Abstract
Oomycetes comprise a diverse group of organisms that morphologically resemble fungi but belong to the stramenopile lineage within the supergroup of chromalveolates. Recent studies have shown that plant pathogenic oomycetes have expanded gene families that are possibly linked to their pathogenic lifestyle. We analyzed the protein domain organization of 67 eukaryotic species including four oomycete and five fungal plant pathogens. We detected 246 expanded domains in fungal and oomycete plant pathogens. The analysis of genes differentially expressed during infection revealed a significant enrichment of genes encoding expanded domains as well as signal peptides linking a substantial part of these genes to pathogenicity. Overrepresentation and clustering of domain abundance profiles revealed domains that might have important roles in host-pathogen interactions but, as yet, have not been linked to pathogenicity. The number of distinct domain combinations (bigrams) in oomycetes was significantly higher than in fungi. We identified 773 oomycete-specific bigrams, with the majority composed of domains common to eukaryotes. The analyses enabled us to link domain content to biological processes such as host-pathogen interaction, nutrient uptake, or suppression and elicitation of plant immune responses. Taken together, this study represents a comprehensive overview of the domain repertoire of fungal and oomycete plant pathogens and points to novel features like domain expansion and species-specific bigram types that could, at least partially, explain why oomycetes are such remarkable plant pathogens.
Oomycetes are a diverse group of organisms that live as saprophytes or as pathogens of plants, insects, fish, vertebrates, and microbes (Govers and Gijzen, 2006). The numerous plant pathogenic oomycete species cause devastating diseases on many different host plants and have a huge impact on agriculture. A prominent example is Phytophthora infestans, the causal agent of late blight of potato (Solanum tuberosum) and tomato (Solanum lycopersicum) and responsible for the Irish potato famine in the 19th century. Plant pathogenic oomycetes include a large number of different species that vary in their lifestyle, from obligate biotrophic and hemibiotrophic to necrotrophic. In addition, they show great differences in host selectivity, ranging from broad to very narrow (Erwin and Ribeiro, 1996; Agrios, 2005). Oomycetes have morphological features similar to filamentous fungi, and the two groups exploit common infection structures and mechanisms (Latijnhouwers et al., 2003). Together with diatoms, brown algae, and golden-brown algae, oomycetes are classified as stramenopiles, a lineage that is united with alveolates in the supergroup of chromalveolates (Baldauf et al., 2000; Yoon et al., 2002). The monophyly of this supergroup, however, is under debate (Baurain et al., 2010). The genomes of oomycetes sequenced so far are variable in size and content, ranging from 65 Mb in Phytophthora ramorum to 240 Mb in P. infestans (Haas et al., 2009), and only include plant pathogenic species. Analysis of these genomes revealed that several gene families facilitating the infection process are expanded (Martens et al., 2008). Extreme examples are gene families encoding cytoplasmic effector proteins such as RXLR effectors, which share the host cell-targeting motif RXLR and suppress defense responses in the host, and the necrosis-inducing proteins classified as Crinklers (Crn; Haas et al., 2009). To date, a few oomycete genomes have been sequenced, and this enables a comprehensive comparison of genomic features present in oomycetes, fungi, and other eukaryotic species such as gene families and protein domains. Experimentally derived functional knowledge of the majority of gene products in oomycetes in a comparable depth as for model species like Saccharomyces cerevisiae and Arabidopsis (Arabidopsis thaliana) will likely not be accessible in the near future. Hence, comparative genomics provides an important framework to functionally characterize oomycete gene products and generate hypotheses on the basic cellular functions as well as the complex interactions of these plant pathogens with their hosts and environment.
In this study, we focus on protein domains because these are the basic functional, evolutionary, and structural units that shape proteins (Rossmann et al., 1974; Orengo et al., 1997; Vogel et al., 2004). Domains function independently in single-domain proteins or synergistically in multidomain proteins (Doolittle, 1995; Vogel et al., 2004; Bashton and Chothia, 2007). Accordingly, some domains always occur with a defined set of functional partners, whereas others are highly versatile and form combinations of two consecutively occurring domains (also called bigrams) with different N- or C-terminal partners (Marcotte et al., 1999; Basu et al., 2008). Here, we analyzed the domain repertoire predicted from the genome sequences of 67 eukaryotic species and compared filamentous plant pathogens with other eukaryotes with a special emphasis on oomycetes. We show how differences in the domain repertoire of oomycetes, especially in the expansion of certain domain families and the formation of species-specific bigram types, can be linked to the biology of this group of organisms. This allowed the generation of candidate sets of proteins and domains that are likely to play roles in the lifestyle of oomycetes or their interaction with plants.
RESULTS
The Domain Repertoire of Oomycete Plant Pathogens and Its Comparison with Other Eukaryotes
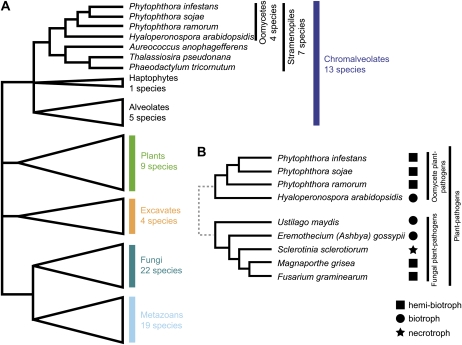
We analyzed the domain architecture of the predicted proteomes in 67 eukaryotes covering all major groups of the eukaryotic tree of life with the exception of the supergroup Rhizaria (Fig. 1A; Supplemental Table S1). We included seven stramenopiles, four of which are plant pathogenic oomycetes, namely the obligate biotrophic downy mildew Hyaloperonospora arabidopsidis and three hemibiotrophic Phytophthora species. The selection also contained five fungal plant pathogens, including rice (Oryza sativa) blast fungus (Magnaporthe grisea) and corn (Zea mays) smut (Ustilago maydis), both species with a (hemi)biotrophic lifestyle comparable to the oomycete plant pathogens used in the analysis (Fig. 1B).
Figure 1.
A, The major eukaryotic groups considered in the analysis and the number of species represented in every group. For the exact species used in the analysis, see Supplemental Table S1. The tree is adapted from Simpson and Roger (2004) and incorporates the phylogeny for the stramenopiles based on Blair et al. (2008). B, Fungal and oomycete plant-pathogenic species used in this analysis. The plant pathogens include species with different lifestyles, indicated by the symbol following the species name. The phylogeny for the fungi is based on James et al. (2006).
The domain architecture of all 1,250,996 predicted proteins in the 67 eukaryotic genomes was analyzed using HMMER (Eddy, 1998) and a local Pfam-A database (Finn et al., 2008). Overall, 59% (737,851) of all proteins have one or more predicted domain. We detected a total of 1,464,807 domains in all species, 80,180 within the stramenopiles and 51,030 in oomycetes.
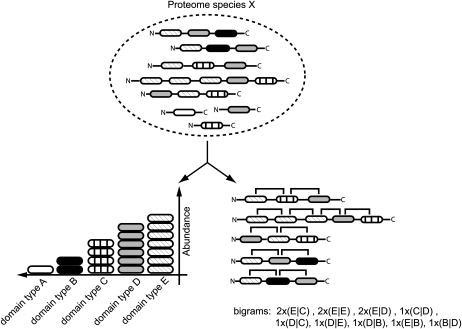
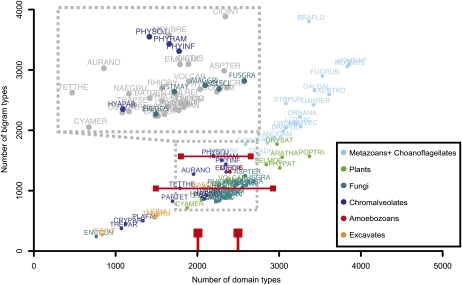
In order to characterize the domain repertoire of eukaryotes, we used two metrics: the number of domain types and the number of different combinations of adjacent domains, also called bigrams (Fig. 2). In total, 13,994 bigram types were identified in the 67 eukaryotic genomes, consisting of 6,356 different domain types. As described by Basu et al. (2008), the number of bigram types increases superlinearly relative to the number of domain types, with the highest numbers in multicellular organisms (Fig. 3). We observed separate clusters for metazoans, fungi, and plants (including land plants and mosses). Oomycetes and fungi have similar numbers of domain types, ranging from 2,000 to 2,500; however, oomycetes, in particular Phytophthora species, contain significantly more bigram types. The three analyzed Phytophthora species appeared to have approximately 50% more bigram types compared with other organisms that have similar numbers of domain types (Fig. 3; P = 0.00019, by one-sided Wilcoxon rank-sum test). This even holds when we apply a more conservative approach by discarding all domain and bigram types that occur once in each predicted proteome (Supplemental Fig. S1A). We observed that the number of domain types as well as the number of bigram types increases with proteome size and reaches saturation for larger proteomes (Supplemental Fig. S1, B and C; Cosentino Lagomarsino et al., 2009). Although oomycetes and in particular Phytophthora species contain a similar number of domain types as fungi, they have a larger predicted proteome (Supplemental Fig. S1B). However, they contain more bigram types than fungi but less than other species with predicted proteomes of similar size (e.g. Drosophila melanogaster; Supplemental Fig. S1C).
Figure 2.
Description of different metrics used in this study. In the example shown, we observe five different domain types. The abundance of a domain type is defined as the number of occurrences of the individual entity within the species (e.g. domain type B has an abundance of two). The versatility is defined as the number of different direct adjacent N- or C-terminal neighbors. We distinguish between N- and C-terminal partners (e.g. the versatility of domain type C is three). A bigram is a set of two directly adjacent domains, and we also consider two entities of the same domain a bigram (e.g. we observe nine different bigram types in the proteome, of which two have an abundance of three [right panel]).
Figure 3.
Dependence of the number of domain and bigram types observed in the analyzed species. The average number of different bigrams of species that have between 2,000 and 2,500 different domain types is indicated with the bottom horizontal red bar. The top horizontal red bar indicates the average number of different bigrams for Phytophthora species. The full species names corresponding to the abbreviations can be found in Supplemental Table S1. A magnification of the area encompassing the oomycete and fungal plant pathogens is shown; the species of interest are highlighted. The dots are colored according to the major eukaryotic groups as indicated in the text box.
Domain Overrepresentation Provides a Snapshot of Pathogen-Host Interaction
Apart from a wide and abundant repertoire of domains related to transposable elements (Haas et al., 2009), the most abundant domain types in oomycetes are similar to those in other eukaryotes (Supplemental Table S2). Hence, absolute domain abundance alone is not indicative enough to correlate domains to the lifestyle of both fungal and oomycete plant pathogens. Instead, we identified domains that are overrepresented in plant pathogens relative to other eukaryotes (Fig. 1B).
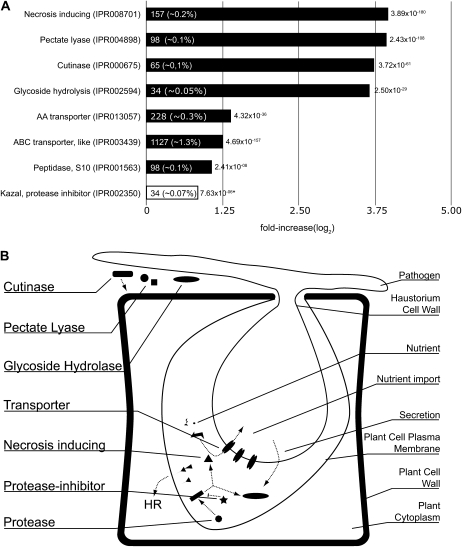
Our analysis inferred 246 overrepresented domains in plant pathogens that are observed in 24,970 proteins (P < 0.001, by Fisher’s exact test; a selection of well-described overrepresented domains is depicted in Fig. 4A; Supplemental Table S3). Since we analyzed the expansion in plant pathogens at the level of a group rather than an individual species, domains that are reported as being expanded in the group are not necessarily expanded in all species of the group or may even be absent (Supplemental Table S3). For example, secreted proteins encoding carbohydrate-binding family 25 domains (IPR005085) are only found in Phytophthora species and not in fungal plant pathogens, whereas secreted proteins containing the Cys-rich domain (CFEM; IPR008427) are only observed in fungal pathogens (Kulkarni et al., 2003).
Figure 4.
Overrepresentation of selected, well-described domains involved in plant-pathogen interaction and establishing or maintaining infection. A, The log2-fold overrepresentation of the domains in plant pathogens is shown in the bar chart. The absolute number of occurrences in plant pathogens and the percentage of all predicted domains in plant pathogens are displayed in the bars, and the corrected P values are shown at the tip of the bars. The fold overrepresentation and the P value for the Kazal protease inhibitor domain were based on the overrepresentation in oomycetes compared with plant pathogens (indicated by the white bar and asterisks). B, The overrepresented domains described in A are depicted in their possible cellular role during infection of the plant host.
Many proteins involved in host-pathogen interaction are secreted in the apoplast or, like the RXLR effector proteins, translocated into host cells following their secretion from the pathogen (Haas et al., 2009). Hence, we also predicted the presence of potential N-terminal signal peptide sequences in the whole proteomes of the analyzed species. The combined secretome encompasses 100,521 potentially secreted proteins, of which 11,352 are predicted in plant pathogens (Supplemental Fig. S2). Approximately 20% (2,478) of these proteins contain overrepresented domains; hence, proteins containing overrepresented domains are 1.85-fold enriched in the predicted secretome of the analyzed plant pathogens (P = 2.57 × 10−231, by Fisher’s exact test).
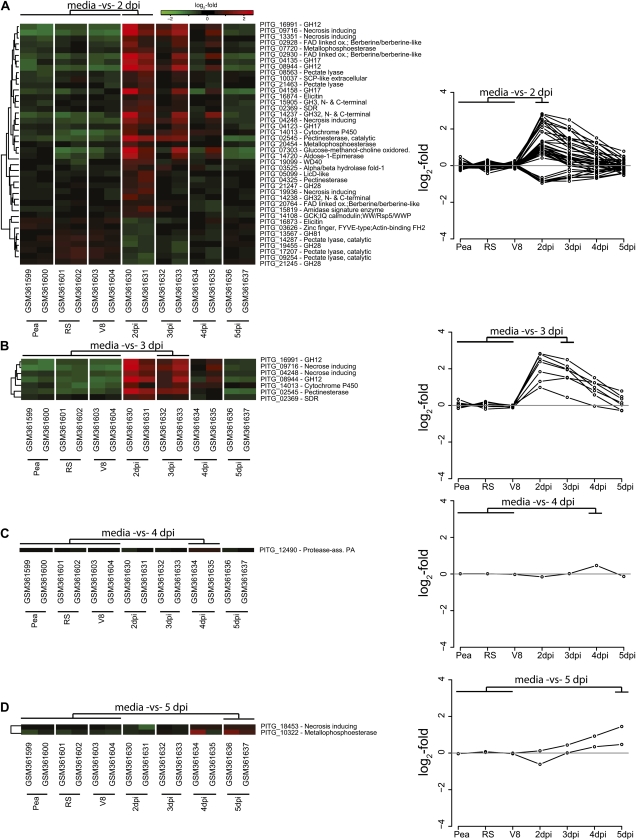
Oomycete proteins with significantly expanded domains are prime candidates for being pathogenicity associated. To assess this hypothesis, we tested if P. infestans genes that are differentially expressed during infection of the potato host are enriched for the aforementioned expanded domains. For this, we utilized NimbleGen microarray data that include genome-wide expression levels of P. infestans genes at different days post inoculation (dpi) of potato leaves as well as from mycelium grown in vitro on different media (Haas et al., 2009). We identified in total 1,584 genes that are significantly induced or repressed in P. infestans during infection (differentially expressed for at least one of the time points 2–5 dpi) compared with those grown in vitro (three different growth media; P < 0.05, q < 0.05, by t test; Supplemental Table S4A; Supplemental File S1). Of the 1,584 differentially expressed genes, 259 encode proteins containing significantly expanded domains (Supplemental Table S4B), which is 1.2-fold more than expected (P = 8.8 × 10−5, by Fisher’s exact test). Moreover, 44 of these 259 genes also encode proteins with a predicted signal peptide, which is a significant enrichment (1.8-fold; P = 4.38 × 10−5, by Fisher’s exact test). The majority (41) of these 44 genes are differentially expressed early in infection (2 dpi; Fig. 5A). All genes differentially expressed at 3 dpi are also differentially expressed at 2 dpi (Fig. 5, A and B). Consequently, the 44 differentially expressed genes coding for proteins with both predicted signal peptides as well as overrepresented domains are promising candidates for pathogenicity-associated proteins, of which several will be discussed in detail below.
Figure 5.
Gene expression analysis of P. infestans genes encoding overrepresented domains and a predicted N-terminal signal peptide. Genes with significant gene expression changes at different time points after infection (2–5 dpi) relative to the expression intensities of different growth media are displayed (P < 0.05, q < 0.05, by t test). Heat maps show the significantly differentially expressed genes at different time points relative to growth media. Genes were clustered using Spearman rank correlation and average linkage clustering. Gene identifiers as well as domain descriptions are displayed. Gene expression profiles are displayed for the expression intensities relative to the average intensities of the growth media for each time point after infection. Heat maps and expression profiles of the significantly differentially expressed genes relative to the growth media are shown for individual time points as follows: 2 dpi (A), 3 dpi (B), 4 dpi (C), and 5 dpi (D).
For several groups of overrepresented domains, a direct or indirect role in host-pathogen interaction and/or plant pathogen lifestyle has already been hypothesized or demonstrated (Dean et al., 2005; Tyler et al., 2006; Haas et al., 2009). Nearly 18% of the 246 overrepresented domains belong to three groups of domains: (1) hydrolase domains; (2) domains involved in substrate transport over membranes, such as the general ATP-binding cassette (ABC) transporter-like domain (IPR003439) but also more specialized transporters of sulfate (IPR011547) and amino acids (IPR004841/IPR013057); and (3) domains present in peptidases, such as the metalloprotease-type M28 domain (IPR007484) found in many secreted proteins. Of the hydrolases, which encompass 9% of the overrepresented domains, the majority are present in enzymes that hydrolyze glycosidic bonds. An example is the glycoside hydrolase (GH) family 12 domain (IPR002594). This domain is observed 34 times in plant pathogens, which overall contain 91,747 domains, and 43 times in all eukaryotes, which have a total of 1,464,807 domains, and hence is 12.62-fold (3.66 log2-fold) enriched in the plant pathogens. This domain is mainly observed in secreted proteins (27 out of 34; SignalP prediction). The majority (79%) of the GH-12 domains are found in oomycete plant pathogens, and the expression of two of these hydrolase genes in P. infestans (PITG_08944 and PITG_16991) is significantly induced during infection of potato (Fig. 5; Supplemental Table S4). In total, 33 differentially expressed genes during plant infection in P. infestans encode proteins that contain GH domains, including GH-17 (IPR000490) in endo-1,3-β-glucosidase and GH-81 (IPR005200) in β-1,3-glucanases as well as several members of GH-28 (IPR000743), a domain involved in soft rotting of host tissues and described in both fungal and bacterial plant pathogens (He and Collmer, 1990; Ruttkowski et al., 1990). Twenty-eight P. infestans genes coding for domains involved in transmembrane transport are differentially expressed during plant infection (Supplemental Table S4). Examples of genes encoding domains involved in substrate transport over the membrane are PITG_04307, which encodes an ABC-2-type transporter (IPR013525), PITG_12808, which encodes an amino acid transporter (IPR013057), as well as PITG_22087, a gene encoding both ABC-like (IPR003439) and ABC-2-type domains (Supplemental Table S4). Extracellular degrading enzymes like cutinases contain an overrepresented domain (IPR000675; P = 3.72 × 10−61). This domain is observed 65 times in plant pathogenic species, corresponding to a 13.3-fold (3.73 log2-fold) enrichment (Fig. 4A). In total, 61 proteins in plant pathogens predicted to possess this domain are potentially secreted (Supplemental File S1). Another overrepresented domain that is present in secreted proteins and involved in maceration and soft rotting of plant tissue is the pectate lyase (IPR004898). This domain is 15.34-fold (3.94 log2-fold) enriched in plant pathogens and mainly found in oomycetes. Five genes in P. infestans encode this domain as well as a predicted N-terminal signal peptide and are differentially expressed (Fig. 5).
Novel Candidate Domains Significantly Expanded in Plant Pathogens
Next to domains that were already directly or indirectly implied in host-pathogen interaction, we identified novel candidates that are also expanded in plant pathogens, several of which are encoded in P. infestans genes differentially expressed during infection of the host. Genes encoding the significantly expanded alcohol dehydrogenase (zinc binding; IPR013149) as well as a GroES-like alcohol dehydrogenase (IPR013154) domains are ubiquitous in all analyzed eukaryotes, and also the combination of these two domains is present in all species with only a few exceptions. Nine of these genes in P. infestans are induced during infection (Supplemental Table S4). Sixty-five genes in plant pathogens encode proteins with FAD-linked oxidase (IPR006094) and berberine/berberine-like (BBE) domains (IPR012951), of which three out of six in P. infestans are induced during infection (PITG_02928, PITG_02930, and PITG_20764). The BBE domain is involved in the biosynthesis of the alkaloid berberine (Facchini et al., 1996). The genes encode a predicted N-terminal signal peptide, although molecular analysis of proteins containing these domains in plants indicated that at least some of these are not secreted but instead are targeted to specialized vesicles (Amann et al., 1986; Kutchan and Dittrich, 1995; Facchini et al., 1996). Moreover, Moy et al. (2004) observed induced expression of a soybean (Glycine max) gene (BE584185) shortly after infection with Phytophthora sojae containing these two domains. A recent analysis from Raffaele et al. (2010) focusing solely on the secretome in P. infestans corroborates our results and also concludes that proteins with BBE and FAD-linked oxidase domains are candidate virulence factors. Three genes encoding secreted metallophosphoesterases (IPR004843; PITG_20454, PITG_07720, and PITG_10322) show induced gene expression. These metallophosphoesterase domains are found in phosphatases and hence are involved in the regulation of protein activity, since they work as antagonists of kinase activity.
For approximately 6% of all overrepresented domains, no or limited functional information is available in Pfam. These are the so-called DUFs: domains of unidentified function. Given their expansion in plant pathogens and the fact that other overrepresented domains are known to function in diverse aspects of plant-pathogen interactions, these DUFs are also likely to play a role in the lifestyle of plant pathogens and hence are promising targets for further experimental validation (Supplemental Table S3). Secreted proteins containing a combination of two overrepresented DUFs, DUF2403 (IPR018807) and DUF2401 (IPR018805), are exclusively found in fungi and in oomycetes, with the majority (approximately 75%) in oomycetes. The N-terminal DUF2403 contains a Gly-rich region without further functional annotation, whereas five highly conserved Cys residues characterize the C-terminal DUF2401. Proteins containing both DUFs have been characterized in S. cerevisiae and in Candida albicans as being covalently linked to the cell wall (Terashima et al., 2002; Yin et al., 2005; Klis et al., 2009). Another overrepresented DUF within plant pathogens and mainly found in oomycetes is DUF953 (IPR010357). This domain is present in several eukaryotic proteins with thioredoxin-like function, and two genes in P. infestans containing this domain are differentially expressed during infection (PITG_07008 and PITG_07010). DUF590 (IPR007632), which is ubiquitous in nearly all eukaryotes, is observed in proteins containing eight putative transmembrane helices. These proteins exhibit calcium-activated ion channel activity and are involved in diverse biological processes (Yang et al., 2008). The P. infestans gene PITG_06653 that contains the DUF590 domain is differentially expressed during infection, and this provides further support for a role in host-pathogen interaction. The exemplified DUFs as well as other overrepresented domains with less or no functional annotation are interesting candidates for further functional studies to decipher their precise role in plant pathogens.
Domain Overrepresentation in Oomycete Plant Pathogens
Since the previous analysis grouped both fungal and oomycete plant pathogens, domains specifically enriched in oomycetes were not directly discernible. Hence, we compared the relative domain abundance predicted in plant pathogens (Fig. 1B) with the aim to identify domains specifically enriched in oomycetes. Of the 75 domains that are overrepresented in oomycetes, 20 are not observed in any fungal plant pathogen and therefore can be considered oomycete specific within plant pathogens (Supplemental Table S5). In general, the abundance of expanded domains in Phytophthora species is higher than in H. arabidopsidis. A well-described example is the NPP1 domain (IPR008701) that is present in secreted (SignalP: 122) necrosis-inducing proteins. It shows a significant overrepresentation in oomycetes (1.68-fold [0.75 log2-fold] enriched), in particular in Phytophthora species, but is also observed 10 times in fungal plant pathogens as well as in a few cases in nonpathogenic fungi as noted before (Gijzen and Nürnberger, 2006). Four P. infestans genes encoding this domain are induced early during infection (2–3 dpi), whereas a single gene (PITG_18453) is induced late (5 dpi). Several peptidases (e.g. containing the peptidase S1/S6 and C1A domains) are overrepresented compared with other plant pathogens. S1/S6 (IPR001254; 1.6-fold [0.74 log2-fold]) is predicted in 91 proteins, of which 67 have a predicted secretion signal, while C1A (IPR000668; 1.79-fold [0.85 log2-fold]) is predicted in 78 proteins, of which 31 are potentially secreted. C1A is present in several eukaryotic species, but within the plant pathogenic group it is exclusively found in oomycetes. Several secreted protease inhibitors of the Kazal family containing the Kazal I1 (IPR002350) and Kazal-type (IPR011497) domains are significantly expanded in oomycetes and are within the group of analyzed plant pathogens specific to oomycetes. This suggests that they provide an increased level of protection of the pathogen against host-encoded defense-related proteases (Tyler et al., 2006). Another domain that is oomycete specific within the plant pathogens is the Na/Pi cotransporter (IPR003841) involved in the uptake of phosphate. Several other transporters that have already been described as being overrepresented in plant pathogens (e.g. the ABC-2-type transporters) are significantly expanded within oomycete plant pathogens, since these species are the major contributors to the overall abundance of this domain in plant pathogens. The abundance of predicted Ser/Thr-like kinase domains (IPR017442) compared with other plant pathogenic species is surprisingly high, and this domain is specifically expanded in the Phytophthora species. Even if several expanded domains are observed in both oomycete as well as fungal plant pathogens, the exploration of domains primarily expanded in oomycetes (e.g. certain transporter families and defense- and signaling-related domains) highlights functional entities that discriminate between these groups of plant pathogens.
Clustering of Abundance Profiles Reveals Additional Potential Pathogenicity Factors
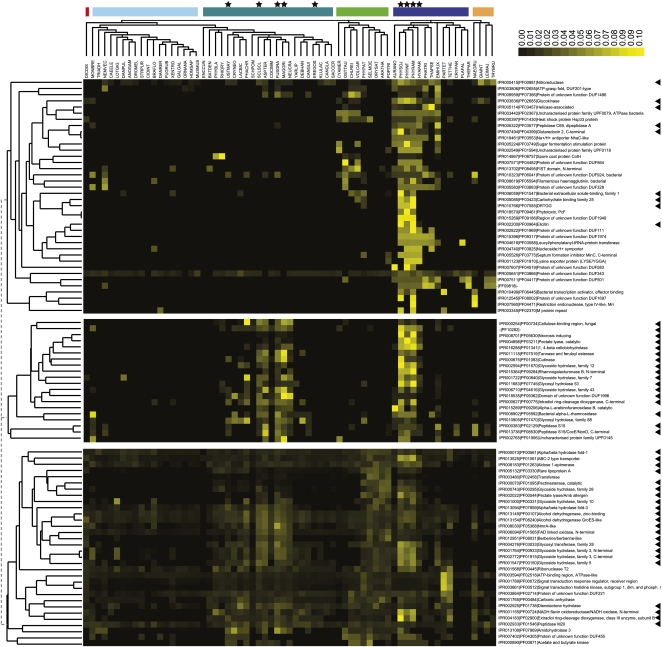
We extended the set of candidate domains that might be important for host-pathogen interaction beyond overrepresented domains by searching for additional domains that show presence, absence, and expansion profiles similar to overrepresented domains, since these domains are likely to be functionally linked or involved in similar biological processes (Pellegrini et al., 1999). We calculated a normalized profile of domain abundance and clustered similar abundance profiles using hierarchical clustering (Supplemental File S1). Several clusters contained a mix of significantly overrepresented domains and domains whose expansion in plant pathogens is not significant. We exemplify this with three clusters that contain 20% of all overrepresented domains in plant pathogens (Fig. 6).
Figure 6.
Average linkage clustering of normalized domain profiles using Spearman rank correlation as a distance measurement. The species tree for all eukaryotic species is depicted on top, with the color code of their supergroup as introduced in Figure 1. Plant pathogens are marked with stars, and the arrowheads highlight domains identified as overrepresented in plant pathogens.
In the first cluster (Fig. 6), domains are mainly expanded in oomycete plant pathogens. The abundance of some domains in plant pathogens is too low to be identified as being overrepresented. For example, the PcF domain (IPR018570), which is present in a small, approximately 50-amino acid necrosis-inducing protein found in various Phytophthora species (Orsomando et al., 2001; Liu et al., 2005), was not identified in the initial overrepresentation analysis. Also in this cluster is the sugar fermentation stimulation domain (IPR005224), which is mainly found in bacteria and involved in the regulation of maltose metabolism (Kawamukai et al., 1991). In this first cluster, we observed a high number (approximately 40%) of domains without functional characterization that are mainly present in bacteria. An example is DUF1949 (IPR015269), a domain that is only found in the three analyzed Phytophthora species. This domain is observed in functional uncharacterized bacterial proteins like YIGZ in Escherichia coli K12 and adopts a ferredoxin-like fold (Park et al., 2004). The Phytophthora and bacterial proteins containing DUF1949 also contain a second, N-terminal uncharacterized protein family, UPF (UPF00029, IPR001498). This domain is also found in the human protein Impact and is conserved from bacteria to eukaryotes (Okamura et al., 2000). The P. infestans gene (PITG_00027) containing both domains is induced early in infection (Supplemental Table S4B). Since these DUFs cluster with overrepresented domains, they are promising candidates for further study.
The domains in the second cluster mainly show an expansion of the abundance in both fungal and oomycete plant pathogens. This cluster contains, for example, cell wall-degrading domains like cutinases, pectate lysases, and other hydrolases and also the NPP1 domain that is found in necrosis-inducing proteins. The glycosyl hydrolase family 88 comprises unsaturated glucoronyl hydrolases thought to be involved in biofilm degradation and is mainly found in bacteria and fungi (Itoh et al., 2006). Interestingly, homologs are also observed in plant pathogenic bacteria (e.g. Pectobacterium atrosepticum), in fungi (e.g. M. grisea), and in all three Phytophthora species.
The third cluster contains domains that are not exclusively found in plant pathogens but have a broader abundance profile. This cluster includes a variety of overrepresented hydrolases, epimerases, and the ABC-2-type transporter domain (IPR013525) that is observed nearly 500 times in plant pathogenic species. Another domain that is found in this cluster is the dienelactone hydrolase domain (IPR002925), observed in all plant pathogens and also in other eukaryotic species, with a high abundance in plants as well as in fungi. This domain hydrolyzes dienelactone to maleylacetate in bacteria (Pathak et al., 1991) and is also detected in a putative 1,3:1,4-β-glucanase from P. infestans that is proposed to be involved in cell wall metabolism (McLeod et al., 2003).
Quantification of Oomycete-Specific Bigrams
Domains generally do not act as single entities in proteins but rather synergistically with other domains in the same protein or with domains in interacting proteins (Park et al., 2001; Vogel et al., 2004). Domains involved in signaling, sensing, and generic interactions are versatile and form combinations with several different partner domains (Supplemental Table S6). As described by others (Vogel et al., 2005), we observed that the versatility of domains is proportional to their abundance (Supplemental Fig. S3). Hence, we applied a weighted bigram frequency that corrects for abundance to detect domains that are promiscuous or prone to form combinations with different partners (Basu et al., 2008). The average number of promiscuous domains in oomycetes is 424 and in Phytophthora is 464. This is higher than the average number of promiscuous domains (357) over all other species (Supplemental Table S7).
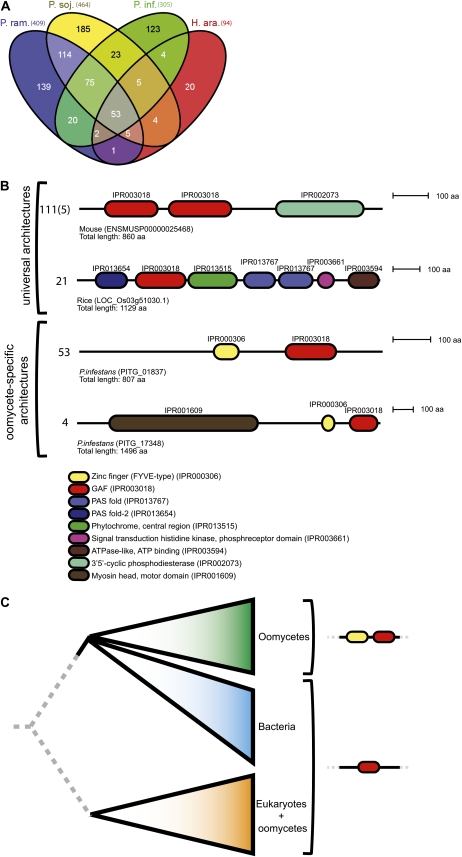
We observed that oomycetes have a higher number of bigram types than species with a comparable number of domain types (Fig. 3). We identified in total 13,994 different bigram types throughout the 67 analyzed species. The majority of these bigram types (i.e. 7,724, or 55.2%) are predicted in only a single species. In oomycetes, bigram types formed by domains that are associated with transposable elements showed a high abundance (Supplemental Tables S8 and S9). We identified 1,107 bigram types occurring exclusively in plant pathogens, the majority of which (773) are only observed in the analyzed oomycetes (Supplemental Table S10). These oomycete-specific bigram types are identified in total 1,511 times in 1,375 predicted proteins. Of the 773 oomycete-specific bigram types, 53 are present in all oomycetes (Fig. 7A). The biggest overlap in oomycete-specific domain types is observed between the Phytophthora species, especially between P. ramorum and P. sojae. A recent analysis of domain combination in P. ramorum and P. sojae already revealed several proteins involved in metabolism and regulatory networks containing novel bigrams (Morris et al., 2009). We additionally observed in total 43 bigram types that are shared either between P. infestans and P. sojae or between P. infestans and P. ramorum. However, the majority of oomycete-specific bigrams (467) are specific for a single species. The number of oomycete-specific bigram types highly exceeds the number of oomycete-specific domain types (41). Interestingly, only six of the oomycete-specific domains participate in forming the specific bigrams. Therefore, common domain types form the majority of the observed species-specific domain combinations, emphasizing the importance of novel domain combinations rather than novel domain types as a source for species-specific functionality. Even when we selectively look at the bigrams that occur at least twice in the same proteome or once in at least two different proteomes, we still observe 320 bigram types that are specific to oomycetes and occur in 982 predicted proteins.
Figure 7.
A, Venn diagram depicting the presence of oomycete-specific bigram types in the analyzed oomycete proteomes and indicating the number of shared bigram types between different proteomes. The total number of oomycete-specific bigram types in each proteome is shown in parentheses. The Venn diagram was produced using Venny (Oliveros, 2007). B, Domain architecture of example proteins containing a GAF domain. The top two architectures resemble common protein architectures: the cGMP-dependent 3′,5′-cyclic phosphodiesterase (observed 111 times in eukaryotes and five times in oomycetes) and phytochrome A (observed 21 times in eukaryotes). The bottom two architectures depict oomycete-specific architectures: the FYVE-GAF fusion is observed 53 times independent of other domains, and the myosin motor head in combination with the FYVE-GAF fusion is observed four times, a single copy in each of the oomycetes included in this study. aa, Amino acids. C, Simplified evolutionary tree based on the phylogenetic analysis of the GAF domain in prokaryotes and eukaryotes. GAF domains from proteins with a FYVE-GAF fusion are exclusively found to be close to bacterial GAF domains. Other oomycete proteins that only contain the GAF domain without the FYVE domain also cluster with other eukaryotic sequences.
Approximately 8% of the proteins containing an oomycete-specific bigram have a predicted secretion signal (9.2% of all oomycete proteins contain a predicted secretion signal). An example that is observed in a secreted putative Cys protease present in all analyzed oomycetes is the combination of the peptidase C1A domain (IPR000668) and the ML domain (IPR003172). The ML domain is known to be involved in lipid binding and innate immunity and has been observed in plants, fungi, and animals (Inohara and Nuñez, 2002). The proteins containing this bigram also have an N-terminal cathepsin inhibitory domain (IPR013201) that is often found next to the peptidase C1A domain and prevents access of the substrate to the binding cleft (Groves et al., 1996). Another bigram that is found in secreted proteins predicted in the analyzed Phytophthora species is the combination of the carbohydrate-binding domain family 25 (IPR005085; CBM25) with a GH-31 domain (IPR000322) as well as the tandem combination of CBM25 domains N terminal to the glycosyl hydrolase domain. The presence of the secreted CBM25 and GH-31 combination has recently been noted in Pythium ultimum (Lévesque et al., 2010). We further tried to elucidate the presence of RXLR or Crn motifs in proteins containing oomycete-specific bigrams. We predicted the presence of one of these motifs using individual HMMER models for both the RXLR and the Crn motif (see “Materials and Methods”). We overall predicted 746 proteins containing an RXLR and 99 proteins with a Crn motif. None of these proteins is predicted to contain an oomycete-specific bigram type.
The most abundant oomycete-specific bigram type that occurs in 64 proteins is a combination of the phosphatidylinositol 3-phosphate-binding zinc finger (FYVE type) and the GAF domain. The presence of this oomycete-specific bigram in P. ramorum and P. sojae has been noted before (Morris et al., 2009). The GAF domain is described as one of the most abundant domains in small-molecule-binding regulatory proteins (Zoraghi et al., 2004). It is present in a large number of different proteins with a wide range of cellular functions, such as gene regulation (Aravind and Ponting, 1997) and light detection and signaling (Sharrock and Quail, 1989; Montgomery and Lagarias, 2002). A typical eukaryotic domain composition involving the GAF domain is N terminal to the 3′5′-cyclic phosphodiesterase domain found in phosphodiesterases that regulate pathways with cyclic nucleotide-monophosphate as second messengers (Sharrock and Quail, 1989; Martinez et al., 2002). This organization is observed in total 111 times, and five times in oomycetes (Fig. 7B). The GAF-FYVE bigram is either observed as a single bigram (in 53 proteins) or in combination with other domains (in 11 proteins), for example with myosin (Richards and Cavalier-Smith, 2005). In P. infestans, two genes (PITG_07627 and PITG_09293) encoding proteins with this combination are induced early during infection of the plant (Supplemental Table S4B). A phylogenetic analysis of the GAF domain in eukaryotes and prokaryotes showed that all GAF domains in oomycetes that are involved in the fusion with FYVE exclusively cluster with prokaryotic GAF domains, whereas other GAFs also cluster with eukaryotes. Hence, this suggests a horizontal gene transfer from bacteria to oomycetes of those GAF domains that are involved in the fusion with FYVE (Fig. 7C; see “Materials and Methods”). The FYVE-type zinc finger is not identified in prokaryotic species; hence, we suggest two independent events, namely a horizontal gene transfer of the GAF domain from bacteria to oomycetes and subsequently a fusion to the zinc finger domain. Horizontal gene transfer seems to play an important role in the evolution of eukaryotes (Keeling and Palmer, 2008), and recent evidence indicates that these events also have a significant contribution to the genome content of protists and oomycetes, as they received genetic material from different sources (Richards and Talbot, 2007; Martens et al., 2008; Morris et al., 2009). Because GAF domains are known to be involved in many different cellular processes, we can only speculate about the biological function of proteins harboring the GAF-FYVE bigram. A possible function is the targeting of proteins to lipid layers by the zinc finger domain in response to second messengers sensed by the GAF domain.
Several domains involved in the phospholipid signaling domain were found to be overrepresented in the filamentous plant pathogens and in particular in oomycetes. These included the phosphatidylinositol 3-/4-kinase, PIK (IPR000403), the phosphatidylinositol 4-phosphate 5-kinase domain, PIPK (IPR002498), as well as the phosphatidylinositol 3-phosphate-binding FYVE. Novel domain compositions in proteins involved in phospholipid signaling and metabolism in Phytophthora species have been reported previously (Meijer and Govers, 2006). Signaling domains like the FYVE and the PIK, as well as domains like the IQ-calmodulin-binding domain (IPR000048) and the phox-like domain (IPR001683), form highly abundant oomycete-specific bigram types (Supplemental Table S10). Moreover, other domains, like the Ser/Thr protein kinase-like (IPR017442), pleckstrin homology (IPR001849), and DEP (IPR000591) domains, are involved in several oomycete-specific bigram types (e.g. the DEP-Ser/Thr protein kinase-like domain fusion is predicted in the proteomes of all analyzed oomycetes). Additionally, domains that are components of the histone acetylation-based regulatory system form oomycete-specific bigrams, such as the AP2 (IPR001471) and the histone deacetylase (IPR000286) domain combination (Iyer et al., 2008), which is observed in P. ramorum as well as in P. sojae.
DISCUSSION
We predicted the domain repertoire encoded in the genomes of four oomycete plant pathogens and compared it with a broad variety of eukaryotes spanning all major groups, including several fungal plant pathogens that have a similar morphology, lifestyle, and ecological niche as oomycete plant pathogens. We quantified and examined domain properties observed in oomycetes and especially emphasized differences and common themes within fungal and oomycete plant pathogens and their probable contribution to a pathogenic lifestyle.
We observed that oomycete plant pathogens, in particular Phytophthora species, have significantly higher numbers of unique bigram types compared with species with a similar number of domain types (Fig. 2A). However, oomycetes also have on average 50% more predicted genes than most of the analyzed fungi, but at the same time they encode a comparable number of domain types and hence exhibit similar domain diversity (Supplemental Fig. S1B). The high number of genes observed in oomycetes suggests enlarged complexity compared with fungi, which is not directly obvious from the domain diversity but instead from the number of unique bigram types (Supplemental Fig. S1C). This observation has two possible explanations: (1) the larger number of genes predicted from oomycete genomes provides the flexibility to form new domain combinations based on a limited set of already existing domains that are in quantities similar to fungi; (2) the domain models that cover specific domains are incomplete and therefore do not provide the required sensitivity for oomycete genomes. Hence, we would underestimate the number of observable domain types (and to a certain extent the number of predicted bigram types). Additionally, oomycetes, especially Phytophthora species, are no longer following the observed trend that organisms with a higher number of genes (proteins) contain a larger number of domain types. Consequently, they are shifted when comparing the number of predicted domain and bigram types. Nevertheless, both possible explanations and the observed numbers allow us to conclude that oomycete genomes, especially Phytophthora species, harbor a large repertoire of genes encoding different bigram types compared with species of comparable complexity and, in the case of filamentous fungi, even similar morphology.
Oomycetes and fungal plant pathogens seem to be very similar to other eukaryotes with respect to absolute domain abundance (Supplemental Table S2), and this metric is hence not sufficiently indicative to correlate domains directly or indirectly with the pathogenic lifestyle. Therefore, we predicted overrepresented domains in plant pathogens and identified 246 domains that are significantly expanded (Supplemental Table S3). Proteins containing overrepresented domains are significantly enriched in the predicted secretome of the analyzed plant pathogens, corroborating the idea that expanded domain families are involved in host-pathogen interaction and that these proteins are mainly acting in the extracellular space. It has to be noted that the presence of a predicted signal peptide does not necessarily mean that these proteins are found extracellularly, since some proteins are retained in the endoplasmic reticulum/Golgi and hence are not secreted (Bendtsen et al., 2004).
Since we anticipate that proteins that are directly involved in host-pathogen interaction are differentially regulated upon infection, we utilized the NimbleGen microarray data of P. infestans (Haas et al., 2009) and identified 259 induced/repressed genes encoding proteins containing overrepresented domains. Genes containing overrepresented domains are significantly enriched within the set of differentially expressed genes containing a predicted domain. Moreover, this subset contains a significantly higher abundance of genes with a predicted N-terminal signal peptide than expected. These observations highlight and corroborate the initially emerging link between domain expansion and host-pathogen interaction.
The majority of the 246 expanded domains are present in proteins that are involved in general carbohydrate metabolism, nutrient uptake, signaling networks, and suppression of host responses and hence might contribute to establishing and maintaining pathogenesis (Fig. 4). The variety of overrepresented domains involved in substrate transport over membranes is of special interest. Filamentous plant pathogens and especially oomycetes exhibit a complex and expanded repertoire of these domains, enabling them to absorb nutrients from their environment and host. The expression of P. infestans genes encoding ABC-2-like transporters, amino acid transporters, and Na/Pi cotransporter is induced early in infection of the plant, suggesting that these proteins act during the biotrophic phase of infection. Several other genes encoding proteins with a predicted extracellular localization are induced during infection and contain overrepresented domains. For example, three P. infestans genes encoding the predicted N-terminal signal peptide as well as FAD-linked oxidase and BBE domains are induced during infection. The BBE domain is involved in the biosynthesis of the alkaloid berberine (Facchini et al., 1996). Moy et al. (2004) showed that a soybean homolog of this gene is inducing after infection with P. sojae. Molecular studies of proteins containing BBE domains in plants have indicated that several proteins containing these domains are in fact not secreted but instead targeted to specific alkaloid biosynthetic vesicles where the proteins accumulate (Amann et al., 1986; Kutchan and Dittrich, 1995; Facchini et al., 1996). The expansion of domain families with potential direct or indirect roles in host-pathogen interaction in filamentous plant pathogens strongly suggests adaptation to their lifestyle at the genomic level.
In addition to known domains, the set of overrepresented domains also revealed domains that, as yet, have not been implicated in pathogenicity nor are functionally characterized. An example is the DUF953 domain, which, within plant pathogens, is mainly found in oomycetes. This domain is observed in eukaryotic proteins with a thioredoxin-like function, and P. infestans genes encoding these domains are differentially expressed during infection. The significant expansion of these domains in plant pathogens, and the fact that other well-described domains with a function in plant pathogenicity are also overrepresented, make proteins encoding poorly described but expanded domains interesting candidates to decipher their role in filamentous plant pathogens in general and oomycetes in particular.
We determined domain overrepresentation on the basis of species groups (plant pathogens and oomycetes) rather than on the level of individual species. We are aware that, as a consequence of this approach, we might have identified domains as being overrepresented in one group even if they do not need to be present or expanded in all the members (Supplemental Tables S3 and S5). Hence, we might falsely extrapolate the functional role of a domain in a subset of species to the whole group (e.g. a domain that is exclusively found in plant pathogenic fungi and not in oomycetes would still be overrepresented in the plant pathogenic group). Especially when comparing oomycete with fungal plant pathogens, the dominant expansion of domain families within Phytophthora species over families in H. arabidopsidis might bias the inferred overrepresented domain (Supplemental Table S5). Since we in general want to identify candidate domains that might be directly or indirectly involved in host-pathogen interaction, either at the level of filamentous plant pathogens or oomycetes, we think our group-based approach is appropriate to establish a set of candidate proteins and domains.
Moreover, the clustering of presence, absence, and expansion patterns of domains known or implicated to be involved in a plant pathogenic lifestyle with domains that have no known or direct connection to host-pathogen interactions aids in expanding this set of novel candidate domains (Fig. 5). For example, DUF1949 is within our species selection exclusively found in Phytophthora species and adopts a ferredoxin-like fold. The N-terminal region of proteins containing this domain shows similarity to another domain (UPF00029) that has been found in the human Impact protein. The P. infestans gene containing both domains is induced early during infection of the plant, providing additional, independent evidence for the possible role of genes encoding this uncharacterized domain in host-pathogen interaction. However, domains that are also abundant in nonpathogenic species (e.g. other stramenopiles) might not be related to or only indirectly involved in pathogenicity. Hence, the exact nature of the contribution of these domains to pathogenesis or to general lifestyle requires more in-depth experimental studies of the candidate domains and genes predicted to contain these functional entities.
Protein domains generally do not act as single entities but in synergy with other domains in the same protein or with other domains in interacting proteins. We identified 773 oomycete-specific bigrams, of which 53 are observed in all analyzed oomycetes (Fig. 7A; Supplemental Table S10). Based on our species selection, we cannot conclude that the oomycete-specific bigrams are common to all oomycetes, since they might only be specific for plant pathogenic oomycetes or even for the selected oomycetes analyzed in this study. The majority of the 773 bigrams, however, are specific for a subset of the tested oomycete species or even a single species. The 320 bigram types that are observed in more than a single species or twice in the same proteome are observed in 982 predicted proteins. These bigrams are less likely to be the result of a wrong gene annotation and include already well-described examples of oomycete-specific domain combinations, such as the FYVE-PIK bigram observed in Phytophthora phosphatidylinositol kinases (Meijer and Govers, 2006), the AP2-histone deacetylase bigram that is specifically found in P. ramorum and P. infestans (Iyer et al., 2008), and the myosin head domain-FYVE bigram as well as the FYVE-GAF bigram found in myosin proteins in all analyzed oomycetes (Richards and Cavalier-Smith, 2005). Still, some of the bigrams could be artificial due to false negatives or false positives in the domain predictions. The remaining, species-specific bigrams could be the result of artificial fusion of genes due to wrong gene annotation or an actual biological signal in one of the analyzed oomycete species. The derived results are not only dependent on the quality of the genome sequences of the analyzed oomycetes but also on that of the other eukaryotes. Wrong predictions of bigrams in these species would lead to false negatives in oomycetes. Hence, the number of derived oomycete-specific bigrams is only an approximation, and the true set of oomycete-specific bigrams needs to be further analyzed. Recent analyses of the underlying molecular mechanisms of domain gain in animals have shown that in fact gene fusion, tightly linked with gene duplication, is the major mechanism that shaped novel protein architecture (Buljan et al., 2010; Marsh and Teichmann, 2010). The contributions of this mechanism in forming lineage- or even species-specific bigrams in oomycetes and the probable role of the flexible genomes have to be further analyzed. The bigrams presented here form a comprehensive starting point for an in-depth bioinformatic and experimental analysis of promising gene families coding novel domain combinations.
Common domain types form the majority of the observed oomycete-specific bigrams, emphasizing the importance of novel combinations rather than novel domain types as a source for species-specific functionality. Only a minority of proteins containing oomycete-specific bigrams are secreted, and none of these proteins is predicted to contain a RXLR or Crn motif. We are aware that the total number of predicted proteins containing the RXLR or Crn motif is lower than reported in other studies where those were predicted using multiple complementary methods (Haas et al., 2009). However, when directly comparing the number of proteins predicted to contain the RXLR motif by HMMER alone, the reported numbers are similar to our predictions. Together with the observation that RXLR proteins do not contain known Pfam-A domains in the C-terminal domain (Haas et al., 2009), our data are not in conflict with RXLR protein predictions from previous studies. Of the known Crn genes in P. infestans, 40% do not encode a secretion signal (Haas et al., 2009); hence, these sequences are not considered in the prediction of Crn motifs in our analysis and explain the discrepancy between the previously reported numbers and our predictions. Haas et al. (2009) have reported a huge number of different C-terminal structures in P. infestans Crns that contained up to 36 different domains, of which 33 are not described in Pfam. Several of these domains induce necrosis in plants. Since we focused in our analysis exclusively on Pfam domains, we did not expect to find these proteins containing specific bigrams.
The majority of proteins containing oomycete-specific bigrams seem to be functional in the pathogen cytoplasm. Moreover, domains involved in mediation between macromolecules or lipids (e.g. the FYYE or the phox-like domain) as well as signaling domains (e.g. Ser/Thr kinase-like or the DEP domain) are highly abundant in oomycete-specific bigrams. Ser/Thr kinase domain-like is overrepresented in oomycetes compared with fungal plant pathogens and is particularly expanded within the Phytophthora species (Supplemental Table S5). This expanded repertoire together with the high abundance of this domain in oomycete-specific bigrams strongly suggests that oomycetes have the capacity to recombine existing signaling pathways in a novel and complicated network that is distinct from other eukaryotes. This might also be true for other interaction networks, since several domains mediating interactions between macromolecules (e.g. DNA-binding zinc finger [IPR007087] or protein-protein interaction like WW/Rsp5/WWP [IPR001202]) are also highly abundant in oomycete-specific bigrams. Whether this reflects a general phenomenon in all oomycetes, specific for the plant pathogenic species analyzed in this study, or only for Phytophthora species, can only be answered when more oomycetes, including saprophytes and pathogens with different hosts, have been sequenced.
We outlined a complex but comprehensive picture of the domain repertoire of filamentous plant pathogens focusing on oomycetes and showed how differences compared with other eukaryotes are reflecting the biology of these groups of organisms. Especially the expansion of certain domain families is directly linked with the lifestyle of oomycete plant pathogens and allowed the generation of a set of candidate domains likely to play important roles in the interaction with the plant host. Proteins containing overrepresented domains are enriched in the predicted secretome of the analyzed species. Moreover, the expression analysis of genes encoding overrepresented domains during infection of the plant revealed a significant enrichment of genes encoding overrepresented domains within the differentially expressed genes. Furthermore, we observed a significantly higher than expected abundance of genes encoding a signal peptide within the set of differentially expressed genes containing expanded domains. This added additional, independent evidence for the biological significance of our observations. Furthermore, oomycete genomes encode a set of proteins containing oomycete-specific domain combinations that are formed by common domain types and include several domains involved in signaling and/or mediation of interactions between macromolecules. Oomycetes, therefore, might possess altered regulatory and signaling networks that differ from other eukaryotes. If the described and discussed differences in the domain repertoire of oomycetes have a direct influence on plant pathogenicity or are generally useful in these organisms needs to be analyzed further. Nevertheless, they provide promising starting points that will aid our understanding of the biology of oomycetes in general and plant pathogens in particular.
MATERIALS AND METHODS
Species Used in the Analysis
In the performed analysis, 67 eukaryotic species representing four of the five eukaryotic supergroups (excluding Rhizaria) were considered (Fig. 1A; for species abbreviations, see Supplemental Table S1). We used the predicted best model proteomes for all subsequent analyses.
Identification of Domain Composition
We predicted the domain repertoire of all proteins encoded in the diverse genomes using hmmpfam (HMMER package version 2.3.2) and a local Pfam-A database (version 23). We applied a domain model-specific gathering cutoff and used HMM models that are optimized to search for full-length entities in the query sequence.
In order to obtain the nonoverlapping domain architecture of multidomain proteins, we resolved overlapping domains according to certain rules. We defined two domains as overlapping if more than 10% of the predicted domain locations were overlapping (based on the relative length of the domains). If, in the case of overlapping domains, the e-value difference was larger than 5 (on a –log10 scale), we kept the domain with the highest e-value. In cases where the difference was smaller, we kept the longest model. If both overlapping models had the same length, we considered differences in e-value and bit score. In the case of the Pfam-based predictions for 15 proteins, the applied rules did not resolve overlapping entities. Therefore, we considered the Conserved Domain Database (version 2.16) superfamily annotation, which automatically clusters domain entities that resemble evolutionarily related domains. If both domains corresponded to the same family, we choose one entity.
Based on the nonoverlapping domain architecture, we derived different metrics for each proteome. We counted the abundance for every domain and the resulting number of different domain types per analyzed proteome. We defined domain bigrams as two consecutively located domains in a single protein. We discriminated between reciprocal domain pairs, so that the bigram (A|B) is not identical to (B|A), and took repeating domains into account, such as (A|A). Based on the set of bigrams, we also determined the versatility of all individual domains in a given proteome, which is defined by the number of different direct N- and C-terminal partners, also including reciprocal and self-repeated pairs.
Prediction of Secreted Proteins
Secreted proteins were predicted using SignalP (version 3.0; Bendtsen et al., 2004) in combination with TMHMM (version 2.0; Krogh et al., 2001). We restricted the analysis to the first 70 amino acids of the protein and accepted signal peptide predictions if both the neural network and the HMM implemented in SignalP predicted the presence of a signal peptide under default parameters. Moreover, we declined predicted signal peptides if TMHMM predicted more than one transmembrane region in the protein. If only a single transmembrane helix was predicted and the predicted region was overlapping with the SignalP prediction for more than 10 amino acids and positioned within the first 35 amino acids from the start, we included the protein in the set of secreted proteins.
Domain Overrepresentation
Domain overrepresentation was calculated using a one-sided Fisher’s exact test. The derived P values were Bonferroni corrected for multiple testing by multiplying the P value with the number of conducted tests. The corrected P values were compared with an α = 0.001 to infer domain overrepresentation. For the overrepresented domains in oomycete plant pathogens compared with fungal plant pathogens, we considered domains that occur at least once in a single plant pathogen but nevertheless could also occur in other eukaryotic species.
Gene Expression Analysis of Phytophthora infestans
We extracted NimbleGen expression data of P. infestans during infection of potato (Solanum tuberosum) 2 to 5 dpi from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). The setup and initial analysis of the NimbleGen data are described by Haas et al. (2009). The log2-transformed and mean-centered array intensities were analyzed for differential expression using Multiexperiment Viewer (Saeed et al., 2006). The t tests were conducted between two groups (group A, different media types; group B, replicates for 1 dpi). The test was applied for each day after inoculation, and significant up-/down-regulated genes were reported applying a P values cutoff of 0.05. False discovery rates were addressed using R and the qvalue package by computing q values for each of the comparisons and subsequently applying a q value cutoff of 0.05 (Storey and Tibshirani, 2003; R Development Core Team, 2010). Visualization of the heat maps was done using R and the Bioconductor package utilizing Spearman correlation as a distance measurement and hierarchical clustering (average linkage; Gentleman et al., 2004). Gene expression intensities relative to the average expression intensities in media types (V8, RS, Pea) were computed in R.
Clustering of Domain Profiles
We created abundance profiles for each domain based on the abundance in each individual proteome. We excluded domains that were only identified in a single species. The rows (domains) were multiplied by a scaling factor so that the sum of squares was 1, and subsequently the columns (species) were normalized in the same way. We performed a hierarchical clustering (average linkage) of the profiles using the Spearman correlation matrix as a distance measurement. The normalization and clustering were performed using Cluster (Eisen et al., 1998), and the visualization was done using TreeView (http://rana.lbl.gov/EisenSoftware.htm).
Domain Promiscuity
We calculated the domain promiscuity for every domain in the analyzed species based on weighted bigram frequency (Basu et al., 2008). We took a relatively moderate cutoff for determining promiscuous domains; every domain with a higher promiscuity score than a domain that is only present once in the genome and is participating in one bigram type is called promiscuous.
Prediction of the RXLR and Crn Motifs in Oomycetes
We identified the presence of the RXLR motif in all predicted proteins in the analyzed oomycetes using three different HMMER models (R.H.Y. Jiang, personal communication). The first model was created using Phytophthora ramorum and Phytophthora sojae RXLRs and included the RXLR motif itself and 10 amino acids downstream and upstream of the motif. The two other models were based separately on RXLRs from P. infestans and Hyaloperonospora arabidopsidis and included 10 amino acids upstream from the RXLR motif and five amino acids downstream of the DEER motif. We used HMMER (hmmsearch) with an e-value cutoff of 10 and subsequently combined all predictions. Furthermore, we demanded the presence of a predicted signal peptide (SignalP) cleavage site within the first 30 amino acids of the protein, the gap between the cleavage site and the start of the motif to be 30 or less, the start of the motif to be within the first 100 amino acids of the protein, and the starting position of the RXLR motif to be downstream of the cleavage site. For the identification of the Crn LFLAK motif, we used a HMMER model of that region (B.J. Haas, personal communication) and the same sequence demands as for the RXLRs.
Phylogenetic Analysis of the GAF Domain
We derived all sequences containing a GAF domain from the selected proteomes and extracted the amino acid sequence of the domain based on the start and end points of the domain model. We conducted a similarity search with the extracted domains using BLASTP (version 2.2.20) with an e-value cutoff of 1 × 10−5 and a low-complexity filter against a set of 295 bacterial predicted proteomes (downloaded from the National Center for Biotechnology Information ftp server on January 27, 2009). In the homologs that were obtained, domains were predicted using hmmpfam as described above. Subsequently, prokaryotic GAF domains were extracted and aligned together with the eukaryotic domains using mafft (version 6.713b) with the local alignment strategy (Katoh et al., 2002). A phylogenetic tree was constructed with RAxML (version 7.0.4) using the GAMMA model of rate heterogeneity and the WAG amino acid substitution matrix (Stamatakis, 2006).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Dependence of the number of domain types, bigram types, and proteome sizes.
Supplemental Figure S2. Size of the predicted secretome of the 67 analyzed eukaryotes.
Supplemental Figure S3. Dependance of versatility and abundance of the analyzed domains.
Supplemental Table S1. Summary of the eukaryotic species analyzed in this study.
Supplemental Table S2. Domain abundance reported for all predicted Pfam domains.
Supplemental Table S3. Overrepresented domains in plant pathogens.
Supplemental Table S4. Differentially expressed genes in P. infestans.
Supplemental Table S5. Overrepresented domains in oomycetes.
Supplemental Table S6. Domain versatility reported for all predicted Pfam domains.
Supplemental Table S7. Domain promiscuity reported for all predicted Pfam domains.
Supplemental Table S8. Bigram abundance reported for all species, plant pathogens, and plant pathogenic oomycetes.
Supplemental Table S9. Bigram abundance (excluding self-repeated domains) reported for all species, plant pathogens, and plant pathogenic oomycetes.
Supplemental Table S10. Summary of the oomycete-specific bigrams.
Supplemental File S1. TreeView file containing the clustered domain abundance profiles.
Supplementary Material
Acknowledgments
We thank Lidija Berke, John van Dam, and Jos Boekhorst for fruitful discussion and comments on the manuscript as well as Rui Peng Wang for support with the P. infestans gene expression data. We also thank Harold J.G. Meijer for discussion of fusion proteins in P. infestans, Rays H.Y. Jiang for providing the RXLR-HMMER model, and Brian J. Haas for the Crn LFLAK-HMMER model. Some of the sequence data and annotation were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov), the Broad Institute of Harvard and the Massachusetts Institute of Technology (http://www.broadinstitute.org), or the Stanford Genome Technology Center (http://med.stanford.edu/sgtc/) in collaboration with the user community (for detailed information, see Supplemental Table S1).
References
- Agrios GN. (2005) Plant Pathology, Ed 5 Academic Press, New York [Google Scholar]
- Amann M, Wanner G, Zenk MH. (1986) Intracellular compartmentation of two enzymes of berberine biosynthesis in plant cell cultures. Planta 167: 310–320 [DOI] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. (1997) The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci 22: 458–459 [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. (2000) A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290: 972–977 [DOI] [PubMed] [Google Scholar]
- Bashton M, Chothia C. (2007) The generation of new protein functions by the combination of domains. Structure 15: 85–99 [DOI] [PubMed] [Google Scholar]
- Basu MK, Carmel L, Rogozin IB, Koonin EV. (2008) Evolution of protein domain promiscuity in eukaryotes. Genome Res 18: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurain D, Brinkmann H, Petersen J, Rodríguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H. (2010) Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol 27: 1698–1709 [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Blair JE, Coffey MD, Park SY, Geiser DM, Kang S. (2008) A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet Biol 45: 266–277 [DOI] [PubMed] [Google Scholar]
- Buljan M, Frankish A, Bateman A. (2010) Quantifying the mechanisms of domain gain in animal proteins. Genome Biol 11: R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino Lagomarsino M, Sellerio AL, Heijning PD, Bassetti B. (2009) Universal features in the genome-level evolution of protein domains. Genome Biol 10: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, et al. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986 [DOI] [PubMed] [Google Scholar]
- Doolittle RF. (1995) The multiplicity of domains in proteins. Annu Rev Biochem 64: 287–314 [DOI] [PubMed] [Google Scholar]
- Eddy SR. (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DC, Ribeiro OK. (1996) Phytophthora Diseases Worldwide. American Phytopathological Society, St. Paul [Google Scholar]
- Facchini PJ, Penzes C, Johnson AG, Bull D. (1996) Molecular characterization of berberine bridge enzyme genes from opium poppy. Plant Physiol 112: 1669–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, et al. (2008) The Pfam protein families database. Nucleic Acids Res 36: D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijzen M, Nürnberger T. (2006) Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67: 1800–1807 [DOI] [PubMed] [Google Scholar]
- Govers F, Gijzen M. (2006) Phytophthora genomics: the plant destroyers’ genome decoded. Mol Plant Microbe Interact 19: 1295–1301 [DOI] [PubMed] [Google Scholar]
- Groves MR, Taylor MA, Scott M, Cummings NJ, Pickersgill RW, Jenkins JA. (1996) The prosequence of procaricain forms an alpha-helical domain that prevents access to the substrate-binding cleft. Structure 4: 1193–1203 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398 [DOI] [PubMed] [Google Scholar]
- He SY, Collmer A. (1990) Molecular cloning, nucleotide sequence, and marker exchange mutagenesis of the exo-poly-alpha-D-galacturonosidase-encoding pehX gene of Erwinia chrysanthemi EC16. J Bacteriol 172: 4988–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Nuñez G. (2002) ML: a conserved domain involved in innate immunity and lipid metabolism. Trends Biochem Sci 27: 219–221 [DOI] [PubMed] [Google Scholar]
- Itoh T, Hashimoto W, Mikami B, Murata K. (2006) Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1. Biochem Biophys Res Commun 344: 253–262 [DOI] [PubMed] [Google Scholar]
- Iyer LM, Anantharaman V, Wolf MY, Aravind L. (2008) Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol 38: 1–31 [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, et al. (2006) Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443: 818–822 [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamukai M, Utsumi R, Takeda K, Higashi A, Matsuda H, Choi YL, Komano T. (1991) Nucleotide sequence and characterization of the sfs1 gene: sfs1 is involved in CRP*-dependent mal gene expression in Escherichia coli. J Bacteriol 173: 2644–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. (2008) Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet 9: 605–618 [DOI] [PubMed] [Google Scholar]
- Klis FM, Sosinska GJ, de Groot PW, Brul S. (2009) Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEM Yeast Res 9: 1013–1028 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kulkarni RD, Kelkar HS, Dean RA. (2003) An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem Sci 28: 118–121 [DOI] [PubMed] [Google Scholar]
- Kutchan TM, Dittrich H. (1995) Characterization and mechanism of the berberine bridge enzyme, a covalently flavinylated oxidase of benzophenanthridine alkaloid biosynthesis in plants. J Biol Chem 270: 24475–24481 [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M, de Wit PJGM, Govers F. (2003) Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol 11: 462–469 [DOI] [PubMed] [Google Scholar]
- Lévesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, Raffaele S, Robideau GP, Thines M, Win J, et al. (2010) Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol 11: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Bos JI, Armstrong M, Whisson SC, da Cunha L, Torto-Alalibo T, Win J, Avrova AO, Wright F, Birch PR, et al. (2005) Patterns of diversifying selection in the phytotoxin-like scr74 gene family of Phytophthora infestans. Mol Biol Evol 22: 659–672 [DOI] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. (1999) Detecting protein function and protein-protein interactions from genome sequences. Science 285: 751–753 [DOI] [PubMed] [Google Scholar]
- Marsh JA, Teichmann SA. (2010) How do proteins gain new domains? Genome Biol 11: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C, Vandepoele K, Van de Peer Y. (2008) Whole-genome analysis reveals molecular innovations and evolutionary transitions in chromalveolate species. Proc Natl Acad Sci USA 105: 3427–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez SE, Beavo JA, Hol WGJ. (2002) GAF domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Mol Interv 2: 317–323 [DOI] [PubMed] [Google Scholar]
- McLeod A, Smart CD, Fry WE. (2003) Characterization of 1,3-beta-glucanase and 1,3;1,4-beta-glucanase genes from Phytophthora infestans. Fungal Genet Biol 38: 250–263 [DOI] [PubMed] [Google Scholar]
- Meijer HJG, Govers F. (2006) Genomewide analysis of phospholipid signaling genes in Phytophthora spp.: novelties and a missing link. Mol Plant Microbe Interact 19: 1337–1347 [DOI] [PubMed] [Google Scholar]
- Montgomery BL, Lagarias JC. (2002) Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci 7: 357–366 [DOI] [PubMed] [Google Scholar]
- Morris PF, Schlosser LR, Onasch KD, Wittenschlaeger T, Austin R, Provart N. (2009) Multiple horizontal gene transfer events and domain fusions have created novel regulatory and metabolic networks in the oomycete genome. PLoS ONE 4: e6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy P, Qutob D, Chapman BP, Atkinson I, Gijzen M. (2004) Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol Plant Microbe Interact 17: 1051–1062 [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagiwara-Takeuchi Y, Li T, Vu TH, Hirai M, Hattori M, Sakaki Y, Hoffman AR, Ito T. (2000) Comparative genome analysis of the mouse imprinted gene impact and its nonimprinted human homolog IMPACT: toward the structural basis for species-specific imprinting. Genome Res 10: 1878–1889 [DOI] [PubMed] [Google Scholar]
- Oliveros JC. (2007) VENNY: An Interactive Tool for Comparing Lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html (October 7, 2010) [Google Scholar]
- Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. (1997) CATH: a hierarchic classification of protein domain structures. Structure 5: 1093–1108 [DOI] [PubMed] [Google Scholar]
- Orsomando G, Lorenzi M, Raffaelli N, Dalla Rizza M, Mezzetti B, Ruggieri S. (2001) Phytotoxic protein PcF, purification, characterization, and cDNA sequencing of a novel hydroxyproline-containing factor secreted by the strawberry pathogen Phytophthora cactorum. J Biol Chem 276: 21578–21584 [DOI] [PubMed] [Google Scholar]
- Park F, Gajiwala K, Eroshkina G, Furlong E, He D, Batiyenko Y, Romero R, Christopher J, Badger J, Hendle J, et al. (2004) Crystal structure of YIGZ, a conserved hypothetical protein from Escherichia coli K12 with a novel fold. Proteins 55: 775–777 [DOI] [PubMed] [Google Scholar]
- Park J, Lappe M, Teichmann SA. (2001) Mapping protein family interactions: intramolecular and intermolecular protein family interaction repertoires in the PDB and yeast. J Mol Biol 307: 929–938 [DOI] [PubMed] [Google Scholar]
- Pathak D, Ashley G, Ollis D. (1991) Thiol protease-like active site found in the enzyme dienelactone hydrolase: localization using biochemical, genetic, and structural tools. Proteins 9: 267–279 [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. (1999) Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA 96: 4285–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Win J, Cano L, Kamoun S. (2010) Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans. BMC Genomics 11: 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2010) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Richards TA, Cavalier-Smith T. (2005) Myosin domain evolution and the primary divergence of eukaryotes. Nature 436: 1113–1118 [DOI] [PubMed] [Google Scholar]
- Richards TA, Talbot NJ. (2007) Plant parasitic oomycetes such as Phytophthora species contain genes derived from three eukaryotic lineages. Plant Signal Behav 2: 112–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Moras D, Olsen KW. (1974) Chemical and biological evolution of nucleotide-binding protein. Nature 250: 194–199 [DOI] [PubMed] [Google Scholar]
- Ruttkowski E, Labitzke R, Khanh NQ, Löffler F, Gottschalk M, Jany KD. (1990) Cloning and DNA sequence analysis of a polygalacturonase cDNA from Aspergillus niger RH5344. Biochim Biophys Acta 1087: 104–106 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. (2006) TM4 microarray software suite. Methods Enzymol 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3: 1745–1757 [DOI] [PubMed] [Google Scholar]
- Simpson AGB, Roger AJ. (2004) The real ‘kingdoms’ of eukaryotes. Curr Biol 14: R693–R696 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Fukuchi S, Nakai K, Arisawa M, Hamada K, Yabuki N, Kitada K. (2002) Sequence-based approach for identification of cell wall proteins in Saccharomyces cerevisiae. Curr Genet 40: 311–316 [DOI] [PubMed] [Google Scholar]
- Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RHY, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, et al. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313: 1261–1266 [DOI] [PubMed] [Google Scholar]
- Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. (2004) Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol 14: 208–216 [DOI] [PubMed] [Google Scholar]
- Vogel C, Teichmann SA, Pereira-Leal J. (2005) The relationship between domain duplication and recombination. J Mol Biol 346: 355–365 [DOI] [PubMed] [Google Scholar]
- Yang YD, Cho H, Koo JY, Tak MH, Cho Y, Shim WS, Park SP, Lee J, Lee B, Kim BM, et al. (2008) TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 455: 1210–1215 [DOI] [PubMed] [Google Scholar]
- Yin QY, de Groot PW, Dekker HL, de Jong L, Klis FM, de Koster CG. (2005) Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via glycosylphosphatidylinositol remnants or mild alkali-sensitive linkages. J Biol Chem 280: 20894–20901 [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Pinto G, Bhattacharya D. (2002) The single, ancient origin of chromist plastids. Proc Natl Acad Sci USA 99: 15507–15512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoraghi R, Corbin JD, Francis SH. (2004) Properties and functions of GAF domains in cyclic nucleotide phosphodiesterases and other proteins. Mol Pharmacol 65: 267–278 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.