Summary
We develop and implement an ensemble-refinement method to study dynamic biomolecular assemblies with intrinsically disordered segments. Data from small angle X-ray scattering (SAXS) experiments and from coarse-grained molecular simulations are combined by using a maximum-entropy approach. The method is applied to CHMP3 of ESCRT-III, a protein with multiple helical domains separated by flexible linkers. Based on recent SAXS data by Lata et al. (J. Mol. Biol. 378, 818, 2008), we construct ensembles of CHMP3 at low and high salt concentration to characterize its closed autoinhibited state and open active state. At low salt, helix α5 is bound to the tip of helices α1 and α2, in excellent agreement with a recent crystal structure. Helix α6 remains free in solution and does not appear to be part of the autoinhibitory complex. The simulation-based ensemble refinement is general and effectively increases the resolution of SAXS beyond shape information to atomically detailed structures.
Introduction
Many important biological functions are carried out by large, multi-protein assemblies. Examples range from DNA transcriptional regulation to signal transduction and the nuclear pore complex. Multi-protein assemblies often form only transiently, and are held together by relatively weak pairwise interactions with dissociation constants Kd > 1 μM. The proteins forming the assemblies frequently contain intrinsically disordered regions, some of which become ordered only in the complex (Sugase et al., 2007,Turjanski et al., 2008). A prominent example is the endosomal sorting complex required for transport (ESCRT), which is built of about a dozen proteins that contain multiple domains connected by long flexible linkers (Hurley, 2010). The ESCRT complexes sort ubiquitin-labeled cargo proteins into the intralumenal vesicles (ILVs) of multivesicular bodies (MVBs) (Williams and Urbe, 2007,Hurley and Hanson, 2010). Also, certain retroviruses such as HIV-1 recruit the ESCRT complexes for budding from host cell membranes since the latter process is topologically equivalent to the formation of ILVs (Zamborlini et al., 2006,Lee et al., 2007). Recent in vitro experiments showed that the ESCRT-I and -II complexes together are capable of inducing membrane budding (Wollert and Hurley, 2010) whereas the ESCRT-III system drives the process of scission of the vesicle buds (Wollert et al., 2009). However, the molecular mechanisms used by the ESCRTs to promote membrane invagination and budding remain unknown (Hurley, 2010,Bassereau, 2010).
Multi-protein assemblies containing disordered, flexible polypeptide segments pose serious experimental challenges. X-ray crystallography is best suited for studies of folded, relatively rigid domains, and tightly bound complexes; and solution nuclear magnetic resonance (NMR) faces challenges because of the large molecular weights. These high-resolution techniques are thus increasingly complemented by lower-resolution methods, in particular cryo-electron microscopy and mass spectrometry. In addition, small angle X-ray scattering (SAXS) offers a promising alternative for the structural characterization of proteins, nucleic acids, and biomolecular complexes in solution (Jacques and Trewhella, 2010; Rambo and Tainer, 2010). Despite the orientational averaging and the typically limited number of features in SAXS intensity curves in the momentum transfer range up to q ≈ 0.3 Å−1, one can extract useful information about the shapes and dimensions of the molecules and their complexes. Indeed, SAXS has been successfully applied to biomolecular systems that range from individual proteins (Heidorn and Trewhella, 1988,Svergun, 1999,Chacon et al., 1998,Svergun et al., 2001,Yang et al., 2009) to protein complexes and biomolecular assemblies (Bernadó et al., 2007,Pelikan et al., 2009,VanOudenhove et al., 2009,Datta et al.,2009,Yang et al.,2009, Yang et al., 2010).
The resolution of the SAXS technique is inherently limited since a complex three-dimensional molecular structure is reduced to a one-dimensional intensity profile. Unlike X-ray crystallography, the SAXS intensity profile is orientationally averaged. Despite the resulting loss in information, SAXS data have proven highly valuable in protein structure determination, especially when supplemented by other techniques. SAXS has been combined with NMR measurements (Grishaev et al., 2005,Grishaev et al., 2008,Mittag et al., 2010), and could in turn be combined with structure prediction (Shen et al., 2008).
SAXS would seem to be particularly useful for the characterization of protein assemblies. Indeed, if there is only a single dominant structure of a protein complex, and high-resolution structures of the complex components are known, then it should be possible to reconstruct the assembly from the SAXS data. In practice, however, this task is often challenging because many of the large protein assemblies, including those of the ESCRT system (Prag et al., 2007,Ren et al., 2009), are dynamic complexes that attain multiple distinct conformations. The problem is further complicated by the presence of disordered linkers that connect the structured domains. In such cases, it is impossible to characterize the typical protein conformations by a single molecular envelope and, thus, the standard regularization methods for molecule shape determination (Svergun, 1999,Svergun et al., 2001) cannot be applied directly. To overcome this problem and quantitatively characterize flexible protein systems in solution using SAXS data, several methods have been developed, including the ensemble optimization method (Bernadó et al., 2007), the minimal ensemble search (Pelikan et al., 2009) and the SAXS module in the integrative modeling platform (Förster et al., 2008). In addition, topology-based (Gō-type) models have been used to create an initial structural ensemble that is then refined against SAXS data (Yang et al., 2010).
Here we develop an alternative, physical approach for the analysis of flexible protein complexes, and apply it to a key protein of the ESCRT system. Our Ensemble-Refinement of SAXS (EROS) method proceeds in two steps: First, we use a coarse-grained model for protein binding to simulate the molecular assembly and, in this way, generate an initial ensemble of protein conformations. Second, we gently refine the simulated ensemble to improve the agreement with SAXS data. The transferable energy function used in our simulations has been shown to reproduce complex structures and binding affinities to within a few kT (Prag et al., 2007,Kim and Hummer, 2008,Ren et al., 2009). The use of an energy function optimized for protein binding constitutes arguably the main difference from approaches that create initial ensembles based primarily on steric exclusion (Bernadó et al., 2007), high-temperature atomistic molecular dynamics simulations (Pelikan et al., 2009), and topology-based Gō-type models (Yang et al., 2010). In contrast, the approach of Förster et al. (2008) employs statistical potentials similar to ours, but performs a gradient-based refinement focused on a single (or few) distinct structures instead of a distributed ensemble. Our coarse-grained simulation model has previously predicted the structures of both specific and transient protein complexes based on NMR paramagnetic relaxation enhancement experiments (Kim et al., 2008). We therefore expect the simulation model to explore the relevant conformation space of the protein complexes. To refine the resulting simulation ensemble in a controlled way, and to prevent data over-fitting, we use a maximum entropy method.
We apply this SAXS refinement formalism to the CHMP3 protein, which is a key component of the ESCRT-III complex (Shim et al., 2007,Saksena et al., 2009). The ESCRT-III proteins are predicted to exhibit transitions between a closed, autoinhibited state in the cytosol and an open, activated conformation on the membrane (Shim et al., 2007). Electrostatic interactions have been implicated in the conformational transitions of CHMP3, based on SAXS measurements that show CHMP3 attaining an extended conformations in a buffer at high salt concentration, and compact conformations at low salt concentration (Lata et al., 2008).
Here we use the SAXS experimental data of Lata et al. (2008), first, to validate our simulation and refinement methodology and then to gain detailed insight into the structures of CHMP3 in solution. For further validation of the simulation methodology, we compare the predicted structures to high-resolution X-ray structures of the CHMP3 core (Bajorek et al., 2009,Muziol et al., 2006). Specifically, we show that the simulations recover the binding of helix α5 to the tip of helices α1 and α2 in agreement with the recently published monomer crystal structure (Bajorek et al., 2009). We also examine the location of helix α6, which has not been crystallized, and show that it does not bind preferentially to the core of CHMP3. Our results shed light on the conformations of CHMP3 in the activated and autoinhibited states.
Results and Discussion
We simulate CHMP3 equilibrium conformations with an energy function optimized previously for protein binding and validated for binary protein-protein complexes (Kim and Hummer, 2008), ESCRT protein assemblies (Kim and Hummer, 2008,Prag et al., 2007,Ren et al., 2009), and transient protein binding (Kim et al., 2008). In the framework of this coarse-grained model, amino acid residues are represented as spherical beads centered at the Cα atoms. The core structure of CHMP3 formed by helices α1 to α4 is simulated as a single rigid domain. Helices α5 and α6 are treated as two separate rigid units connected by flexible linkers. Therefore, helices α5 and α6 can attain different positions relative to the CHMP3 core structure and to each other. For details on our simulations see Methods.
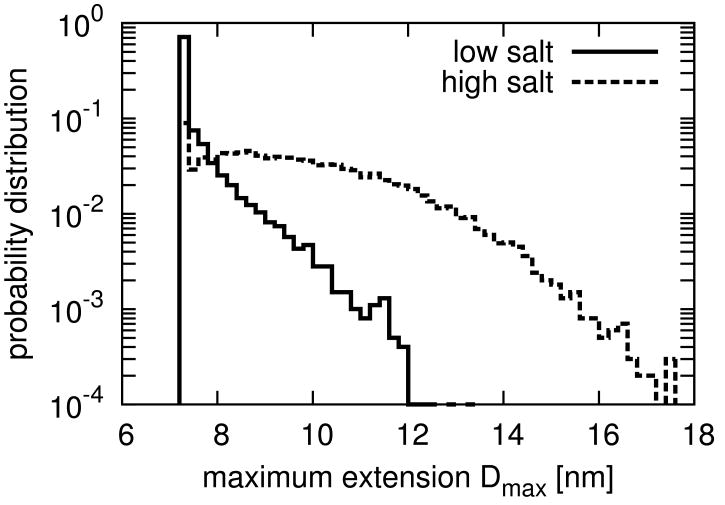
To capture the effect of salt in the buffer, we varied the Debye length λD. In our simulations we used λD = 30 Å and λD = 3 Å, which correspond to salt concentrations of about 10 mM and 1 M, respectively. To probe the spatial extent of CHMP3 at low and high salt, we measured its maximum extension Dmax (see Figure 1). At low salt concentration, CHMP3 attains closed conformations with Dmax ranging from 70 Å to 120 Å and helices α5 and α6 in close vicinity of helices α1 and α2. At high salt concentration, in contrast, CHMP3 predominantly attains open conformations with Dmax up to 180 Å and helices α5 and α6 away from the CHMP3 core. The dependence of the maximum extension Dmax on salt concentration is in qualitative agreement with the earlier interpretation of the SAXS experiments (Lata et al., 2008).
Figure 1. Distribution of the Maximum Extension of CHMP3.
Dmax (in units of nm) at low (solid line) and high salt concentration (dashed line). Potentials of mean force for other structural quantities are shown in Figure S1.
To compare our simulation results quantitatively to experiment, we calculated SAXS intensity profiles for the simulated ensemble of CHMP3 conformations. As shown in Figure 2A, the calculated SAXS profiles for our simulated CHMP3 ensemble agree well with the scattering curves obtained from experiments (Lata et al., 2008). As a matter of fact, the agreement with experimental data is sufficiently good that we could infer from the comparison that the SAXS curves in Figure 3a of Lata et al. (2008) were mislabeled (personal communication, Lata and Weissenhorn). There is, however, some noticeable disagreement between our calculations and the experimental data in the interval 0.1 < q < 0.15 Å−1 in the low salt concentration case, and in the interval 0.15 < q < 0.2 Å−1 in the case of high salt concentration (Figure 2A).
Figure 2. SAXS Intensity Profiles of CHMP3.
SAXS intensities at low salt (LS) and high (HS) salt concentration from simulations (lines) and experiment (symbols, taken from Figure 3a of Lata et al. (2008)).
(A) SAXS intensities before cluster reweighting.
(B) SAXS intensities after cluster reweighting.
See also Figure S2 for details on SAXS profile calculations.
To improve the agreement with experiment and gain insight into the relevant CHMP3 conformations, we refine the simulated ensemble as described in Methods. In brief, the simulated structures are clustered and the cluster weights are varied to decrease the discrepancy between simulations and experiments. In order to minimally deform the set of initial cluster weights, and to prevent data over-fitting, the maximum-entropy method is used in the cluster reweighting algorithm (see Methods). As a result, the cluster weights are changed on average by less than 2 kT in free energy (or ≈1.2 kcal/mol, as determined from the ratio of cluster weights before and after reweighting, ; see Methods), which is within the expected error of the energy function used in our simulations (Kim and Hummer, 2008).
The SAXS profiles for the refined ensembles agree with the experimental data to within the estimated experimental error (Figure 2B). At low salt concentration, the refined ensemble is dominated by only a few well-defined structures (Figure 3 top). The six clusters with the largest weights suffice to account for the SAXS data (see Methods and Figure 4C). These clusters constitute about 40% of the refined ensemble (Figure 4C) and do not differ significantly in terms of their maximum extension Dmax and their radius of gyration Rg (see Figure S3 A and B). At high salt concentration, in contrast, the refined ensemble contains many distinct conformations that jointly contribute to the SAXS intensity profile (Figure 3 bottom). In this case, about 60 of the most populated clusters are required to account for the SAXS data (see Methods and Figure 4D). These 60 clusters constitute about 50% of the refined ensemble (Figure 4D), and jointly cover a broad range of Dmax and Rg values (Figure S3 C and D).
Figure 3. Structures of CHMP3 in Solution.
Most populated clusters at low (top) and high (bottom) salt concentration, together with their relative weights. Blue: helices α1-α4. Red: helix α5. Magenta: helix α6. Green: helix α5 in crystal structure (PDB code 3FRT).
Figure 4. Maximum-Entropy Refinement of Cluster Weights.
(A) Discrepancy χ2 between experimental and simulated SAXS data versus relative entropy S. The red and blue curves correspond to simulations at low and high salt concentration, respectively. The black dots indicate the entropy threshold S0 below which the simulated data are considered over-fitted.
(B) Optimized weights wk at the entropy threshold S0 versus initial weights wk(0). The black dashed line indicates wk=wk(0). The dashed gray lines indicate a cluster free energy change of kT log(wk/wk(0) = ±2 kT.
(C) Discrepancy χ2 as a function of the cumulative weight of the clusters at low salt. Clusters were rank ordered by their optimized weights. The discrepancy χ2 was calculated for a truncated ensemble consisting of the n top-ranked clusters with cumulative weight . The relative weights were maintained in the calculation of χ2.
(D) Discrepancy χ2 at high salt.
See also Figure S3 for variations in the structures of the clusters.
With the refined ensembles fully matching the experiment, we can now examine the conformations of CHMP3 at low and high salt concentrations (Figure 3 top and bottom, respectively). At high salt concentration, the ensemble consists of a large number of clusters, with a maximum relative population of 4 %. In the most highly populated clusters, helices α5 and α6 are both dissociated from the core. The ensemble refinement of the SAXS data thus indicates that at high salt concentrations CHMP3 attains open, extended conformations.
In contrast, CHMP3 populates closed conformations at low salt concentration. Helix α5 is then predominantly bound to the core. In the two most populated clusters that account for 9 % and 8 % of the refined ensemble, respectively, helix α5 is bound close to the tip of helices α1 and α2 (Figure 3 top). These structures are in excellent agreement with the recently published crystal structure (Bajorek et al., 2009) (PDB code 3FRT). Even though we do not observe helix α5 at the opposite end of helix α2, as suggested by an earlier crystal structure of CHMP3 dimers (Muziol et al., 2006) (PDB code 2GD5) mimicking CHMP3 assemblies, the local binding mode of α5 with respect to helices α1 and α2 is again nearly identical to our simulation structures.
Helix α6 does not interact strongly with the rest of the protein. We find that α6 tends to be located close to the core of CHMP3, but without preferential binding (Figure 3). The weak interaction of helix α6 with the CHMP3 core might be a reason why constructs including helix α6 have so far not been crystallized.
Our findings of distinct binding by α5 to the core and of unbound α6 are consistent with biochemical evidence. Zamborlini et al. (2006) showed that a shortened CHMP3 construct, with helices α5 and α6 removed, bound to the C-terminal domain and inhibited HIV-1 release. In contrast, if α5 was left intact and only α6 was removed, inhibition of HIV-1 release and CHMP3 binding were both reduced. These results are consistent with our simulation ensemble, with α5 being bound in the autoinhibited state, and α6 unbound. Helix α6 therefore likely serves other functions that may include binding to other parts of the ESCRT system (Tsang et al., 2006) or to the membrane. The corresponding helix in Vps2 is known to bind to Vps4 (Obita et al., 2007,Stuchell-Brereton et al., 2007).
Our simulations show that at low salt helix α5 is bound to the core in a significant fraction of structures, both before and after refinement. Before (after) refinement, we find that about 28 % (25 %) of the ensemble is within distance root-mean-square DRMS < 5 Å (see Methods) from the crystal structure, and about 57 % (55 %) is within DRMS < 7 Å. Nevertheless, even at low salt concentration a significant fraction of the ensemble assumes conformations far from the crystal structure. About 20 % of all structures in the refined ensemble have DRMS > 10 Å from the crystal structure.
To quantify the amount of helix α5 that is bound to the core, we have performed an additional refinement of the SAXS data using only bound structures. Specifically, we used a restricted set of structures from our simulation ensemble at low salt concentration, requiring that they are close to the crystal structure (Bajorek et al., 2009) with DRMS < 3 Å. Remarkably, in refining this restricted ensemble, we achieved the same level of agreement with the SAXS curve up to q=0.35 Å−1 as with the full simulation ensemble, at comparable values of the relative entropy S, i.e., without overfitting (see Methods). In effect, helix α6 unbound in solution and the connecting linkers both contribute significantly to the X-ray scattering, and substantially blur the SAXS profile of the complex of the core and α5. We therefore cannot tell with certainty from the low-resolution SAXS data alone, with q < 0.35 Å−1, whether the crystallographic complex is fully populated at low-salt conditions, or whether it has only a fractional population.
Conclusions
The ESCRT-III system is involved in multivesicular body biogenesis (Williams and Urbe, 2007,Hurley, 2010). It can also be engaged by HIV-1 to promote viral budding (Zamborlini et al., 2006). Over the last several years, substantial progress has been made in understanding the assembly and disassembly of the ESCRT-III complex (Hanson et al., 2008,Saksena et al., 2009) and the interactions of its components with other proteins (Zamborlini et al., 2006,Lata et al., 2008). In particular, in vivo experiments have provided evidence of a regulatory role of the C-terminal domains of ESCRT-III proteins (Shim et al., 2007). It has been suggested that ESCRT-III proteins cycle between a default closed state and an activated open state under the control of their C-terminus (Shim et al., 2007,Williams and Urbe, 2007). Also SAXS data indicate that CHMP3 in vitro attains open, extended conformations in a buffer at high salt concentration, and closed, more compact conformations at low salt concentration (Lata et al.,2008).
Our simulation results agree very well with these experimental predictions and accurately account for the measured SAXS data (Figure 2). The maximum extension Dmax of CHMP3 increases dramatically with the increase of salt concentration (Figure 1). Moreover, we correctly recover the interaction of helix α5 with the core of CHMP3 at low salt concentrations (Figure 3). In the most populated clusters of CHMP3 conformations, helix α5 binds close to the tip of helices α1 and α2 in agreement with the recently published crystal structure (Bajorek et al., 2009). We also find that helix α6, which has not been crystallized, does not bind preferentially to the core of CHMP3. This finding is consistent with evidence from an assay probing the inhibition of HIV-1 release by CHMP3 fragments (Zamborlini et al., 2006). Helix α6 is thus a candidate for interactions with other components of the ESCRT system.
The differences in binding of helix α5 to the core as well as the weak interactions of helix α6 with the core indicate that relatively small changes in the environment can suffice to drive CHMP3 from closed, autoinhibited states to open, activated conformations. The sensitivity to the ion concentration (Lata et al., 2008), reproduced here in simulations, suggests an important role of electrostatics and possibly the membrane environment.
Refinement of low-resolution structural data poses a major challenge in structural studies of large and dynamic biomolecular assemblies (Schr der et al., 2010). These challenges are highlighted by the original refinement of the CHMP3 SAXS data (Lata et al., 2008). As a first step, a molecular envelope was determined with the program DAMMIN (Svergun, 1999). The CHMP3 core was then manually fitted into the envelope. The parts of the envelope not accounted for by the core gave some broad indications concerning possible locations of helices α5 and α6, but without the detailed structural information provided by our ensemble refinement. Because of the disorder, in particular under high salt conditions, the ensemble of structures is not easily captured by a single envelope. In contrast, the EROS approach avoids the construction of envelopes as intermediates by directly calculating the low-resolution SAXS data from an ensemble of structures obtained by molecular simulations. This ensemble refinement methodology is general and immediately applicable to other multi-protein complexes with flexible linkers. The combination of experiment and theory effectively increases the resolution down to the level of residues, allowing us to go beyond the conventional modeling of SAXS data that is focused first on shape information, followed by docking of structures into the shapes.
In practice, the two approaches of simulation and envelope-based refinement can be complementary. In EROS, we create an initial ensemble of structures from simulations with an energy function that can predict both structures and binding affinities of protein complexes (Prag et al., 2007,Kim and Hummer, 2008,Kim et al., 2008,Ren et al., 2009). In the refinement, we implicitly assume that this initial ensemble is already “close” to the actual ensemble. With the maximum-entropy method, we then minimally modify the relative weights of the structures to match the measured SAXS curve. This procedure allows us to obtain relative weights for possibly a large number of clusters from a combination of low-resolution experimental data and simulation output, by striking a balance between the two. In contrast, refinement methods based on molecular envelopes do not normally use molecular structures as input. At least in the cases where only a few structures dominate the ensemble, the resulting unbiased envelopes can therefore be used to validate the simulations.
An approach similar to ours was recently used to assemble Hck tyrosine kinase with the help of SAXS data (Yang et al., 2010), demonstrating the power of combining a low-resolution experiment with coarse-grained simulations. The main difference between the two approaches is that Yang et al. (2010) simulated their system with a multi-state topology-based (Gō-type) energy function (Best et al., 2005) that used information about the complex structures and its motions as input. In contrast, here we used a fully transferable energy function optimized for protein binding (Kim and Hummer, 2008), but treated the folded protein domains as rigid. Additional differences are in the refinement procedure. Yang et al. (2010) grouped the ensemble into a small number of clusters (9 for Hck kinase) based on similarity of structures and scattering functions, and then refined the cluster weights. Here, we refined the entire simulation ensemble with a maximum entropy method.
As we demonstrated for CHMP3, and for the dimeric complex PF0863 of P. furiosus, our simulation refinement produces atomically detailed structures that recover the high-resolution crystal structures of the complexes. In the case of CHMP3, a key interaction between helix α5 and the core is accurately captured. The ensemble of refined structures is consistent with the SAXS experiments, the topology and stereochemistry of the biomolecular constructs, and the energetics of biomolecular interactions. Here we validated the ensemble against high-resolution crystal structures. In general, additional low-resolution information can be used, in particular distances from spin labeling, fluorescence, or cross-linking experiments. Simulation-based ensemble refinement should therefore prove useful not only for ESCRT proteins, but also for other complexes that include intrinsically disordered linkers.
Methods
SAXS Intensity Calculation for Coarse-Grained Protein Models
In the coarse-grained model we use to simulate CHMP3, amino acid residues are represented as spherical beads centered at corresponding Cα atoms. The “crysol” software package used to evaluate SAXS intensity curves from protein models (Svergun et al., 1995) requires all-atom structures as input. Following the crysol methodology, we developed a simple method that allows us to calculate SAXS intensity profiles for protein models in the Cα representation. As in crysol, a multipole expansion is used for fast calculations of the scattering profile. For the systems studied here, and the q range, an expansion up to order 20 was found to be sufficient. However, there are two main differences between our algorithm and the one implemented in crysol. First, the form factors fj(q) of the 20 types j of amino acids are assumed to be constant in our calculation, fj = Ne(j) − ρVj. Here, Ne(j) is the number of electrons of residue j and ρ is the electron density of solvent. We assume ρ = 0.334 e Å−3, as in (Svergun et al. (1995). The solvent volume Vj displaced by residue j is calculated as a sum of displaced solvent volumes of atoms forming residue j. Values from Table 1 in (Svergun et al. (1995) are used. Second, we use a different hydration shell model that is built on a cubic lattice with a spacing of 3 Å. Grid points that are further than a distance Å from the Cα atoms of all residues j, but closer than Å from at least one residue Cα atom are assigned an electron density w. The radii rj of amino acids are consistent with their excluded volumes used to calculate the amino acid form factors, . As with amino acids, we use uniform form factors for the water grid points. Although the hydration shell construction used in our calculation is slower than the algorithm implemented in crysol (Svergun et al., 1995), it avoids the problem of nonuniform distributions of water at the irregular surfaces of multi-domain proteins.
One relevant parameter that enters this calculation is the electron density w of the hydration shell around the protein (Svergun et al., 1998). The latter quantity depends on the buffer condition as well as on physical properties of the protein surface. Consistent with previous SAXS refinement protocols (Svergun et al., 1995,Yang et al., 2009), we treat the hydration shell electron density w as a free parameter in our calculation that is varied between w=0 (meaning no hydration shell) and w=0.045 e Å−3.
In Figure S2A, we compare the results of our method to crysol output for CHMP3 of ESCRT-III (PBD code 3FRT), ESCRT-II (PBD code 1U5T) and ESCRT-I (PBD code 2P22) crystal structures. The crysol calculations are performed with the default program parameters, with a hydration-shell electron density of w=0.03 e Å−3 in both calculations. Since the ESCRT-I core structure is thin and elongated, the hydration shell built by our algorithm and crysol differ locally, which leads to small differences in calculated intensities in the high q regime.
In Figure S2B, SAXS experimental data for lysozyme (Svergun et al., 1995) and for P. furiosus dimeric product PF1026 (BioIsis ID 37; Hura et al., 2009; http://www.bioisis.net) are compared to results both from atomistic crysol calculations and from our coarse-grained calculations. In the scattering intensity calculation we used the crystal structures with PDB codes 6LYZ (lysozyme) and 2DVM (PF1026). In both cases, we obtain agreement with the experimental data for reduced hydration shell electron densities of w≈0.01 e Å−3. Moreover, the SAXS profiles from atomistic crysol calculations and from the coarse-grained calculations agree if the default crysol hydration shell density of w=0.03 e Å−3 is used. This ambiguity suggests that the hydration shell electron density, which is a phenomenological parameter in models that do not include the solvent explicitly, should be treated as a variable in the SAXS intensity calculation. For this reason we compared the CHMP3 SAXS data with our simulation results for different values of hydration shell density ranging from w=0 to w=0.045 e Å−3. We obtained the best agreement for w=0.03 e Å−3, which is the default value in crysol (Svergun et al., 1995,Svergun et al., 1998) and in our calculations (Figure 2).
Simulation Methodology
To simulate CHMP3 at different buffer conditions, we used a coarse-grained model for protein binding (Kim and Hummer, 2008). In this model, folded protein domains are represented as rigid bodies. Here, the CHMP3 core structure formed by helices α1 to α4 is treated as one rigid body and helices α5 and α6 are modeled as two additional rigid bodies. Possible partial disorder in these helices should not substantially affect the SAXS curves for q < 0.3 Å−1 and the binding equilibria. The interactions between the domains are treated at the residue level with amino-acid-dependent pair potentials and Debye-Hückel-type electrostatic interactions. Flexible linker peptides connecting the three rigid domains are represented as amino acid beads on a polymer with appropriate stretching, bending, and torsion-angle potentials. We perform replica exchange Monte Carlo (REMC) simulations with 2 108 Monte Carlo (MC) sweeps and, after the initial equilibration of 108 MC sweeps, consider 104 configurations chosen every 104 MC sweeps for analysis. With 20 replica temperatures between 300 and 485 K, the MC production run took ∼2 weeks on a parallel PC architecture. Different buffer conditions are captured by varying the Debye length λD that enters the Debye-H ckel potential. We used λD = 30 Å and λD = 3 Å, which corresponds to salt concentrations of about 10 mM and 1 M, respectively, to simulate CHMP3 at low and high salt conditions.
Cluster Reweighting
We use the standard QT-clustering method (Heyer et al., 1999) with DRMS as a metric to cluster the simulated structures. The distance between two structures g and h is
| (1) |
where is the Cartesian distance of the α-carbon atoms of residues n and m in two different rigid domains of structure g, and N2 is the number of residue pairs over which the sum is performed.
In the QT-clustering, for every structure the number of neighboring structures in a sphere of radius Rcutoff is calculated. The structure with the maximal number of neighbors is removed from the structure pool together with all its neighbors. Then the procedure is iterated until the structure pool is emptied. We choose Rcutoff = 5 Å and 7 Å to cluster structures form the low and high salt concentration simulations, respectively. With this choice we obtain about 550 and 1400 clusters of structures at low and high salt concentrations, respectively. In both cases, about 40 % of all clusters contain only a single structure. At low salt concentration, they mostly correspond to structures with helix α6 dissociated from the CHMP3 core. At high salt concentration, the single-structure clusters mainly represent conformations with unbound helices α5 and α6.
The SAXS intensity Ik(q) assigned to a cluster k is the arithmetic mean of SAXS intensities resulting from all structures in this cluster. The average SAXS intensity from the whole ensemble of simulated structures is
| (2) |
where Nc is the total number of clusters, and wk is the weight of cluster k, normalized to Σkwk = 1. The discrepancy between the computed, average SAXS intensity Isim(qi) and experimental data Iexp(qi) can be quantified by
| (3) |
with a scale factor c that results from. Here Nq is the number of data points on the SAXS curve. We assume shot noise as the dominant source of the experimental error, σ2(qi) ≈ AIexp(qi), thus ignoring additional uncertainties from, e.g., the buffer subtraction. From the magnitude of the noise in the experimental curves, we estimate amplitudes of A=0.025 and A=0.25 at low and high salt concentration, respectively.
To improve the agreement with experimental data, we vary the cluster weights wk. We prevent over-fitting by using the maximum entropy method (Jaynes, 1957). First, we define the relative entropy
| (4) |
where the are the initial cluster weights. Note that S = 0 if . Second, we introduce a “free energy” function G = χ2 −θS with a temperature-like control parameter θ. Third, we minimize the function G numerically with respect to the normalized weights wk by using simulated annealing. In this way we obtain the optimal set of weights {wk}. For large θ, when the relative entropy term dominates over χ2, minimizing G leads to small perturbations of the initial weights and thus for the majority of clusters k. In contrast, minimization of G with θ = 0 produces the best possible agreement with experiment, but possibly large changes in the weights as a result of over-fitting.
Figure 4A shows how the discrepancy χ2({wk}) varies with the relative entropy S({wk}) for optimal weights wk that minimize function G. We chose an entropy threshold S0 below which the data might be over-fitted. The entropy threshold S0 corresponds to the average change in cluster weights by 2 kT, which is the estimated accuracy of the energy function we use in our CHMP3 simulations (Kim and Hummer, 2008). Figure 4B shows the optimal weights wk that correspond to this entropy threshold versus the initial weights . Reflective of the fact that our initial simulation ensembles are already very close to the SAXS experiments in the case of CHMP3 (Figure 2A), we find that the reweighting procedure only minimally deforms the ensemble. As shown in Figure S1 A-C, reweighting results in only small changes of a few kT in the potentials of mean force for the maximum extension Dmax, the radius of gyration Rg, and the root-mean-square deviation (RMSD) from the crystal structure. (The potential of mean force for quantity X is defined as -kT ln p(X), where p(X) is the probability that the quantity of interest attains the value X.) Similarly, the pair distance distribution functions change only minimally, as shown in Figure S1 D.
We also estimated the minimum number of clusters that account for the SAXS data. First, we rank ordered the clusters by their optimized weights. Second, we calculated χ2 for truncated ensembles containing only the n top-ranked clusters, with their relative weights maintained. Figures 4C and D show the discrepancy χ2 versus the cumulative weight of these truncated ensembles at low and high salt concentration, respectively. At low salt concentration, the six clusters with the largest weights fully account for the SAXS data (χ2 ≈0.5). These clusters together constitute about 40% of the reweighted ensemble. In contrast, at high salt concentration the ∼60 most populated clusters are required to account for the SAXS data (χ2 ≈2), reflecting the disorder in the relatively open high salt structures.
The maximum-entropy refinement is locally stable and robust. We checked that small variations of the control parameter θ lead to small changes of the optimal weights {wk} for all clusters k. We also minimized the function G using simulated annealing with different seeds for the random number generator. The calculation was repeated 7 times at the entropy threshold. The average relative error of cluster weights was 21% and 31% at low and high salt concentration, respectively. The largest standard deviation for any of the clusters was 0.0004, which means that only very small weights bear noticeable uncertainty. To test the robustness of the ensemble refinement method, we (i) divide the simulated ensemble into four blocks, (ii) cluster the structures in each block separately and (iii) repeat the reweighting calculation for each block. We find that the resulting cumulative distributions of the DRMS to the crystal structure (PDB code 3FRT) do not differ significantly between the blocks. At low salt concentration, the probability of finding a structure within DRMS < 5 Å from the crystal structure varies between 0.24 and 0.27 in different blocks.
Validation
As an additional validation of the EROS methodology, we performed simulations of the protein complex PF0863 of P. furiosus, for which both SAXS data and a crystallographic structure have been reported (Hura et al., 2010; BioIsis ID 36; http://www.bioisis.net). The two proteins in this complex contain flexible chains, and the SAXS profile calculated from the crystal structure results in a marginal fit of the measured profile (χ2 =5.8). We performed simulations of this system in which each of the two proteins could translate and rotate independently inside a periodic simulation box. Their structured parts were kept rigid and the flexible chains were treated as polymers. In these simulations, we found that ∼10% of the complexes were close to the crystal structure (within ∼6 Å DRMS), the rest populating different structures (Figure S2 C and D). The simulation ensemble was then clustered and the weights refined, again with an entropy threshold of S0=2 and the default hydration shell density of w=0.03 e Å−3. This refinement not only reduced the χ2 from 10.8 to 3.7 (compared to 5.8 of the model in the BioIsis database; Hura et al., 2010), but also greatly increased the weight of structures consistent with the high-resolution X-ray crystal. After refinement, the fraction of structures within ∼6 Å DRMS from the crystal structures increased from ∼10 to ∼80%, with ∼40% of the structures within ∼3 Å DRMS (Figure S2 D). We conclude from this test that the EROS methodology can also be useful in cases where a single structure dominates the ensemble, but flexible tails “blur” the SAXS profile and interfere with a conventional analysis.
Supplementary Material
Acknowledgments
We thank Dr. James Hurley and Dr. Alexander Grishaev for many helpful discussions. This work used the Biowulf computing cluster at the NIH. The research was supported by the Intramural Research Program of the NIH, NIDDK, and by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Programme (to B.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WMB, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nature Struct Mol Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassereau P. Division of labour in ESCRT complexes. Nature Cell Biol. 2010;12:422–423. doi: 10.1038/ncb0510-422. [DOI] [PubMed] [Google Scholar]
- Bernadó P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- Best RB, Chen YG, Hummer G. Slow protein conformational dynamics from multiple experimental structures: The helix/sheet transition of arc repressor. Structure. 2005;13:1755–1763. doi: 10.1016/j.str.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Chacon P, Moran F, Diaz JF, Pantos E, Andreu JM. Low-resolution structures of proteins in solution retrieved from X-ray scattering with a genetic algorithm. Biophys J. 1998;74:2760–2775. doi: 10.1016/S0006-3495(98)77984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta AB, Hura GL, Wolberger C. The structure and conformation of Lys63-linked tetraubiquitin. J Mol Biol. 2009;392:1117–1124. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster F, Webb B, Krukenberg KA, Tsuruta H, Agard DA, Sali A. Integration of small-angle X-ray scattering data into structural modeling of proteins and their assemblies. J Mol Biol. 2008;382:1089–1106. doi: 10.1016/j.jmb.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishaev A, Tugarinov V, Kay LE, Trewhella J, Bax A. Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints. J Biomol NMR. 2008;40:95–106. doi: 10.1007/s10858-007-9211-5. [DOI] [PubMed] [Google Scholar]
- Grishaev A, Wu J, Trewhella J, Bax A. Refinement of multidomain protein structures by combination of solution small-angle X-ray scattering and NMR data. J Am Chem Soc. 2005;127:16621–16628. doi: 10.1021/ja054342m. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidorn DB, Trewhella J. Comparison of the crystal and solution structures of calmodulin and troponin-C. Biochemistry. 1988;27:909–915. doi: 10.1021/bi00403a011. [DOI] [PubMed] [Google Scholar]
- Heyer LJ, Kruglyak S, Yooseph S. Exploring expression data: Identification and analysis of coexpressed genes. Genome Research. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, II, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nature Meth. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010 doi: 10.3109/10409238.2010.502516. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol. 2010 doi: 10.1038/nrm2937. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques DA, Trewhella J. Small-angle scattering for structural biology-expanding the frontier while avoiding the pitfalls. Protein Sci. 2010;19:642–657. doi: 10.1002/pro.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes ET. Information theory and statistical mechanics. Phys Rev. 1957;106:620–630. [Google Scholar]
- Kim YC, Hummer G. Coarse-grained models for simulations of multiprotein complexes: application to ubiquitin binding. J Mol Biol. 2008;375:1416–1433. doi: 10.1016/j.jmb.2007.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Tang C, Clore GM, Hummer G. Replica exchange simulations of transient encounter complexes in protein-protein association. Proc Natl Acad Sci USA. 2008;105:12855–12860. doi: 10.1073/pnas.0802460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Roessle M, Solomons J, Jamin M, Göttlinger HG, Svergun DI, Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. J Mol Biol. 2008;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nature Struct Mol Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T, Marsh J, Grishaev A, Orlicky S, Lin H, Sicheri F, Tyers M, Forman-Kay JD. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure. 2010;18:494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Göttlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase vps4. Nature. 2007;449:735–740. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prag G, Watson H, Kim YC, Beach BM, Ghirlando R, Hummer G, Bonifacino JS, Hurley JH. The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Dev Cell. 2007;12:973–986. doi: 10.1016/j.devcel.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr Opin Struct Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XF, Kloer DP, Kim YC, Ghirlando R, Saidi LF, Hummer G, Hurley JH. Hybrid structural model of the complete human ESCRT-0 complex. Structure. 2009;17:406–416. doi: 10.1016/j.str.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schr der GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu GH, Eletsky A, Wu YB, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by vps4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystal. 1995;28:768–773. [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Richard S, Koch MHJ, Sayers Z, Kuprin S, Zaccai G. Protein hydration in solution: Experimental observation by X-ray and neutron scattering. Proc Natl Acad Sci USA. 1998;95:2267–2272. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang HTH, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88:333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Turjanski AG, Gutkind JS, Best RB, Hummer G. Binding-induced folding of a natively unstructured transcription factor. PLoS Comp Biol. 2008;4:e1000060. doi: 10.1371/journal.pcbi.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanOudenhove J, Anderson E, Krueger S, Cole JL. Analysis of PKR structure by small-angle scattering. J Mol Biol. 2009;387:910–920. doi: 10.1016/j.jmb.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nature Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–870. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–178. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SC, Park S, Makowski L, Roux B. A rapid coarse residue-based computational method for X-ray solution scattering characterization of protein folds and multiple conformational states of large protein complexes. Biophys J. 2009;96:4449–4463. doi: 10.1016/j.bpj.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Blachowicz L, Makowski L, Roux B. Multidomain assembled states of Hck tyrosine kinase in solution. Proc Natl Acad Sci USA. 2010;107:15757–15762. doi: 10.1073/pnas.1004569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Göttlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci USA. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.