Abstract
Soybean (Glycine max) is a self-pollinating species that has relatively low nucleotide polymorphism rates compared with other crop species. Despite the low rate of nucleotide polymorphisms, a wide range of heritable phenotypic variation exists. There is even evidence for heritable phenotypic variation among individuals within some cultivars. Williams 82, the soybean cultivar used to produce the reference genome sequence, was derived from backcrossing a Phytophthora root rot resistance locus from the donor parent Kingwa into the recurrent parent Williams. To explore the genetic basis of intracultivar variation, we investigated the nucleotide, structural, and gene content variation of different Williams 82 individuals. Williams 82 individuals exhibited variation in the number and size of introgressed Kingwa loci. In these regions of genomic heterogeneity, the reference Williams 82 genome sequence consists of a mosaic of Williams and Kingwa haplotypes. Genomic structural variation between Williams and Kingwa was maintained between the Williams 82 individuals within the regions of heterogeneity. Additionally, the regions of heterogeneity exhibited gene content differences between Williams 82 individuals. These findings show that genetic heterogeneity in Williams 82 primarily originated from the differential segregation of polymorphic chromosomal regions following the backcross and single-seed descent generations of the breeding process. We conclude that soybean haplotypes can possess a high rate of structural and gene content variation, and the impact of intracultivar genetic heterogeneity may be significant. This detailed characterization will be useful for interpreting soybean genomic data sets and highlights important considerations for research communities that are developing or utilizing a reference genome sequence.
Intracultivar genetic heterogeneity refers to the genetic variation present from plant to plant within a named cultivar or variety. Although the phenomenon of intracultivar heterogeneity has long been recognized in crop species (Byth and Weber, 1968), it is oftentimes ignored, as most researchers assume that elite cultivars are composed of relatively homogenous genetic pools (Fasoula and Boerma, 2007). However, a small number of studies have documented the phenotypic consequences of intracultivar genetic heterogeneity in inbred crop accessions, including studies in tobacco (Nicotiana tabacum; Gordon and Byth, 1972), maize (Zea mays; Higgs and Russell, 1968; Tokatlidis, 2000), wheat (Triticum aestivum; Tokatlidis et al., 2004), and cotton (Gossypium hirsutum; Tokatlidis et al., 2008).
The segregation of parental loci during the breeding process is one source of intracultivar heterogeneity. For self-pollinating species, new cultivars are typically derived from either intermating elite lines or backcrossing traits into elite lines, followed by several rounds of single-seed descent via self-mating and subsequent seed increase generations. At the termination of the single-seed descent generations, any remaining heterozygous loci will segregate in subsequent generations. Assuming that the population remains intact, each plant lineage will eventually fix almost all of the segregating loci in the homozygous state of either parent. Therefore, the population will maintain some degree of plant-to-plant variation due to the heterogeneity at these loci.
Genetic heterogeneity may also be generated de novo by spontaneous mutation (Shaw et al., 2000; Ossowski et al., 2010), novel recombination events, DNA transposition, or epigenetic processes (Rasmusson and Phillips, 1997). Recent studies in yeast and fungi have reported striking genomic structural variation, such as large-scale duplications, deletions, and rearrangements, induced de novo in response to drug treatments or nutrient-stressed conditions (Gresham et al., 2008; Selmecki et al., 2009). Furthermore, a recent study has reported striking genomic structural variation derived de novo in Arabidopsis (Arabidopsis thaliana) lineages within five or fewer generations when individuals are grown in stressful conditions (DeBolt, 2010).
Studies that have investigated the genetic basis of intracultivar heterogeneity have primarily reported on the rates of molecular marker polymorphisms within cultivars and inbred lines of crops such as barley (Hordeum vulgare), maize, rice (Oryza sativa), sunflower (Helianthus annuus), and wheat (Zhang et al., 1995; Olufowote et al., 1997; Gethi et al., 2002; Röder et al., 2002; Sjakste et al., 2003; Soleimani et al., 2005; Giarrocco et al., 2007). However, the number and types of markers applied in these studies limited their throughput and ability to resolve major features of structural variation, including large-scale deletions, duplications, and more complicated genomic rearrangements. Consequently, little is known about the origins and mechanisms of intracultivar heterogeneity.
In soybean (Glycine max), Fasoula and Boerma (2005, 2007) have reported on the impact of intracultivar variation on several traits, including seed composition, seed weight, maturity, plant height, and lodging. They noted that this remnant variation could be used to select elite individuals from within existing cultivars, and they recently registered a total of 18 lines directly selected from within the cvs Benning, Cook, and Haskell (Fasoula et al., 2007a, 2007b, 2007c).
The recent sequencing of the soybean genome (Schmutz et al., 2010) has enabled the development of genomic tools and methodologies that can address the question of soybean intracultivar heterogeneity in great detail. Williams 82, the sequenced accession, was derived from a composite of four individual plants selected from a Williams × Kingwa BC6F3 generation (Bernard and Cremeens, 1988). This implies that Williams 82 experienced one generation of single-seed descent following the six back-cross generations. Residual heterozygous loci in the BC6F2 generation may have differentially segregated among the four BC6F3 individuals and in subsequent generations. In theory, this process would fix genetic heterogeneity into the Williams 82 population after several rounds of self-pollination. Furthermore, as seed is propagated and distributed throughout the scientific community, founder effects from genetic bottleneck events may give rise to distinct Williams 82 subpopulations at different locations, resulting in disparate Williams 82 lines among researchers.
Detailed genomic comparisons of different Williams 82 individuals should resolve the regions of intracultivar genomic heterogeneity. Further comparisons of each Williams 82 individual with the Williams and Kingwa parents would then trace the ancestry of the heterogeneous regions to either parent; different Williams 82 individuals would match different parents within these regions. Additionally, genomic variation derived de novo after the split of the Williams 82 lineages may also contribute novel heterogeneous loci. In this case, the genomic comparisons of different Williams 82 individuals would still resolve the regions of intracultivar genomic heterogeneity. However, the ancestry of these loci would not specifically trace back to either parent. We would expect to observe novel genomic compositions at such loci.
In this study, we have utilized high-density single nucleotide polymorphism (SNP) genotyping, comparative genomic hybridization (CGH), and exome resequencing data to obtain an unprecedented resolution of the genetic heterogeneity that is extant in Williams 82. The SNP genotyping resolved the parental origins of Williams 82 genetic heterogeneity. Furthermore, the CGH and exon resequencing analyses from more than 203,000 loci revealed the consequences of this heterogeneity in terms of structural and gene content variants between the Williams 82 individuals. Collectively, these findings demonstrate that intracultivar genetic heterogeneity can be pervasive in soybean. Implications on the interpretation of the Williams 82 reference genome and the potential of applying similar approaches to interspecific comparative genomics are discussed.
RESULTS
Origins of Williams 82 Genomic Heterogeneity
In the course of performing preliminary CGH experiments, we noted several soybean cultivars, including Minsoy, Archer, and Williams 82, that showed evidence of structural genomic variation among different individual plants within each cultivar (data not shown). We postulated that the regions of structural variation may have arisen by one of two mechanisms: (1) differential segregation of the parental genetic material among individuals during the breeding process, or (2) variation generated de novo by mutation and genome rearrangements, such as large deletions and DNA transposition. Importantly, these events are molecularly distinguishable. Variation based on differential segregation should be identifiable in the parental lines, while variation generated de novo would be expected to be unique to the cultivar and not preferentially shared with either parent.
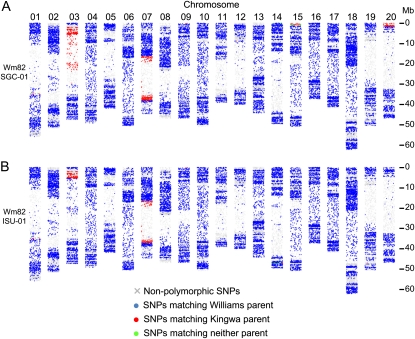
To test these hypotheses and dissect the origin of the Williams 82 genomic heterogeneity, Williams 82 individuals and parental lines were genotyped using the Illumina Infinium iSelect SoySNP50 chip consisting of 44,299 informative SNPs specifically designed for soybean (Fig. 1). We isolated DNA from a single Williams 82 plant from two different seed sources and a single plant each of Williams and Kingwa, the parental lines for Williams 82 (Bernard and Cremeens, 1988). The two Williams 82 individuals were respectively derived from seed stocks held at Iowa State University and the U.S. Department of Agriculture Soybean Germplasm Collection in Urbana, Illinois. These individuals were named Wm82-ISU-01 and Wm82-SGC-01, respectively, for this analysis.
Figure 1.
SNP genotyping reveals the parental origins of Williams 82 genetic heterogeneity. The Infinium SNP genotypes of the Wm82-SGC-01 and Wm82-ISU-01 individuals are shown in A and B, respectively. Blue spots indicate SNP positions that match the Williams genotype. Red spots indicate SNP positions that match the Kingwa genotype. Green spots indicate SNP positions that match neither Williams nor Kingwa. Gray X indicates SNPs that were nonpolymorphic between Wm82, Williams, and Kingwa. Data were jittered along the x axis of each chromosome to better resolve individual data points.
As expected, most of the Williams 82 SNPs match the Williams genotype, with several introgressions derived from Kingwa (Fig. 1). Interestingly, the regions of introgressed Kingwa haplotypes are different for the two Williams 82 individuals. Approximately 52.8 and 24.9 Mb of Kingwa appear to have been introgressed into Wm82-SGC-01 and Wm82-ISU-01, respectively (Fig. 1; Table I).
Table I. Approximate positions of the Kingwa DNA introgression into Wm82-SGC-01 and Wm82-ISU-01.
Regions need to show a minimum of two contiguous Kingwa SNPs to be included in this list.
| Chromosome | Start Position | End Position | Approximate Size |
| kb | |||
| Genotype Wm82-SGC-01 | |||
| Gm01 | 34,668,255 | 35,080,457 | 412 |
| Gm03 | 2,116,324 | 29,593,761 | 27,477 |
| Gm07 | 16,189,219 | 38,036,244 | 21,847 |
| Gm09 | 1 | 313,275 | 313 |
| Gm15 | 1 | 1,007,132 | 1,007 |
| Gm20 | 1 | 1,733,347 | 1,733 |
| Total | 52,789 | ||
| Genotype Wm82-ISU-01 | |||
| Gm01 | 34,668,255 | 35,080,457 | 412 |
| Gm03 | 2,056,210 | 5,356,747 | 3,301 |
| Gm07 | 16,615,822 | 37,107,849 | 20,492 |
| Gm09 | 1 | 313,275 | 313 |
| Gm14 | 47,133,164 | 47,475,993 | 343 |
| Total | 24,861 | ||
Small, conserved Kingwa introgressions are evident in both Wm82-SGC-01 and Wm82-ISU-01 at position approximately 35 Mb on chromosome 1 and the top approximately 300 kb on chromosome 9 (Fig. 1; Table I). The remaining Kingwa introgressions, however, are polymorphic between these individuals. For example, three relatively small introgressions are present in one individual and absent in the other. Wm82-ISU-01 appears to carry an approximately 300-kb introgression at position approximately 47 Mb on chromosome 14; Wm82-SGC-01 does not (Fig. 1; Table I). Conversely, Wm82-SGC-01 appears to carry introgressions on the top approximately 1.0 Mb and approximately 1.7 Mb of chromosomes 15 and 20, respectively; Wm82-ISU-01 does not (Fig. 1; Table I).
Large, polymorphic Kingwa introgressions are evident on chromosomes 3 and 7 (for details, see Supplemental Fig. S1). These introgressions span approximately 28 and 22 Mb, respectively, on chromosomes 3 and 7. However, Wm82-SGC-01 and Wm82-ISU-01 exhibit different borders for these introgressions. The more striking example is on chromosome 3 (Fig. 1; Supplemental Fig. S1). Wm82-SGC-01 chromosome 3 carries an introgression of the Kingwa haplotype from positions approximately 2.0 to 29.6 Mb. Wm82-ISU-01 chromosome 3 carries the Kingwa haplotype from positions approximately 2.0 to 5.4 Mb; the rest of the chromosome matches the Williams parent. This indicates that Wm82-ISU-01 and Wm82-SGC-01 carry different haplotypes for approximately 24 Mb of chromosome 3, which is nearly half of the chromosome.
The large Kingwa introgression on chromosome 7 appears to have slightly different recombination points in Wm82-SGC-01 and Wm82-ISU-01 (Supplemental Fig. S1). The Wm82-SGC-01 introgression spans from approximately 16.2 to 38.0 Mb. The Wm82-ISU-01 introgression spans from approximately 16.6 to 37.1 Mb. Consequently, the Wm82-SGC-01 introgression is approximately 1.5 Mb larger than the Wm82-ISU-01 introgression.
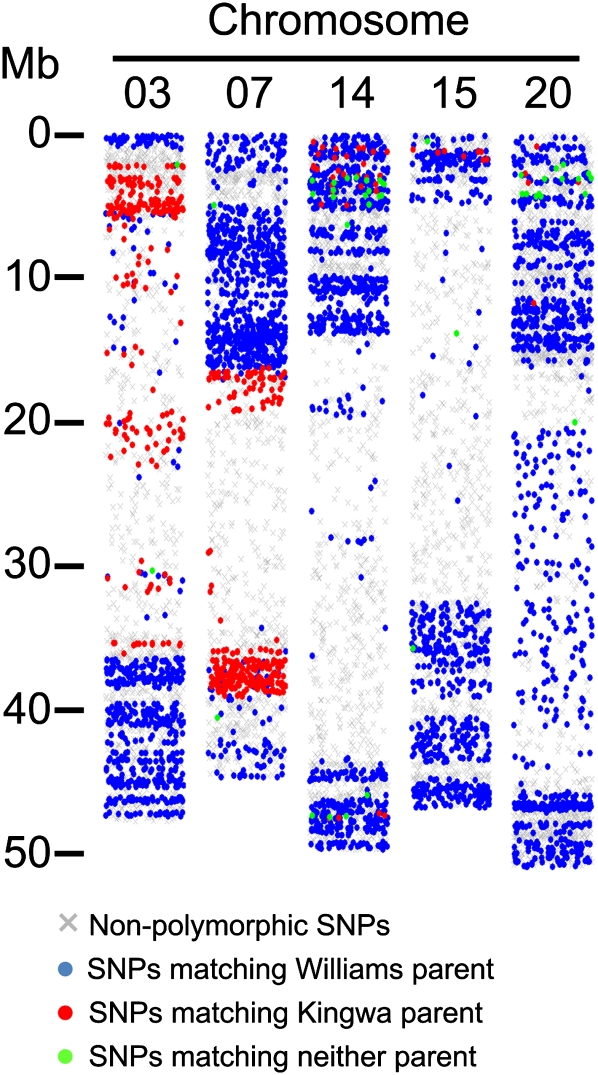
The genomic heterogeneity observed between Wm82-SGC-01 and Wm82-ISU-01 presented an interesting new question: within the regions of heterogeneity, which haplotypes are represented by the published genome sequence (Schmutz et al., 2010)? To address this question, we compared the Williams and Kingwa SNP profiles with the published Williams 82 genome sequence (Fig. 2). The distribution of Williams and Kingwa SNPs appeared to be interspersed with one another throughout the regions of heterogeneity, indicating that these sequences are presumably assembled from a pool of heterogeneous Williams 82 individuals. Figure 2 shows the mixed parentage of SNPs along regions of chromosomes 3, 7, 14, 15, and 20. The large (approximately 4.8 Mb) Williams-Kingwa mosaic at the top of chromosome 14 (Fig. 2) was not identified as an introgressed region in either Wm82-SGC-01 or Wm82-ISU-01. All other noticeable mosaics were identified as Kingwa introgressions in either Wm82-SGC-01 and/or Wm82-ISU-01.
Figure 2.
SNP genotyping reveals the parental origins of the Williams 82 reference sequence. The Infinium genotypes of Williams and Kingwa were compared with the Williams 82 reference sequence to identify which haplotypes are represented in the reference sequence within regions of Wm82 heterogeneity. The genotype of the Williams 82 reference sequence is shown for chromosomes 3, 7, 14, 15, and 20. Blue spots indicate SNP positions that match the Williams genotype. Red spots indicate SNP positions that match the Kingwa genotype. Green spots indicate SNP positions that match neither Williams nor Kingwa. Gray X indicates SNPs that were nonpolymorphic between Wm82, Williams, and Kingwa. Regions of heterogeneity appear to be mosaics of Williams and Kingwa sequences in the Williams 82 reference sequence, as evidenced by the interspersion of blue and red spots throughout the regions of heterogeneity. Data were jittered along the x axis of each chromosome to better resolve individual data points.
Structure of Intercultivar Variation and Intracultivar Genomic Heterogeneity in Soybean
The Williams 82 reference sequence was used to develop a NimbleGen CGH custom microarray. CGH platforms are useful for comparative studies of soybean genomes, particularly the detection of structural variation between different genotypes. Structural variants that are detected between two genomes are commonly referred to as copy number variants (CNV) and are thought to arise from differential duplication, deletion, or insertion of DNA sequences at a given locus. A subclass of CNV, termed presence/absence variants (PAV), describe sequences that are present in one genome but absent in the other (Springer et al., 2009).
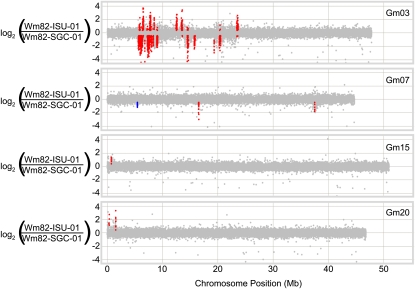
Direct CGH comparisons between Wm82-SGC-01 and Wm82-ISU-01 were conducted to reveal significant CNV within regions of known genetic heterogeneity and to look for possible structural variants generated de novo within or outside of heterogeneous regions. Figure 3 shows the CNV profile of the four chromosomes with greater than 1 Mb of heterogeneous loci. Nearly all of the structural variants observed between these two genotypes occur within known regions of SNP heterogeneity; Supplemental Figure S2 shows the alignment between the region of heterogeneity and the structural variation. Only one significant CNV, on chromosome 7, was located outside of the known regions of heterogeneity (Fig. 3). It is unclear if this small CNV is associated with a small pocket of heterogeneity or was derived de novo since the split of Wm82-SGC-01 and Wm82-ISU-01.
Figure 3.
Structural variation within regions of heterogeneity between Wm82-ISU-01 and Wm82-SGC-01. A detailed view of CNV on chromosomes 3, 7, 15, and 20 reveals major structural polymorphism within known regions of heterogeneity (Fig. 1; Table I). Each data point represents the log2 ratio of the hybridization for a given microarray probe. Colored data points represent probes within significant CNV segments that exceeded the significance threshold value. Red data points are CNV located within known regions of heterogeneity based on SNP genotyping. Blue data points are CNV outside of known regions of heterogeneity. Gray data points indicate probes that are not located in significant segments. All significant CNV are located within known regions of heterogeneity between the genotypes, except for the left-most feature on chromosome 7.
We proceeded to compare the genome structures of Williams and Kingwa to gauge the level of structural variation between the two parent lines. CGH comparisons of the Williams and Kingwa individuals revealed a surprisingly high amount of structural variation throughout the genomes, including instances of significant CNV on all 20 chromosomes and several conspicuous CNV hotspots (Supplemental Fig. 3).
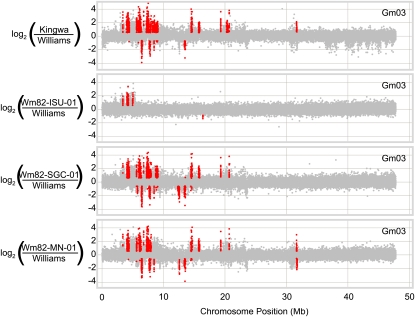
A series of hybridizations were then conducted to determine the origins of the differential CNV profiles of the Wm82 individuals. The Williams genotype was used as the common reference for hybridizations with four different Wm82 individuals: Wm82-SGC-01, Wm82-ISU-01, Wm82-MN-01, and Wm82-PU-01 (for the last two samples, DNA was isolated from a single Williams 82 plant obtained from seed lots at the University of Minnesota and Purdue University, respectively). The results of the chromosome 3 comparisons are shown in Figure 4. The Wm82-SGC-01, Wm82-ISU-01, and Wm82-MN-01 individuals all exhibited differential CNV patterning relative to one another (the Wm82-PU-01 pattern essentially matched the Wm82-SGC-01 pattern). Therefore, these three individuals each exhibit distinct chromosome 3 haplotypes. Importantly, the vast majority of significant CNV observed between Williams and the Wm82 individuals were also observed in the Williams-Kingwa comparison (Fig. 4), indicating that these CNV were directly inherited from the Kingwa introgressions and are structurally unchanged since the original introgression. There were a few regions in which the significant peaks of the Wm82 individuals were not called significant in the Williams-Kingwa comparison; however, these regions (e.g. the DownCNV at position approximately 12.5 Mb in the Wm82-SGC-01 and Wm82-MN-01 comparison with Williams) typically appeared to have similar structural variation patterns in the Williams-Kingwa comparison. For chromosome 3, there is little evidence for significant CNV outside of the introgressed regions or novel structural variation within the introgressed regions that are not observed in the comparison between Kingwa and Williams. Thus, the differential CNV patterning of Wm82-SGC-01, Wm82-ISU-01, and Wm82-MN-01 chromosome 3 appears to be caused by different Kingwa introgressions within these three individuals. The largest of these introgressions appears to be in the Wm82-MN-01 individual (30 Mb or more).
Figure 4.
A detailed view of CNV between Wm82 individuals reveals three distinct structural compositions for chromosome 3 based on differential introgressions from Kingwa. Each data point represents the log2 ratio of the hybridization for a given microarray probe for each genotype versus the Williams reference. In the top panel, CNV were compared between Kingwa and Williams as a reference for differences between the Wm82 parents. Red data points represent probes within significant CNV segments that exceeded the significance threshold value. The other panels display the CNV patterns of Wm82-ISU-01, Wm82-SGC-01, and Wm82-MN-01 versus the Williams reference. For all panels, gray data points indicate probes that are not located in significant segments. (Wm82-PU-01 exhibited a similar chromosome 3 structure to Wm82-SGC-01 and thus is not included here.)
The Wm82-Williams CGH data for all 20 chromosomes are shown in Supplemental Figure S4. CNV within the known regions of introgression match the Kingwa-Williams patterns, as was observed on chromosome 3 (see chromosome 7 in Supplemental Fig. S4). Additionally, there are numerous significant small CNV throughout the genome that are located outside of regions of known introgressions. Several of these small CNV resemble the Kingwa-Williams CNV patterns, indicating that these may be structural variant introgressions that were not represented on the Infinium platform or were too small to be resolved by the SNP introgression analyses. However, a small number of these CNV do not match the Kingwa-Williams CNV patterns (e.g. the UpCNV peak on the end of chromosome 12 in Supplemental Fig. S4). This suggests that these features may be pockets of de novo structural variation. Alternatively, these loci may be heterogeneous within the Williams and/or Kingwa lines, such that the loci inherited by the Wm82 individuals are structurally different from the Williams and Kingwa individuals used in the CGH analyses.
Evidence for Pervasive Presence/Absence Gene Content Variation within Soybean Haplotypes
The SNP genotyping analysis resolved the parental origins of Williams 82 intracultivar heterogeneity. Furthermore, the CGH analysis revealed extensive structural variation associated with this heterogeneity. Therefore, the extensive structural variation between Williams 82 individuals primarily represents the structural variation between the Williams and Kingwa haplotypes, which have been differentially maintained in the respective Williams 82 individuals. Next, we utilized a NimbleGen custom soybean exon-capture microarray to perform exome resequencing of the Wm82-ISU-01 and Wm82-SGC-01 individuals to molecularly validate the fine structure of this variation and investigate any impacts on gene content variation.
We aligned the exome resequencing reads with the soybean genome sequence version 4.1. The data revealed an abundance of SNPs between Wm82-ISU-01 and Wm82-SGC-01. A total of 52,837,460 reads from Wm82-ISU-01 and 38,192,508 reads from Wm82-SGC-01 were uniquely aligned to the reference genome sequence. A total of 1,838 SNPs were found between Wm82-ISU-01 and Wm82-SGC-01 (SNPs that were heterozygous in either genotype are not included in this list). The newly discovered intracultivar SNPs were overwhelmingly located in the genomic regions defined as heterogeneous based on both the CGH and Infinium SNP analyses (Supplemental Table S1). The vast majority (approximately 94%) of the SNPs mapped to chromosomes 3, 7, 15, and 20. The regional distributions of these data are in agreement with the genomic heterogeneity observed between these individuals based on the Infinium SNP genotyping and CGH analyses.
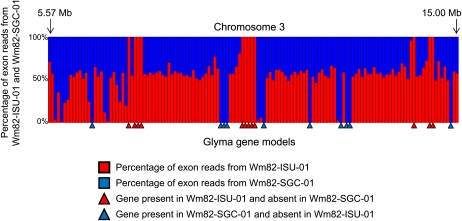
We also defined the gene content variation based on the exome resequencing read counts. Specifically, we aligned the reads with the Glyma version 5.0 annotation file, which defines the exon space of the predicted soybean gene models. The vast majority of genes exhibited similar read counts between Wm82-SGC-01 and Wm82-ISU-01 (Supplemental Table S2). We classified a gene as a putative PAV if we observed a minimum of 30 counts among exons in one individual and zero counts in the other individual. We identified 25 genes that satisfied these stringent criteria (Supplemental Tables S1 and S3); only one of these genes does not reside within a known region of Williams 82 heterogeneity (Table I; Supplemental Table S3). The PAV genes were primarily located within an approximately 10-Mb region of chromosome 3 (22 of 25 genes; Supplemental Table S3). The PAV genes were discontinuously located throughout the region, alternating between present Wm82-ISU-01 and present Wm82-SGC-01 genes or gene clusters (Fig. 5). This region is noteworthy for hosting a relative abundance of Leu-rich repeat genes; approximately 23% (five of 22) of the present-absent gene models within this region were defined as Leu-rich repeat genes (Supplemental Table S3). The exome resequencing data indicate that Williams and Kingwa likely possess extensive PAV gene content variation within their chromosome 3 haplotypes, which is thereby reflected in the gene content variation of heterogeneous Williams 82 individuals.
Figure 5.
Exome resequencing reveals gene content variation between two Williams 82 lines. Genomic DNA for Wm82-ISU-01 and Wm82-SGC-01 was captured on a soybean exome microarray and then sequenced via the Illumina IIX system. The relative frequency of reads matching the soybean Glyma gene models is shown for the two Williams 82 lines; 134 gene models are shown. Colored triangles indicate gene models that exhibited presence in one line and absence (no captured exon reads) in the other line. Nearly 90% of the presence-absence gene content variants identified between Wm82-ISU-01 and Wm82-SGC-01 reside within the 10-Mb region of chromosome 3 shown here.
DISCUSSION
Origin of Intracultivar Genomic Heterogeneity in Williams 82
Soybean is thought to possess relatively limited genetic diversity due to self-fertilization and successive genetic bottleneck events during the course of domestication (Hyten et al., 2006). However, soybean exhibits a wide range of phenotypic variation, including variation observed within established cultivars (Fasoula and Boerma, 2005, 2007). In this study, we used comparative genomics analyses within soybean cv Williams 82 to show substantial genetic heterogeneity between individuals that could be traced to variable Kingwa introgressions. Comparisons of individuals from different Williams 82 seed stocks revealed genomic identity among most chromosomes, with small pockets of variation interspersed. However, certain regions, most notably chromosome 3, displayed extensive SNP and structural heterogeneity between individuals.
Williams 82 was originally released as a composite of four resistant lines selected from a Williams × Kingwa BC6F3 generation (Bernard and Cremeens, 1988). Kingwa was used as the donor parent to introgress Phytophthora root rot resistance into the recurrent parent Williams (Bernard and Cremeens, 1988). Collectively, the SNP and CGH analyses show that Williams 82 intracultivar variation is primarily derived from the segregation and fixation of residual heterozygosity in the BC6F2 generation of Williams × Kingwa. Therefore, the polymorphic regions observed between Williams 82 individuals may be a consequence of heterogeneity between and/or within the four lines originally selected at the BC6F3 stage.
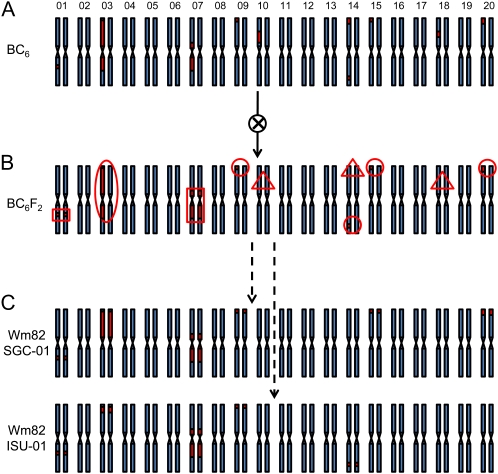
A genetic model of how this may have occurred is shown in Figure 6. One can presume that several small and perhaps large Kingwa introgressions were maintained in the heterozygous state into the BC6 generation. Thereafter, one generation of single-seed descent should have fixed approximately one-half of these loci into the homozygous stage of either Williams or Kingwa origin. However, any heterozygous loci remaining in the BC6F2 generation would be subject to segregation and differential fixation among the four selected BC6F3 lineages.
Figure 6.
A model for the origin of genomic heterogeneity in two Williams 82 lines. A, The Williams × Kingwa BC6 generation, in which contributions from Williams are shown in blue and contributions from Kingwa are shown in red. In this example, 10 loci are heterozygous; the Kingwa Rps1k locus has been selected near the top of chromosome 3. B, The BC6F2 plant after one generation of selfing. In this example, loci that fix the Williams type are shown in red triangles, loci that fix the Kingwa type are shown in red rectangles, and loci that remain heterozygous are shown in red circles. C, The heterozygous loci from the BC6F2 have segregated and fixed homozygosity within each individual plant after several rounds of selfing. The resulting individuals, Wm82-SGC-01 and Wm82-ISU-01, fix heterogeneous types for four of these loci. On chromosome 3, the Rps1k locus is fixed for the Kingwa type in both individuals; however, differential recombination below this locus fixes heterogeneous types for much of the chromosome.
Most of the heterogeneous loci appear to be small genomic regions, with the exception of the large blocks of heterogeneity along chromosomes 3 and the conspicuous approximately 1-Mb regions on chromosomes 7 (differential introgression points; Supplemental Fig. S1), 15, and 20. Interestingly, the Rps1k locus, which confers the Phytophthora root rot resistance, is located approximately at position 4 Mb on chromosome 3 (Gao and Bhattacharyya, 2008). This position is conserved among the Wm82 individuals, as they carry the Kingwa version of this locus. However, this region is adjacent to the strongest region of structural variation (including profound CNV clusters and gene content variation) in the Wm82/Wm82 comparisons. During the series of six back-crosses, the Rps1k locus was necessarily recovered in the heterozygous condition in every generation. Our data indicate that the Rps1k locus remained linked to a large (greater than 30 Mb) genomic region derived from Kingwa, possibly into the BC6F2 generation. Our data suggest that this large Kingwa-derived region recombined and segregated either among the four BC6F3 individuals or in subsequent generations prior to homozygous fixation within each line. This region appears to have recombined into at least three different forms among Williams 82 samples, as Wm82-SGC-01, Wm82-ISU-01, and Wm82-MN-01 all maintain different forms of this chromosome (Fig. 4). It is unknown how many different forms of chromosome 3 (and 7) might be extant in the various stocks of Williams 82 and its derived cultivars, as this has been a popular line utilized in breeding programs (Mikel et al., 2010).
Selection and Intracultivar Heterogeneity
Soybean breeding does not currently utilize a reliable haploid induction system (Ravi and Chan, 2010); therefore, breeding is primarily accomplished by single-seed descent or back-cross strategies. These breeding methods impose selection on plants that maintain variable levels of heterozygosity during the early generations of the breeding cycle. Following the single-seed descent generations, heterozygous loci may segregate, resulting in genetic heterogeneity within a released accession (Fig. 6). Heterozygous loci may be preferentially maintained during the early rounds of breeder selection, as individuals with higher rates of heterozygosity may exhibit greater yields or other advantageous traits due to heterosis (Birchler et al., 2010). Genetic theory predicts, on average, a halving of heterozygous loci with every self-pollination following a given cross. However, heterozygosity may be retained at higher rates if loci confer desirable and selectable phenotypes. In fact, comparative genotyping of maize recombinant inbred lines has identified excess residual heterozygosity maintained in the highly diverse pericentromeric regions (Gore et al., 2009; McMullen et al., 2009), which have been presumably maintained due to phenotypic advantages. Soybean also exhibits heterosis (Palmer et al., 2001; Burton and Brownie, 2006); thus, early selections during the breeding process may preferentially maintain lines with greater heterozygosity, as these lines would exhibit phenotypic superiority. Preferential maintenance of heterozygous loci would result in more segregating loci during the seed increase generations, ultimately leading to greater than expected rates of intracultivar heterogeneity among individuals.
Selective advantages of heterozygosity can theoretically increase the likelihood of establishing heterogeneity during the breeding process; however, there are also possible advantages to maintaining the genetic heterogeneity once the cultivar is established. For example, increased genetic diversity within a cultivar may stabilize a stand against pathogen invasion or spread (Burdon et al., 2006).
Clearly, the genetic heterogeneity on Williams 82 chromosome 3 was influenced by the heterozygous selection of the Rps1k locus during the back-crossing process. However, it is unclear if the genetic heterogeneity observed elsewhere in the genome was influenced by selective advantages of heterozygous loci during the backcross or single-seed descent generations. Excluding the chromosome 3 introgressions, both Wm82-SGC-01 and Wm82-ISU-01 maintained substantial Kingwa introgressions, particularly the large introgression on chromosome 7. The sample size used in this study is too small to allow for speculation on how common this type of donor retention is in soybean breeding. However, it remains an intriguing question whether introgressed loci are typically retained at higher than expected rates during traditional soybean back-cross breeding.
Implications for Soybean Comparative Genomics
The high rates of intracultivar structural variation observed within the Williams 82 regions of heterogeneity are primarily a consequence of structural variation between the Williams and Kingwa parental lines (Fig. 4; Supplemental Fig. S4). Presumably, the gene content variation within these regions is also directly inherited from these parental lines.
The relatively high levels of structural variation within regions of Williams 82 heterogeneity appear to be somewhat representative of the genome-wide structural variation observed between the Williams and Kingwa parents (Supplemental Fig. S3). The Williams-Kingwa CGH comparison indicates that there may be a great deal of genomic variation between soybean lines, more so than is generally assumed. Moreover, this variation may include substantial differences in gene structure and gene content, as was observed in the gene presence/absence variation analysis of the exome resequencing data in this study.
In addition to the 25 genes we defined as PAV in this analysis, there were also several genes that were nearly called PAV. In these cases, one Williams 82 individual exhibited an abundance of read counts while the other individual exhibited trace levels of read counts. Seven such genes, each exhibiting greater than 95% of their reads from either Wm82-SGC-01 or Wm82-ISU-01, are observed in Figure 5. The trace read counts could be explained by technical or biological causes, including exon reshuffling, gene truncation, and resequencing misalignments to the reference genome. Additionally, our analysis only focused on the gene set defined by the annotation of the published soybean genome sequence; there are possibly additional PAV genes that are missing from the genome sequence, not identified by the current annotation, or not successfully captured by the exome microarray. Collectively, these data suggest that the true number of PAV and rearranged genes between Wm82-SGC-01 and Wm82-ISU-01 may be substantially greater than the 25 genes we identified using the stringent criteria described above.
Gene content differences in the form of PAV have been extensively documented in maize inbred line comparisons, and there is speculation that these may significantly contribute to maize phenotypic variation (Springer et al., 2009; Beló et al., 2010; Swanson-Wagner et al., 2010). Likewise, it will be critical to evaluate the extent and consequences of such structural variation and presence/absence gene variants in providing phenotypic plasticity in soybean lines and breeding populations. Although there were no obvious phenotypic dissimilarities between the Wm82 individuals in this study, there was an apparent enrichment of Leu-rich repeat annotations observed within the Wm82 PAV genes. Comparative sequencing of maize inbred lines recently identified an abundance of Leu-rich repeat genes with large-effect SNPs (Lai et al., 2010). No transcription factors were identified within the Wm82 PAV gene list; it may be of great interest to identify and investigate the effect of transcription factor PAV between soybean cultivars.
Implications for the Williams 82 Genome Sequence
The vast majority of the Williams 82 genome appears to be homogeneous among different Williams 82 individuals and subpopulations. However, a comparison of the Williams and Kingwa SNP genotypes with the reference soybean genome sequence (Schmutz et al., 2010) revealed a surprising result: within regions of genetic heterogeneity, the reference sequences consist of a mosaic of the Williams and Kingwa haplotypes. We assume that no Williams 82 individual plant will match the soybean reference genome sequence throughout these regions of heterogeneity. Therefore, researchers investigating comparative studies of soybean that include Williams 82 as a reference genotype must factor in the inherent differences between each Wm82 individual and the reference genome sequence.
For example, the CGH tiling microarray used in this study was designed based on the reference genome sequence. Ideally, one would hope to identify a Wm82 individual that is a perfect match to the soybean genome sequence; such an individual would be useful as a common reference in CGH experiments. If this were the case, the interpretation of UpCNV and DownCNV would be relatively straightforward: UpCNV peaks would indicate increased copy number relative to Williams 82, and DownCNV peaks would indicate an absent or polymorphic sequence relative to Williams 82. Applying a reference Wm82 individual that is polymorphic to the CGH microarray is a slightly different matter: the interpretation of UpCNV and DownCNV will be the same as described above for chromosomal regions that are not heterogeneous between the genome sequence and the Wm82 individual (this, fortunately, is the case for the majority of the genome sequence). However, within the regions of known heterogeneity, UpCNV would now have to be interpreted as either an increased copy number relative to Williams 82 or sequences that are absent in the particular Wm82 individual that was used as a reference for the given experiment. We imagine that similar considerations will need to be made for a variety of comparative methodologies and platforms (e.g. interpretations of RNA-Seq data, analysis of the Affymetrix soybean GeneChip, etc.), including phenotypic characters in which Williams 82 serves as the experimental control.
Similar circumstances may apply to the utility of other plant genome sequences. Several presumably homogenous accessions were used as the DNA source for the genome sequences of Arabidopsis (Arabidopsis Genome Initiative, 2000), rice (International Rice Genome Sequencing Project, 2005), maize (Schnable et al., 2009), and other species. To our knowledge, it is not known if there is persistent genetic heterogeneity within the respective sequenced accessions, nor is it known whether a single individual or a pool of heterogeneous individuals was used to construct the sequence assemblies for each species. Nevertheless, for future genome sequencing projects, it would be advisable to sequence the genome of a single individual (perhaps a double haploid individual when possible). It would also be preferable that seeds or clones from the reference individual be stored in a repository, so that they could be used in future experiments and analyses.
MATERIALS AND METHODS
Plant Material and Nucleic Acid Extraction
Seed for soybean (Glycine max ‘Williams 82’) was obtained from the laboratories of Dr. James Orf at the University of Minnesota, Dr. Randy Shoemaker at Iowa State University, Dr. Scott Jackson at Purdue University, and the U.S. Department of Agriculture Soybean Germplasm Collection in Urbana, Illinois. Seed for cv Williams and Kingwa was obtained from the U.S. Department of Agriculture Soybean Germplasm Collection.
Seeds were planted in individual four-inch pots containing a 50:50 mix of sterilized soil and Metro Mix. Growth chambers contained a mixture of fluorescent and incandescent light bulbs set to 16 h of light per day. Young trifoliate leaves from 3-week-old plants were harvested and immediately frozen in liquid nitrogen. Frozen leaf tissue was ground with a mortar and pestle with liquid nitrogen. DNA was extracted from approximately 100 mg of powderized tissue using the Qiagen Plant DNeasy Mini Kit according to the manufacturer’s protocol (including an RNase degradation step). DNA was quantified on a NanoDrop spectrophotometer.
Illumina Infinium Genotyping
The Illumina Infinium iSelect SoySNP50 chip (Q. Song, C.V. Quigley, G. Jia, P.B. Cregan, and D.L. Hyten, unpublished data) was used to obtain genotyping data for four individual plants: Wm82-SGC-01, Wm82-ISU-01, Williams, and Kingwa. Illumina GenomeStudioV2010.2 software was used to identify polymorphic SNPs among samples in both the homozygous and heterozygous states; only homozygous calls were used for this analysis. Any ambiguous or otherwise uninformative data points were not used in this analysis. Visual displays showing the distribution of Williams and Kingwa contributions to the Wm82 lines were generated using Spotfire DecisionSite software.
Comparative Genomic Hybridizations and Analyses
A 696,139-feature oligonucleotide microarray was designed and built by Roche NimbleGen to perform soybean array comparative genome hybridization. Unique probes of varying lengths (maximum 75 bp, minimum 50 bp, median 55 bp) were designed based on the soybean genome version 4.0 assembly (Schmutz et al., 2010). Probes were spaced at a median interval length of 1,120 bp across the entire anchored genome. This array may be ordered from Roche NimbleGen by requesting the design 091113_Gmax_RS_CGH_HX3.
Total genomic DNA (isolated as described above) was labeled according to the Roche NimbleGen CGH User’s Guide (version 5.1). Briefly, 1 μg of genomic DNA was labeled with either Cy3- or Cy5-labeled random nonamers via incubation with exo-Klenow enzyme and 10 mm deoxyribonucleotide triphosphates at 37°C for 2 h in a total volume of 100 μL. Labeling reactions were stopped with the addition of 10 μL of 0.5 m EDTA and 11.5 μL of 5 m NaCl. DNA was precipitated with 0.9 volumes of isopropanol, washed with 500 μL of ice-cold 80% ethanol, and dried in an Eppendorf Vacufuge on low heat for 5 to 10 min. The labeled samples were resuspended in 25 μL of nuclease-free water and quantified on a NanoDrop spectrophotometer. Thirty-one micrograms each of the Cy3- and Cy5-labeled samples were combined in a 1.5-mL tube and dried in a Centri-Vac. Samples were resuspended in 5.6 μL of sample tracking control and 14.4 μL of hybridization solution supplied by Roche NimbleGen. Samples were heat denatured at 95°C for 5 min, followed by incubation at 42°C for at least 5 min prior to loading. Samples were hybridized to the arrays for 60 to 72 h at 42°C with mixing. Microarrays were washed with Roche NimbleGen wash buffers and dried by centrifugation. Arrays were scanned with a GenePix4000B scanner (Axon Instruments) at 5-μm resolution. Automated image gridding, alignment, and data extraction were performed using NimbleScan software version 2.5.
For each CGH comparison, the segMNT algorithm in the NimbleScan software (version 2.5) was used to extract the raw data and make segmentation calls. The parameters of the algorithm were as follows: minimum segment difference = 0.1, minimum segment length (number of probes) = 2, acceptance percentile = 0.99, number of permutations = 10, nonunique probes were included, and spatial correction and qspline normalization were applied. The list of resulting segments was then processed to identify significant segments. A segment was called significant if the log2 ratio mean of the probes within the segment was above the upper threshold or below the lower threshold for that given array comparison. The upper threshold for each comparison was determined to be the log2 ratio value of the 95th percentile of all data points. The lower threshold for each comparison was determined to be the log2 ratio value of the 5th percentile of all data points. Visual displays of the CGH data were generated using Spotfire DecisionSite software.
The comparative genomic hybridization data from this study have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE25294.
Exome Resequencing and Analyses
Wm82-ISU-01 and Wm82-SGC-01 genomic DNA samples were isolated using the Qiagen Plant DNeasy system. Illumina Paired End libraries (Illumina) were constructed for Wm82-ISU-01 and Wm82-SGC-01 using Illumina’s PE Kit (part no. PE-102-1001). The mean library fragment size was found to be 328 bp. The details of library preparation and prehybridization amplification are provided in Supplemental Materials and Methods S1.
A custom microarray was built for soybean exome sequence capture based on the soybean gene annotation (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v4.0/Gmax/annotation/initialRelease/Glyma1.gff2.gz). This array may be ordered from Roche NimbleGen by requesting design 100310_Gmax_public_exome_cap_HX3. The details of this microarray design are included in Supplemental Materials and Methods S1. The library exon sequences were captured by hybridizing to the microarray for 72 h at 42°C in the presence of 20 μL of plant capture enhancer per subarray. Slide washing and sample library elution were performed as published previously (Fu et al., 2010). Posthybridization amplification consisted of 16 cycles of PCR. Following the completion of the amplification reaction, the samples were purified using a Qiagen Qiaquick column using the manufacturer’s recommended protocol, and the DNA was quantified spectrophotometrically using the NanoDrop-1000 and electrophoretically evaluated with an Agilent Bioanalyzer 2100 using a DNA1000 chip. The resulting postcapture enriched sequencing libraries were diluted to 10 nm and used in cluster formation on an Illumina cBot, and paired-end sequencing was done using Illumina’s Genome Analyzer IIX. Both cluster formation and 76-bp paired-end sequencing were performed using the Illumina protocols. The full methodology of library capture, capture array processing, posthybridization amplification, and sequencing are described in Supplemental Materials and Methods S1.
Software SOAP2 (Li et al., 2009b) and SOAPsnp (Li et al., 2009a) were used for SNP discovery between the Wm82-ISU-01 and Wm82-SGC-01 exon reads. A customized pipeline was developed for this analysis (Severin et al., 2010). Briefly, a total of 56,010,828 76-base read sequences of Wm82-ISU-01 and 40,599,004 reads of Wm82-SGC-01 from Illumina Solexa sequencing were aligned to the soybean genome sequence version 4.1 (Gmax.main_genome.scaffolds assembly; ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v4.1/Gmax/assembly/sequences/) using SOAP2. The paired alignment was set to a maximum mismatch of two, and only the unique alignment hits were selected. After imposing these filters, 52,837,460 reads from Wm82-ISU-01 and 38,192,508 reads from Wm82-SGC-01 were uniquely aligned to the reference genome sequence. The alignments were screened for SNPs by SOAPsnp analyses; we only screened for SNPs that were homozygous in both genotypes. Potential SNPs were selected using a minimum base-call quality of 10, average quality of 20, and minimum best hits of four. The SNP was not allowed to be an ambiguous base (e.g. SNP ≠ ≢“N”).
We used the exome resequencing data to identify gene content variation between Wm82-ISU-01 and Wm82-SGC-01 based on read counts. The Glyma version 5.0 annotation file, Glyma1_highConfidence.gff3 (February 8, 2010), was used for exon annotations. A perl script was designed to count the number of paired-end reads that mapped to each exon. To identify genes that are present in one Williams 82 line and absent in the other (present-absent genes), we first summed the number of reads among exons for each Glyma gene model. Genes were categorized as “present-absent” if they had a minimum of 30 reads in one genotype and zero in the other.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SNP genotyping reveals the fine structure of differential Kingwa introgression in Wm82-SGC-01 and Wm82-ISU-01 on chromosomes 3 (A) and 7 (B).
Supplemental Figure S2. Structural variation between Wm82-SGC-01 and Wm82-ISU-01 corresponds to regions of heterogeneity.
Supplemental Figure S3. Genome-wide CGH analysis reveals extensive copy number variations between the Kingwa and Williams genotypes.
Supplemental Figure S4. Structural variation between different Wm82 individuals and Williams.
Supplemental Table S1. Chromosomal abundance of nucleotide and gene content variants between Wm82-ISU-01 and Wm82-SGC-01 based on exome resequencing.
Supplemental Table S2. Resequencing read counts following exome capture for 43,442 Glyma gene models.
Supplemental Table S3. Presence-absence genes between Wm82-ISU-01 and Wm82-SGC-01 based on exon resequencing counts.
Supplemental Materials and Methods S1. A detailed description of the exome resequencing methods.
Supplementary Material
Acknowledgments
We are grateful to Drs. Jim Orf, Randy Shoemaker, Scott Jackson, and the U.S. Department of Agriculture-Agricultural Research Service Soybean Germplasm Collection for providing the seeds used in this study. We thank Ruth Swanson-Wagner for CGH training and support. We thank Dawn Green and Tracy Millard for sequencing support.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Beló A, Beatty MK, Hondred D, Fengler KA, Li B, Rafalski A. (2010) Allelic genome structural variations in maize detected by array comparative genome hybridization. Theor Appl Genet 120: 355–367 [DOI] [PubMed] [Google Scholar]
- Bernard RL, Cremeens CR. (1988) Registration of Williams 82 soybean. Crop Sci 28: 1027–1028 [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA. (2010) Heterosis. Plant Cell 22: 2105–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon JJ, Thrall PH, Ericson AL. (2006) The current and future dynamics of disease in plant communities. Annu Rev Phytopathol 44: 19–39 [DOI] [PubMed] [Google Scholar]
- Burton JW, Brownie C. (2006) Heterosis and inbreeding depression in two soybean single crosses. Crop Sci 46: 2643–2648 [Google Scholar]
- Byth DE, Weber CR. (1968) Effects of genetic heterogeneity within two soybean populations. I. Variability within environments and stability across environments. Crop Sci 8: 44–47 [Google Scholar]
- DeBolt S. (2010) Copy number variation shapes genome diversity in Arabidopsis over immediate family generational scales. Genome Biol Evol 2: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasoula VA, Boerma HR. (2005) Divergent selection at ultra-low plant density for seed protein and oil content within soybean cultivars. Field Crops Res 91: 217–229 [Google Scholar]
- Fasoula VA, Boerma HR. (2007) Intra-cultivar variation for seed weight and other agronomic traits within three elite soybean cultivars. Crop Sci 47: 367–373 [Google Scholar]
- Fasoula VA, Boerma HR, Yates JL, Walker DR, Finnerty SL, Rowan GB, Wood ED. (2007a) Registration of five soybean germplasm lines selected within the cultivar ‘Benning’ differing in seed and agronomic traits. J Plant Regist 1: 156–157 [Google Scholar]
- Fasoula VA, Boerma HR, Yates JL, Walker DR, Finnerty SL, Rowan GB, Wood ED. (2007b) Registration of seven soybean germplasm lines selected within the cultivar ‘Cook’ differing in seed and agronomic traits. J Plant Regist 1: 158–159 [Google Scholar]
- Fasoula VA, Boerma HR, Yates JL, Walker DR, Finnerty SL, Rowan GB, Wood ED. (2007c) Registration of six soybean germplasm lines selected within the cultivar ‘Haskell’ differing in seed and agronomic traits. J Plant Regist 1: 160–161 [Google Scholar]
- Fu Y, Springer NM, Gerhardt DJ, Ying K, Yeh CT, Wu W, Swanson-Wagner R, D’Ascenzo M, Millard T, Freeberg L, et al. (2010) Repeat subtraction-mediated sequence capture from a complex genome. Plant J 62: 898–909 [DOI] [PubMed] [Google Scholar]
- Gao H, Bhattacharyya MK. (2008) The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences. BMC Plant Biol 8: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gethi JG, Labate JA, Lamkey KR, Smith ME, Kresovich S. (2002) SSR variation in important US maize inbred lines. Crop Sci 42: 951–957 [Google Scholar]
- Giarrocco LE, Marassi MA, Salerno GL. (2007) Assessment of the genetic diversity in Argentine rice cultivars with SSR markers. Crop Sci 47: 853–860 [Google Scholar]
- Gordon IL, Byth DE. (1972) Comparisons among strains of the tobacco cultivar Hicks illustrating variability within a single cultivar. Queensl J Agric Anim Sci 29: 255–264 [Google Scholar]
- Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, et al. (2009) A first-generation haplotype map of maize. Science 326: 1115–1117 [DOI] [PubMed] [Google Scholar]
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. (2008) The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet 4: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs RL, Russell WA. (1968) Genetic variation in quantitative characters in maize inbred lines. I. Variation among and within Corn Belt seed sources of six inbreds. Crop Sci 8: 345–348 [Google Scholar]
- Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB. (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA 103: 16666–16671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Lai J, Li R, Xu X, Jin W, Xu M, Zhao H, Xiang Z, Song W, Ying K, Zhang M, et al. (2010) Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42: 1027–1030 [DOI] [PubMed] [Google Scholar]
- Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, Wang J. (2009a) SNP detection for massively parallel whole-genome resequencing. Genome Res 19: 1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. (2009b) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, et al. (2009) Genetic properties of the maize nested association mapping population. Science 325: 737–740 [DOI] [PubMed] [Google Scholar]
- Mikel MA, Diers BW, Nelson RL, Smith HH. (2010) Genetic diversity and agronomic improvement of North American soybean germplasm. Crop Sci 50: 1219–1229 [Google Scholar]
- Olufowote JO, Xu Y, Chen X, Park WD, Beachell HM, Dilday RH, Goto M, McCouch SR. (1997) Comparative evaluation of within-cultivar variation of rice (Oryza sativa L.) using microsatellite and RFLP markers. Genome 40: 370–378 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M. (2010) The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RG, Gai J, Sun H, Burton JW. (2001) Production and evaluation of hybrid soybean. Plant Breed Rev 21: 263–307 [Google Scholar]
- Rasmusson DC, Phillips RL. (1997) Plant breeding progress and genetic diversity from de novo variation and elevated epistasis. Crop Sci 37: 303–310 [Google Scholar]
- Ravi M, Chan SW. (2010) Haploid plants produced by centromere-mediated genome elimination. Nature 464: 615–618 [DOI] [PubMed] [Google Scholar]
- Röder MS, Wendehake K, Korzun V, Bredemeijer G, Laborie D, Bertrand L, Isaac P, Rendell S, Jackson J, Cooke RJ, et al. (2002) Construction and analysis of a microsatellite-based database of European wheat varieties. Theor Appl Genet 106: 67–73 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. (2009) Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet 5: e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin AJ, Peiffer GA, Xu WW, Hyten DL, Bucciarelli B, O’Rourke JA, Bolon YT, Grant D, Farmer AD, May GD, et al. (2010) An integrative approach to genomic introgression mapping. Plant Physiol 154: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RG, Byers DL, Darmo E. (2000) Spontaneous mutational effects on reproductive traits of Arabidopsis thaliana. Genetics 155: 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjakste TG, Rashal I, Röder MS. (2003) Inheritance of microsatellite alleles in pedigrees of Latvian barley varieties and related European ancestors. Theor Appl Genet 106: 539–549 [DOI] [PubMed] [Google Scholar]
- Soleimani VD, Baum BR, Johnson DA. (2005) Genetic diversity among barley cultivars assessed by sequence-specific amplification polymorphism. Theor Appl Genet 110: 1290–1300 [DOI] [PubMed] [Google Scholar]
- Springer NM, Ying K, Fu Y, Ji T, Yeh CT, Jia Y, Wu W, Richmond T, Kitzman J, Rosenbaum H, et al. (2009) Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet 5: e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Eichten SR, Kumari S, Tiffin P, Stein JC, Ware D, Springer NM. (2010) Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res 20: 1689–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokatlidis IS. (2000) Variation within maize lines and hybrids in the absence of competition and relation between hybrid potential yield per plant with line traits. J Agric Sci 134: 391–398 [Google Scholar]
- Tokatlidis IS, Tsialtas JT, Xynias IN, Tamoutsidis E, Irakli M. (2004) Variation within a bread wheat cultivar for grain yield, protein content, carbon isotope discrimination and ash content. Field Crops Res 86: 33–42 [Google Scholar]
- Tokatlidis IS, Tsikrikoni C, Tsialtas JT, Lithourgidis AS, Bebeli PJ. (2008) Variability within cotton cultivars for yield, fibre quality and physiological traits. J Agric Sci 146: 483–490 [Google Scholar]
- Zhang YX, Gentzbittel L, Vear F, Nicolas P. (1995) Assessment of inter- and intra-inbred line variability in sunflower (Helianthus annuus) by RFLPs. Genome 38: 1040–1048 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.