Abstract
Circadian oscillations in clock components are central to generation of self-sustained 24-h periodicity. In the Drosophila molecular clock, accumulation, phosphorylation, and degradation of PERIOD (PER) and TIMELESS (TIM) proteins govern period length. Yet little is known about the kinases that phosphorylate TIM in vivo. It has been shown previously that the protein kinase CK2 phosphorylates TIM in vitro. Here, we identify a role for CK2 in TIM regulation in vivo. Induction of a dominant-negative CK2α, CK2αTik (Tik), increases TIM protein and tim transcript levels, reduces oscillation amplitude, and results in persistent cytoplasmic TIM localization. Exposure to light and subsequent TIM degradation results in an increase in the fraction of the transcriptional repressor PER that is nuclear and suppression of per and tim RNA levels. TIM protein, but not tim transcript, levels are elevated in Tik mutants in a per01 background. In contrast, Tik effects on PER are undetectable in a tim01 background, suggesting that TIM is required for CK2 effects on PER. To identify potential CK2 target sites, we assayed TIM phosphorylation rhythms in a deletion mutant that removes a conserved serine-rich domain and found that TIM protein does not show robust rhythmic changes in mobility by Western blotting, a hallmark of rhythmic phosphorylation. The period lengthening effects in Tik heterozygotes are reduced in a timUL mutant that disrupts a putative CK2 phosphorylation site. Together, these data indicate that TIM is an important mediator of CK2 effects on circadian rhythms.
Keywords: circadian, phosphorylation, CK2, TIMELESS, PERIOD, Drosophila
Introduction
Circadian (from the Latin circa dia, ∼1 d) rhythms are daily oscillations in gene expression, physiology, and behavior driven by biological clocks. These clocks can be entrained to the sun-imposed light/dark (LD) cycle (DeCoursey, 2004). Circadian clocks enable organisms to anticipate daily environmental changes. Even in the absence of an LD cycle, circadian rhythms persist, reflecting a functional clock. Circadian rhythms have been found in all the kingdoms of life, suggesting they are a fundamental property (DeCoursey, 2004).
Molecularly, circadian clocks consist of interlocked transcriptional feedback loops that drive self-sustained oscillations in gene expression (Allada et al., 2001). The structure of the molecular clock is highly conserved between mammals and insects. In the fruit fly Drosophila, transcription of the clock components period (per) and timeless (tim) is activated by the CLOCK (CLK)/CYCLE (CYC) heterodimer (Allada et al., 1998; Rutila et al., 1998). PERIOD (PER) and TIMELESS (TIM) proteins heterodimerize in the cytoplasm (Lee et al., 1996; Zeng et al., 1996; Meyer et al., 2006) and translocate to the nucleus to repress CLK/CYC-driven activation (Curtin et al., 1995; Darlington et al., 1998; Chang and Reppert, 2003; Nawathean and Rosbash, 2004). Several studies have suggested that the PER/TIM heterodimer may dissociate before these proteins translocate to the nucleus (Shafer et al., 2002; Meyer et al., 2006), where PER may act as a repressor (Rothenfluh et al., 2000; Chang and Reppert, 2003; Nawathean and Rosbash, 2004) by facilitating an interaction between DOUBLETIME (DBT) and CLK (Kim and Edery, 2006; Yu et al., 2006; Kim et al., 2007; Nawathean et al., 2007). In wild-type flies, it takes ∼24 h to complete this cycle of transcriptional activation and repression, thus giving rise to 24 h periodicity.
PER and TIM phosphorylation is critical for regulating period length. Both proteins demonstrate a circadian rhythm in phosphorylation (Edery et al., 1994; Myers et al., 1996; Zeng et al., 1996); the appearance of hyperphosphorylated PER and TIM precedes their disappearance, suggesting hyperphosphorylation signals degradation (Edery et al., 1994; Myers et al., 1996; Zeng et al., 1996; Ko et al., 2002). PER is phosphorylated by the protein kinases DBT (Kloss et al., 1998; Price et al., 1998; Ko et al., 2002; Muskus et al., 2007), the fly ortholog of CK1ε, and CK2 (formerly known as casein kinase 2) (Lin et al., 2002, 2005). DBT activity promotes PER degradation (Kloss et al., 1998; Grima et al., 2002; Ko et al., 2002), whereas CK2 activity promotes PER and TIM nuclear translocation in pacemaker neurons (Lin et al., 2002; Atken et al., 2003; Smith et al., 2008). Both DBT and CK2 enhance PER repression in cultured Schneider 2 (S2) cells (Nawathean and Rosbash, 2004). SHAGGY, the fly ortholog of mammalian glycogen synthase kinase 3 β (GSK3β), phosphorylates TIM in vitro and promotes TIM nuclear translocation (Martinek et al., 2001). PER–TIM interaction blocks DBT phosphorylation (Kloss et al., 1998, 2001; Price et al., 1998; Ko et al., 2002), stabilizing PER. PER and TIM stability is promoted by the phosphatases PP2A and PP1, respectively (Sathyanarayanan et al., 2004; Fang et al., 2007). During light/dark cycling, TIM is degraded by light through its interaction with a blue-light photoreceptor, CRYPTOCHROME (CRY) (Ceriani et al., 1999; Busza et al., 2004). GSK3β also phosphorylates CRY, which could explain in vivo TIM effects (Stoleru et al., 2007). Analysis of tim mutants suggests regulation of TIM stability during constant conditions is distinct from regulation during light (Rothenfluh et al., 2000). Nonetheless, little is known about the mechanisms that regulate TIM stability in constant darkness. Here, we present evidence suggesting that CK2 regulates TIM independently of light.
Materials and Methods
Western blotting.
TIM and PER Western blotting was performed as described previously (Lin et al., 2002, 2005). To ensure equivalent amounts of protein were loaded in each lane, we used equal numbers of heads and confirmed equal loading by Ponceau staining as done in previous studies (Lin et al., 2002, 2005). Western blot data were quantified using Scion Image software. Data shown graphically in Figure 7 were normalized such that per01 = 100 or tim01 = 100.
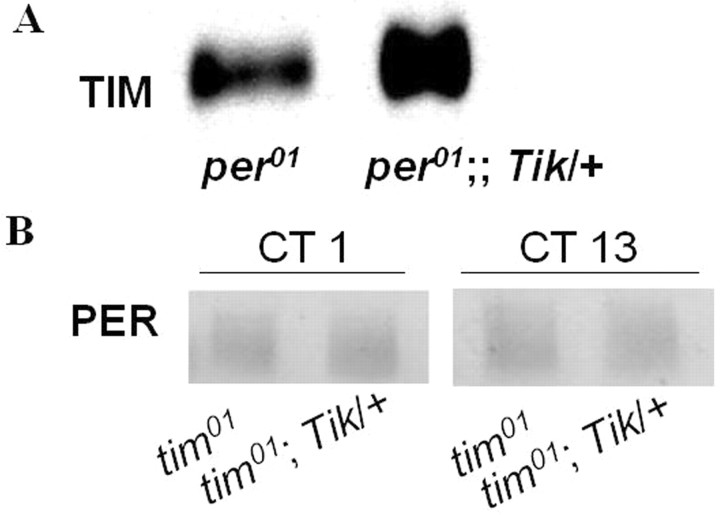
Figure 7.
In the absence of PER, TIM is increased by Tik. Tik has no effect on PER in the absence of TIM. A, Tik can increase TIM protein abundance by ∼40% in the absence of PER. In A, male flies were collected at circadian time (CT) 14 on DD3. A, Representative TIM Western blot of whole head extracts. B, Tik has no effect on PER in the absence of TIM. Flies were collected at CT 1 and CT13 on DD1. Representative PER Western blot of whole head extracts are shown.
Quantitative real-time reverse transcriptase-PCR.
Total RNA was isolated from frozen whole heads using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. DNA was removed from RNA extracts using RQ1 DNase from Promega. Real-time PCRs were run using the Applied Biosystems 7900HT fast real-time PCR instrument. Data were collected using SDS software version 2.2.1. Data were analyzed using the 2−ΔCt method using RP49 expression values to normalize for differences in RNA amount among samples. Student's t tests were used to determine statistically significant differences between samples. Transcript data for per01 and per01;; Tik/+ were normalized such that per01 = 100.
For PCRs, ∼100 ng of RNA were used per reaction. We used the Qiagen SYBR green RT-PCR kit to set up reactions. Total reaction volume was 25 μl and reactions were run in 96-well plates. Primer sets used were ordered from Integrated DNA Technologies. Primer sequences are as follows: per forward primer is 5′-CAGCAGCAGCCTAATCG-3′, and the reverse primer is 5′-GAGTCGGACACCTTGG-3′; tim forward primer is 5′-CTGGGGAGTGACCATGG-3′, and the reverse primer is 5′- GCTGGAATCGCCACTG-3′; RP49 forward primer is 5′-CGACGCTTCAAGGGACAGTATC-3′, and the reverse primer is 5′-TCCGACCAGGTTACAAGAACTCTC-3′. RT-PCR cycling parameters were as follows: 30 min at 50°C, 15 min at 95°C, and 30 cycles of 15 s at 94°C, 30 s at 55°C, and 30 s at 72°C.
Fly genetics and husbandry.
We published previously the UAS-Tik transgene (Smith et al., 2008). It was made using the following strategy. We cut the BglII and XhoI fragment containing Tik from pET-Tik, and cloned it into pUAST to generate transgenic flies. All transgenic DNA constructs were sequenced (Applied Biosystems) and injected in y w embryos at the CBRC Transgenic Drosophila Core (Charlestown, MA). Stable stocks of timGAL4–62; UAS-Tik, pdfGAL4; UAS-Tik, and tim01; Tik/TM6 were made using standard fly genetics. timUL, timL1, timL3, and timL4 mutants were a kind gift from Michael Young (The Rockefeller University, New York, NY). TimΔ260–292 1–8 and TimΔ260–292 1–8 tim01 flies were a kind gift from Amita Sehgal. The published TimΔ260–292 transgenic line used in our study is identified as 1–8 (Ousley et al., 1998). All fly stocks and crosses were maintained in standard cornmeal-molasses-agar food at 25°C.
Fly locomotor behavior and analysis.
Fly locomotor behavior was assayed using the Drosophila Activity Monitor System as described previously (Lin et al., 2005). To analyze behavior during light/dark entrainment, we used the Counting Macro 5.11 software developed by the Allada laboratory. Counting Macro 5.11 was also used to generate the average activity traces shown in Figure 4 (right column).
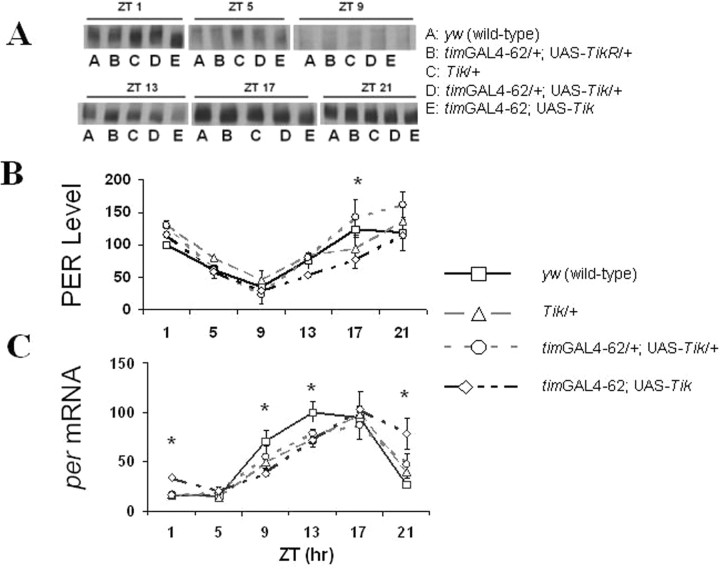
Figure 4.
Light corrects the weak oscillations and abnormally high trough levels of PER protein and transcript observed in Tik-expressing flies during DD1 (Smith et al., 2008). A–C, PER protein (A, B) and per transcript (C) were measured using methods described in Figure 1. PER protein levels are normalized such that in y w flies, ZT1 = 100. per transcript levels are normalized such that in y w flies, ZT17 = 100. Flies were collected at 4 h intervals during LD. ZT here refers to time during a 12 h light/dark cycle. ZT 0 is lights on. Error bars are SEM. Time-point-specific protein and transcript levels were compared between y w and timGAL4–62; UAS-Tik; *p < 0.05 for y w versus timGAL4–62; UAS-Tik (Student's t test).
Phosphorylation site predictions.
Potential phosphorylation sites in TIM260–292 were identified using an on-line server, NetPhosK, that makes kinase-specific phosphorylation site predictions based on the sequences of known phosphorylation sites (NetPhosK 1.0 server; http://www.cbs.dtu.dk/services/NetPhosK/) (Blom et al., 2004).
Immunohistochemistry.
Male flies were entrained for at least 2 d to 12 h light/dark cycles. Flies were dissected at the indicated time points and fixed in 4% formalin in PBS for 40 min. After rinsing in 0.1% Triton in PBS (PBT) brains were stained with mouse anti-pigment-dispersing factor (anti-PDF; 1:800) and either rabbit anti-PER (1:4000; gift from M. Rosbash, Brandeis University, Waltham, MA) or rat anti-TIM (1:1000; gift from M. Rosbash) overnight in blocking solution (10% goat-normal-serum-PBT). After PBT rinses, Invitrogen secondary antibodies were applied at 1:500 in block for at least 3 h at room temp or overnight (for PER/PDF double labels, goat anti-mouse Alexa 488 with goat anti-rabbit Alexa 594; for TIM/PDF, goat anti-rat Alexa 488 with goat anti-mouse Alexa 594). After PBT and PBS rinse, brains were mounted in glycerol.
Slides were coded and processed blind through quantification and analysis. Tissue was imaged at 60× on a Nikon C1 confocal microscope. Images were analyzed with NIH ImageJ. Briefly, small ventral lateral pacemaker neurons (sLNvs) were identified by location, size, and PDF staining. From a single section containing the middle of the nucleus, the cytoplasm and nucleus of each cell were separately circled from the PDF staining. Average intensity of each compartment was quantified from the PER or TIM staining. Nearby background staining intensity was subtracted from each. Measurements were scaled to the average peak within each experiment to combine experiments. Then, the average nuclear intensity for each cell was divided by cytoplasmic intensity to get the nuclear/cytoplasmic ratio. Averages and SEM were calculated in Excel and t tests were used to test for significant differences. A minimum of 22 cells were measured for TIM and 26 for PER.
Results
Expression of a dominant-negative CK2 increases trough levels of TIM protein and mRNA in constant darkness
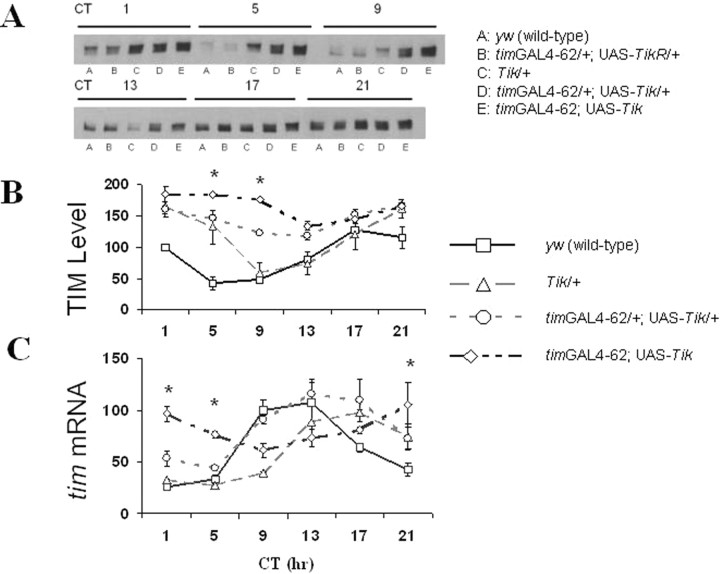
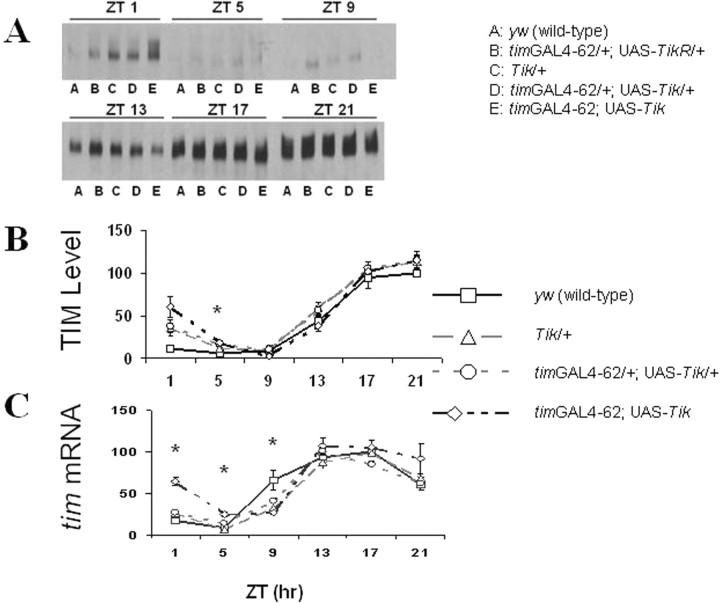
Tik behaves as a dominant-negative CK2α allele and expression of Tik using the GAL4/UAS system (Brand and Perrimon, 1993) can generate strikingly long behavioral rhythms (>30 h) or behavioral arrhythmia (Smith et al., 2008). To express a UAS-driven Tik, we crossed UAS-Tik flies to timGAL4–62 flies which express GAL4 in all circadian cells (Kaneko and Hall, 2000). To determine whether tim expression is altered by UAS-Tik expression, we measured TIM protein and transcript levels in Tik-expressing flies as well as Tik mutants during the first day of constant darkness (DD1) (Fig. 1). Whereas wild-type flies show robust oscillations in tim expression, heterozygous progeny (timGAL4–62/+; UAS-Tik/+) show delayed reduced amplitude molecular oscillations (Fig. 1). TIM protein and tim transcript levels weakly oscillate in timGAL4–62; UAS-Tik homozygotes. In addition, timGAL4–62; UAS-Tik homozygotes have elevated trough levels of TIM protein and tim transcript during the first half of the circadian cycle (subjective day) (Fig. 1). We observed similar results for PER (Smith et al., 2008). Elevated tim transcript in the context of elevated PER levels suggests reductions in CK2 activity may disrupt PER negative feedback, consistent with S2 cell data (Nawathean and Rosbash 2004).
Figure 1.
Tik-expressing flies display weak oscillations in and elevated trough levels of TIM protein and tim transcript during DD1 after a 12 h LD cycle. A–C, TIM protein (A, B) and tim transcript (C) were measured in the heads of y w (wild-type, squares), Tik/+ (triangles), timGAL4–62/+; UAS-Tik/+ (circles), and timGAL4–62; UAS-Tik (diamonds) flies. timGAL4–62/+; UAS-TikR/+ flies express TikR and were used as a control for UAS-CK2α expression. TikR is a recessive allele of CK2α that was derived spontaneously from the Tik stock. In addition to the two point mutations (M161K and E165D) found in Tik, TikR contains an additional insertion and deletion resulting in deletion of amino acids 234–240 and an additional amino acid change, R242E (Lin 2002). Flies were collected at 4 h intervals during DD1. TIM protein was assayed by Western blotting (A) and quantified using Scion Image software (B). TIM protein levels are normalized such that in y w flies, circadian time 1 (CT1) = 100. tim transcript (C) was assayed by quantitative real-time RT-PCR and normalized to RP49 transcript levels. tim transcript levels are normalized such that in y w flies, CT13 = 100. CT here refers to time during the first day of constant darkness after entrainment in LD cycling. CT 0 is “subjective dawn” during constant darkness. Error bars are SEM. Time-point-specific protein and transcript levels were compared between y w and timGAL4–62; UAS-Tik; *p < 0.05 for y w vs timGAL4–62; UAS-Tik (Student's t test).
TIM demonstrates persistent cytoplasmic localization in Tik-expressing flies
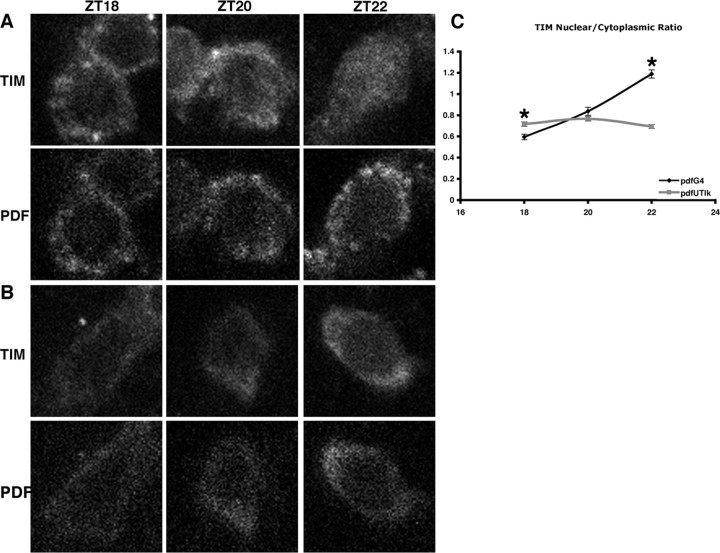
We and others have shown that CK2 is important for the transition of PER from cytoplasm to nucleus in the core sLNvs (Lin et al., 2002; Atken et al., 2003; Smith et al., 2008). The sLNvs are critical drivers of morning anticipation under LD conditions and are the master pacemaker neurons that drive circadian behavior under DD conditions (Renn et al., 1999; Grima et al., 2004; Stoleru et al., 2004, 2005). After observing effects of Tik on PER nuclear translocation, we wondered whether Tik would alter the ability of TIM to localize to the nucleus. Previous studies had shown that TIM nuclear localization is delayed in mutants of the regulatory β subunit of CK2 (Atken et al., 2003). We examined TIM localization under light/dark conditions to reduce the influence of free-running period differences on the timing of nuclear localization. Indeed, during late night [Zeitgeber time (ZT) 22], TIM translocates to the nucleus in sLNvs of pdfGAL4 control flies but remains mostly cytoplasmic in sLNvs of pdfGAL4; UAS-Tik homozygotes, as quantified by the nuclear:cytoplasmic ratio (Fig. 2). This result, together with our previous results, shows that CK2 function controls the proper timing of TIM and PER nuclear translocation.
Figure 2.
TIM persists in the cytoplasm in pacemaker neurons of Tik-expressing flies. A, B, TIM nuclear entry in the sLNvs of pdfGAL4 controls (A) and pdfGAL4; UAS-Tik homozygotes (B). Top panels are TIM; bottom panels are neuropeptide PDF that labels cytoplasm. C, Average of two experiments, quantified as the ratio of average nuclear/cytoplasmic intensities. *p < 0.05. Error bars indicate SEM.
Light/dark cycling can restore robust oscillations in TIM levels in Tik-expressing flies
Tik-expressing flies display elevated levels of tim expression during the subjective day of DD1 (Fig. 1), a time of day during which TIM protein is rapidly degraded by light in wild-type flies subjected to light/dark entrainment (Hunter-Ensor et al., 1996; Lee et al., 1996; Myers et al., 1996; Zeng et al., 1996). We wondered whether light would be able to reduce TIM levels in Tik-expressing flies, thus “correcting” TIM molecular oscillations. In light/dark entrainment, Tik-expressing flies display TIM protein and transcript levels and oscillations that are very similar to those observed in wild-type flies (Fig. 3). Whereas, during DD1, TIM protein shows less than a twofold change in levels in homozygous timGAL4–62; UAS-Tik flies, TIM protein oscillates with an ∼10-fold amplitude when these flies are subjected to environmental light/dark cycling. Interestingly, higher amplitude protein oscillations are accompanied by higher amplitude RNA oscillations, specifically by suppressing transcript levels after TIM degradation (Fig. 3C). These data suggest that light degradation of TIM allows PER feedback repression of tim transcription (Fig. 3C). It should be noted that TIM levels are not entirely corrected, as elevated levels can be observed at ZT1, including lower mobility forms (Fig. 3A). We also examined PER protein and per RNA in LD and observed a similar pattern (Fig. 4). Reductions of PER protein and RNA in the light phase are also consistent with enhanced negative feedback (Fig. 4). Although oscillations in LD are more like wild-type, per and tim transcript oscillations are phase-delayed in Tik-expressing flies during LD entrainment (Figs. 3C, 4C), suggesting that CK2 function may be necessary for the proper timing of PER repression.
Figure 3.
Light corrects the weak oscillations and abnormally high trough levels of TIM protein and transcript observed in Tik-expressing flies during DD1 (Fig. 1). A–C, TIM protein (A, B) and tim transcript (C) were measured as described in Figure 1. TIM protein levels are normalized such that in y w flies, ZT1 = 100. tim transcript levels are normalized such that in y w flies, ZT17 = 100. Flies were collected at 4 h intervals during LD. ZT here refers to time during a 12 h light/dark cycle. ZT 0 is lights on. Error bars are SEM (SEM). Time-point-specific protein and transcript levels were compared between y w and timGAL4–62; UAS-Tik; *p < 0.05 for y w vs timGAL4–62; UAS-Tik (Student's t test).
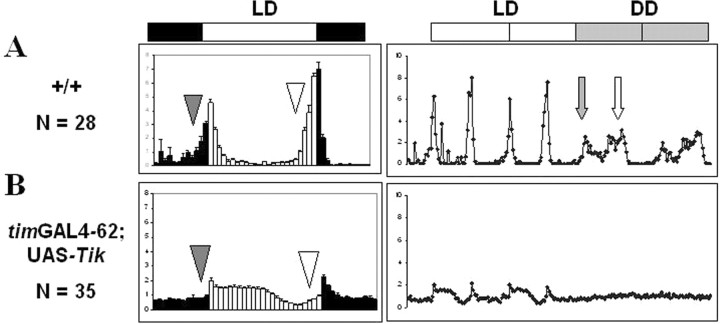
Light can induce evening anticipation in Tik-expressing flies
Because light/dark cycling can restore quasi-normal oscillations in TIM protein in Tik-expressing flies, we hypothesized that behavioral rhythms may also be restored in this condition, particularly in homozygous timGAL4–62; UAS-Tik expression flies, which are behaviorally arrhythmic in constant darkness (Fig. 5B) (Smith et al., 2008). During light/dark cycling, wild-type flies display anticipatory increases in locomotor activity before lights-on (morning anticipation) and lights-off (evening anticipation) and these peaks persist early in constant darkness (Fig. 5A). Strikingly, homozygous Tik expression flies show moderate evening anticipation during LD, but during DD1, these flies show neither morning nor evening peaks, consistent with the behavioral arrhythmia we observe over multiple days in DD (Fig. 5B) (Smith et al., 2008). These observations suggest light is sufficient to generate evening anticipation without normal CK2 activity. In addition to altered anticipatory activity, Tik expression flies show striking changes in the distribution of activity during light/dark cycles with a sustained increase in activity after lights-on. Together, our data suggest that clock defects as a result of CK2 inhibition can be partially compensated by the presence of light, consistent with our molecular data.
Figure 5.
Tik-expressing flies exhibit evening anticipatory behavior in LD cycling. The light phase is indicated with white bars, and dark phase is indicated by black bars. A, Wild-type flies (+/+) exhibit an increase in locomotor activity before the lights-on and lights-off transitions; these increases in activity levels are known as morning and evening anticipation, respectively. The gray triangle on each LD plot (left column) indicates the time at which morning anticipation ought to occur, and the white triangle indicates the time at which evening anticipation ought to occur. These peaks of activity persist in constant conditions in wild-type flies. The right column shows the average activity traces for a population of flies during 2 d of LD cycling (white boxes) and 2 d of DD (gray boxes). For the first day of DD, the gray arrow indicates the morning peak of activity, and the white arrow indicates the evening peak of activity. B, Tik-expressing flies anticipate the lights-off transition but fail to anticipate the lights-on transition. In DD, homozygous Tik-expressing flies show neither morning nor evening peaks (as indicated by the lack of arrows). In this assay, locomotor activity of male flies was tested with Drosophila activity monitors, and behavior data were analyzed using the Counting Macro 5.11 software developed by the Allada laboratory.
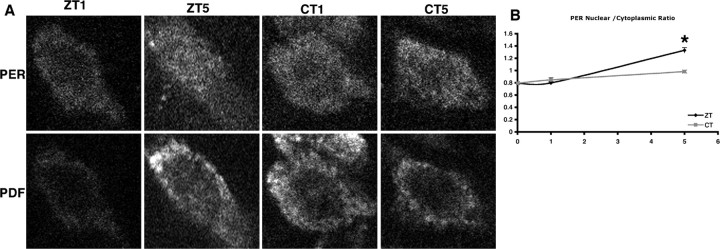
Light induces PER nuclear translocation in pacemaker neurons of Tik-expressing flies
The ability of light to rescue quasi-normal molecular oscillations and evening anticipation in UAS-Tik expression flies prompted us to examine the effect of light on PER subcellular localization in the pacemaker sLNvs. Previously we showed that PER nuclear translocation is delayed in Tik heterozygotes or Tik-expressing flies (Lin et al., 2002; Smith et al., 2008). We asked whether light could correct this defect in pdfGAL4; UAS-Tik flies. The presence of light increases the proportion of nuclear PER by ZT5 relative to flies not exposed to light (circadian time 5) (Fig. 6) (p < 0.05). Importantly, this result shows that when CK2 activity is reduced, light can enhance PER nuclear translocation, consistent with the notion that in the presence of reduced CK2 activity, TIM may retain PER in the cytoplasm. This effect of light on PER may work directly on PER through TIM. We cannot rule out the possibility that it may work indirectly through light effects on the phase of the clock. The fact that changes in PER localization are not observed until after TIM is degraded suggests that TIM degradation may gate nuclear localization but that additional processes, such as progressive phosphorylation, may be necessary for nuclear entry.
Figure 6.
Light increases the proportion of nuclear PER in pacemaker neurons of Tik-expressing flies. A, PER subcellular localization in the sLNvs of pdfGAL4; UAS-Tik homozygotes in the hours after lights on (ZT1, ZT5) and in the first hours of constant darkness (CT1, CT5). PER is significantly more nuclear at ZT5 than at CT5 (*p < 0.05). Top panels are PER; bottom panels are PDF. CT, Circadian time. B, Graph is as in Figure 2.
Tik increases TIM abundance independently of PER
We have previously reported data supporting the notion that PER is an in vivo substrate of CK2 (Lin et al., 2005). To determine whether PER is required for the ability of Tik to increase TIM levels, we examined TIM protein in per01;; Tik/+ mutants during constant darkness. By DD3, TIM levels are increased in Tik flies by ∼40% compared with per01 controls (Fig. 7A) [per01 = 100 ± 0 (SEM) arbitrary units; per01;; Tik/+ = 140.3 ± 4.5, three replicates, p < 0.05]. tim transcript levels are not increased by Tik, indicating that this effect is posttranscriptional [per01 = 100 ± 0 (SEM) arbitrary units, per01;; Tik/+ = 102.0 ± 4.2, two replicates, p = 0.7].
TIM is required for Tik effects on PER abundance
Having established that Tik can increase TIM abundance in the absence of PER protein, we sought to determine whether TIM is required for the ability of Tik to increase PER levels. We found that PER protein levels are unaltered by Tik in tim01; Tik/+ mutants compared with tim01 controls (Fig. 7B) [tim01 = 99.4 ± 0.9 (SEM) arbitrary units; tim01; Tik/+ = 98.1 ± 1.4, two replicates, p = 0.5], suggesting that Tik requires TIM to affect PER levels. We have confirmed that PER levels in tim01 are detectable and within the linear range to detect any differences (supplemental text and supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Together, our data suggest that CK2 effects on PER levels require TIM, although CK2 may still alter PER repressor activity.
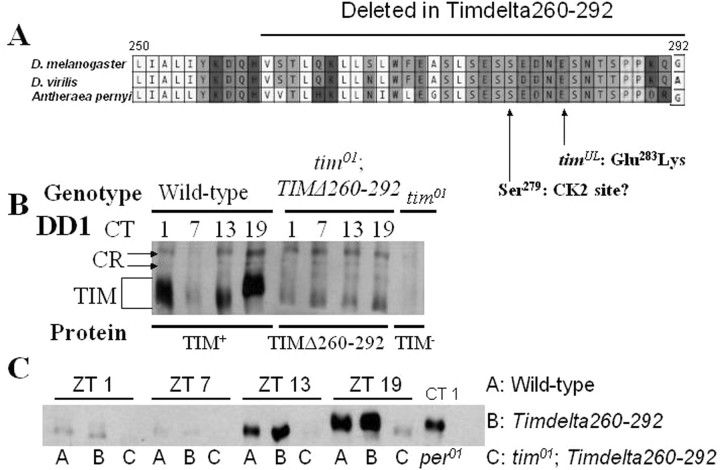
Deletion of a serine/threonine-rich domain in TIM severely reduces rhythms in TIM abundance and mobility
To our knowledge, no specific phosphorylation sites have been identified in TIM. To identify such regions, we searched for potential kinase target sites that are conserved among insect TIMs. We identified an N-terminal domain (260–292) rich in serines (Fig. 8A), which we have named TIM serine-rich domain (SRD). This domain contains seven predicted phosphorylation sites, including four CK2 sites and two DBT sites. Intriguingly, a previous study found that this serine/threonine-rich domain in TIM is required for normal circadian behavior: a tim transgene in which amino acids 260–292 (TimΔ260–292) have been deleted can poorly rescue tim01 rhythmicity, but these rhythms have very long periods of >30 h, suggesting this region of TIM is also critical for normal period length (Ousley et al., 1998). To test the effect of this deletion on TIM oscillations, we examined TIM protein by Western blotting during DD1 and LD conditions. In TimΔ260–292 tim01 flies during DD1, TIM protein levels do not show a significant change in mobility, remaining in a high mobility (likely hypophosphorylated) state (Fig. 8B). High mobility TIM does not appear to be a general consequence of weak molecular rhythms, as arrhythmic per01 flies express TIM that is not high mobility (Fig. 8C). We cannot exclude the possibility, however, that the lack of low-mobility forms of TIM may be attributable to reduced TIM expression or detection in TimΔ260–292 tim01 flies. During LD entrainment, TIMΔ260–292 cycles modestly (Fig. 8C). The lack of TIMΔ260–292 protein during and immediately after the light phase indicates that light can degrade this form of TIM. This observation suggests TIMΔ260–292 retains the ability to interact with CRY. Our results suggest TIM SRD is particularly important for light-independent regulation of TIM hyperphosphorylation.
Figure 8.
A TIM domain important for phosphorylation and CK2 action. A, Drosophila TIM amino acids 250–292 are highly conserved in insects. Shown here is an alignment of TIM 250–292 from Drosophila melanogaster, Drosophila virilis, and Antheraea pernyi (silkmoth). Amino acids 260–292 are deleted in the TimΔ260–292 transgene (Ousley et al., 1998). The timUL mutation, Glu283Lys, is also found within this region of TIM. timUL is hypothesized to inhibit CK2 phosphorylation of a predicted CK2 site at Ser279. B, Representative Western blot of whole head extracts from flies collected during DD1 demonstrating that TIM fails to show significant mobility shifts in TimΔ260–292 tim01 flies. Cross-reacting (CR) bands are indicated as markers for linearity of samples across the blot. CT, Circadian time. C, Representative Western blot of whole head extracts demonstrating that TIM cycles weakly in TimΔ260–292 tim01 flies under LD entrainment conditions. ZT 0, lights on; ZT 12, lights off. Western blots shown in B and C are representative of results from at least two independent experiments.
Ability of Tik to lengthen behavioral period is partially suppressed in timUL mutants
The behavioral period lengthening and misregulation of tim gene expression observed in Tik mutants and Tik-expressing flies are reminiscent of behavioral and molecular phenotypes observed in timUL mutants. TIMUL protein is unusually stable in constant darkness yet is rapidly degraded by light, suggesting the timUL mutation alters light-independent TIM degradation. Interestingly, light likely restores negative feedback in timUL mutants (Rothenfluh et al., 2000) as we propose for Tik flies (Figs. 3C, 4C). Based on the phenotypic similarities, we hypothesized that Tik and timUL might affect the same molecular pathways that regulate TIM stability. We confirmed the presence of the point mutation in timUL originally reported as a Glu260Lys mutation (Rothenfluh et al., 2000; Tauber et al., 2007). This residue is Glu283 in the long tim isoform (Tauber et al., 2007). Interestingly, Glu283 is a key acidic residue near a potential CK2 site at Ser279. CK2 targets serines and threonines with acidic amino acids C-terminal to the target site, as is the case for this site (Meggio and Pinna, 2003) (Fig. 8A). To test for interactions between Tik and timUL, we examined circadian behavior in timUL/tim01; Tik/+ flies (Table 1). timUL/tim01 flies display ∼34 h periods. Tik mutants normally lengthen period by ∼2–3 h (Lin et al., 2002); in contrast, Tik only lengthens period by ∼1 h in timUL/tim01; Tik/+ flies. Rhythmicity is robust, indicating that period assessments are reliable. This suppression of period lengthening is unlikely to be a result of having reached an upper limit for period length: perL; timUL flies reportedly exhibit ≥40 h periods and robust rhythms (Rothenfluh et al., 2000), and we have observed periods ≥38 h in timGAL4–62/+; UAS-Tik flies (E. Smith, J. Lin, R. Meissner, and R. Allada, unpublished observations). This suppression of period lengthening is allele-specific: Tik has an additive effect on period length (≥2 h) in combination with other long-period tim alleles (Table 1). Together, these results suggest that mutation of a putative CK2 site in TIM reduces CK2 effects on circadian period.
Discussion
Here, we present evidence that CK2 operates through TIM to control circadian clock function in vivo. Expression of the dominant-negative CK2α allele, Tik, elevates trough levels of TIM protein and RNA during constant darkness and alters TIM subcellular localization. Tik effects on PER are undetectable in a tim01 mutant, whereas Tik effects on TIM are evident in a per01 background, suggesting direct TIM effects. Behavioral period effects of Tik are reduced in the timUL mutant that disrupts a putative CK2 site. The effects on TIM metabolism as well as the genetic requirement of tim for CK2 effects indicate CK2 primarily operates through TIM to regulate the circadian clock. Importantly, we demonstrate these effects in vivo.
One potential model consistent with our data is that CK2 regulates TIM abundance and in turn, promotes negative feedback. Elevated trough TIM levels are accompanied by elevated trough tim transcript levels in DD, suggesting impaired turnover, negative feedback, and/or tim transcriptional regulation. Effects on TIM are evident in per01 flies but are not accompanied by changes in tim transcript levels, suggesting a direct effect on TIM protein, perhaps by regulating TIM stability, although we cannot rule out a translational effect. Importantly, the per01 data argue strongly against a direct effect of CK2 on CLK/CYC-driven transcription of tim. The ability of light to degrade TIM in Tik-expressing flies implies that TIM levels can be regulated by two distinct pathways, a light/CRY-dependent pathway and a CK2-dependent pathway. During light/dark entrainment, light robustly degrades TIM in Tik-expressing flies. This degradation is accompanied by a sharp suppression in per and tim transcript levels, suggesting that it is excessive TIM levels, rather than direct PER effects, that abrogate negative feedback in Tik-expressing flies. per and tim RNA oscillations remain phase delayed during LD in homozygous Tik-expressing flies, suggesting that Tik also affects the timing of PER repression.
Some CK2 effects on PER may be mediated by CK2 effects on TIM. CK2 effects on PER levels require TIM (Fig. 7). CK2 is localized to the cytoplasm where PER/TIM dimers are likely present (Lin et al., 2002; Shafer et al., 2002; Meyer et al., 2006). CK2 regulates PER and TIM nuclear entry (Fig. 2) (Lin et al., 2002; Atken et al., 2003; Smith et al., 2008), a process that likely depends on the PER/TIM dimer (Fig. 6) (Saez and Young, 1996; Lin et al., 2002; Shafer et al., 2002; Atken et al., 2003; Meyer et al., 2006). Although effects on PER require TIM, PER is likely a direct in vivo CK2 substrate (Lin et al., 2005). CK2 robustly and specifically phosphorylates PER in vitro and in-vitro-defined sites have clear in vivo functions (Lin et al., 2002, 2005). Tik mutants can also strongly effect PER mobility on Western blots (Fig. 4A) (Smith et al., 2008); PER mobility is highly dependent on phosphorylation (Edery et al., 1994; Myers et al., 1996; Zeng et al., 1996). These data are most consistent with the idea that CK2 targets the PER/TIM dimer in the cytoplasm and phosphorylates PER to promote nuclear entry. Additional experiments will be required to test the dimer hypothesis. Based on S2 cell experiments, it has been proposed that CK2 phosphorylation of PER promotes its intrinsic repressor activity independent of its effects on nuclear localization (Nawathean and Rosbash, 2004). Our observation that light-induced TIM degradation results in a suppression of per and tim RNA in Tik-expressing flies suggests that elevated TIM levels block PER repression. Nonetheless, remaining alterations in transcript levels suggest that the freed PER repressor may not be entirely functional, consistent with additional CK2 effects on PER.
TIM may be an in vivo CK2 substrate. We and others have shown that CK2 phosphorylates TIM in vitro (Zeng et al., 1996; Lin et al., 2002). In vivo, Tik increases TIM levels in the absence of PER, suggesting CK2 effects on TIM may be direct. Importantly, increases in TIM protein are not accompanied by increases in transcript, indicating CK2 regulates TIM posttranscriptionally. We favor the notion that CK2 acts to phosphorylate TIM in vivo, consistent with published in vitro phosphorylation experiments (Zeng et al., 1996).
One potential argument against the hypothesis that CK2 phosphorylates TIM is the finding that low-mobility TIM accumulates to high levels in timGAL4–62; UAS-Tik homozygotes compared with wild-type controls (Fig. 3A). Interestingly, this result is similar to the observation that in Dbtg mutants, low mobility forms of PER accumulate that are even lower in mobility than those in wild-type (Suri et al., 2000), although PER is widely established as a DBT substrate (Kloss et al., 1998; Price et al., 1998; Ko et al., 2002; Muskus et al., 2007). In both cases, phosphorylation-induced mobility changes cannot be attributed solely to a single kinase. It is also possible that elevated TIM levels may render low mobility forms more visible in Tik-expressing flies. Thus, the presence of low-mobility forms does not exclude CK2 as an in vivo kinase for TIM.
Although no phosphorylation sites have been identified in TIM, deletion of a small TIM SRD (TIM 260–292) reduces or eliminates significant circadian mobility changes (Fig. 8B). This domain is conserved among insects and may contain multiple phosphorylation sites that are responsible for regulating period length and rhythms in TIM levels and mobility. CK2 may be one of several kinases that phosphorylates TIM SRD. In addition, although this domain is critical for shifts in TIM mobility, there are likely phosphorylation sites for CK2 outside of this region. When we sequenced the bacterial expression construct used previously to generate TIM protein for our CK2 in vitro phosphorylation assay, we discovered that the construct lacks amino acids 260–292 (data not shown). We also cannot exclude the possibility that loss of rhythmic mobility changes may be secondary to reduced TIM levels in SRD mutants or may not be linked to CK2. Nonetheless, the in vivo data and sequence conservation raise the possibility that the SRD may be a kinase substrate.
The TIM SRD contains four predicted CK2 sites. One of these sites, Ser279, is potentially altered in timUL mutants by the Glu283Lys point mutation. CK2 preferentially phosphorylates serine or threonine residues located 2–5 residues N-terminal to acidic amino acid residues such as glutamate or aspartate (Meggio and Pinna, 2003). The placement of basic residues, such as lysine as in the timUL mutant, near CK2 sites inhibits CK2 phosphorylation of the target Ser/Thr residue (Marin et al., 1986, 1992). timUL and Tik show allele-specific genetic interactions such that in timUL mutants, Tik period lengthening is partially suppressed. These results are consistent with our hypothesis that timUL is a CK2 site mutant. This prediction suggests that Ser279 plays a very important role in regulating circadian period length in flies. It will be of interest to test the hypothesis that CK2 promotes TIM degradation.
It has been claimed that timUL shows a late-night specific defect principally based on the phase response curve to light and persistent nuclear TIM levels (Rothenfluh et al., 2000). CK2, however, is mostly restricted to the cytoplasm and thus would be predicted to act on cytoplasmic PER and/or TIM proteins during the early night. Although it is true that timUL shows profound late-night defects, on closer examination, there are alterations in the phase response curve to light during the early night (Rothenfluh et al., 2000) (Fig. 1, 2 h in darkness). In addition, TIM protein was not examined in pacemaker neurons during the early night in timUL, so we cannot rule out the possibility that there is also an early-night defect in timUL. Therefore it is possible that timUL also displays early-night defects, as we propose for CK2 effects. Second, it is possible that phosphorylation of TIM by CK2 in the cytoplasm could influence the function of phospho-TIM in the nucleus. Third, although we principally observe CK2 in the cytoplasm (Lin et al., 2002), we cannot exclude the possibility that small but functional levels of CK2 are present in the nucleus. Regardless, the allele-specific, nonadditive genetic interaction between Tik and timUL argue that timUL is important for CK2 effects on circadian behavior.
Our data are consistent with the speculation that Tik expression reduces CK2 phosphorylation of TIM. This results in retention of PER in the cytoplasm, thus reducing PER-mediated repression. Light-mediated degradation of TIM can then liberate PER (pending some additional events) to enter the nucleus and repress CLK/CYC. Although additional experiments will be needed to test this model, our data demonstrate that it is likely that CK2 effects through TIM will play an important role in circadian clock function.
Footnotes
This work was supported by National Institutes of Health Grant R01 MH067870. R.-M.M. was supported by a Vision Training Grant awarded to Northwestern University. We thank Michael Young and Amita Sehgal for flies and Michael Rosbash for antibodies. We thank Fred Turek for use of his Applied Biosystems 7900HT fast real-time PCR instrument. We thank Elaine Smith for helpful discussions.
References
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: The genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- Atken B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR. A role for CK2 in the Drosophila circadian oscillator. Nature Neuroscience. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- Ceriani MF, Darlington TK, Staknis D, Más P, Petti AA, Weitz CJ, Kay SA. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- Chang DC, Reppert SM. A novel C-terminal domain of Drosophila PERIOD inhibits dCLOCK:CYCLE-mediated transcription. Curr Biol. 2003;13:758–762. doi: 10.1016/s0960-9822(03)00286-0. [DOI] [PubMed] [Google Scholar]
- Curtin KD, Huang ZJ, Rosbash M. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron. 1995;14:365–372. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- DeCoursey PJ. Overview of biological timing from unicells to humans. In: Dunlap JC, Loros JJ, DeCoursey PJ, editors. Chronobiology: biological timekeeping. Sunderland, MA: Sinauer; 2004. pp. 2–24. Chap 1. [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci U S A. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Sathyanarayanan S, Sehgal A. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21:1506–1518. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Chélot E, Papin C, Limbourg-Bouchon B, Rouyer F. The F-box protein Slimb controls the levels of clock proteins Period and Timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein Timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kim EY, Edery I. Balance between DBT/CK1epsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci U S A. 2006;103:6178–6183. doi: 10.1073/pnas.0511215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu W, Hardin PE, Edery I. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol. 2007;27:5014–5028. doi: 10.1128/MCB.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein kinase closely related to human casein kinase Ie. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Kloss B, Rothenfluh A, Young MW, Saez L. Phosphorylation of PERIOD is influenced by cycling physical associations of DOUBLE-TIME, PERIOD, and TIMELESS in the Drosophila Clock. Neuron. 2001;30:699–706. doi: 10.1016/s0896-6273(01)00320-8. [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting of the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- Lin JM, Schroeder A, Allada R. In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J Neurosci. 2005;25:11175–11183. doi: 10.1523/JNEUROSCI.2159-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Meggio F, Marchiori F, Borin G, Pinna LA. Site specificity of casein kinase-2 from rat liver cytosol. A study with model peptide substrates. Euro J Biochem. 1986;160:239–244. doi: 10.1111/j.1432-1033.1986.tb09962.x. [DOI] [PubMed] [Google Scholar]
- Marin O, Meggio F, Draetta G, Pinna LA. The consensus sequences for cdc2 kinase and for casein kinase-2 are mutually incompatible. A study with peptides derived from the beta-subunit of casein kinase-2. FEBS Letters. 1992;301:111–114. doi: 10.1016/0014-5793(92)80221-2. [DOI] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- Meyer P, Saez L, Young MW. PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science. 2006;311:226–229. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- Muskus MJ, Preuss F, Fan YJ, Bjes ES, Price JL. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol. 2007;27:8049–8064. doi: 10.1128/MCB.00680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Wagner-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Rosbash M. The Doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell. 2004;13:213–223. doi: 10.1016/s1097-2765(03)00503-3. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Stoleru D, Rosbash M. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol Cell Biol. 2007;27:5002–5013. doi: 10.1128/MCB.02338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousley A, Zafarullah K, Chen Y, Emerson M, Hickman L, Sehgal A. Conserved regions of the timeless (tim) clock gene in Drosophila analyzed through phylogenetic and functional studies. Genetics. 1998;148:815–825. doi: 10.1093/genetics/148.2.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Tagher PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A, Young MW, Saez L. A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron. 2000;26:505–514. doi: 10.1016/s0896-6273(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Saez L, Young MW. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Rosbash M, Truman JW. Sequential nuclear accumulation of the clock proteins Period and Timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Lin JL, Meissner RA, Allada R. Dominant-negative CK2alpha induces potent effects on circadian rhythmicity. PLoS Genetics. 2008;4:99–109. doi: 10.1371/journal.pgen.0040012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Nawathean P, de la Paz Fernández M, Menet JS, Ceriani MF, Rosbash M. The Drosophila circadian network is a seasonal timer. Cell. 2007;129:207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Suri V, Hall JC, Rosbash M. Two novel doubletime mutants alter circadian properties and eliminate the delay between RNA and protein in Drosophila. J Neurosci. 2000;20:7547–7555. doi: 10.1523/JNEUROSCI.20-20-07547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, Breda C, Daga A, Selmin A, Monger K, Benna C, Rosato E, Kyriacou CP, Costa R. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science. 2007;316:1895–1898. doi: 10.1126/science.1138412. [DOI] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]