Summary
The miR-17-92 microRNA cluster is often activated in cancer cells, but the identity of its targets remains elusive. Using SILAC and quantitative mass spectrometry, we examined the effects of activation of the miR-17-92 cluster on global protein expression in neuroblastoma cells. Our results reveal cooperation between individual miR-17-92 miRNAs and implicate miR-17-92 in multiple hallmarks of cancer, including proliferation and cell adhesion. Most importantly, we show that miR-17-92 is a potent inhibitor of TGFβ-signaling. By functioning both upstream and downstream of pSMAD2, miR-17-92 activation triggers downregulation of multiple key effectors along the TGFβ-signaling cascade as well as through direct inhibition of TGFβ-responsive genes.
Introduction
MicroRNAs (miRNAs) belong to a regulatory class of small non-coding RNAs with a fundamental role in numerous aspects of cell biology, such as cell cycle regulation, apoptosis, differentiation and maintaining stemness (reviewed in (Bartel, 2004)). Only 20–25 nucleotides in length, miRNAs function as key molecules in the post-transcriptional repression of gene expression. Upon miRNA assembly in the RNA induced silencing complex (RISC), binding between the miRNA seed (nucleotides 2 – 7 counted from the 5’ end of the miRNA) and complementary sites in the 3’ untranslated region (3’UTR) of target mRNAs results in degradation of the mRNA or inhibition of translation (reviewed in (Bartel, 2009)). Based on the 3’UTR site context, algorithms predict that up to 60% of all coding genes are under the control of one or more miRNAs (Friedman et al., 2009). However, these predictions suffer from a high degree of false positives, and to date, only a fraction of miRNA-mRNA interactions have been experimentally validated.
In cancer, miRNAs function both as oncogenes or tumor-suppressors (reviewed in (Calin and Croce, 2006; Esquela-Kerscher and Slack, 2006)). Some of these miRNAs were identified as essential components of known cancer pathways, such as the p53-induced miR-34 family (He et al., 2007; Raver-Shapira et al., 2007) or the c-MYC/MYCN-induced miR-17-92 cluster (O'Donnell et al., 2005). The oncogenic miR-17-92 cluster consists of six individual miRNAs (miR-17, miR-18a, miR-19a, miR-19b, miR-20a and miR-92a) located within a polycistronic transcript on human chromosome 13. Gene duplications and deletions eventually resulted in two miR-17-92 paralogs, the miR-106b-25 cluster on chromosome 7 and the miR-106a-363 cluster on chromosome X. Of these clusters, miR-17-92 is the most frequently activated one in cancer. MiRNA expression profiling studies revealed miR-17-92 overexpression, both in hematopoietic malignancies, such as B-cell lymphomas (He et al., 2005), and solid tumors, including breast, colon and lung cancer (Castellano et al., 2009; Hayashita et al., 2005; Lanza et al., 2007) and neuroblastoma (Mestdagh et al., 2009a). Overexpression can result from amplification of the miR-17-92 locus (He et al., 2005) or direct miR-17-92 transactivation by c-MYC/MYCN (Dews et al., 2010; Fontana et al., 2008; Mestdagh et al., 2009a; O'Donnell et al., 2005). The oncogenic nature of miR-17-92 activation is supported by the identification of miR-17-92 targets with key roles in cell cycle control and cell death. In particular, miR-17 and miR-20a target the cyclin dependent kinase inhibitor CDKN1A (p21), a negative regulator of the G1-S transition (Fontana et al., 2008), and miR-17 targets the pro-apoptotic BCL2L11 (Bim) (Fontana et al., 2008). In gastric cancer, downregulation of p21 by the miR-17 and miR-20a paralogs miR-106b and miR-93 renders the cells insensitive to TGFβ-induced cell cycle arrest whereas miR-25 (a miR-92a paralog) inhibits TGFβ-dependent apoptosis through the repression of BCL2L11 (Petrocca et al., 2008).
Thus far, the number of identified miR-17-92 targets remains relatively limited thus precluding a comprehensive understanding of the full oncogenic potential of this miRNA cluster. In a first step towards this goal, we examined the effects of miR-17-92 cluster activation on the proteome of neuroblastoma cancer cells. Using quantitative mass spectrometry, we analyzed the response of thousands of proteins upon miR-17-92 activation in neuroblastoma cells. Neuroblastoma is an excellent model to study the effects of miR-17-92 activation because high-risk neuroblastoma tumors are characterized by increased MYCN/c-MYC activity either through MYCN amplification or increased c-MYC expression, both resulting in elevated miR-17-92 levels (Mestdagh et al., 2009a). Our results demonstrate that miR-17-92 is implicated in multiple hallmarks of the tumorigenic program, including proliferation and cell adhesion. Most importantly, we dissect the role of miR-17-92 as a potent inhibitor of TGFβ-signaling acting on multiple levels along the signaling cascade.
Results
miR-17-92 cluster activation is a marker for poor survival
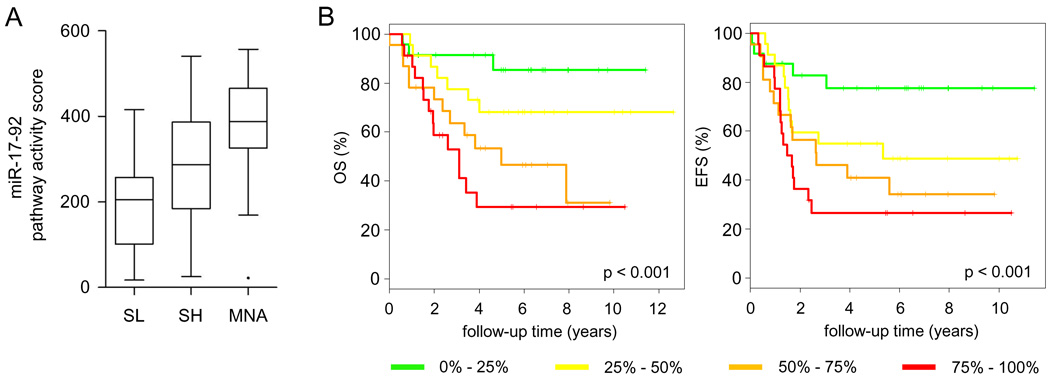
In neuroblastoma (NB), miR-17-92 expression is activated through direct MYCN/c-MYC promoter binding (Fontana et al., 2008; Mestdagh et al., 2009b). We quantified miR-17-92 expression on a cohort of 95 primary untreated NB tumor samples (dataset D1, Supplemental Table 1, GEO accession: GSE21713) (Mestdagh et al., 2009a). The activation of the entire miR-17-92 cluster was evaluated by means of a pathway activity score (Fredlund et al., 2008; Mestdagh et al., 2009a). NB tumors were divided into three cohorts, MYCN single copy low-risk tumors (SL), MYCN single copy high-risk tumors (SH) and MYCN amplified tumors (MNA). The miR-17-92 pathway activity was highest in the MNA tumors, followed by the SH tumors and the SL tumors (Figure 1A). Each individual miRNA is upregulated in the MNA samples suggesting that the entire miR-17-92 cluster, rather than a subset of miRNAs, is of potential relevance (Mann Whitney, p < 0.05) (Supplemental Figure 1A). We next evaluated miR-17-92 pathway activation with respect to NB patient survival. Kaplan-Meier analysis demonstrated that miR-17-92 activity was proportional to overall and event-free survival (log-rank, p < 0.001), underscoring the importance of miR-17-92 activation in NB tumor biology (Figure 1B). Except for miR-19b, expression of the other miRNAs within the miR-17-92 cluster showed similar correlations (Supplemental Figure 1B).
Figure 1. miR-17-92 cluster activation is a marker for poor prognosis.
(A) miR-17-92 pathway activity is scored in three clinicogenetic subsets of neuroblastoma tumors (dataset D1, Supplemental Table 1), MYCN amplified tumors (MNA), MYCN single copy high-risk tumors (SH) and MYCN single copy low risk tumors (SL) (whiskers: Tukey). miR-17-92 pathway activity score is significantly higher in MNA vs. SH (Mann Whitney, p < 0.05), MNA vs. SL (p < 0.0001) and SH vs. SL (p < 0.01). (B) Kaplan Meier plots for overall (OS) and event free survival (EFS) based on the pathway activity score of miR-17-92, represented as quartiles. Increased activity of miR-17-92 is proportionally correlated to both poor overall and event-free survival.
Impact of miR-17-92 activation on protein output
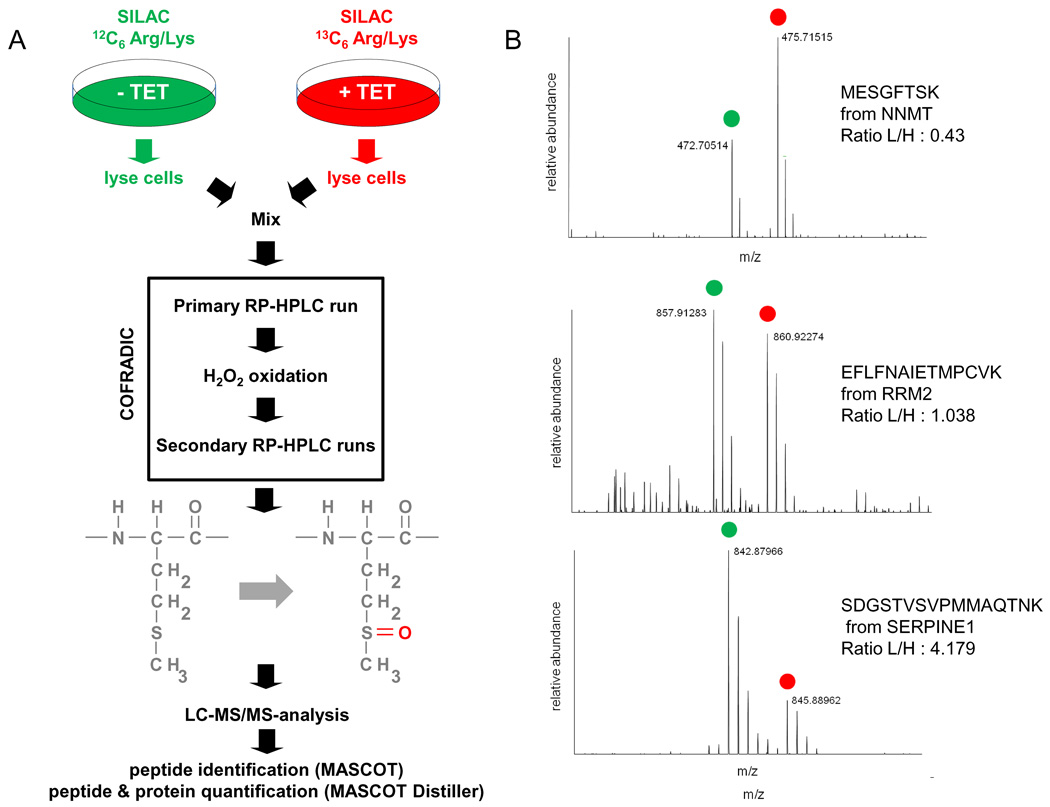
To study the regulatory effects of miR-17-92 activation, quantitative mass spectrometry was applied to measure protein response in a cellular model (SHEP-TR-miR-17-92) with tetracycline-inducible miR-17-92 expression (Mestdagh et al., 2009a). This approach provides the most relevant readout as it directly measures the impact of a miRNA on protein output (Baek et al., 2008; Selbach et al., 2008). Average miR-17-92 induction upon tetracycline treatment was in the range of miR-17-92 fold changes between MNA and SL tumours (Supplemental Figure 2A and data not shown). Profiling of 430 miRNAs revealed no significant effects on global miRNA expression suggesting that miR-17-92 induction does not affect the processing of other miRNAs (data not shown). SHEP-TR-miR-17-92 cells were differentially labeled using SILAC (stable isotope labeling with amino acids in cell culture) (Ong et al., 2002) and then either treated with tetracycline for 72 h or left untreated, followed by methionine COFRADIC isolation of methionyl peptides (Gevaert et al., 2002) and identification of these peptides by LC-MS/MS (Figure 2A). Only proteins that were quantified by at least two different peptides over two different proteome analyses (n = 3249) were selected for further analysis (Colaert et al.). Most proteins were in fact quantified by more than two peptides (Supplemental Figure 3B). Differential protein expression was determined as the average protein ratio of the differentially labeled fractions across the biological replicates (Supplemental Table 2, Figure 2B, Supplemental Figure 3C). Based on a fold change expression cutoff of 0.5 log2 units (see supplemental materials for cutoff definition), 144 proteins were downregulated upon miR-17-92 activation.
Figure 2. Analysis of global protein expression upon miR-17-92 activation.
(A) Tetracycline treated (+TET) and untreated (−TET) SHEP-TR-miR-17-92 cells were metabolically labeled using SILAC. Methionine-containing peptides were isolated using COFRADIC technology and subsequently analyzed using LC-MS/MS. (B) Representative LC-MS/MS spectra for an upregulated protein (NNMT), unchanged protein (RRM2) and downregulated protein (SERPINE1).
To assess whether the measured protein response reflects regulatory miR-17-92 effects, we performed an unbiased search for all possible 7mer motifs (n = 16,384) in the 3’UTR of the downregulated proteins (15th percentile) and compared these to motif occurrence in the 3’UTR of the remaining proteins. We found seven motifs to be overrepresented in the 3’UTR of the downregulated proteins, with the five most significant motifs belonging to the miR-17-92 miRNAs: miR-17, miR-19a, miR-19b, miR-20a and miR-92a (Fisher Exact, p < 0.05, Bonferoni multiple testing correction) (Figure 3A). Strikingly, there was no enrichment for miR-18a seeds, suggesting that miR-18a does not substantially contribute to protein repression upon miR-17-92 activation. Analyses using the 20th percentile gave similar results (data not shown). Analyses for the 5’UTR and coding sequence (CDS) did not reveal significant enrichments for miR-17-92 miRNA seed sequences. However, we did observe an enrichment for the 7mer-m8 seed of miR-17* in the CDS of the downregulated proteins, suggesting that miR-17*-mediated protein repression might depend on CDS binding.
Figure 3. miR-17-92 activation induces widespread repression of targeted proteins.
(A) Overview of all significantly enriched heptamer motifs in the 3’UTR of transcripts from repressed proteins. The top five significantly enriched motifs correspond to miR-17-92 target sites. One motif corresponds to the 7mer-m8 seed of the miR-302/miR-372 family, which differs in only one base with the 7mer-m8 seed of miR-17/miR-20a. The last motif did not correspond to any known miRNA nor did it show any overlap with miR-17-92 seeds. (B) The cumulative distribution of protein fold changes upon miR-17-92 activation, calculated for five different protein subsets: proteins with at least one miR-17-92 8mer site (red), 7mer-m8 site (blue), 7mer-A1 site (yellow), 6mer site (green) and no site (black). (C) Identical analysis as in (B) but for each individual miRNA from the miR-17-92 cluster. The miR-17/miR-20a and the miR-19a/miR-19b were analyzed together as they share identical seeds.
To evaluate miR-17-92 seed efficiency with respect to protein repression, we plotted the cumulative distribution of protein fold changes for proteins with at least one miR-17-92 3’UTR 6mer, 7mer-A1, 7mer-m8 or 8mer seed and compared these to proteins without miR-17-92 seeds (Figure 3B). As expected, protein repression was highest in the presence of a 8mer seed (Kolmogorov-Smirnov, p = 2.20 × 10−16) followed by 7mer-m8 (p = 1.11 × 10−6), 7mer-A1 (p = 0.00011) and 6mer seeds (p = 0.0027). When evaluating each miR-17-92 miRNA separately, we observed similar results for miR-17/miR-20a, miR-19a/miR-19b and miR-92a (miR-17/mir-20a and miR-19a/miR-19b were analyzed together as they share identical seeds) (Figure 3C). For miR-18a, the relation between seed occurrence and protein fold change was less pronounced, further supporting our observation that the contribution of miR-18a to miR-17-92 mediated protein repression is limited.
The fraction of proteins containing at least one miR-17-92 7-8mer seed was highest for proteins that were downregulated at least 2-fold (82%) and decreased to background levels (45%) for unchanged proteins (Supplemental Figure 3A). Robust protein repression was also characterized by the presence of multiple miR-17-92 3’UTR sites per protein (Supplemental Figure 3B), suggesting that individual miR-17-92 miRNAs cooperate to achieve target repression. This correlation was only observed for 3’UTR sites and not for 5’UTR or CDS sites (Supplemental Figure 3C, D). To further evaluate miRNA cooperation, we analyzed co-occurrence of individual miR-17-92 sites in the 3’UTR of downregulated proteins and compared this to co-occurrence in the 3’UTR of upregulated proteins (used as a reference control set). We identified significant co-occurrence for miR-17/miR-20a sites and miR-19a/miR-19b sites confirming cooperation between individual miRNAs (Supplemental Figure 3E). miR-18a sites almost never occurred in the absence of other miR-17-92 sites (8.33%) and were significantly associated with miR-17/miR-20a sites (Supplemental Figure 3E).
miR-17-92 affects multiple cancer pathways
To gain insight into the pathways affected by oncogenic miR-17-92 activation, we performed gene set enrichment analysis (GSEA) (Subramanian et al., 2005) using all measured proteins, ranked according to their fold change. Thirty-six gene sets were significantly enriched in the positive phenotype (i.e., downregulated proteins) while nine were enriched in the negative phenotype (i.e., upregulated proteins). Of the latter, six were related to increased metabolic activity of the mitochondrial oxidative phosphorylation energy production pathway (Supplemental Figure 4). In neuroblastoma, miR-17-92 expression is activated by MYCN/c-MYC transcription factors that have been shown to regulate genes involved in the biogenesis of mitochondria and metabolism (Zhang et al., 2007). Our results now provide evidence that this, at least in part, is mediated through miR-17-92 activation.
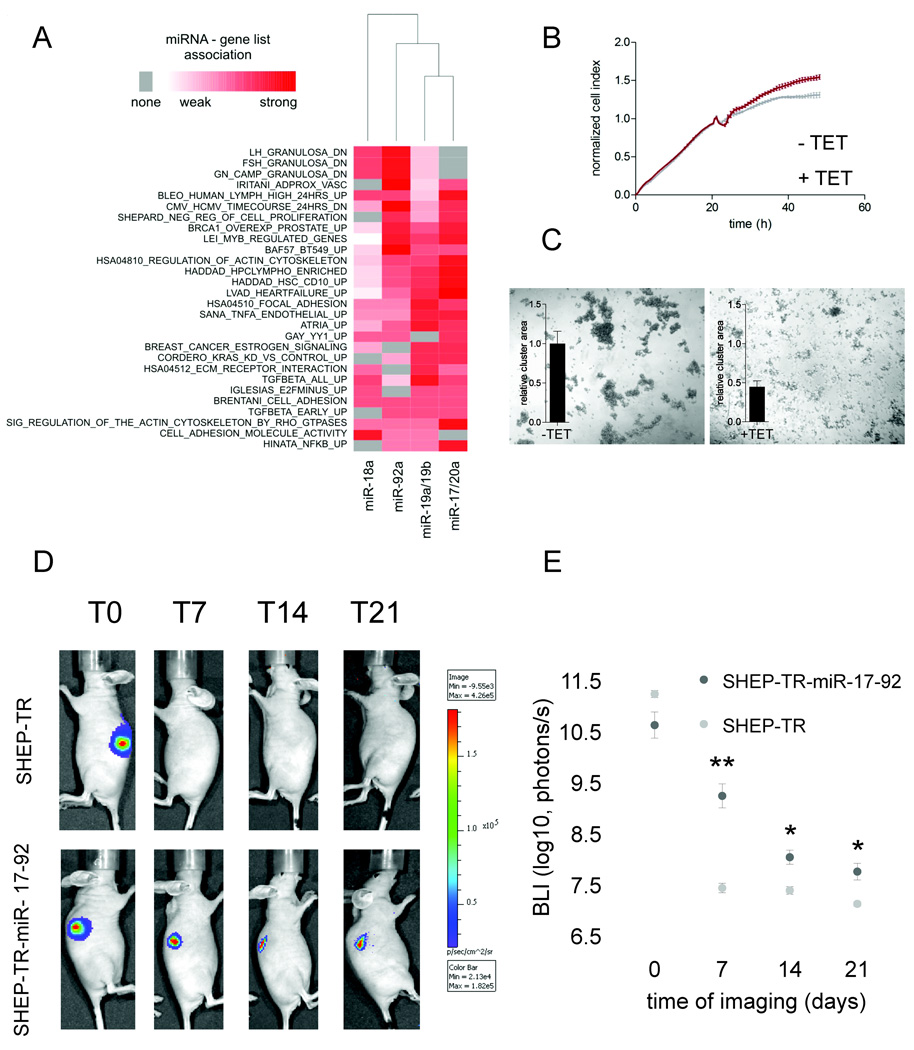
The contribution of each individual miRNA to the significant gene lists in the positive phenotype was calculated and visualized as a heatmap (Figure 4A). Among the gene lists enriched in the positive phenotype, which reflect direct miR-17-92 regulated pathways, we identified multiple cancer-related processes such as cell proliferation, cell adhesion, TGFβ-signaling, estrogen-signaling and RAS-signaling (Figure 4A). Hierarchical clustering reveals a close association between miR-17/miR-20a and miR-19a/miR-19b regulated pathways, reflecting the previously observed co-occurrence of these sites. Again, miR-18a clusters further away from the remaining miR-17-92 miRNAs and is characterized by weak gene list associations.
Figure 4. miR-17-92 activation regulates multiple cancer pathways.
(A) Heatmap of significant miRNA-pathway associations, identified through gene set enrichment analysis. The intensity of the association is based on the fraction of genes with at least one 7mer or 8mer 3’UTR site. (B) Normalized cell index (mean ± standard deviation) as a measure for proliferation of tetracycline treated (+TET) and untreated (−TET) SHEP-TR-miR-17-92 cells. Treatment was initiated 20 hours post seeding. (C) Evaluation of the cell-cell adhesion of tetracycline treated and untreated SHEP-TR-miR-17-92 cells. Measurements of the relative cluster area for three independent experiments using ImageJ are displayed as bar plots. Upon miR-17-92 activation, the area of the clusters dropped by >50% resulting in more but smaller aggregates. (D) Representative analyses of Bioluminescence imaging (BLI) of luciferase positive SHEP-TR-miR-17-92 and SHEP-TR cells injected etherotopically and subcutaneously in the right and left flank of nude athymic mice. Bar scale color indicates the number of photons/s measured by IVIS 3D imaging instrumentation. SHEP miR17-92 cells in vivo are still alive after 21 days post cell subcutaneous implantation. (E) Bioluminescence imaging (BLI) of luciferase positive SHEP-TR-miR-17-92 and SHEP-TR cells injected etherotopically and subcutaneously in the right and left flank of nude athymic mice. Luciferase signals were measured at 0, 7, 14 and 21 days post engraftment and are shown as the mean ± SEM of 5 mice. Significant differences between SHEP-TR and SHEP-TR-miR-17-92 cells are indicated by * (Student t-test, p < 0.05) and ** (Student t-test, p < 0.01).
In neuroblastoma, the oncogenic nature of miR-17-92 has been ascribed to its ability to promote cell proliferation through the regulation of CDKN1A and BCL2L11 (Fontana et al., 2008). GSEA results indicate that miR-17-92 has a much broader influence and targets different oncogenic pathways. As a proof of concept, we tried to validate the association with increased proliferation and decreased cell adhesion in the SHEP-TR-miR-17-92 cells. Cell proliferation was evaluated in real-time using the xCELLigence system. Upon miR-17-92 activation, proliferation of SHEP-TR-miR-17-92 cells increased (Figure 4B) and intercellular cell adhesion significantly decreased (Figure 4C). To evaluate the effect of miR-17-92 activation in vivo we performed etherotopic injection of SHEP-TR-miR-17-92 and SHEP-TR (control) cells in the right and left flanking site respectively of atymic nude mice that were given tetracyclin and visualized tumour cells using bioluminescence imaging. For SHEP-TR cells, the luciferase signal dropped to background levels after 7 days of engraftment which is in line with previous findings demonstarting that SHEP cells are not tumourigenic in vivo (Schweigerer et al., 1990) (see Figure 4D and 4E of the revised manuscript). In contrast, SHEP-TR-miR-17-92 cells persisted much longer and showed statistically higer luciferase signals at 7, 14 and 21 days indicating that, although tumourigenesis decreases, miR-17-92 activation significantly prolongs the engraftment of SHEP cells, probably through increased proliferation and decreased apoptosis, activities previously ascribed to miR-17-92 overexpression (Fontana et al., 2008). Together, these results confirm the relation between miR-17-92 activation and cell proliferation and reveal a role for miR-17-92 in the regulation of cell adhesion, hereby confirming the GSEA results.
miR-17-92 impairs TGFβ activity
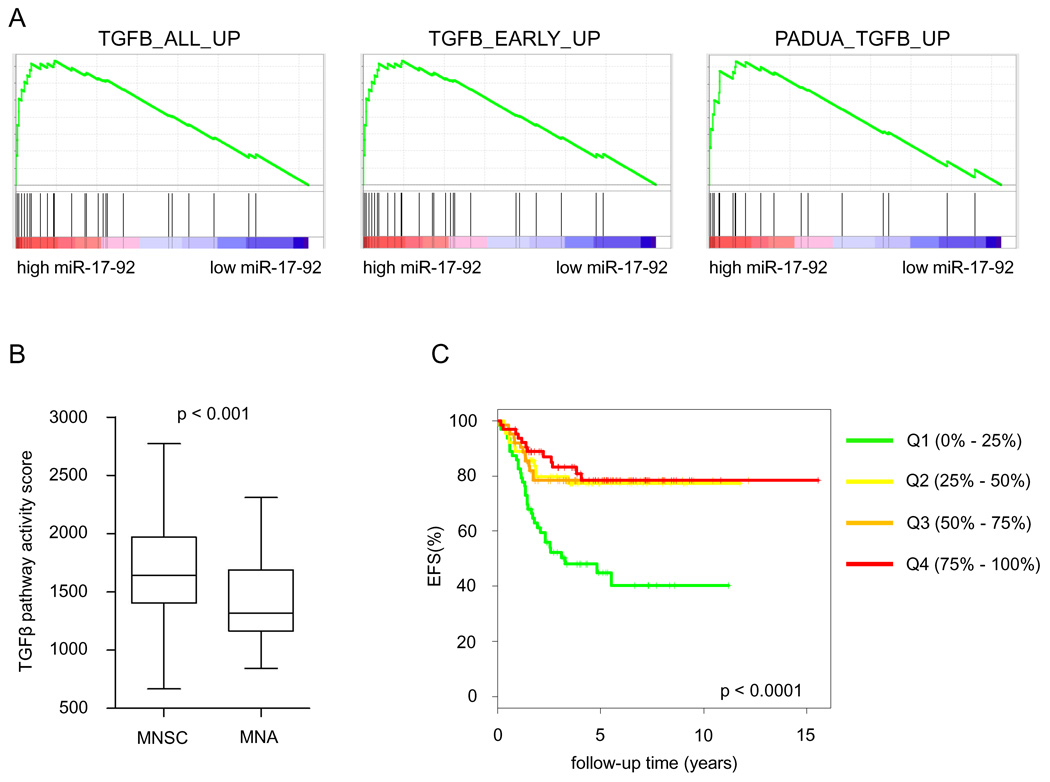
GSEA-analysis identified three TGFβ responsive gene sets (Padua et al., 2008; Verrecchia et al., 2001) among the proteins downregulated upon miR-17-92 activation in the SHEP-TR-miR-17-92 cells (Figure 5A). To exclude the possibility that repression of TGFβ responsive genes is an artifact of miRNA overexpression, we analyzed 8 published protein expression datasets of miRNA overexpression (Baek et al., 2008; Selbach et al., 2008) using GSEA. None of the TGFβ-gene lists were significantly enriched in any of the datasets suggesting the observed effect to be related to miR-17-92. For a subset of the TGFβ responsive genes, the measured protein repression was confirmed on the mRNA level using RT-qPCR (Supplemental Figure 5). We next evaluated this TGFβ-signature in neuroblastoma tumor samples using the pathway activity score of all genes that significantly contributed to the GSEA results (n = 21). For this purpose, we used the larger Oberthuer dataset (Oberthuer et al., 2006) (dataset D2, Supplemental Table 1) to increase the power of our analysis. TGFβ pathway activity was significantly downregulated in MNA neuroblastoma tumors, that are characterized by high miR-17-92 expression (Mann Whitney, p < 0.001) (Figure 5B), and showed a negative correlation to MYC pathway activity (Spearman’s Rank p < 0.01, rho = −0.460). In addition, Kaplan-Meier survival analysis indicates that tumors with low TGFβ pathway activity are characterized by poor event-free survival (log-rank, p < 0.0001) (Figure 5C). To further substantiate the inverse relation between TGFβ-target gene expression and miR-17-92 expression, we performed an expression correlation analysis in a subset of 40 of the 95 neuroblastoma tumors for which also mRNA expression was available (dataset D3, Supplemental Table 1) (Mestdagh et al., 2009a). Hierarchical clustering of the correlation coefficients revealed that, indeed, miR-17-92 expression inversely correlates to TGFβ-target gene expression (Supplemental Figure 6A). These results confirm that TGFβ-signaling is downregulated in aggressive neuroblastoma tumors with high miR-17-92 expression and underscore the potential importance of TGFβ-activity in neuroblastoma tumor biology.
Figure 5. miR-17-92 activation represses the TGFβ-pathway.
(A) Gene set enrichment analysis plots for three different TGFβ gene sets showing significant enrichment among the miR-17-92 repressed proteins. (B) TGFβ pathway activity score in MYCN amplified neuroblastoma tumors (MNA) and MYCN single copy neuroblastoma tumors (MNSC) (dataset D2, Supplemental Table 1). MNA tumors show significantly lower TGFβ pathway activity (Mann Whitney, p<0.001) (whiskers: Tukey). (C) Kaplan Meier plot for event free survival (EFS) based on the TGFβ pathway activity score, represented as quartiles (dataset D2, Supplemental Table 1). Increased activity of miR-17-92 is proportionally correlated to event-free survival.
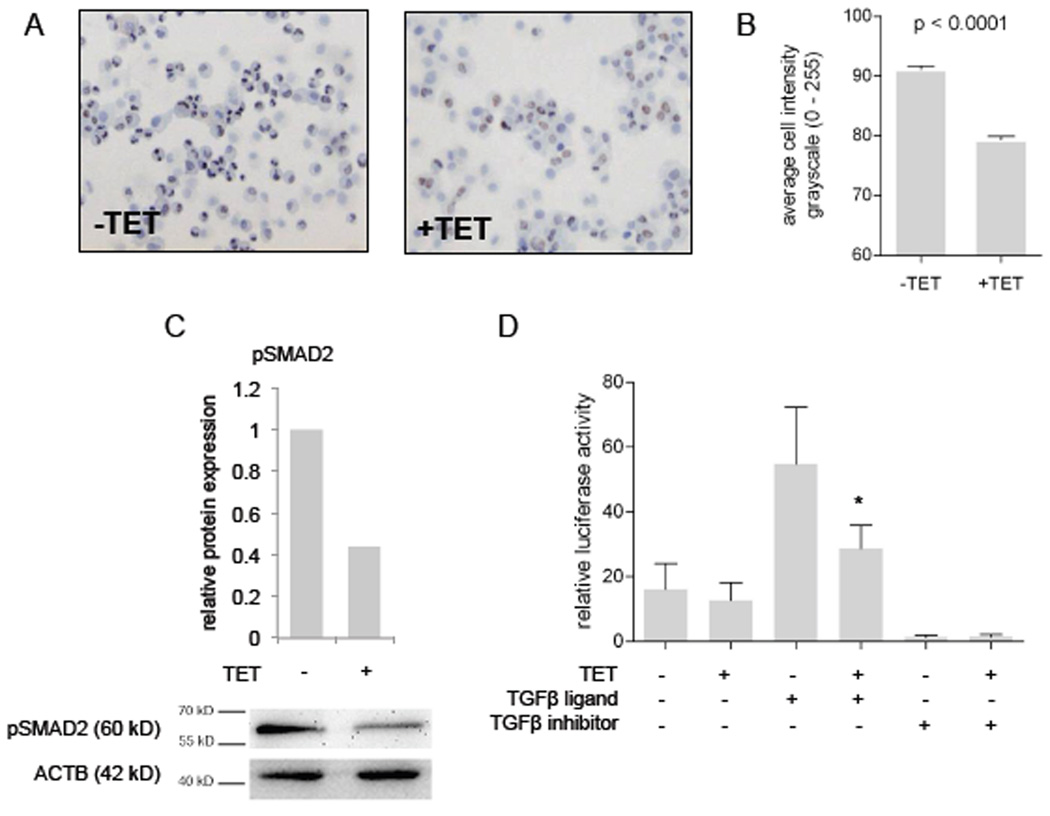
We next evaluated which components of the TGFβ-signaling cascade are controlled by miR-17-92 miRNAs. One important effector of active TGFβ-signaling is phosphorylated SMAD2 protein (pSMAD2) that translocates to the nucleus to induce gene transcription. Upon tetracycline treatment of SHEP-TR-miR-17-92 cells, we observed a significant decrease in nuclear pSMAD2 levels (Mann Whitney, p < 0.0001) (Figure 6A, B, C). A similar decrease was observed for pSMAD3 levels (data not shown). When SHEP-TR-miR-17-92 cells were transfected with a plasmid containing a SMAD regulated luciferase reporter ((CAGA)12-Luc) and treated with TGFβ1, a strong activation of the reporter gene was observed (Figure 6D). However, when miR-17-92 expression was activated through tetracycline treatment, reporter gene activation was substantially attenuated (Mann Whitney, p < 0.001) (Figure 6D). When the SHEP-TR-miR-17-92 cells were cultured in the presence of the potent TGFBR1 inhibitor SB431542 (Laping et al., 2002), the SMAD reporter gene activity was completely abrogated (Figure 6D). These results suggest that miR-17-92 activation impairs the TGFβ signaling cascade by acting upstream of pSMAD2.
Figure 6. miR-17-92 inhibits pSMAD2 levels and activity.
(A)(B) Immunohistochemical detection of phosphorylated SMAD2 protein (pSMAD2) in tetracycline treated (+TET) and untreated (−TET) SHEP-TR-miR-17-92 cells. The cell intensity measurement (mean ± SEM) reveals a significant decrease in pSMAD2 levels in tetracycline treated cells (Mann Whitney, p<0.0001). (C) Western blot analysis indicates a strong decrease (2.3-fold) in pSMAD2 levels upon miR-17-92 induction (+TET). (D) The relative luciferase activity of a pSMAD2 reporter construct (mean ± SEM). Activation of miR-17-92 expression through tetracycline treatment (+TET) results in a significant (*) decrease in reporter activity after stimulation of the TGFβ-pathway with TGFβ1 (TGFβ-ligand). TGFβ-inhibitor treatment completely abrogates the reporter activity.
miR-17-92 affects multiple levels of the TGFβ pathway
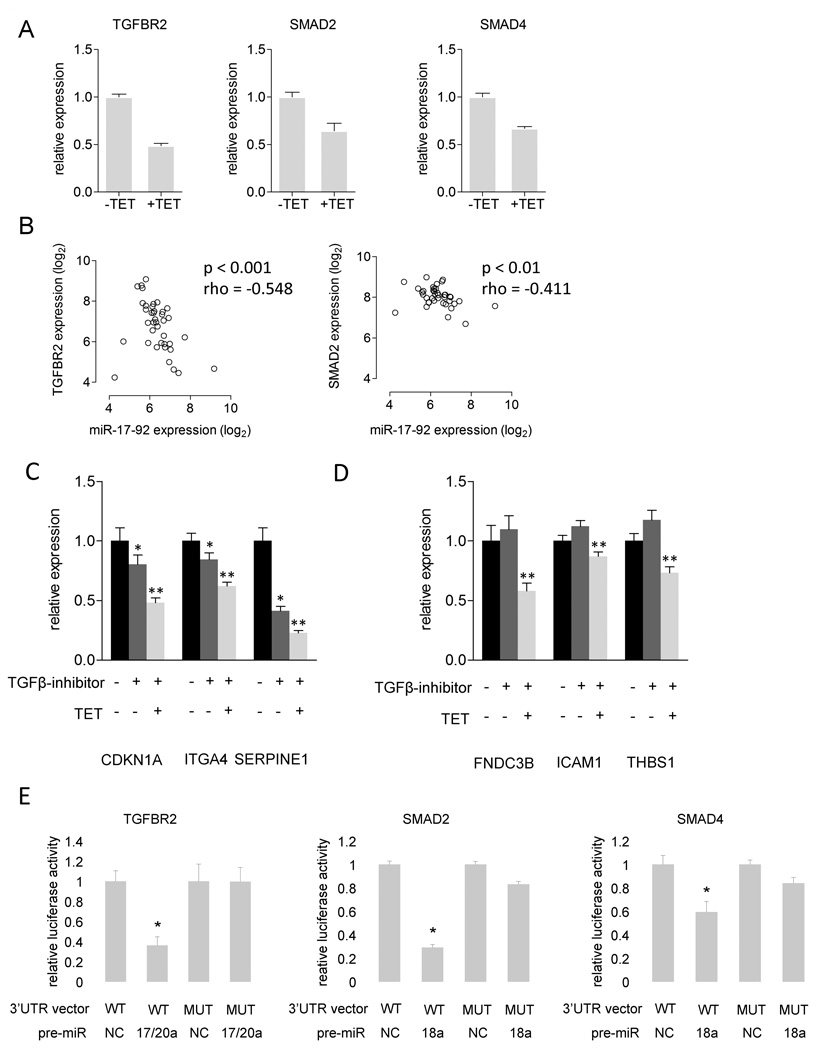
As decreased pSMAD2 levels are either caused by reduced receptor activity or reduced SMAD2 expression, we quantified TGFBR2 and SMAD2 mRNA expression in the SHEP-TR-miR-17-92 cells. Both TGFBR2 and SMAD2 expression levels decreased by at least 1.5-fold upon miR-17-92 activation (Figure 7A). SMAD4, the binding partner of pSMAD2, also displayed a decrease in expression upon miR-17-92 activation (Figure 7A). This negative correlation with miR-17-92 expression could be confirmed in primary neuroblastoma tumor samples for SMAD2 and TGFBR2 (Spearman’s Rank, p < 0.01) (Figure 7B), suggesting that miR-17-92 regulates their expression. Indeed, both genes contain miR-17-92 binding sites in their 3’UTR and a direct interaction between TGFBR2 and miR-20a has been established (Volinia et al., 2006). This miR-17-92 mediated silencing of TGFBR2 ultimately results in decreased pSMAD2 levels and decreased transcription of the TGFβ-target genes. In total we identified 13 TGFB-target genes to be downregulated on the protein level with a log2 fold-change < −0.5 (7 out of 20 proteins in the PADUA_TGFB_UP gene set, 3 out of 16 proteins from the TGFB_EARLY_UP gene set and 7 out of 28 proteins from the TGFB_ALL_UP gene set) (Supplemental Table 3). As 10 of these genes harbor miR-17-92 binding sites in their 3’UTR (Supplemental Table 3), we wondered whether they might also be targeted directly by miR-17-92. To exclude the effects of miR-17-92 directed inactivation of TGFβ-signaling on the expression of TGFβ-responsive genes, we first treated SHEP-TR-miR-17-92 cells for 4 h with the TGFBR1 inhibitor SB431542, which completely abrogates TGFβ-signaling (Figure 6C). Cells were subsequently treated with tetracycline to activate miR-17-92 expression and harvested at 24 h and 48 h after tetracycline treatment. From the six genes that were evaluated, three (CDKN1A, ITGA4 and SERPINE1) were downregulated after 24h of TGFβ-inhibitor treatment (t-test, p < 0.05), confirming that they are regulated by TGFβ (Figure 7C). The remaining three genes (FNDC3B, ICAM1 and THBS1) did not show any differential expression after 24h however, FNDC3B and THBS1 did respond to TGFB-inhibitor treatment after 48h (data not shown). This suggests that, in neuroblastoma, these are either not or indirectly responsive to TGFβ-signaling (Figure 7D). Upon miR-17-92 activation, the TGFβ-responsive genes were further downregulated (t-test, p < 0.001) (Figure 7C), supporting our hypothesis that miR-17-92 also influences the expression of these genes, independent of its ability to inactivate TGFβ-signaling. As expected, the genes that were not responsive to TGFβ inhibition did show decreased expression upon miR-17-92 activation (t-test, p < 0.001) (Figure 7D).
Figure 7. miR-17-92 targets multiple components of the TGFβ-pathway.
(A) The relative mRNA expression of TGFβ-responsive genes in tetracycline treated (+TET) and untreated SHEP-TR-miR-17-92 cells (+TET) (mean ± SEM). (B) Significant negative correlation between TGFBR2 mRNA expression and miR-17-92 expression and SMAD2 mRNA expression and miR-17-92 expression in primary neuroblastoma tumors. Spearman’s rank rho-values and p-values are listed. (C)(D) The relative mRNA expression (mean ± SEM) of a representative set of genes responsive to TGFβ (C) and genes not or (indirectly) responsive to TGFβ (D) in SHEP-TR-miR-17-92 cells that were either untreated, treated with TGFβ-inhibitor or treated with TGFβ-inhibitor followed by miR-17-92 activation with tetracycline (TET) for 24h. (C) Genes respond to TGFβ-inhibitor treatment (t-test, p < 0.05, indicated by *) and show an additional decrease in expression upon combined TGFβ-inhibitor treatment and miR-17-92 activation (t-test, p < 0.001, indicated by *). (D) Genes only respond to mR-17-92 treatment (t-test, p < 0.001, indicated by **). (E) Relative 3’UTR luciferase reporter activity for TGFBR2, SMAD2 and SMAD4, measured in DLD1DICERhypo cells (mean ± SEM). Plasmids with a wild type seed site for the active miRNA were introduced in DLD1DICERhypo cells in combination with a pre-miR negative control (NC) or miR-17-92 pre-miR. Luciferase activity is decreased significantly in the presence of the active miRNA (*) (t-test, p < 0.01) and increases significantly when the seed for the active miRNA is mutated (MUT) (t-test, p < 0.01).
To investigate which specific miRNAs contribute to the repression of the TGFβ-pathway, we overexpressed each miRNA from the miR-17-92 cluster separately and measured the expression of TGFβ-pathway components and target genes. Interestingly, we found that each miRNA contributes to the repression of one or more genes from the TGFβ-pathway suggesting that the entire miR-17-92 cluster, rather than a subset of miRNAs, mediates the repression of TGFβ-signaling in neuroblastoma cells (Supplemental Figure 6B). Downregulation (log2 fold change < −0.5) upon miRNA transfectection was almost exclusively observed for those genes harboring a 3’UTR seed sites for the respective miRNA (Fisher Exact, p < 0.001).
We next evaluated whether the miR-17-92-induced downregulation of TGFβ-pathway components is caused by direct binding between miR-17-92 miRNAs and miR-17-92 seed sites in the 3’UTR of TGFBR2, SMAD2 and SMAD4. To this purpose, DLD1DICERhypo cells were transfected with 3’UTR luciferase reporter plasmids in combination with a pre-miR negative control or a miR-17-92 pre-miR for which one or multiple seed sites were present in the 3’UTR of the respective genes. We identified a direct interaction between TGFBR2 and miR-17/20, SMAD2 and miR-18a and SMAD4 and miR-18a as evidenced by the significant decrease in luciferase activity compared to the pre-miR negative control (t-test, p < 0.01, Figure 7E). Other putative miR-17-92 sites in the 3’UTR of TGFBR2 (miR-19a/miR-19b), SMAD2 (miR-19a/miR-19b, miR-92a) and SMAD4 (miR-19a/miR-19b) did not affect luciferase signals (data not shown). Mutagenesis of the active miRNA seed sites resulted in a significant rescue of the luciferase signal (t-test, p < 0.01) suggesting that the observed effects depend on the presence of the 3’UTR seed site. These results confirm TGFBR2 as a direct miR-17-92 target gene and identify 2 additional TGFβ-pathway components, SMAD2 and SMAD4, as miR-17-92 target genes.
To assess the importance of TGFβ-pathway inhibition in the proliferation phenotype observed upon miR-17-92 activation we overexpressed SMAD2 and SMAD4 in the presence of activated miR-17-92. SMAD2/SMAD4 overexpression resulted in a 25% decrease in cell growth (t-test, p < 0.05) indicating that miR-17-92 accelerated proliferation is, at least in part, depending on the downregulation of the TGFβ-pathway. The relatively modest decrease in cell growth is probably explained by the fact that miR-17-92 directly regulates TGFB-target genes in a SMAD2/SMAD4 independent manner.
In conclusion, our data demonstrate that miR-17-92 activation triggers a targeted clampdown of TGFβ-signaling by acting on multiple key effectors along the signaling cascade, as well as through the direct inhibition of TGFβ-responsive genes, hereby repressing the cytostatic effects of active TGFβ-signaling (Supplemental Figure 7).
Discussion
Transcriptional activation of the miR-17-92 miRNA cluster by MYC/MYCN transcription factors occurs in multiple tumor entities, including neuroblastoma (Hayashita et al., 2005; Mestdagh et al., 2009a; O'Donnell et al., 2005). Although the oncogenic nature of miR-17-92 activation is well established, the underlying targets and signaling cascades that are deregulated remain largely elusive. In addition, studies aimed at determining miR-17-92 targets have focused on individual members of the cluster, despite the observation that the entire cluster is activated (Mestdagh et al., 2009b; O'Donnell et al., 2005). Here we have used an unbiased proteomics approach to identify miR-17-92 targeted pathways in a neuroblastoma tumor model. Direct quantitative measurement of protein expression is preferred over the more straightforward mRNA profiling as a high-throughput method for miRNA target identification (Baek et al., 2008; Selbach et al., 2008).
Computational analysis of miR-17-92 seeds in the 3’UTR of transcripts from proteins supported the expected enrichment of direct miR-17-92 targets within the list of down regulated proteins detected using mass spectrometry. Moreover, a proportional relationship between seed frequency and fold downregulation was noted. This relationship not only holds for multiple seeds from an individual miR-17-92 miRNA but also for multiple seeds from different miR-17-92 miRNAs, suggesting cooperation between individual miRNAs from the cluster towards target protein repression. MiR-17-92 miRNAs have indeed been shown to function in a cooperative and additive manner amongst others in the regulation of PTEN by miR-17 and miR-19 (Xiao et al., 2008). Our results further indicate that miR-19a/miR-19b and miR-17/miR-20a sites significantly co-occur in the 3’UTR of transcripts from several downregulated proteins. As these co-occurring sites were not observed for every possible combination of individual miR-17-92 miRNAs, we hypothesize that in neuroblastoma, the miRNA components of the miR-17-92 cluster can regulate target expression either individually or in certain combinations with additive effects. However, miR-17-92 function might be highly context and cell-type specific as miR-19 was shown to be both necessary and sufficient to promote MYC-induced lymphomagenesis in the Eµ-myc mouse B-cell lymphoma model (Olive et al., 2009).
While the fraction of downregulated proteins was enriched for seeds of miR-17/miR-20a, miR-19a/miR-19b and miR-92a, enrichment for the miR-18a seed was not detected. Strikingly, miR-18a seeds rarely occur as the only seed(s) in the 3’UTR of a downregulated target and showed little or no correlation to protein fold change. Although this suggests that miR-18a is not substantially contributing to target deregulation, it does not imply that miR-18a lacks functionality, as miR-18a has been shown to regulate important cancer genes such as CTGF in colon cancer and estrogen receptor-α (ESR1) in neuroblastoma (Dews et al., 2006; Loven et al.). Interestingly, we found miR-18a to regulate both SMAD2 and SMAD4, 2 key components of the TGFβ-signaling cascade, suggesting that miR-18a substantially contributes to pathway deregulation by regulating a selected set of target genes.
When all cluster components were combined, we identified a large number of targeted proteins belonging to diverse cancer-related pathways. Notably, estrogen receptor signaling was also among the targeted pathways. The fact that we identified such a wide variety of functions in neuroblastoma cells suggests that miR-17-92 pleiotropy is not only related to different targets in different cell types but also occurs within cell types. The molecular basis for this observation likely lies within the multiple components of the cluster and the complex interplay between them.
Mir-17-92-directed regulation of the TGFβ-responsive genes CDKN1A and BCL2L11 in neuroblastoma cells has been described by previously (Fontana et al., 2008). In gastric cancer, members of the miR-106b-25 cluster have also been shown to target CDKN1A and BCL2L11 (Petrocca et al., 2008). Here we comprehensively demonstrate that miR-17-92 dampens TGFβ-signaling in a multifaceted way by acting both upstream and downstream of pSMAD2/SMAD4, further underscoring its ability to regulate multiple components of the same pathway. This ability to simultaneously target the components of the signaling cascade as well as the downstream effectors through multiple miRNAs, allows for tight control of the TGFβ-transcriptional program. Moreover, it offers the cells enormous flexibility and plasticity for regulation of different subsets of TGFβ-target genes. In neuroblastoma, enhanced TGFβ-signaling, through increased TGFBR2 expression, results in reduced cell growth in vitro and disables the ability of the cells to form tumors in vivo (Turco et al., 2000). Instead, cells assume a terminally differentiated neuronal phenotype and display increased expression of axonal growth-associated protein (GAP43) and neurofilaments (Turco et al., 2000). Treatment of neuroblastoma cells with TGFβ1 induces a similar phenotype (Scarpa et al., 1996). In addition, retinoic acid (RA) induces differentiation of neuroblastoma cells, known to down regulate MYCN, accompanied by the increased expression of TGFβ1, TGFBR1, TGFBR2 and TGFBR3, resulting in the induction of a negative autocrine TGFβ1 growth regulatory loop (Cohen et al., 1995). We have shown that aggressive neuroblastoma tumors evade the cytostatic TGFβ-pathway through miR-17-92 directed targeting of key components of the pathway as well as downstream effectors. Reactivation of TGFβ-signaling through miR-17-92 inhibition could be a promising therapeutic approach as it would not only result in reactivation of TGFBR2 expression but also relieve the direct miR-17-92-mediated repression of TGFβ responsive genes.
Experimental Procedures
Cell culture
SHEP-TR-miR-17-92 cells (Mestdagh et al., 2009a) were cultured in RPMI (Invitrogen) supplemented with 10% fetal calf serum unless stated otherwise. SHEP-TR-miR-17-92 cells were treated with 2 µg/ml tetracycline (Sigma-Aldrich) to induce miR-17-92 expression (Supplemental Figure 2A). TGFβ1 (PeproTech) and TGFBR1 inhibitor (SB431542, Sigma-Aldrich) were used at a concentration of 0.25 ng/ml and 2 µM, respectively, unless stated otherwise.
COFRADIC analysis
SHEP-TR-miR-17-92 cells were metabolically labeled by growing them in DMEM medium supplemented with dialyzed fetal calf serum and with either heavy lysine and arginine (both 13C6) or with natural, light lysine and arginine (12C6). This stable isotope labeling (SILAC (Ong et al., 2002)) ensures that following trypsin digestion, all generated peptides can be quantified by mass spectrometry (MS, see Supplemental Materials). Mass spectrometry data for the forward en reverse experiment are available in Supplemental Table 4 and 5 and in the PRIDE database (www.ebi.ac.uk/pride, accession 14860).
mRNA and miRNA expression quantification
See supplemental materials for details on mRNA and miRNA quantification and data normalization. miRNA expression data are available in rdml-format (Supplemental File 1) (Lefever et al., 2009).
Immunohistochemistry and Western blot
Briefly, SHEP-TR-miR-17-92 cells, tetracycline treated or untreated, were stimulated with TGFβ1 for 4 h. pSMAD2 activity was evaluated by immunochemistry on cytopreparations or by Western blot. See supplemental materials for detailed experimental procedures.
Cell adhesion and proliferation assays
Details on cell adhesion and proliferation assays are described in the supplemental materials.
Xenografts
SHEP-TR-miR-17-92 and SHEP-TR (control) cells were transfected with a luciferase expressing mammalian vector. Etherotopic xenografts were established in atymic nude mice (n=5) by injection of 106 SHEP-TR cells subcutaneosly in the left flanking site and 106 SHEP-TR-miR-17-92 cells in the rigth flanking site of each individual animal. See supplemental materials for detailed experimental procedures.
CAGA-Luciferase reporter assay
For luciferase experiments, tetracycline or control treated SHEP-TR-miR-17-92 cells were transfected with the (CAGA)12-Luc luciferase reporter vector and assayed for luciferase and renilla activity. See supplemental materials for detailed experimental procedures.
3’UTR reporter assay
DLD1Dicerhypo cells were seeded in DMEM (Invitrogen) supplemented with fetal calf serum (10%) at a density of 10000 cells per well in an opaque 96-well plate. Twenty-four hours after seeding, using DharmaFECT Duo (Dharmacon), cells were cotransfected, either with a combination of a 3’UTR containing pGL4.11[luc2p] vector (Switchgear Genomics), a pRL-TK vector (Promega) for normalization and a miR-17-92 pre-miR (Ambion) (10 nM) or with a combination of a psi-check2 vector (Promega) containing only part of the 3’UTR and a miR-17-92 pre-miR. Forty-eight hours after transfection, luciferase reporter gene activity was measured using the Dual-Glo Luciferase Assay System (Promega) and a FLUOstar OPTIMA microplate reader (BMG LABTECH). See Supplemental Materials for details on plasmid construction and miRNA binding site mutation.
Statistics
See supplemental materials for details on all statistical procedures and gene set enrichment analysis.
Supplementary Material
Acknowledgements
This research was funded by the Fund for Scientific Research (grant number: G.0198.08 and 31511809), the Belgian Kid’s Fund and the Stichting tegen Kanker. P.M. is supported by the Ghent University Research Fund (BOF 01D31406). A-K. B., K.S. and H.A. are supported by grants from the Swedish Childhood Cancer Foundation and the Swedish Cancer Society. E.F. is supported by The Royal Swedish Physiographic Society and the American Cancer Society. B.G. is a Postdoctoral Research Fellow for the Fund for Scientific Research – Flanders (Belgium). The VIB/UGent lab further acknowledges support by a research grant from the Fund for Scientific Research – Flanders (Belgium) (project numbers G.0077.06), the Concerted Research Actions (project BOF07/GOA/012) from the Ghent University and the Inter University Attraction Poles (IUAP06). NCI grants R01 CA122334 and P30 CA016520 to A.T-T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman J, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci U S A. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PS, Letterio JJ, Gaetano C, Chan J, Matsumoto K, Sporn MB, Thiele CJ. Induction of transforming growth factor beta 1 and its receptors during all-trans-retinoic acid (RA) treatment of RA-responsive human neuroblastoma cell lines. Cancer Res. 1995;55:2380–2386. [PubMed] [Google Scholar]
- Colaert N, Helsens K, Impens F, Vandekerckhove J, Gevaert K. Rover: a tool to visualize and validate quantitative proteomics data from different sources. Proteomics. 2010;10:1226–1229. doi: 10.1002/pmic.200900379. [DOI] [PubMed] [Google Scholar]
- Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, Enders GH, El-Deiry W, Schelter JM, et al. The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 2010;70:8233–8246. doi: 10.1158/0008-5472.CAN-10-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V, Giacomini P, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredlund E, Ringner M, Maris JM, Pahlman S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proc Natl Acad Sci U S A. 2008;105:14094–14099. doi: 10.1073/pnas.0804455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert K, Van Damme J, Goethals M, Thomas GR, Hoorelbeke B, Demol H, Martens L, Puype M, Staes A, Vandekerckhove J. Chromatographic isolation of methionine-containing peptides for gel-free proteome analysis: identification of more than 800 Escherichia coli proteins. Mol Cell Proteomics. 2002;1:896–903. doi: 10.1074/mcp.m200061-mcp200. [DOI] [PubMed] [Google Scholar]
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza G, Ferracin M, Gafa R, Veronese A, Spizzo R, Pichiorri F, Liu CG, Calin GA, Croce CM, Negrini M. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer. 2007;6:54. doi: 10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laping NJ, Grygielko E, Mathur A, Butter S, Bomberger J, Tweed C, Martin W, Fornwald J, Lehr R, Harling J, et al. Inhibition of transforming growth factor (TGF)-beta1-induced extracellular matrix with a novel inhibitor of the TGF-beta type I receptor kinase activity: SB-431542. Mol Pharmacol. 2002;62:58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- Lefever S, Hellemans J, Pattyn F, Przybylski DR, Taylor C, Geurts R, Untergasser A, Vandesompele J. RDML: structured language and reporting guidelines for real-time quantitative PCR data. Nucleic Acids Res. 2009;37:2065–2069. doi: 10.1093/nar/gkp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Zinin N, Wahlstrom T, Muller I, Brodin P, Fredlund E, Ribacke U, Pivarcsi A, Pahlman S, Henriksson M. MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc Natl Acad Sci U S A. 107:1553–1558. doi: 10.1073/pnas.0913517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Fredlund E, Pattyn F, Schulte JH, Muth D, Vermeulen J, Kumps C, Schlierf S, De Preter K, Van Roy N, et al. MYCN/c-MYC-induced microRNAs repress coding gene networks associated with poor outcome in MYCN/c-MYC-activated tumors. Oncogene. 2009a doi: 10.1038/onc.2009.429. [DOI] [PubMed] [Google Scholar]
- Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009b;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, Ernestus K, Konig R, Haas S, Eils R, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, Massague J. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Scarpa S, Coppa A, Ragano-Caracciolo M, Mincione G, Giuffrida A, Modesti A, Colletta G. Transforming growth factor beta regulates differentiation and proliferation of human neuroblastoma. Exp Cell Res. 1996;229:147–154. doi: 10.1006/excr.1996.0352. [DOI] [PubMed] [Google Scholar]
- Schweigerer L, Breit S, Wenzel A, Tsunamoto K, Ludwig R, Schwab M. Augmented MYCN expression advances the malignant phenotype of human neuroblastoma cells: evidence for induction of autocrine growth factor activity. Cancer Res. 1990;50:4411–4416. [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco A, Scarpa S, Coppa A, Baccheschi G, Palumbo C, Leonetti C, Zupi G, Colletta G. Increased TGFbeta type II receptor expression suppresses the malignant phenotype and induces differentiation of human neuroblastoma cells. Exp Cell Res. 2000;255:77–85. doi: 10.1006/excr.1999.4750. [DOI] [PubMed] [Google Scholar]
- Verrecchia F, Chu ML, Mauviel A. Identification of novel TGF-beta /Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J Biol Chem. 2001;276:17058–17062. doi: 10.1074/jbc.M100754200. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.