Abstract
The SLEEPY1 (SLY1) F-box gene is a positive regulator of gibberellin (GA) signaling in Arabidopsis (Arabidopsis thaliana). Loss of SLY1 results in GA-insensitive phenotypes including dwarfism, reduced fertility, delayed flowering, and increased seed dormancy. These sly1 phenotypes are partially rescued by overexpression of the SLY1 homolog SNEEZY (SNE)/SLY2, suggesting that SNE can functionally replace SLY1. GA responses are repressed by DELLA family proteins. GA relieves DELLA repression when the SCFSLY1 (for Skp1, Cullin, F-box) E3 ubiquitin ligase ubiquitinates DELLA protein, thereby targeting it for proteolysis. Coimmunoprecipitation experiments using constitutively expressed 35S:hemagglutinin (HA)-SLY1 and 35S:HA-SNE translational fusions in the sly1-10 background suggest that SNE can function similarly to SLY1 in GA signaling. Like HA-SLY1, HA-SNE interacted with the CULLIN1 subunit of the SCF complex, and this interaction required the F-box domain. Like HA-SLY1, HA-SNE coimmunoprecipitated with the DELLA REPRESSOR OF GA1-3 (RGA), and this interaction required the SLY1 or SNE carboxyl-terminal domain. Whereas HA-SLY1 overexpression resulted in a decrease in both DELLA RGA and RGA-LIKE2 (RGL2) protein levels, HA-SNE caused a decrease in DELLA RGA but not in RGL2 levels. This suggests that one reason HA-SLY1 is able to effect a stronger rescue of sly1-10 phenotypes than HA-SNE is because SLY1 regulates a broader spectrum of DELLA proteins. The FLAG-SLY1 fusion protein was found to coimmunoprecipitate with the GA receptor HA-GA-INSENSITIVE DWARF1b (GID1b), supporting the model that SLY1 regulates DELLA through interaction with the DELLA-GA-GID1 complex.
This study examines the roles of the SLEEPY1 (SLY1) F-box gene and its homolog SNEEZY (SNE)/SLY2 in GA hormone signaling. GA is required for several important transitions in Arabidopsis (Arabidopsis thaliana) development, including seed germination, stem elongation, fertility, and the transition to flowering (for review, see Sun and Gubler, 2004; Ueguchi-Tanaka et al., 2007; Aya et al., 2009). GA is also required for normal fertility and flower development and plays an important role in adaptations to cold, drought, and anoxia (Achard et al., 2006, 2008; Fukao and Bailey-Serres, 2008). During the green revolution, GA-insensitive semidwarf mutations in the DELLA genes provided resistance to lodging as well as increased yield for biomass (Allan, 1986). The mechanisms of GA signaling are highly conserved between Arabidopsis, rice (Oryza sativa), barley (Hordeum vulgare), and tomato (Solanum lycopersicum; for review, see Sun and Gubler, 2004; Jasinski et al., 2008). Thus, elucidating the fundamental mechanisms of GA signaling will be important in developing future strategies for crop improvement.
Previous research has shown that GA stimulates GA responses through destruction or deactivation of DELLA repressors of GA responses (McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004). The DELLA gene family is a subset of the GRAS family of putative transcription factors defined by the presence of a conserved N-terminal DELLA regulatory domain and a C-terminal GRAS functional domain. The term DELLA refers to the signature conserved amino acid sequence Asp-Glu-Leu-Leu-Ala (D-E-L-L-A). There are five DELLA genes in Arabidopsis with partly overlapping functions defined based on the capacity of each DELLA mutation to suppress the phenotypes of the severe GA biosynthesis mutant ga1-3. DELLAs REPRESSOR OF GA1-3 (RGA) and GA-INSENSITIVE (GAI) are the main DELLA genes repressing stem elongation (Dill and Sun, 2001), but the DELLA RGA-LIKE1 (RGL1) also contributes (Wen and Chang, 2002). DELLAs RGA, RGL2, and RGL1 repress flowering and fertility (Cheng et al., 2004). Finally, the DELLA RGL2 is the main repressor of seed germination (Lee et al., 2002; Peng and Harberd, 2002; Tyler et al., 2004; Ariizumi and Steber, 2007), although DELLAs RGA, GAI, and RGL3 also function in seed germination (Cao et al., 2005; Piskurewicz and Lopez-Molina, 2009). The resemblance of the GRAS domain to STAT transcription factors and the fact that DELLA proteins localize to the nucleus initially suggested that DELLAs might function in transcriptional control. Chromatin immunoprecipitation experiments showed that DELLA RGA localizes to promoter elements and appears to activate the expression of downstream negative regulators of GA responses (Zentella et al., 2007). Nevertheless, Zentella et al. (2007) suggested that the enrichment for DELLA RGA at these promoters is weak (2- to 3.5-fold), because DELLA appears to interact indirectly with these promoters rather than through protein-protein interaction. DELLA also appears to repress hypocotyl elongation in the dark by direct protein-protein interaction with the phytochrome-interacting factors PIF3 and PIF4 (de Lucas et al., 2008; Feng et al., 2008). DELLA binding prevents PIF3 and PIF4 transcriptional activators from binding to their promoter elements, thus blocking their transcription. Thus, it appears that DELLA may repress GA responses both with and without association with promoter elements.
The SCFSLY1 E3 ubiquitin ligase complex lifts DELLA repression by targeting DELLA for destruction by the ubiquitin-proteasome pathway. The SCF complex is composed of a Skp1 homolog termed ASK (for Arabidopsis Skp1), a Cullin homolog, an Rbx1 homolog, and an F-box protein that binds a specific target. There are 23 ASK genes, five Cullins (CULs), two Rbx1 homologs, and 694 F-box proteins in Arabidopsis (Gagne et al., 2002; Gray et al., 2002; Risseeuw et al., 2003). The F-box protein binds to the Skp1 homolog of the complex through direct protein-protein interaction via the F-box domain. The F-box protein generally binds the substrate protein via the C-terminal domain. Yeast two-hybrid data indicate that the SLY1 protein can bind to DELLA protein via the C-terminal domain (Dill et al., 2004; Fu et al., 2004). GA binding stimulates the ability of the GA receptor GA-INSENSITIVE DWARF1 (GID1) to bind DELLA proteins. Based on yeast three-hybrid data, Arabidopsis GID1-GA binding to DELLA appears to increase the affinity of the F-box protein SLY1 for DELLA (Griffiths et al., 2006). Thus, GA stimulates SCFSLY1 binding to DELLA proteins, thereby allowing SCFSLY1 to catalyze the polyubiquitination of DELLA protein. Addition of four ubiquitin moieties to a target protein triggers its recognition and proteolysis by the 26S proteasome (Smalle and Vierstra, 2004). It appears that DELLA is destroyed via the ubiquitin-proteasome pathway, because both mutations in SLY1 and 26S proteasome inhibitors result in stabilization and increased accumulation of DELLA protein in the presence of GA (McGinnis et al., 2003; Dill et al., 2004; Fu et al., 2004).
We first identified MIF21.6 as a homolog of SLY1 in Arabidopsis by BLAST database search (At5g48170; McGinnis et al., 2003), and this predicted protein was subsequently named SNE (Strader et al., 2004) or SLY2 (Fu et al., 2004). Functional analysis showed that SNE/SLY2 overexpression can rescue the sly1 mutant phenotype (Fu et al., 2004; Strader et al., 2004). We chose to refer to this gene as SNE rather than as SLY2 because it is not yet clear whether SNE/SLY2 normally shows functional overlap with SLY1. Moreover, it is still possible that SNE/SLY2 may have some functions that do not overlap with those of SLY1. This study examines the relative roles of the two F-box proteins, SLY1 and SNE/SLY2, first through a careful analysis of their ability to complement the sly1-10 mutation, then by determining which domains are required for function, and finally by examining the ability of these proteins to bind to other proteins involved in GA signaling, including the CUL1 subunit of the SCF complex and the DELLA protein RGA in Arabidopsis. The ability of SNE to regulate the accumulation of DELLAs RGA and RGL2 is examined. Finally, we established the interaction of the SLY1 protein with the GA receptor GID1 through coimmunoprecipitation.
RESULTS
Sequence Homology of SLY1 and SNE/SLY2
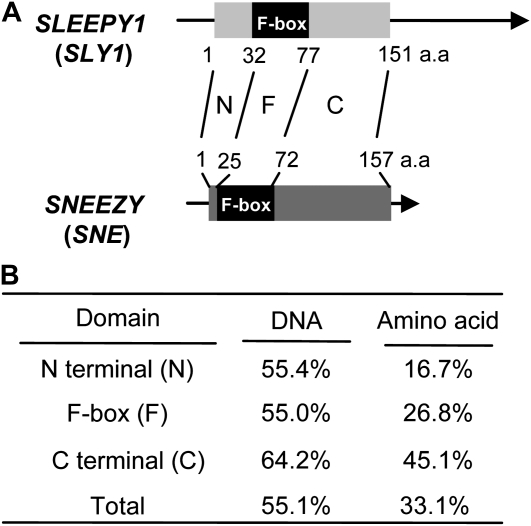
The homology of SLY1 to SNE is diagrammed in Figure 1 at both the level of mRNA and predicted protein sequence. The 453-bp SLY1 gene encodes a predicted 151-amino acid protein, whereas the 471-bp SLY2/SNE open reading frame (ORF) encodes a predicted protein of 157 amino acids. Both gene sequences contain no introns (McGinnis et al., 2003). Based on InterPro motif analysis (http://www.ebi.ac.uk/Tools/InterProScan/; Hunter et al., 2009), the predicted SNE protein contains an F-box motif composed of amino acids 25 to 72. The presence of this functional domain suggests that SNE encodes an F-box subunit protein of an SCF E3 ubiquitin ligase (Fig. 1; Supplemental Fig. S1). The homology of SLY1 and SNE was considered by dividing the proteins into three domains: the F-box domain, the N-terminal domain, and the C-terminal domain. Previously published yeast two-hybrid studies indicated that the C-terminal domain of SLY1 is required for interaction with DELLA proteins (Dill et al., 2004; Fu et al., 2004). The full-length SLY1 and SNE genes have 55.1% DNA and 33.1% amino acid sequence homology. The N-terminal domains before the F-box domain (SLY1, amino acids 1–32; SNE, 1–24) have 55.4% DNA/16.7% amino acid homology; the F-box domain itself (SLY1, amino acids 32–77; SNE, 25–72) has 55.0% DNA/26.8% amino acid homology; and the C-terminal domain (SLY, amino acids 78–151; SNE, 73–157) has 64.2% DNA/45.1% amino acid homology. Thus, the C-terminal region, which is believed to be involved in SLY1 interaction with its DELLA target protein, contains the highest degree of sequence homology with SNE (Fig. 1B).
Figure 1.
Domain structure and homology of the SLY1 and SNE genes and predicted protein sequences. A, Gene structure of two F-box proteins, SLY1 and SNE, consisting of N-terminal (N), F-box (F), and C-terminal (C) domains. The number below the SLY1 and above the SNE gene shows the amino acid (a.a.) number, where the first amino acid is +1. B, DNA and amino acid homology between SLY1 and SNE in the N-terminal, F-box, and C-terminal domains.
Functional Analysis of SLY1 and Its Homolog SNE in Arabidopsis
Previous research showed that the SNE gene sequence was able to partly suppress the sly1 mutant phenotypes when expressed under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter (Fu et al., 2004; Strader et al., 2004). This study used deletion analysis to further explore the functional importance of each of the three domains in controlling plant height, fertility, and DELLA destruction. An N-terminal hemagglutinin (HA) epitope tag was fused in-frame to SLY1 and SNE full-length ORFs and ORFs containing deletions of each of the three domains defined in Figure 1 and placed under the control of the 35S promoter. The full-length constructs are referred to as HA-SLY1 and HA-SNE, whereas the constructs lacking the N-terminal, F-box, and C-terminal domains are referred to as ΔN, ΔF, and ΔC alleles, respectively (Supplemental Table S1). These chimeric constructs and an HA vector-only control were then transformed into the sly1-10 mutant to determine whether they were able to rescue the sly1-10 dwarfism and fertility phenotypes. Supplemental Table S1 shows the number of transgenic plants obtained for each construct and the number of transformed plants that appeared to rescue the sly1-10 dwarf phenotype in the T2 generation. Based on initial observations, the HA-SLY1 construct fully complemented the sly1-10 dwarfism, whereas HA-SNE, HA-sly1ΔN, and HA-sneΔN partially complemented the dwarfism. The remaining constructs failed to complement.
When the expression of each chimeric protein was examined by protein-blot analysis using the HA antibody, bands corresponding to the predicted full-length and truncated HA fusion proteins were observed (Supplemental Fig. S2). For each construct, two lines showing similar levels of protein expression were used for further analysis. However, it was noted that HA-SNE protein levels were on the whole lower than those of the HA-SLY1 constructs and that loss of the C-terminal domain led to some decrease in protein accumulation.
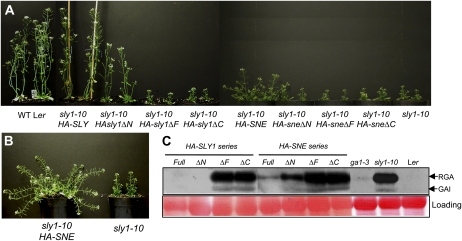
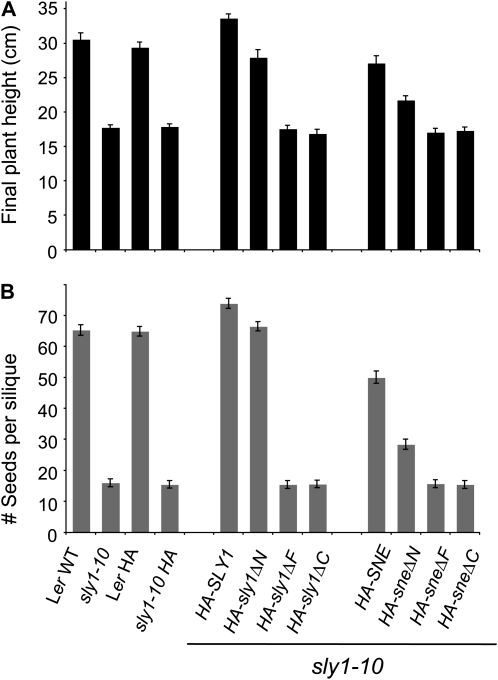
The degree to which each HA-SLY1 and HA-SNE construct was able to complement the sly1-10 mutation was determined by observing 30-d-old plants (Fig. 2A) and 45-d-old plants (Supplemental Fig. S3) and by measuring final plant height and the number of seeds per silique (Fig. 3). The full-length HA-SLY1 construct fully complemented the sly1-10 mutant phenotypes, including dwarfism, infertility, and delayed flowering (Fig. 2A; Supplemental Table S1). The HA-sly1ΔN construct appeared to fully rescue the final plant height and fertility phenotypes; however, these plants were shorter than the wild-type and the sly1-10 HA-SLY1 lines at 30 d due to slower plant growth (Figs. 2A and 3). HA-sly1ΔF and HA-sly1ΔC resulted in no apparent rescue of sly1-10 phenotypes compared with the untransformed and the vector-only controls. These results indicated that the F-box and C-terminal domains of SLY1 are required for SLY1 function, whereas the N-terminal region of SLY1 is required for full functionality of the SLY1 protein. The HA-SNE construct strongly complemented the sly1-10 mutation, resulting in a final plant height approximately 89% of the wild-type plant height and in fertility approximately 84% of the wild type (Figs. 2B and 3; Supplemental Fig. S3B). The sly1-10 HA-sneΔN did not show significant restoration of plant height in 30-d-old plants but appeared to result in some restoration of plant height and fertility in 45-d-old plants (Figs. 2A and 3; Supplemental Fig. S3C). The sly1-10 HA-sneΔF and the sly1-10 HA-sneΔC showed no suppression of sly1-10 phenotypes (Figs. 2A and 3). Thus, the F-box and C-terminal domains are required for SLY1 function and for SNE rescue of the sly1-10 plant height and fertility phenotypes. These results are consistent with previous reports indicating that SNE/SLY2 overexpression rescued sly1 mutant phenotypes (Fu et al., 2004; Strader et al., 2004).
Figure 2.
The effect of N-terminal, C-terminal, and F-box domain deletions on the ability of HA-SLY1 and HA-SNE to rescue sly1-10. A, Shown are 30-d-old wild-type (WT) Ler, sly1-10, and sly1-10 transformed with the indicated HA fusion constructs. B, Shown are 45-d-old sly1-10 and sly1-10 HA-SNE. The HA-SNE construct partially rescued the dwarfism. C, DELLA RGA and GAI protein accumulation in wild-type Ler, ga1-3, sly1-10, and sly1-10 transformed with constructs described above was determined by anti-RGA immunoblot analysis of 40 μg of total protein extracted from 21-d-old rosette leaves.
Figure 3.
Final plant height and fertility in the wild-type (WT) Ler, sly1-10, and sly1-10 transformed with the indicated constructs. A, Final plant height (cm) was determined after 120 d of incubation in a growth chamber. B, Fertility was determined based on the number of seeds per silique. Wild-type Ler and sly1-10 plants transformed with the HA vector were used as controls.
The Effect of HA-SLY1 and HA-SNE Constructs on DELLA Protein Accumulation
To determine if the restoration of sly1-10 plant height was associated with a decrease in the accumulation of DELLA repressors of stem elongation, protein-blot analysis was performed to detect DELLAs RGA and GAI (Fig. 2C). Expression of HA-SLY1 and HA-sly1ΔN was associated with a considerable decrease in DELLA protein accumulation in 21-d-old sly1-10 seedlings, whereas expression of HA-sly1ΔF and HA-sly1ΔC did not result in decreased DELLA protein accumulation. This suggests that the HA-SLY1 and HA-sly1ΔN constructs relieve DELLA repression through ubiquitination and proteolysis and that the F-box and C-terminal regions are required for this E3 ubiquitin ligase activity. Expression of HA-SNE also resulted in decreased DELLA protein accumulation in sly1-10 seedlings, suggesting that the HA-SNE fusion protein can partly replace SLY1 function in DELLA protein ubiquitination and destruction during stem elongation (Fig. 2C). Only a slight decrease in DELLA protein accumulation was observed in sly1-10 seedlings transformed with HA-sneΔN, suggesting that the N terminus may be needed for full function. Alternatively, the fact that HA-SLY1ΔN and HA-SNEΔN proteins accumulate at lower levels than HA-SLY1 and HA-SNE may explain their lesser effect on DELLA protein accumulation (Supplemental Fig. S2). The HA-sneΔF and HA-sneΔC constructs resulted in no significant decrease in DELLA protein accumulation in sly1-10 seedlings, suggesting that the F-box and C-terminal domains are required for SNE/SLY2 to regulate DELLA protein accumulation.
Because the infertility phenotype of the sly1-10 mutant was also fully suppressed by the HA-SLY1 construct and partially suppressed by the HA-sly1ΔN, HA-SNE, and HA-sneΔN constructs (Fig. 3B), we examined whether this suppression is associated with decreased DELLA protein accumulation. Protein-blot analysis was performed to examine the accumulation of DELLA proteins RGA, GAI, and RGL2 in flower bud tissue from 30-d-old plants. Transformation of sly1-10 with the HA-SLY1 and HA-SNE constructs resulted in a dramatic decrease in DELLA RGA and GAI accumulation compared with untransformed sly1-10 (Supplemental Fig. S4); transformation with HA-sly1ΔN resulted in a large decrease; transformation with HA-sneΔN resulted in a slight decrease; and transformation with HA-sly1ΔF, HA-sly1ΔC, HA-sneΔF, and HA-sneΔC resulted in no decrease in DELLA RGA and GAI protein accumulation. These results suggested that the suppression of infertility by SNE gene overexpression was, at least in part, the result of decreased RGA and GAI protein accumulation. Interestingly, a decrease in RGL2 protein levels was observed when sly1-10 was transformed with HA-SLY1 and HA-sly1ΔN (Supplemental Fig. S4A), but no decrease in RGL2 was observed when sly1-10 was transformed with HA-SNE (Supplemental Fig. S4B). This suggests that SNE is able to direct the degradation of DELLAs RGA and GAI but not of the DELLA RGL2.
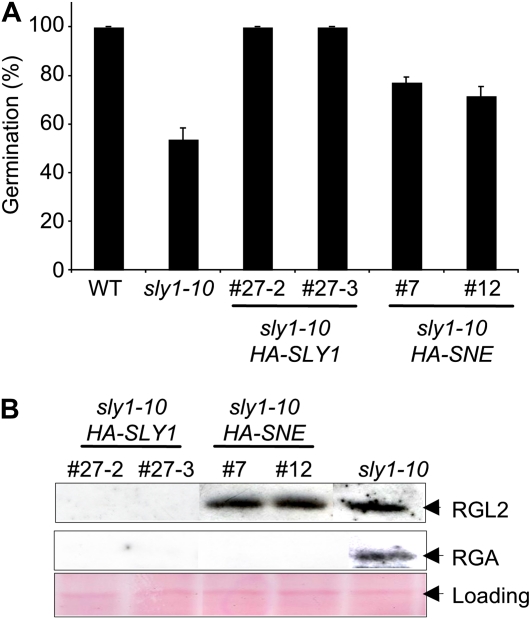
Because RGL2 is the major DELLA repressing seed germination, we next examined the effect of HA-SNE and HA-SLY1 overexpression on DELLA accumulation and on seed germination efficiency in sly1-10. The sly1-10 mutant has increased seed dormancy and when after-ripened germinates more slowly than the wild type (Ariizumi and Steber, 2007). The sly1-10 seed germination phenotype was fully rescued by HA-SLY1 and partially rescued by HA-SNE overexpression after 4 d of incubation (Fig. 4A). Protein-blot analysis showed that HA-SLY1 overexpression was associated with the disappearance of both RGA and RGL2. In contrast, the rescue of seed germination by HA-SNE overexpression was associated with a decrease in RGA protein but not in RGL2 protein levels (Fig. 4B). This suggests that an SCFSNE E3 complex may be able to ubiquitinate and target RGA but not RGL2 for destruction.
Figure 4.
SNE overexpression partial rescue of the sly1-10 seed germination phenotype is associated with decreased DELLA RGA but not RGL2 protein accumulation. A, Germination of sly1-10 and two independent sly1-10 lines transformed with HA-SLY1 (#27-2 and #27-3) and HA-SNE (#7 and #12). Seeds were incubated on MS agar plates at 4°C for 3 d, followed by 22°C for 4 d. WT, Wild type. B, Protein-blot analysis of RGA and RGL2 in sly1-10 HA-SLY1 and sly1-10 HA-SNE seeds imbibed on MS agar plates at 4°C for 3 d, followed by 22°C for 24 h. Protein was detected with anti-RGA and anti-RGL2. Sixty micrograms of total protein was loaded. [See online article for color version of this figure.]
Evidence for the Formation of SCFSLY1 and SCFSNE Complexes and for Protein Interaction with DELLA RGA
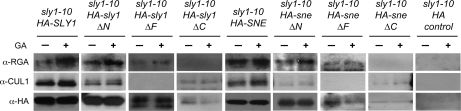
The fact that sly1-10 rescue by overexpression of full-length and N-terminal deletions of SLY1 and SNE is associated with a decrease in DELLA protein accumulation suggested that these two F-box proteins form SCF complexes that interact with and regulate DELLA proteins by ubiquitination. If this is true, we would expect SLY1 and SNE to interact with DELLA RGA protein and with the SCF CUL1 subunit via the F-box domain. To examine in planta protein-protein interactions, we performed coimmunoprecipitation experiments using an HA antibody matrix to affinity purify full-length and deletion alleles of HA-SLY1 and HA-SNE proteins from seedling extracts. Protein-blot analysis of these extracts showed that CUL1 coimmunoprecipitated with the HA-SLY1, HA-sly1ΔN, HA-sly1ΔC, HA-SNE, HA-sneΔN, and HA-sneΔC fusion proteins but not with the HA-vector control or with the HA-sly1ΔF and HA-sneΔF proteins (Fig. 5; Supplemental Fig. S5). This result suggests that SLY1 and SNE proteins form SCF complexes that include CUL1 and that complex formation requires the F-box domain. Protein-blot analysis also showed that the DELLA protein RGA coimmunoprecipitated with the HA-SLY1, HA-sly1ΔN, HA-sly1ΔF, HA-SNE, HA-sneΔN, and HA-sneΔF fusion proteins but not with the HA-sly1ΔC and HA-sneΔC proteins and not with the HA-vector control. The addition of GA to the protein extract increased the quantity of RGA coimmunoprecipitated with HA-SLY1, but the interaction was still observed without GA addition due to the presence of endogenous GA in seedling extracts. This result indicates that both SLY1 and SNE can interact with RGA protein in planta and that the C-terminal domain of SLY1 and SNE are required. Taken together, these data indicate that SCFSNE and SCFSLY1 E3 ubiquitin ligases form in vivo and regulate DELLA RGA by direct protein interaction.
Figure 5.
HA-SLY1 and HA-SNE fusion proteins interact with DELLA RGA and CUL1 protein in planta. Protein was extracted from 12-d-old sly1-10 plants transformed with the indicated constructs and incubated with HA matrix agarose in the presence of 0.1% ethanol (mock) or 100 μm GA3. Immunoprecipitated protein was loaded on an SDS-PAGE gel and detected with anti-HA antibody. Coimmunoprecipitated protein was detected with anti-RGA and anti-CUL1.
The Effect of SNE Overexpression on Plant Growth Habit
In addition to partly suppressing the sly1-10 dwarf and infertility phenotypes, we observed that HA-SNE plants show an aberrant plant growth habit consisting of decreased apical dominance and a prone growth habit (Fig. 2, A and B). All 17 sly1-10 HA-SNE plants showed a similar phenotype where secondary lateral shoots formed at a wide angle to the primary shoot (Supplemental Fig. S3). To determine whether this HA-SNE overexpression phenotype was dependent on the sly1-10 background, wild-type ecotype Landsberg erecta (Ler) and the GA biosynthesis mutant ga1-3 were transformed with the HA-SNE overexpression construct. It appeared that wild-type Ler HA-SNE plants also showed the decreased apical dominance and prone growth habit (Supplemental Fig. S6A). Thus, the aberrant growth phenotype is not dependent on the sly1-10 background. The HA-SNE overexpression construct did not rescue the dwarfism of the ga1-3 plants. GA treatment of the ga1-3 HA-SNE plants rescued the ga1-3 dwarf and fertility phenotypes but resulted in the same aberrant prone growth habit seen in the wild-type Ler transformants (Supplemental Fig. S6B). These results suggest that the aberrant growth phenotype is a direct result of SNE overexpression and does not result from an interaction between SNE overexpression and the sly1 mutant background.
Expression Analysis of SNE/SLY2
Previous reverse transcription-PCR and northern-blot analyses showed that the SLY1 mRNA is present throughout the plant, whereas the SNE mRNA is mainly present in flowers and to a lesser extent in stems (Strader et al., 2004). The meta-analysis tool Genevestigator (https://www.genevestigator.com/gv/index.jsp) and the Arabidopsis microarray database were used to compare the spatiotemporal pattern of SLY1 and SNE gene expression (Hruz et al., 2008). The level of SLY1 mRNA accumulation appears to be 3- to 5-fold higher than that of SNE in most tissues, with the two genes showing similar expression trends in a developmental analysis (Supplemental Fig. S7A). SNE transcript appeared highest in callus, shoot apex, and root endodermis (Supplemental Fig. S8) and lowest in germinating seeds and seed tissues (endosperm, seed coat). SLY1 expression appeared highest in the abscission zone, hypocotyl, shoot apex, and petals and lowest in pollen and stigma.
GUS transcriptional fusions were used to further compare the expression patterns of these F-box genes. A construct containing the SLY1 promoter region fused to the GUS reporter gene was transformed into wild-type Ler and the expression pattern analyzed by histochemical staining for GUS activity (Ariizumi et al., 2002a). A total of six independent SLY1p-GUS lines were examined; representative GUS expression patterns are shown in Supplemental Figure S9A. Consistent with the previous reverse transcription-PCR analysis (McGinnis et al., 2003), SLY1p-GUS expression was found in most parts of the plant. Strong expression was seen in the cotyledons and hypocotyls of 6-d-old seedlings, in the vasculature of seedlings, leaves, and roots, in the primary root tip, and in the anthers, filaments, petals of flowers, and the receptacle of siliques. In contrast, the expression of SNE-GUS appears to be much less widespread. An enhancer trap line in which a T-DNA containing the GUS reporter was inserted just before the SNE translational start site was used to examine expression in germinating seeds, seedlings and seedling roots, mature leaves, and flowers. Low-level SNE-GUS expression was evident only in flower anthers (Supplemental Fig. S9B).
Protein Interaction between SLY1 and the GID1b GA Receptor
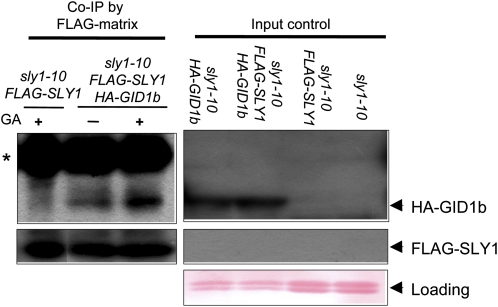
Based on our results, SLY1 most likely is the predominant F-box protein functioning in GA signaling because it is expressed at higher levels and because it can target more DELLA proteins for destruction. In the current model of GA signaling, the formation of the GID1-GA-DELLA complex allows SCFSLY1 to bind to and ubiquitinate DELLA protein (Hirano et al., 2008). This model predicts that SLY1 should form a complex that includes not only DELLA but also GID1 protein. Coimmunoprecipitation was used to examine whether or not the SLY1 protein interacts with GID1b in planta using sly1-10 plants transformed with an HA-GID1b and FLAG-SLY1 epitope-tagged fusion proteins expressed on the 35S promoter. When the FLAG-SLY1 protein was affinity purified from 12-d-old seedling extracts using a FLAG antibody matrix, protein-blot analysis showed that HA-GID1b coimmunoprecipitated with FLAG-SLY1 and that this interaction increased with the addition of GA to the plant protein extract (Fig. 6). This indicates that SCFSLY1 interacts with a complex that includes GID1b.
Figure 6.
Interaction of SLY1 and GID1 in coimmunoprecipitation (Co-IP) assays. The coimmunoprecipitation experiment was performed using protein extracted from 12-d-old sly1-10 plants transformed with HA-GID1a and/or FLAG-SLY1. The protein extract was incubated with FLAG matrix agarose in the presence of 0.1% ethanol (mock) or 10 μm GA3 and loaded on an SDS-PAGE gel. Protein-blot analysis was performed using anti-FLAG and anti-HA antibodies. The asterisk indicates a background band. Forty micrograms of total protein was loaded (input). [See online article for color version of this figure.]
DISCUSSION
Previous work suggested that the SLY1 homolog SNE/SLY2 may encode a second F-box protein functioning in GA signaling, since overexpression of SNE on the 35S promoter partly rescued the sly1-10 dwarf phenotype and resulted in reduced DELLA RGA accumulation (Fu et al., 2004; Strader et al., 2004). This paper examined the mechanism by which SNE substitutes for SLY1 in the regulation of GA signaling, whether the SNE protein could form an SCF E3 ubiquitin ligase complex in planta, and the relative roles of SLY1 and SNE/SLY2 in GA signaling. Finally, we examined the interaction of SLY1 with the GA receptor GID1.
Functional Analysis of SLY1 and SNE F-Box Proteins in GA Signaling
A side-by-side comparison was conducted to determine the domains required for HA-SLY1 and HA-SNE to rescue sly1-10 and to interact with the SCF subunit CUL1 and the DELLA RGA proteins in planta (Fig. 5). Consistent with previous results, HA-SLY1 gave complete rescue of the germination, fertility, and plant height phenotypes of sly1-10, whereas HA-SNE only partly rescued these phenotypes (Figs. 2–4; Supplemental Fig. S3). In both cases, the F-box and C-terminal domains were required for function. Deletion of the N-terminal domains of HA-SLY1 and HA-SNE decreased the capacity of these proteins to complement the sly1-10 phenotype (Figs. 2 and 3; Supplemental Fig. S3). Thus, it appears that although the N terminus is not essential, it does play some role. Consistent with the functional data, it was observed that the C terminus was required for HA-SLY1 and HA-SNE to interact with the DELLA RGA protein in planta in coimmunoprecipitation experiments (Fig. 5). Previous work demonstrated the interaction of SLY1 with DELLA protein in vitro, by yeast two-hybrid assay, and by pull-down assay (Dill et al., 2004; Fu et al., 2004; Wang et al., 2009) as well as the association of SLY1 with the SCF complex protein CUL1 in vitro (Wang et al., 2009). In this study, both HA-SLY1 and HA-SNE coimmunoprecipitated with CUL1 in planta, and this interaction required the F-box domain (Fig. 5), indicating that it is required for SCF complex formation. The F-box domain is required for the F-box protein to interact with the Skp1 subunit during SCF complex formation (Gagne et al., 2002, 2004). Both DELLA RGA and CUL1 proteins coimmunoprecipitate with the HA-SLY1ΔN and HA-SNEΔN proteins, indicating that the N-terminal domain is not absolutely essential for these protein-protein interactions (Fig. 5). It is possible that less RGA coimmunoprecipitates with HA-SLY1ΔN than with HA-SLY1 protein because the N-terminal deletion accumulates at lower levels (Supplemental Fig. S2). Taken together, these results indicate that SNE can function in a manner very similar to SLY1, forming an SCF complex in planta that can function in GA signaling through interaction with the DELLA RGA.
SNE Function Partly Overlaps with SLY1 Function in GA Signaling
Overexpression of HA-SNE partially rescued the dwarfism, infertility, and germination phenotypes of the sly1-10 mutant (Figs. 2–4; Supplemental Fig. S3). This rescue was correlated with decreased levels of the DELLA proteins RGA and GAI, suggesting that an SCFSNE E3 ubiquitin ligase can regulate these DELLA proteins via ubiquitination and destruction by the 26S proteasome (Figs. 2 and 4; Supplemental Fig. S4). Several observations support this theory, including that HA-SNE interacts with DELLA RGA and forms an SCF complex in planta (Fig. 5). Both interaction with CUL1 and rescue of the sly1-10 phenotypes required the F-box domain, indicating that formation of an SCFSNE complex is required for its function in GA signaling (Figs. 2, 3, and 5). This is consistent with previously published data showing that SNE protein can interact with Arabidopsis Skp1 homologs in the yeast two-hybrid system (Fu et al., 2004). Finally, HA-SNE overexpression does not rescue the dwarfism or flowering phenotypes of the GA biosynthesis mutant ga1-3 in the absence of GA (Supplemental Fig. S6C). This suggests that SNE acts through GA signaling rather than via a parallel pathway and that the SCFSNE E3 ubiquitin ligase targets DELLA for destruction via a GA-dependent mechanism similar to the SCFSLY1 (McGinnis et al., 2003; Dill et al., 2004; Fu et al., 2004).
The function of SNE/SLY2 in GA signaling appears to only partly overlap with that of SLY1. Whereas overexpression of HA-SLY1 caused a decrease in DELLA RGA, GAI, and RGL2 protein accumulation in seeds and flowers of the sly1-10 mutant, overexpression of HA-SNE caused a decrease in DELLA RGA and GAI but not in DELLA RGL2 protein accumulation (Fig. 4; Supplemental Fig. S4). This suggests that SNE can only regulate a subset of DELLA proteins. Future work will need to examine whether SNE protein shows stronger binding to RGA and GAI than to RGL2 or whether SNE binds to RGL2 but fails to ubiquitinate RGL2. RGL2 is a key DELLA protein controlling seed germination as well as flowering and fertility (Lee et al., 2002; Cheng et al., 2004; Tyler et al., 2004; Cao et al., 2005, 2006). Thus, failure to regulate RGL2 may partly explain why HA-SNE overexpression only partly rescues the fertility and germination of sly1-10 (Figs. 3 and 4). This, taken together with the facts that the SNE mRNA accumulates at lower levels than the SLY1 mRNA (Supplemental Figs. S7–S9) and that the sly1 mutant shows strong dwarfism and infertility phenotypes in the presence of the normal SNE gene, supports the conclusion that SLY1 is the major F-box protein contributing to GA signaling in Arabidopsis.
The SNE gene can regulate DELLA proteins, but might it also regulate genes that are not part of GA signaling? The overexpression of HA-SNE not only rescued the GA-insensitive phenotypes of sly1-10 but also resulted in changes in growth habit, including loss of apical dominance and a prone growth habit where stems were angled downward (Fig. 2, A and B; Supplemental Fig. S3). The phenotype was not dependent on the sly1-10 background, as these phenotypes were also observed when HA-SNE was transformed into wild-type Ler and ga1-3 (Supplemental Fig. S6). Digital northern analysis shows that the SNE mRNA is expressed in the shoot apex and in callus cells (Supplemental Fig. S8). It is not clear whether the prone phenotype does or does not result from changes in GA signaling. Thus, future research will need to examine whether SNE regulates an alternative target involved in meristem function and/or GA signaling. Previous work demonstrated that SNE mRNA is expressed in the endodermis and the quiescent center of the root, whereas SLY1 is only expressed in the stele (Cui and Benfey, 2009). Cui and Benfey (2009) hypothesized that SNE may be important for DELLA regulation in the root cells that do not show SLY1 expression.
SLY1 Physical Interactions with the GID1 GA Receptor
This study next examined the interaction of the SCFSLY1 E3 ubiquitin ligase with the GA receptor, GID1. The current model of GA signaling proposes that the F-box protein SLY1 of Arabidopsis and the orthologous F-box GID2 of rice trigger DELLA destruction only when DELLA protein is bound by the GA receptor GID1. GID1 protein affinity for DELLA protein increases when GID1 binds GA hormone (Griffiths et al., 2006; Nakajima et al., 2006; Willige et al., 2007). Yeast three-hybrid analysis has demonstrated that the rice F-box GID2 only binds to DELLA protein when it is in the GID1-GA-DELLA complex (Hirano et al., 2010). It appears that GA may also stimulate SLY1-GID1 interaction in Arabidopsis, as coimmunoprecipitation demonstrated that FLAG-SLY1 forms a complex with HA-GID1b (Fig. 6). Based on the work of Hirano et al. (2010), the GID1-SLY1 complex likely includes a DELLA protein. This is, to our knowledge, the first in planta demonstration that the F-box protein forms a complex that includes the GA receptor, thus supporting the rice model derived from yeast three-hybrid data.
Implications for GA Signaling
The research presented here has several broader implications for GA signaling. This study and others indicate that SLY1 is the major F-box protein regulating DELLA protein in GA signaling (Dill et al., 2004; Fu et al., 2004; Ariizumi and Steber, 2007). It appears that SNE can regulate DELLAs RGA and GAI but not RGL2 (Fig. 4; Supplemental Fig. S4). However, based on the unusual growth phenotype of SNE overexpression lines and the apparently unique role of SNE in the control of root elongation, it appears that SCFSNE may play a specialized role that will need to be considered in future studies (Supplemental Fig. S3; Cui and Benfey, 2009). This study demonstrated that the SLY1 protein exists in complex with the GID1b receptor protein in planta. This result supports the model that SCFSLY1 binding to and ubiquitination of DELLA protein is stimulated when DELLA is in complex with GID1 and GA hormone (Fig. 6; Hirano et al., 2010), resulting in GA-stimulated ubiquitination and destruction of DELLA (McGinnis et al., 2003; Dill et al., 2004; Fu et al., 2004).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana) Ler, as well as sly1-10 and ga1-3 mutations in the Ler background, were obtained as described previously (Koornneef and van der Veen, 1980; Ariizumi et al., 2008). The germination of ga1-3 seed was stimulated by imbibing in 10 μm GA4 for 3 d at 4°C and then washing with sterile water. Seeds were sterilized and imbibed in sterile water for 3 d at 4°C to break dormancy, and then all seeds were transferred to half-strength Murashige and Skoog (MS) salts (Sigma-Aldrich)/0.8% agar (MS agar) and incubated at 22°C under constant fluorescent light for 10 to 14 d. Seedlings were transferred to soil and grown at 22°C under fluorescent light (16-h day; McGinnis et al., 2003) for growth rate and fertility comparisons. To determine the effect of GA treatment on plant growth, plants grown in soil were sprayed every 3 d with 10 μm GA4.
A sly1-10 line transformed with the FLAG-SLY1 and HA-GID1b translation fusions was constructed by crossing the sly1-10 FLAG-SLY1 line (described below) to the previously constructed sly1-10 HA-GID1b line (Ariizumi et al., 2008). F1 and F2 seeds were sown on MS agar containing 20 mg L−1 hygromycin. F3 seeds from each F2 individual were harvested. Among several independent F3 plants, a FLAG-SLY1 F3 individual expressing HA-GID1b protein at a level similar to the original sly1-10 HA-GID1b line was selected based on protein-blot analysis (Fig. 5).
Plasmid Construction and Plant Transformation
In-frame N-terminal fusions of the full-length or truncated SLY1 (At4g24210; GenBank accession no. NM_118554) and SNE (At5g48170; NM_124191) coding regions to the HA epitope tag were constructed under the control of the constitutive CaMV 35S promoter. Fragments containing the full-length, N-terminal, and C-terminal deletions of the SLY1 and SNE genes were obtained by PCR using gene-specific primer pairs (Supplemental Table S2). N-terminal deletions of the SLY1 and SNE genes were generated using the SLY-Ndel-F/SLYorf-R and SNE-Ndel-F/SNEorf-R primers, respectively. C-terminal deletions were generated using SLYorf-F/SLY-Cdel-R and SNEorf-F/SNE-Cdel-R primers. Internal deletions of the SLY1 and SNE F-box domains were created using the following strategy. (1) The SLYorf-F and SLY-Fbox-R primers were used to amplify fragment A, while the SLY-Fbox-Fr and SLYorf-R primers were used to amplify fragment B. (2) These PCR products were gel purified and then diluted in water (1:1,000). (3) A reaction was set up using 0.5 μL of the A and B fragments as a PCR template, generating a deletion of the F-box by amplification with the SLY1orf-F and SLYorf-R primers (SNEorf-F and SNEorf-R). The proofreading enzyme KOD Hotstart DNA polymerase (Novagen) was used, and all constructs were confirmed by sequencing. These amplified PCR fragments were then phosphorylated and directly cloned as blunt-end fragments into the SmaI site of HA/pBluescript (Ariizumi et al., 2008) to obtain the HA-SLY1, HA-SLY1ΔN, HA-SLY1ΔF, HA-SLY1ΔC, HA-SNE, HA-SNEΔN, HA-SNEΔF, and HA-SNEΔC translational fusions. The HindIII-SacI fragments from these HA-fused full-length and truncated SLY1/pBluescript plasmids were excised and cloned into the HindIII-SacI site of T-DNA binary vector pBI101, while the HindIII-SacI fragments from HA-SNE/pBluescript and HA-SNEΔC/pBluescript plasmids were excised and cloned into the HindIII-SacI site of T-DNA binary vector pBI101H (Ariizumi et al., 2002b).
To construct the N-terminal FLAG-tagged SLY1 construct, the DNA sequence for three repeats of the FLAG epitope was amplified using FLAG-F and FLAG-R primers (Supplemental Table S2). The PCR fragment was phosphorylated with T4 polynucleotide kinase (Fermentas) and then blunt ligated into the EcoRV site of pBluescript II KS− vector to generate FLAG/pBluescript. PCR fragments containing the full-length SLY1 ORF were directly cloned as a blunt-end fragment into the SmaI site of FLAG/pBluescript to obtain FLAG-SLY1/pBluescript. The HindIII-SacI fragment from the FLAG-SLY1/pBluescript plasmids was cloned into the HindIII-SacI site of T-DNA binary vector pGTV-HPT.
The constructs were transformed into Agrobacterium tumefaciens GV3101 by the freeze-thaw method (An et al., 1988). These constructs were transformed into sly1-10, wild-type Ler, or ga1-3 and selected on MS agar with 20 mg L−1 hygromycin (Clough and Bent, 1998). The expression of each chimeric protein of appropriate size was confirmed by protein-blot analysis using HA antibody (Supplemental Fig. S2). Two independent lines showing similar expression of each fusion protein were used for further analysis.
To create the GUS transcriptional fusion to the SLY1 promoter, the 2.0-kb region upstream of the SLY1 ORF was amplified with KOD Hotstart DNA polymerase using the SLY1pro-F and SLY1pro-R primers, phosphorylated, and cloned as a blunt-end fragment into the SmaI site of pBI121 (Clontech). These constructs were transformed into Ler and selected on MS agar plus kanamycin (30 mg L−1). Expression of the SNE gene was examined using the sne-t2 line, which contains an enhancer trap T-DNA insertion 1 bp before the translational start site. The sne-t2 line was isolated by PCR screening of the Sussman Basta line pools using the T-DNA left border primer JL202 (5′-CATTTTATAATAACGCTGCGGACATCTAC-3′) and the SNE-specific primer T-sly2F (5′-AAGAAACAGGAGTGGGAAAAAAATCACG-3′) to obtain a 1.2-kb product (Krysan et al., 1999; Sussman et al., 2000). The pD991 T-DNA binary vector contains the −60 CaMV minimal promoter fused to the uidA GUS gene (http://www.dartmouth.edu/∼tjack/et.html#pD991). This construct shows no expression until in the vicinity of an enhancer, allowing histochemical detection of SNE-GUS expression.
Gene Expression Analysis
GUS activity was examined according to Ariizumi et al. (2002a). Two-day-old, 6-d-old, 14-d-old, and 30-d-old plants were soaked in GUS solution and incubated for 2 to 24 h at 37°C. After incubation, plants were incubated in 70% ethanol for 24 h to bleach the pigments. In silico expression analysis was performed using the online tool Genevestigator (Hruz et al., 2008).
Protein Expression Analysis
Protein-blot analysis was used to examine DELLA protein (RGA, GAI, and RGL2) accumulation in nonsegregating T3 seedlings, flower buds, and seeds. Seeds were germinated under kanamycin selection and imbibed for 3 d at 4°C, followed by incubation at 22°C for 10 to 14 d prior to tissue collection. Flower buds were collected from 44-d-old plants. Total plant protein was extracted according to Silverstone et al. (2001) with a modified extraction buffer X (50 mm Tris, pH 6.8, 10% glycerol, and 1% SDS). Total protein was extracted from seed tissue as described by Ariizumi and Steber (2007). Forty micrograms of total protein from seedlings and flower buds, or 60 μg of total protein from seeds, was separated by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Immobulon). Protein concentration was determined using the Bio-Rad protein assay, and even loading was confirmed by Ponceau membrane staining. Protein detection was performed using an enhanced chemiluminescence system (GE Healthcare) according to the manufacturer’s protocol except that primary antibody incubations were conducted overnight. RGA and GAI proteins were detected using the RGA polyclonal antibody (1:25,000; Silverstone et al., 2001) and RGL2 using an RGL2 polyclonal antibody (1:25,000; Hussain et al., 2005) using controls described previously (Ariizumi and Steber, 2007). HA fusion proteins were detected using monoclonal HA antibody (1:25,000; Sigma-Aldrich). CUL1 protein was detected using CUL1 antibody (1:25,000; Chen et al., 2006). The anti-rabbit IgG-horseradish peroxidase (GE Healthcare) was used as a secondary antibody (1:250,000).
Germination Experiments
For germination experiments, 30 to 60 seeds from each genotype grown in the same incubator were sterilized with 10% bleach for 15 to 20 min and plated on MS agar or MS agar including 0 to 1.2 μm (+)-abscisic acid (PBI58; gift of S. Abrams). Percentage germination based on radicle emergence was determined following 3 d of incubation at 4°C followed by incubation under constant fluorescent light at 22°C. The average germination rate was calculated using three independent replicates.
Coimmunoprecipitation Experiment
For coimmunoprecipitation experiments, 10-d-old sly1-10 plants transformed with the indicated constructs were incubated as a suspension of 100 μm MG132 in half-strength MS buffered with 5 mm MES, pH 5.5, for 2 h on ice. The cross-linking reagent dithiobis-succinimidyl propionate (Pierce) was added to a final concentration of 1 mm and further incubated for 30 min on ice. The cross-linking reaction was stopped by adding 2 mm Gly for 15 min on ice. Seedlings were washed twice with chilled phosphate-buffered saline (pH 7.6), blotted dry on Kimwipes, and ground under liquid N2. Ground tissue was transferred to buffer A (100 mm Tris, pH 7.5, 150 mm NaCl, 0.5% Triton X-100, and protease inhibitor cocktail [Roche]) on ice, then centrifuged at 21,000g for 15 min. Protein concentration of the supernatant was determined, and 5 mg of protein extract was incubated with 40 μL of anti-HA matrix (Roche) in the presence of 100 μm GA3 or no GA3 (mock; 0.1% ethanol) for 16 h at 4°C. After the anti-HA matrix was washed three times with buffer A, it was resuspended in 1× SDS sample buffer, boiled for 3 min, and pelleted for 2 min at 14,000 rpm, and 20 μL of the supernatant was loaded for SDS-PAGE separation.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_118554 (SLY1), NM_124191 (SNE/SLY2), NM_126218 (RGA), NM_101361 (GAI), NM_111216 (RGL2), and NM_116166 (GID1b).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of predicted SLY1 and SNE proteins.
Supplemental Figure S2. Expression of HA fusion proteins in sly1-10.
Supplemental Figure S3. Vegetative phenotype of sly1-10 HA-SNE lines.
Supplemental Figure S4. DELLA protein expression in flower buds.
Supplemental Figure S5. Input control for Figure 5.
Supplemental Figure S6. Vegetative phenotype of Ler and ga1-3 HA-SNE lines.
Supplemental Figure S7. Digital northern of SNE and SLY1.
Supplemental Figure S8. Developmental pattern of SNE and SLY1 mRNA expression.
Supplemental Figure S9. GUS histochemical staining showing the pattern of expression from the SLY1 and SNE promoters.
Supplemental Table S1. Complementation of sly1-10.
Supplemental Table S2. Primers used.
Supplementary Material
Acknowledgments
We thank T.-P. Sun for the gift of RGA antibody and J. Peng for the gift of the RGL2 antibody. We are also grateful to X.W. Deng for providing the CUL1 antibody. We appreciate S. Abrams for providing (+)-abscisic acid (PBI58). Finally, we thank members of the Steber laboratory and L. Strader for lively discussion and critical comments on the manuscript.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P. (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan RE. (1986) Agronomic comparison among wheat lines nearly isogenic for three reduced-height genes. Crop Sci 26: 707–710 [Google Scholar]
- An G, Ebert RR, Mitra A, Ha SB. (1988) Binary vectors. Gelvin SB, Schilperoort RA, , Plant Molecular Biology Manual. Kluwer, Dordrecht, The Netherlands, pp 1–19 [Google Scholar]
- Ariizumi T, Amagai M, Shibata D, Hatakeyama K, Watanabe M, Toriyama K. (2002a) Comparative study of promoter activity of three anther-specific genes encoding lipid transfer protein, xyloglucan endotransglucosylase/hydrolase and polygalacturonase in transgenic Arabidopsis thaliana. Plant Cell Rep 21: 90–96 [Google Scholar]
- Ariizumi T, Kishitani S, Inatsugi R, Nishida I, Murata N, Toriyama K. (2002b) An increase in unsaturation of fatty acids in phosphatidylglycerol from leaves improves the rates of photosynthesis and growth at low temperatures in transgenic rice seedlings. Plant Cell Physiol 43: 751–758 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Murase K, Sun TP, Steber CM. (2008) Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20: 2447–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Steber CM. (2007) Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 19: 791–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M. (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Cheng H, Wu W, Soo HM, Peng J. (2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Hussain A, Cheng H, Peng J. (2005) Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223: 105–113 [DOI] [PubMed] [Google Scholar]
- Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, Zhang Y, Zhang H, Feng S, Strickland E, et al. (2006) Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J. (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui H, Benfey PN. (2009) Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J 58: 1016–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T. (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP. (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J. (2008) Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc Natl Acad Sci USA 105: 16814–16819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD. (2004) Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA 101: 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Hellmann H, Dharmasiri S, Estelle M. (2002) Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14: 2137–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Asano K, Tsuji H, Kawamura M, Mori H, Kitano H, Ueguchi-Tanaka M, Matsuoka M. (2010) Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22: 2680–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Ueguchi-Tanaka M, Matsuoka M. (2008) GID1-mediated gibberellin signaling in plants. Trends Plant Sci 13: 192–199 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37: D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A, Cao D, Cheng H, Wen Z, Peng J. (2005) Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J 44: 88–99 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Tattersall A, Piazza P, Hay A, Martinez-Garcia JF, Schmitz G, Theres K, McCormick S, Tsiantis M. (2008) PROCERA encodes a DELLA protein that mediates control of dissected leaf form in tomato. Plant J 56: 603–612 [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J. (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM. (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Peng J, Harberd NP. (2002) The role of GA-mediated signalling in the control of seed germination. Curr Opin Plant Biol 5: 376–381 [DOI] [PubMed] [Google Scholar]
- Piskurewicz U, Lopez-Molina L. (2009) The GA-signaling repressor RGL3 represses testa rupture in response to changes in GA and ABA levels. Plant Signal Behav 4: 63–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL. (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J 34: 753–767 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP. (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Strader LC, Ritchie S, Soule JD, McGinnis KM, Steber CM. (2004) Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc Natl Acad Sci USA 101: 12771–12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F. (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S. (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP. (2004) DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. (2007) Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol 58: 183–198 [DOI] [PubMed] [Google Scholar]
- Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan LM, Deng XW. (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen CK, Chang C. (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. (2007) Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.