [Google Scholar]
- van Alphen RJ, Wiemer EA, Burger H, Eskens FA. The spliceosome as target for anticancer treatment. Br J Cancer. 2009;100:228–232. doi: 10.1038/sj.bjc.6604801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- Kirschbaum-Slager N, Lopes GM, Galante PA, Riggins GJ, de Souza SJ. Splicing factors are differentially expressed in tumors. Genet Mol Res. 2004;3:512–520. [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaustein M, Pelisch F, Srebrow A. Signals, pathways and splicing regulation. Int J Biochem Cell Biol. 2007;39:2031–2048. doi: 10.1016/j.biocel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Adams JM, Cory S. Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Curr Opin Immunol. 2007;19:488–496. doi: 10.1016/j.coi.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Gillespie SK, Borrow JM, Hersey P. The histone deacetylase inhibitor suberic bishydroxamate regulates the expression of multiple apoptotic mediators and induces mitochondria-dependent apoptosis of melanoma cells. Mol Cancer Ther. 2004;3:425–435. [PubMed] [Google Scholar]
- Johansson P, Pavey S, Hayward N. Confirmation of a BRAF mutation-associated gene expression signature in melanoma. Pigment Cell Res. 2007;20:216–221. doi: 10.1111/j.1600-0749.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- Cartlidge RA, Thomas GR, Cagnol S, Jong KA, Molton SA, Finch AJ, et al. Oncogenic BRAF(V600E) inhibits BIM expression to promote melanoma cell survival. Pigment Cell Melanoma Res. 2008;21:534–544. doi: 10.1111/j.1755-148X.2008.00491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DL, Wang Y, Liu M, Martinka M, Li G. Bim expression is reduced in human cutaneous melanomas. J Invest Dermatol. 2008;128:403–407. doi: 10.1038/sj.jid.5700989. [DOI] [PubMed] [Google Scholar]
- Park JW, Graveley BR. Complex alternative splicing. Adv Exp Med Biol. 2007;623:50–63. doi: 10.1007/978-0-387-77374-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter N, Herrlich P, König H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- Patel NA, Kaneko S, Apostolatos HS, Bae SS, Watson JE, Davidowitz K, et al. Molecular and genetic studies imply Akt-mediated signaling promotes protein kinase CbetaII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. J Biol Chem. 2005;280:14302–14309. doi: 10.1074/jbc.M411485200. [DOI] [PubMed] [Google Scholar]
- Weg-Remers S, Ponta H, Herrlich P, König H. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 2001;20:4194–4203. doi: 10.1093/emboj/20.15.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YF, Jiang CC, Kiejda KA, Gillespie S, Zhang XD, Hersey P. Apoptosis induction in human melanoma cells by inhibition of MEK is caspase-independent and mediated by the Bcl-2 family members PUMA, Bim, and Mcl-1. Clin Cancer Res. 2007;13:4934–4942. doi: 10.1158/1078-0432.CCR-07-0665. [DOI] [PubMed] [Google Scholar]
- Jiang CC, Chen LH, Gillespie S, Wang YF, Kiejda KA, Zhang XD, et al. Inhibition of MEK sensitizes human melanoma cells to endoplasmic reticulum stress-induced apoptosis. Cancer Res. 2007;67:9750–9761. doi: 10.1158/0008-5472.CAN-07-2047. [DOI] [PubMed] [Google Scholar]
- Liu H, Jiang CC, Lavis CJ, Croft A, Dong L, Tseng HY, et al. 2-Deoxy-D-glucose enhances TRAIL-induced apoptosis in human melanoma cells through XBP-1-mediated up-regulation of TRAIL-R2. Mol Cancer. 2009;8:122. doi: 10.1186/1476-4598-8-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CC, Wroblewski D, Yang F, Hersey P, Zhang XD. Human melanoma cells under endoplasmic reticulum stress are more susceptible to apoptosis induced by the BH3 mimetic obatoclax. Neoplasia. 2009;11:945–955. doi: 10.1593/neo.09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.