Abstract
The F-box protein superfamily represents one of the largest families in the plant kingdom. F-box proteins phylogenetically organize into numerous subfamilies characterized by their carboxyl (C)-terminal protein-protein interaction domain. Among the largest F-box protein subfamilies in plant genomes are those with C-terminal kelch repeats. In this study, we analyzed the phylogeny and evolution of F-box kelch proteins/genes (FBKs) in seven completely sequenced land plant genomes including a bryophyte, a lycophyte, monocots, and eudicots. While absent in prokaryotes, F-box kelch proteins are widespread in eukaryotes. Nonplant eukaryotes usually contain only a single FBK gene. In land plant genomes, however, FBKs expanded dramatically. Arabidopsis thaliana, for example, contains at least 103 F-box genes with well-conserved C-terminal kelch repeats. The construction of a phylogenetic tree based on the full-length amino acid sequences of the FBKs that we identified in the seven species enabled us to classify FBK genes into unstable/stable/superstable categories. In contrast to superstable genes, which are conserved across all seven species, kelch domains of unstable genes, which are defined as lineage specific, showed strong signatures of positive selection, indicating adaptational potential. We found evidence for conserved protein features such as binding affinities toward A. thaliana SKP1-like adaptor proteins and subcellular localization among closely related FBKs. Pseudogenization seems to occur only rarely, but differential transcriptional regulation of close relatives may result in subfunctionalization.
The process of protein degradation is an important posttranslational regulatory mechanism. It is integral to cellular homeostasis by removing nonfunctional and misfolded proteins and allows living organisms to adapt to changing environments by providing fast responses to intracellular signals. A major player in this process is the ubiquitin/26S proteasome system, which is responsible for selective degradation of many intracellular proteins (Stone and Callis, 2007; Vierstra, 2009). Proteins destined for degradation are modified by covalent attachment of multiple ubiquitin moieties. The polyubiquitinated substrates are recognized and degraded by the 26S proteasome, while the ubiquitin molecules are recycled. The ubiquitination process includes three steps. First, the ubiquitin is activated by a ubiquitin-activating enzyme (E1). Subsequently, the ubiquitin is transferred to the ubiquitin-conjugating enzyme (E2). Finally, the transfer of activated ubiquitin to the target protein is catalyzed by a ubiquitin ligase (E3; Stone and Callis, 2007). The selective components of this cascade are the E3 ubiquitin ligases. These structurally diverse enzymes occur as monomers or in multimeric complexes. The various E3 families are classified according to their mode of action and subunit composition (Mazzucotelli et al., 2006). The most prevalent E3 ubiquitin ligases in plants are the Skp1-Cullin-F-box (SCF) protein complexes. The SCF complex is formed by an S-phase kinase-associated protein 1 (SKP1), Cullin 1 (CUL1), RING-box 1 (RBX1), and an F-box protein. While CUL1 functions as a scaffold, SKP1 mediates the connection between CUL1 and the F-box subunit. RBX1 serves as a docking port for the ubiquitin-conjugating E2 enzyme. The F-box protein mediates the specificity of the SCF complex by selectively recruiting target proteins via a protein-protein interaction domain (Cardozo and Pagano, 2004). The Arabidopsis thaliana genome (The Arabidopsis Information Resource, TAIR9 version) contains a single CUL1 gene (Risseeuw et al., 2003), 19 functional genes coding for A. thaliana SKP1-like proteins (ASKs; Takahashi et al., 2004), and approximately 700 F-box genes (Gagné et al., 2002). The huge amount of combinatorial possibilities is evident and seems to be dominated by the hundreds of F-box proteins within the plant genomes assessed so far. Whereas human, Drosophila melanogaster, and Schizosaccharomyces pombe genomes contain 68 (Jin et al., 2004), 33 (Ou et al., 2003), and 18 (Hermand, 2006) F-box genes, respectively, this number is much higher in plants. The only species for which numbers comparable to plants were reported seems to be Caenorhabditis elegans, with approximately 520 F-box proteins (Thomas, 2006). To date, for A. thaliana, Oryza sativa, Populus trichocarpa, and Vitis vinifera, 692, 779, 337 (Xu et al., 2009), and 156 (Yang et al., 2008) F-box genes were identified, respectively. Even for the model plant A. thaliana, less than 5% of the F-box proteins have been functionally characterized, but F-box proteins with known functions suggest that they play prominent roles in multiple physiological processes in plants, such as responses to various hormones (Ruegger et al., 1998; Xu et al., 2002; Guo and Ecker, 2003; Dill et al., 2004; Binder et al., 2007), the circadian clock and photomorphogenesis (Han et al., 2004; Fukamatsu et al., 2005; Kim et al., 2007; Sawa et al., 2007), flower development (González-Carranza et al., 2007; Chae et al., 2008), senescence (Woo et al., 2001), and defense responses (Kim and Delaney, 2002).
F-box proteins share a well-conserved approximately 50-amino acid F-box motif at their N terminus (Kipreos and Pagano, 2000). Furthermore, most F-box proteins contain a C-terminal protein-protein interaction domain, such as leucine-rich repeats (LRRs), kelch repeats, or F-box associated domains (Gagné et al., 2002; Xu et al., 2009). Phylogenetically, F-box proteins cluster according to their protein-protein interaction domains (Gagné et al., 2002), which putatively mediate binding to the corresponding target. One of these subfamilies is constituted by the F-box kelch proteins (FBKs), which contain C-terminal kelch repeats in addition to the N-terminal F-box. In plants, the subfamily of FBKs includes members with one to five kelch repeats. Proteins containing kelch repeat domains in the absence of an F-box are widespread and have been described for A. thaliana, human, D. melanogaster, C. elegans, and S. pombe (Prag and Adams, 2003; Leung et al., 2004; Mora-García et al., 2004). Such proteins typically include five to seven kelch repeats, which may form β-propellers, as shown by the crystal structure of the human kelch protein KEAP1 (Li et al., 2004). Given their rare occurrence in nonplant organisms and the fact that only a single nonplant FBK has been functionally described so far (Sun et al., 2009), FBK proteins seem to be rather plant specific. The A. thaliana genome codes for approximately 100 FBKs, four of which have been functionally characterized. ATTENUATED FAR-RED RESPONSE (AFR) is a positive regulator of phytochrome A-mediated light signaling (Harmon and Kay, 2003). ZEITLUPE (ZTL), FLAVIN-BINDING KELCH-REPEAT F-BOX1 (FKF1), and LOV KELCH PROTEIN2 (LKP2), which contain N-terminal PAS/LOV domains in addition to the F-box motif, are involved in photomorphogenesis and regulation of the circadian clock (Sawa et al., 2007; Demarsy and Fankhauser, 2009; Kim et al., 2010).
Representing one of the biggest protein families in A. thaliana, the F-box protein superfamily has been studied on a phylogenetic and evolutionary scale (Gagné et al., 2002; Yang et al., 2008; Xu et al., 2009). However, the analysis of up to 700 genes/proteins can only provide a glimpse on the evolution, phylogenetics, and functional divergence of the various distinct subfamilies. Several of these subfamilies are by themselves larger than most other known gene families in plants. To get a better understanding of how single subfamilies evolved, we focused our attention on one of the biggest yet largely uncharacterized F-box subfamilies in A. thaliana, the FBKs. Due to nearly completely sequenced and annotated genomes, we were able to examine FBK families in the eudicots A. thaliana, P. trichocarpa, and V. vinifera, the monocots O. sativa and Sorghum bicolor, the lycophyte Selaginella moellendorffii, and the bryophyte Physcomitrella patens. In this study, we first de novo identified FBKs by a combined approach using BLASTP and hidden Markov model (HMM)-based searches in the respective genomes. We then constructed phylogenetic trees and calculated Ka/Ks values to explore the evolutionary and selective forces acting on FBKs in plants. Lastly, expression patterns of FBK genes, subcellular localization, and ASK-binding patterns of selected FBK proteins in A. thaliana were investigated to assess potential functional conservation/diversification of closely related FBKs in land plants. Besides the evolutionary insights gained by this study, these data also provide a scaffold for future functional analysis of this large family of F-box proteins.
RESULTS
FBKs Expanded in Land Plant Genomes
To identify FBKs, we performed a BLASTP search against the annotated genomes of A. thaliana (At), P. trichocarpa (Pt), and O. sativa (Os) using species-specific consensus sequences derived from the full-length protein sequences of previously published FBKs (Gagné et al., 2002; Jain et al., 2007; Xu et al., 2009). Furthermore, we used an HMM-based search to identify additional FBKs in the aforementioned species as well as in the genomes of V. vinifera (Vv), S. bicolor (Sb), S. moellendorffii (Sm), and P. patens (Pp). In the eudicots At, Vv, and Pt, we identified 103, 36, and 68 FBKs, respectively. Thirty-nine and 44 FBKs were detected in the monocots Os and Sb. Forty-six and 71 FBKs were identified in the nonseed embryophytes Sm and Pp (Table I; Supplemental Table S1), which we subsequently refer to as lower land plants. Whereas animal model organisms and the single-celled green alga Chlamydomonas reinhardtii contain only a single FBK each (Supplemental Table S2), this subfamily of F-box genes has apparently dramatically expanded in land plants. The observation that FBKs are prevalent in high numbers not only in monocots and eudicots but also in the lower land plants indicates an early expansion of this F-box subfamily in land plant history.
Table I. Number of F-box kelch proteins in A. thaliana, V. vinifera, P. trichocarpa, O. sativa, S. bicolor, S. moellendorffii, and P. patens.
| Species | No. of FBKs (K) | No. of F-Box Genes (F)a | No. of Protein-Coding Genes (G)b | Percent F/Gc | Percent K/Gd | Percent K/Fe |
| A. thaliana | 103 | 692 | 27,379 | 2.53 | 0.38 | 14.88 |
| V. vinifera | 36 | 156 | 30,434 | 0.51 | 0.12 | 23.08 |
| P. trichocarpa | 68 | 337 | 41,377 | 0.81 | 0.16 | 20.18 |
| O. sativa | 39 | 779 | 40,838 | 1.91 | 0.10 | 5.01 |
| S. bicolor | 44 | 549 | 34,496 | 1.59 | 0.13 | 8.01 |
| S. moellendorffii | 46 | 247 | 22,273 | 1.11 | 0.21 | 18.62 |
| P. patens | 71 | 165 | 39,727 | 0.42 | 0.18 | 43.03 |
Number of F-box genes (At, Os, Pt [Xu et al., 2009]; Vv [Yang et al., 2008]; Sb, Pp, Sm [superfamily 1.73, release August 2010; Gough et al., 2001]).
Number of predicted protein-coding genes (At [TAIR9]; Vv, Pt, Os, Sb, Sm, Pp [Phytozome version 5.0]).
Percentage of genes that encode F-box proteins.
Percentage of genes that encode FBKs.
Percentage of F-box genes that contain kelch repeats.
The presence of the F-box and kelch domains for each of the proteins implemented in the following analyses was verified by the Pfam database (version 24.0, release October 2009; Sonnhammer et al., 1997). While we are not aware that FBKs for Vv, Sb, Sm, and Pp have been described before, we generally identified more FBKs than previously reported for the remaining three species. Initially, Gagné et al. (2002) identified 98 FBKs in At. We recovered all of these FBKs, but three of them (AT2G03460, AT2G29610, and AT4G39750) were not confirmed as FBK proteins by Pfam and therefore were excluded from further analysis. In addition to the remaining 95 genes/proteins identified by Gagné et al. (2002), we were able to identify eight novel FBKs. In Pt and Os, we recovered all previously published FBKs and were able to significantly increase these numbers (Os, 39 in this study versus 25 by Jain et al. [2007]; Pt, 68 in this study versus 40 by Xu et al. [2009]; Table I). Hence, the combination of BLASTP and HMM-based search algorithms efficiently identified previously known and numerous novel FBKs. Since the majority of the novel AtFBKs, for example, were present in the category “F-box proteins with unknown C-terminal domains” in the other studies, the better recognition of the kelch domain by our approach most likely contributes significantly to the identification of such novel FBKs. However, since the kelch motif is rather weakly conserved at the sequence level (Supplemental Fig. S1), the existence of additional yet undetected FBKs is likely. Furthermore, cases of complete loss of the kelch domain would not have been detected in this study. In contrast to plants, FBKs are absent from prokaryotes and occur only rarely in nonplant eukaryotic genomes, usually as single-copy genes with three conserved kelch repeats (Supplemental Table S2). This indicates a possible single common ancestor of FBKs in eukaryotes and a dramatic expansion in land plants. The ratio of genes encoding the F-box protein superfamily (F) and the FBK subfamily (K) relative to the whole protein-coding genome (G) is highest for At among the seven analyzed land plant species (F of G, approximately 2.5%; K of G, approximately 0.4%; Table I). However, due to the lower total number of F-box genes, the eudicots Vv and Pt show a higher ratio of FBK-encoding genes in relation to the total number of F-box genes (K of F, approximately 23.1% and approximately 20.2%, respectively). Since F-box proteins have been shown to be involved in numerous developmental processes (Lechner et al., 2006), one could expect lower land plants to reveal decreased numbers of F-box proteins relative to the complete genome. But this is obviously not the case, and interestingly, roughly half of the F-box proteins in Pp are FBKs (Table I).
FBKs and FBAs Are Closely Related
With both consensus sequence and HMM-based searches, numerous F-box proteins with so-called F-box associated domains were identified in addition to the FBKs (Table II; Supplemental Table S3). Despite its name, the F-box associated domain also occurs in the absence of the F-box domain (Jaso-Friedmann et al., 2002). Although FBKs and F-box proteins with C-terminal F-box associated domains (FBAs) are annotated as different F-box subfamilies in the Pfam database and previous publications (Xu et al., 2009), FBKs and FBAs are quite similar on the amino acid sequence level (Supplemental Fig. S2). Using our consensus sequences, we did not detect members of any F-box protein subfamily other than FBKs and FBAs. This indicates that the similarity is not due to the similarity of the F-box domain but rather to C-terminal regions of the proteins, most likely the similarity between the F-box associated domains and kelch repeats (Supplemental Fig. S2). Therefore, we speculate that the F-box associated domain might also form a tertiary propeller-like structure. Although FBAs represent the largest F-box protein subfamily, with 206 members in At (Xu et al., 2009), little functional information is available. So far, five AtFBAs have been characterized in detail and are involved in lateral root formation (Dong et al., 2006), pathogen responses (Kim and Delaney, 2002; Gou et al., 2009), and ethylene signaling (Qiao et al., 2009). Some additional FBAs are related to proteins that are part of the self-incompatibility system (Wang et al., 2004). It is unclear why this subfamily expanded to this extent in the self-fertile species At.
Table II. Number of F-box proteins with FBAs in A. thaliana, V. vinifera, P. trichocarpa, O. sativa, S. bicolor, S. moellendorffii, and P. patens.
| Species | No. of FBAs | No. of F-Box Proteins with FBA and Kelch Domains | Previously Published Nos. of FBAsa |
| A. thaliana | 205 | 1 | 193 |
| V. vinifera | 3 | 2 | – |
| P. trichocarpa | 47 | 4 | 32 |
| O. sativa | 38 | 1 | 2 |
| S. bicolor | 29 | 5 | – |
| S. moellendorffii | 2 | 1 | – |
| P. patens | 5 | 2 | – |
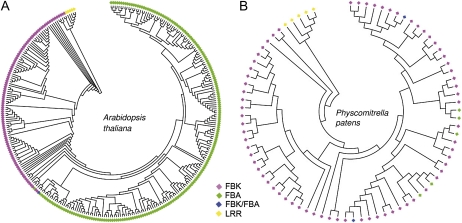
To get an insight into the phylogenetic relationship of FBKs and FBAs, we created phylogenetic trees including FBKs and FBAs of At and Pp, respectively (Fig. 1). For the At tree, we used the 103 FBKs that were confirmed by us and 206 FBAs, of which 193 were previously published (Xu et al., 2009). To construct the Pp tree, 71 FBKs and seven FBAs (five FBAs + two FBK/FBAs, both identified in this study) were used (Supplemental Tables S1 and S3). While the number of FBKs in At and Pp is rather similar in both species (103 in At versus 71 in Pp), the number of FBAs varies substantially (206 in At versus seven in Pp; Table II). Apparently, FBAs have dramatically expanded in At. Furthermore, in At, FBKs and FBAs form closely related but clearly distinct groups within the phylogenetic tree (Fig. 1). The same is true for Pt, Vv, Os, and Sb (Supplemental Fig. S3). In contrast to that, such an obvious phylogenetic distinction of FBKs and FBAs is not visible in the Pp tree (Fig. 1). In both At and Pp, the common ancestors of FBKs and FBAs contain kelch domains, indicating that FBKs may be evolutionary precursors of the FBAs. Taken together, our data indicate that kelch and F-box associated domains share a common evolutionary history.
Figure 1.
FBKs and FBAs are closely related to each other. A, NJ tree of 103 FBK proteins and 205 FBAs of A. thaliana. B, NJ tree of 71 FBKs and 5 FBAs of P. patens. Alignment of full-length protein sequences and construction of the NJ trees were performed in MEGA using a bootstrap value of 1,000. The A. thaliana tree was rooted with the auxin receptors (F-box LRRs) TIR1, AFB1, AFB2, AFB3, and AFB5. The P. patens tree was rooted with the respective homologous genes identified by BLASTP searches with AtTIR1. FBK/FBA proteins are characterized by overlapping positions of an F-box associated domain and individual kelch repeats.
Evolution of FBKs in Seven Land Plant Species
To compare FBKs in different land plant species, we created a phylogenetic tree including all identified FBKs of the seven analyzed species by using neighbor-joining (NJ) methods (Fig. 2A; Supplemental Fig. S4). Since previous studies generally analyzed the whole F-box protein superfamily, phylogenetic reconstructions were most feasible by using only the F-box domain. Analyzing F-box proteins with the same C-terminal interaction domains enabled us to include the amino acid sequence of the full-length proteins. Therefore, this tree reflects the evolution of both the F-box and kelch domains. Since we were unable to identify FBKs in Charophyceaes, a small group of predominantly freshwater green algae that represent the most recent common ancestor of land plants (Kenrick and Crane, 1997), we rooted the tree with the previously mentioned human FBK (Sun et al., 2009) and the only FBK that we identified in the annotated genome of the single-celled green alga C. reinhardtii. To confirm the NJ tree, three additional representative trees were constructed by using maximum likelihood, Bayesian, and NJ methods (Supplemental Figs. S5 and S6). The representative trees with the best likelihood support the NJ tree in Figure 2A (Supplemental Table S4). The significance of this finding was supported by comparing the topologies using p-SH (Shimodaira and Hasegawa, 1999) and 1sKH (Kishino and Hasegawa, 1989) tests. To further evaluate the quality of the NJ tree, we performed an exemplary search for orthologous genes using the 39 Os FBKs (Supplemental Table S1) as queries. The search was carried out in the ENSEMBL tool using the GRAMENE (http://www.gramene.org) and GEVO (http://synteny.cnr.berkeley.edu/CoGe) databases. Nearly all proteins annotated as orthologous in ENSEMBL also formed orthologous clusters in our NJ tree. Therefore, we conclude that the NJ tree topology accurately reflects the phylogenetic relations between FBKs in the analyzed species.
Figure 2.
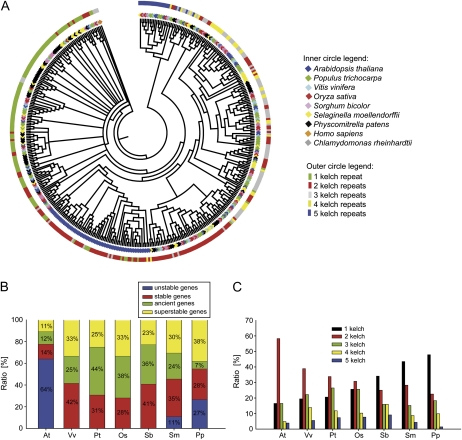
FBKs cluster according to their number of kelch repeats. A, NJ tree created with the full-length protein sequences of A. thaliana, V. vinifera, P. trichocarpa, O. sativa, S. bicolor, S. moellendorffii, and P. patens FBKs. The tree was rooted with FBKs of C. reinhardtii and Homo sapiens. The inner colored circle corresponds to the different species. The outer colored circle indicates the number of kelch repeats. B, Ratio of unstable, stable, ancient, and superstable FBKs in the seven analyzed land plant species. C, Ratio of FBKs with one, two, three, four, or five conserved kelch repeats according to Pfam in the seven analyzed land plant species.
We identified a total of 40 well-supported clades ranging from species-specific clades to clades containing FBKs from all seven analyzed species. For the evolutionary classification of the genes in the all-species tree (Supplemental Fig. S4), we adopted the “stable/unstable” terminology coined by Thomas (2006) in a slightly modified manner and extended it with “superstable” and “ancient” categories (Supplemental Fig. S4). Unstable genes are lineage specific without clear orthologs in the other species analyzed. Stable genes are conserved across species with orthologs in at least one additional species. Superstable genes have orthologs in all analyzed species and, therefore, exhibit the highest degree of evolutionary conservation. It is conceivable that superstable genes perform functions in developmental or physiological processes conserved in all land plant species. The distinction between stable and superstable genes cannot be directly translated into the evolutionary age of a certain gene, because possible gene losses in individual species have to be taken into account. Therefore, we classified genes as ancient if they contain orthologs in at least one lower land plant, one monocot, and one eudicot species.
We identified eight clades of superstable genes (Supplemental Fig. S4). Depending on the species and the number of paralogs within a species, the ratio of genes in this category varies from 11% to 38% (Fig. 2B). While all species likewise contained various numbers of stable genes, we identified unstable genes only in At and the two lower land plant species (Fig. 2B; Supplemental Table S5). We are aware that the identification of unstable genes is strongly affected by the set of species included in the analysis. Since the two Poaceae species Os and Sb, for example, are closely related, the existence of unstable genes as we defined them is less likely than for species with no close relatives analyzed. However, if we account for this bias by screening for clades that contain either monocot-specific or Pt-Vv-specific FBKs, we find only three genes specific for Os and Sb and none specific for Pt and Vv. Therefore, most FBKs in these species (Pt, Vv, Os, and Sb) likely perform functions conserved across species. In agreement with this, we find that the majority of FBKs in these species are of ancient origin (ancient + superstable; Fig. 2B). In At, on the other hand, we identified a large lineage-specific clade of unstable genes containing 64% of all FBKs in this species. This indicates that most FBKs in At result from lineage-specific duplication events. All unstable AtFBKs clustered in clade 22 (Supplemental Fig. S4). To assess how recent this clade evolved/expanded, we screened the genome of Arabidopsis lyrata, a close relative within the same genus, for orthologous genes. We identified A. lyrata orthologs for most of the At members of this clade (Supplemental Fig. S7), indicating that the expansion of this clade predates the split of At and A. lyrata. Furthermore, orthologs of this clade can also be identified in EST databases of other related Brassicaceae species (data not shown) suggesting that this clade originated after the separation of Pt and the Brassicaceae. Several of the Sm and Pp FBKs (20% and 25%, respectively) fell into clades containing only lower land plants (Supplemental Fig. S4). These genes either duplicated after the separation of lower land plants and angiosperms or were completely lost in the ancestor of monocots and eudicots. As in At, several FBKs of Sm and Pp (11% and 27%) are lineage specific (Fig. 2B). Taken together, while most eudicots and both monocots contain only FBKs that are conserved across species, significant portions of At and lower land plant FBKs are lineage specific/unstable.
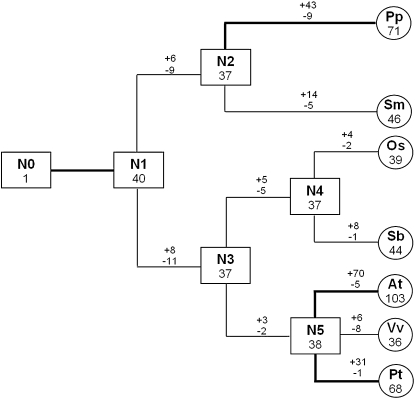
We next estimated the number of FBKs in the most recent common ancestor (MRCA) of the seven species analyzed in this study and determined the number of gained and lost genes. Reconciliation of the all-species tree identified 40 clades containing orthologous genes that were present in the potential MRCAs of the seven analyzed species (N1; Fig. 3). Furthermore, we identified 37 orthologous genes in the lower land plant MRCA (N2), the angiosperm MRCA (N3), and the monocot MRCA (N4) and 38 in the MRCA of eudicots (N5). When we compared the number of ancestral genes with those in the extant species, it appeared that the FBK family has expanded in several of the analyzed species. In At, for example, the number of FBKs increased approximately 2.5-fold since the divergence of the various eudicot species from their respective MRCA. This is consistent with the situation of the whole F-box superfamily in At, which has increased in size as much as 3-fold since the divergence of eudicots and monocots 145 million years ago (Xu et al., 2009). In Vv and Os, the number of FBKs remained largely unchanged since the emergence of angiosperms and the divergence of eudicots and monocots, respectively. In summary, while C. reinhardtii and animal species usually contain a single-copy FBK (Supplemental Table S2), the number of FBKs increased dramatically in land plants (N0–N1; Fig. 3). The number of FBKs then remained relatively stable through evolutionary history from the land plant MRCA (N1) to the MRCAs of the lower land plants (N2) and monocots/eudicots (N4/N5; Fig. 3). Only after the separation of the various eudicot species did FBKs once more expand significantly. Such inconsistent rates of gene gains over time had previously been demonstrated by Hanada et al. (2008), who showed that gain rates for branches linked together by older ancestral nodes are smaller than those linked by younger branches.
Figure 3.
Evolutionary change in the number of FBK proteins in land plants. The numbers in squares and circles represent the maximum numbers of genes in ancestral and extant species, respectively. The numbers with plus and minus indicate the gene gains and losses, respectively, for each branch. Thick lines represent branches with high gene expansion rate. N0, Eukaryotic ancestor; N1, land plant ancestor; N2, lower land plant ancestor; N3, angiosperm ancestor; N4, monocot ancestor; N5, eudicot ancestor. Branch lengths are not in proportion to evolutionary time.
FBKs Cluster According to Their Number of Kelch Repeats
The all-species tree shows that FBK proteins cluster according to the number of kelch repeats (Fig. 2A, outer circle). In principle, the tree shows large clades including mainly FBKs with one and two kelch repeats. FBKs with higher numbers of kelch repeats are most common in an additional clade, whereby the FBKs with five kelch repeats are clearly concentrated in a distinct subclade. As the consensus sequences (Supplemental Fig. S1) indicated, kelch repeats may be more similar when found at the same position in different proteins compared with repeats within the same protein. To substantiate this observation, we performed a permutation test to estimate whether the similarity of kelch repeats within a protein differs from the similarity between proteins. And indeed, kelch repeats at the same position between proteins are significantly more similar to each other than the repeats within the same protein (P = 2 × 10−6; Supplemental Fig. S8). This indicates that multiple kelch repeats did not arise independently in each protein but were already present in “the” ancestral FBK. In numerous proteins, we detected conserved amino acid residues downstream of the last Pfam-confirmed kelch repeats that were quite similar to a kelch motif structure but not conserved well enough to be considered a kelch motif by Pfam. We assume that these conserved regions are rudimentary kelch repeats that decayed over time. The only FBK we identified in C. reinhardtii contains three kelch repeats. Likewise, the FBKs identified in other animal model species also contain three conserved kelch repeats recognized by Pfam (Supplemental Table S2). Hence, we hypothesize that the MRCA of eukaryotic FBKs was formed by the combination of an F-box and three conserved kelch repeats.
Unstable FBKs Are Organized in Clusters of Tandem Repeats
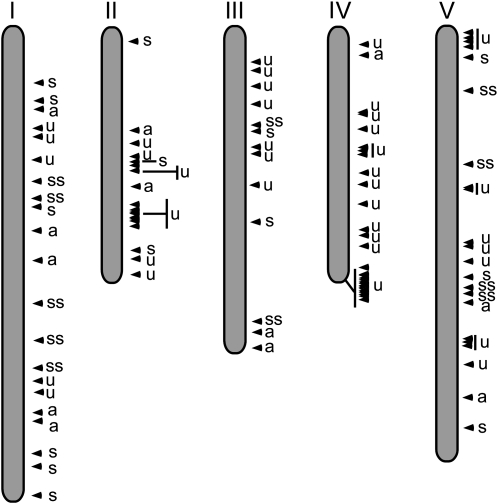
The hypothesis that correlates stable/superstable FBKs with conserved and potentially ancient functions in plant development and physiology likewise suggests that unstable genes constantly continue to evolve by birth-death evolution (Thomas, 2006). Since significant portions of F-box genes are arranged in tandem repeats, this hypothesis would predict that such tandemly arranged genes are primarily unstable genes. Indeed, this prediction is supported by the chromosomal localization of FBKs in At (Fig. 4), the species with the highest ratio of unstable genes (64%; Fig. 2B). Unstable genes are strongly clustered, while stable and superstable genes are evenly scattered over the chromosomes (Fig. 4). Substantiating this, the majority of unstable genes in At (62%) emerged after the most recent whole genome duplication event (Blanc et al., 2003; Bowers et al., 2003; http://wolfe.gen.tcd.i.e./athal/dup). In contrast to At, where 35% of the FBKs are arranged in tandem repeats, considerably fewer FBKs are arranged in tandem repeats in Pt (3%), Vv (17%), Os (5%), and Sb (14%), indicating that in these species, FBKs mainly emerged by mechanisms other than tandem duplication. Analysis of the Pt genome, for example, revealed evidence of a recent whole genome duplication event (8–13 million years ago) that affected approximately 92% of the Pt genome (Sterck et al., 2005; Tuskan et al., 2006). In agreement with the low ratio of tandem repeat FBKs in Pt, Vv, Os, and Sb, they do not contain unstable genes (Fig. 2B). Furthermore, with the exception of Pt, the total number of FBKs in these species is rather similar to that of the respective MRCAs (Fig. 3). In summary, our results suggest that after the dramatic expansion of the FBK gene family in land plants that followed the divergence from C. reinhardtii/single-celled green algae, a second wave of expansion can largely be explained by tandem duplications.
Figure 4.
Chromosomal distribution of A. thaliana FBKs. Approximate positions of FBKs are displayed on the respective chromosome (black triangles). Letters denote evolutionary classification of FBKs according to Supplemental Figure S4. a, Ancient; s, stable; ss, superstable; u, unstable.
Superstable and Unstable FBKs Carry Different Signatures of Selection
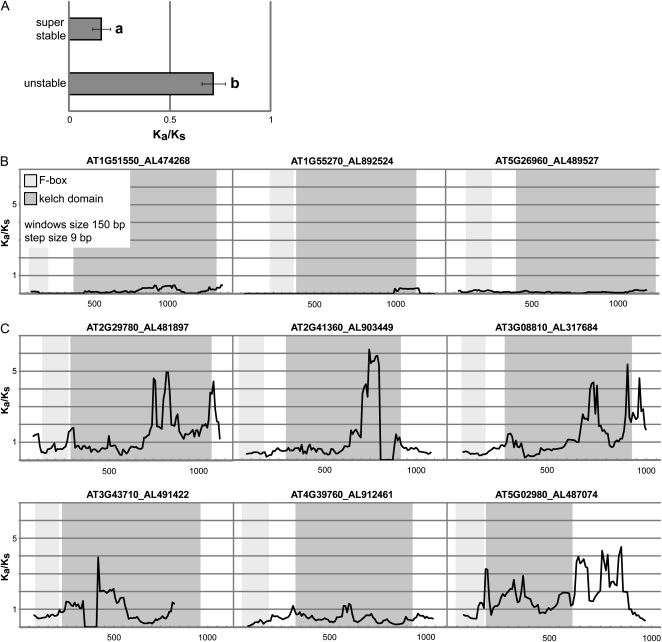
If stable and superstable genes have evolved to recognize similar targets across species, one would expect purifying selection to act on such genes. Genes that confer adaptational properties, on the other hand, typically are under positive selective pressure. To examine whether these expectations also hold for FBKs, we determined Ka/Ks ratios for superstable and unstable genes of At. For the calculation of Ka/Ks ratios, we first identified the closest orthologs for each gene in the genome of the close relative A. lyrata (Supplemental Fig. S7) and included only those At genes that had a single ortholog in A. lyrata. Ka/Ks ratios of 0.16 for the complete coding regions for superstable FBKs (Fig. 5A) strongly indicate purifying selective pressures. In contrast to that, unstable genes seem to be more close to neutral selection, as inferred by significantly higher Ka/Ks ratios (0.72; P < 0.0001) for the complete coding region (Fig. 5A).
Figure 5.
Divergence levels of FBKs (A. thaliana versus A. lyrata). A, Mean Ka/Ks ratios of superstable (n = 10) and unstable (n = 37) FBKs. Only A. thaliana genes with a single A. lyrata ortholog were included (Supplemental Fig. S7). Error bars denote se. a and b denote significant differences according to Student’s t test (P < 0.0001). B, Sliding window plots of representative superstable FBKs. C, Sliding window plots of representative unstable FBKs. For sliding window analysis, nucleotide sequences of A. thaliana (indicated by Arabidopsis Genome Initiative identifier) and homologous nucleotide sequences of A. lyrata (indicated by protein identifier according to the Joint Genome Institute) were used. Window size was 150 bp, and step size was 9 bp. For A. lyrata protein 491422, only a partial coding sequence could be analyzed. Light gray boxes highlight the F-box domain, and dark gray boxes highlight the kelch domain positions.
We then randomly selected representative genes for sliding window analysis to identify possible differences between F-box and Kelch domains (Fig. 5, B and C). For unstable genes, sliding window analysis clearly shows numerous sites/regions under positive selection, with Ka/Ks >> 1 (Fig. 5C). Furthermore, for unstable genes, we observed that the F-box domain had generally lower Ka/Ks values in comparison with the C-terminal part of the proteins/genes including the kelch domain (Fig. 5C). This suggests that as a consequence of natural selection, ASK-binding patterns remain conserved while the target-recruiting kelch domain has evolved to recognize new target proteins.
Closely Related FBKs Are Differentially Expressed in At
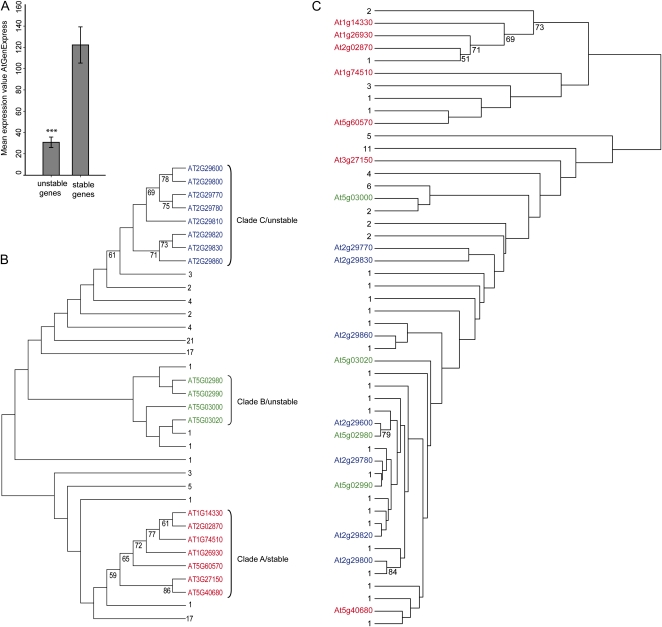
The recent expansion of FBKs especially in At raises two interesting questions. First, are unstable genes generally expressed, or have many of them already become pseudogenes? Second, are phylogenetically closely related FBKs functionally redundant, or have duplicated genes subfunctionalized? To address these questions, we made use of the publicly available AtGenExpress data (Schmid et al., 2005; Toufighi et al., 2005) and additionally selected a small clade consisting of seven closely related AtFBKs (clade A in Fig. 6B) for further molecular characterization.
Figure 6.
Differential transcription profiles of phylogenetically closely related FBKs. A, Mean expression values for stable (including ancient and superstable) and unstable A. thaliana FBKs extracted from the AtGenExpress_Plus extended tissue series (Schmid et al., 2005). Error bars represent se. Statistical significance was assessed using Student’s t test (*** P < 0.001). B, NJ tree of 103 A. thaliana FBKs based on amino acid sequence homology. C, Phylogeny based on coexpression data from the AtGenExpress_Plus extended tissue series including 81 FBKs of A. thaliana. Discrepancy in the number of FBKs between the trees results from 22 missing FBKs on the ATH1 microarray. Noninformative clades were collapsed and labeled according to the number of underlying FBKs. Numbers at nodes display bootstrap values greater than 50%. Classification of clades as stable and unstable is according to the categorization of FBKs in Supplemental Figure S4.
Although both stable (including also ancient and superstable) and unstable genes were expressed at rather low levels in the AtGenExpress extended tissue series (Schmid et al., 2005), the average expression levels across tissues were significantly higher for stable than for unstable genes (P < 0.001; Fig. 6A). Fifty-eight percent (26 of 45 that were present on the ATH1 array) of the unstable genes had mean absolute expression levels of less than 25 across the 86 tissues/developmental stages assessed (Supplemental Data Set S1). However, only four of these 26 had absolute expression levels of less than 50 for all tissues/developmental stages assessed, demonstrating that the vast majority of these unstable FBKs were significantly higher expressed (greater than 50) in at least a few selected tissues/developmental stages (Supplemental Data Set S1). This indicates temporal and/or spatial specialization of unstable genes, which argues against widespread pseudogenization.
To investigate possible functional diversity of AtFBKs in general, we again used the AtGenExpress data. Both members of phylogenetically closely related unstable FBKs organized in tandem repeats (e.g. clades B + C in Fig. 6B) and clades of stable FBKs (clade A in Fig. 6B) grouped into different clades when clustered according to their coexpression profiles (Fig. 6C). These array data could be confirmed by quantitative PCR data of the seven clade A genes (Supplemental Fig. S9). Together with the above-described temporal and/or spatial specialization of unstable genes, this indicates that FBKs within a common phylogenetic clade can be differentially regulated at the mRNA level and therefore could have evolved different functions or spatiotemporal specificities.
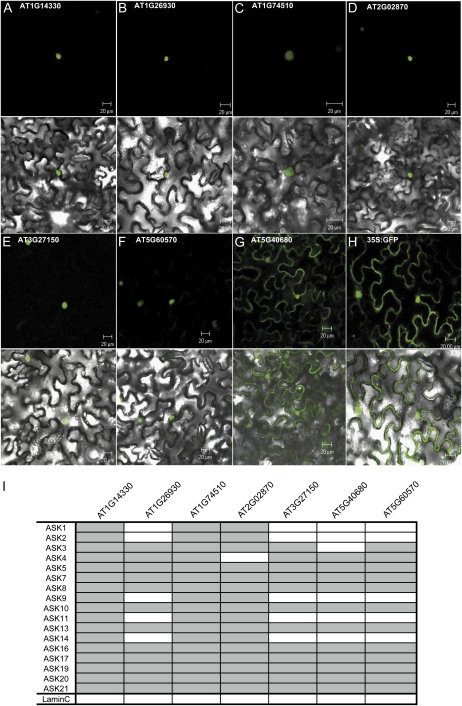
Posttranslational mechanisms that might create functional diversity between family members are subcellular localization and the ASK-binding patterns that define the specific SCF complexes in which a certain F-box protein may be incorporated. Subcellular localization was examined with GFP-FBK fusion proteins of the seven clade A FBKs (Fig. 6B), which were transiently transformed into epidermal cells of Nicotiana benthamiana (Fig. 7, A–H). Six of them localized exclusively to the nucleus. AT5G40680 was additionally located in the cytoplasm. In agreement with this, the 26S proteasome is present in both the cytoplasm and the nucleus (Book et al., 2009). The specific interaction of the FBKs with 17 of the 21 ASK adaptor proteins was examined in the yeast two-hybrid system. ASK6 and ASK15 were excluded from the interaction studies because they were shown to be pseudogenes (Seki et al., 2002; Takahashi et al., 2004). ASK12 and ASK18 were excluded because they showed autoactivity in our system. Figure 7I indicates that the ASK-binding pattern seems to be rather unspecific for the seven selected FBKs. Similar unspecific binding had been reported previously for other F-box proteins such as the kelch domain-containing ZTL (Risseeuw et al., 2003). The seven analyzed FBKs interacted with 11 to 17 of the tested ASK proteins. However, a pattern is visible in which AT1G26930, AT3G27150, AT5G60570, and AT5G40680 interact with nearly the same ASK proteins. The same is true for AT2G02870, AT1G74510, and AT1G14330. Taken together, while the selected FBKs showed rather similar ASK interaction patterns and subcellular localizations, they differed considerably in their expression profiles, indicating that possible functional diversification may be achieved by a combination of transcriptional regulation and positive selection acting on the kelch domain.
Figure 7.
Protein features of selected A. thaliana FBKs. A to H, Subcellular localization of 35S::GFP-FBK constructs in leaf epidermal cells of N. benthamiana. I, Yeast two-hybrid analysis of specific interactions of the selected FBK proteins with ASK adaptor proteins. BD fusions of ASK proteins were tested with AD fusions of FBK proteins. Gray and white boxes indicate positive and negative interactions, respectively. Human LaminC served as a negative control.
DISCUSSION
Comparison of gene family content across species may provide insight into evolutionary mechanisms that have shaped adaptation and diversity (Rubin et al., 2000). The F-box gene superfamily represents one of the largest and fastest evolving gene families in the plant kingdom (Clark et al., 2007). While this superfamily in its entirety had previously been characterized on a phylogenetic and evolutionary scale (Gagné et al., 2002; Yang et al., 2008; Xu et al., 2009), a detailed characterization of one of its subfamilies allows us to turn the focus from the F-box domain to a specific protein-protein interaction domain. The F-box protein subfamily we chose for this study was composed of an N-terminal F-box with various numbers of C-terminal kelch repeats.
The Kelch Repeat Domain
In yeast, Drosophila, and mammalian F-box proteins, WD40 repeats and LRRs are the predominant substrate recruitment domains (Skaar et al., 2009a, 2009b). Interestingly, both kelch and WD40 repeats adopt the stereotypical topology of a β-propeller. As kelch and WD40 repeats have no similarity at the sequence level, it was speculated that convergent evolution of a subset of F-box proteins has originated a common tertiary structure specialized in protein-protein interactions (Andrade et al., 2001).
Kelch repeat proteins have become widespread in evolution. Typically, five to seven kelch repeats form a β-propeller with the blades arranged around a central axis. Intrablade and interblade loops of varying lengths protrude above, below, or at the sides of the β-sheets and contribute variability to the binding properties of individual β-propellers (Fülöp and Jones, 1999; Jawad and Paoli, 2002; Prag and Adams, 2003). The entire kelch β-propeller forms a functional unit that can be found in combination with other conserved protein domains (Adams et al., 2000). In plants, kelch motifs have been identified in proteins without additional conserved domains (Prag and Adams, 2003) as well as in combination with various other N- and/or C-terminal domains, such as C-terminal phosphatase domains (Mora-García et al., 2004) or N-terminal acyl-CoA-binding domains (Suzui et al., 2006; Du et al., 2010). However, the combination with N-terminal F-box domains seems to be most prevalent. With the exception of AtAFR (AT2G24540), all functionally characterized kelch proteins in plants contain a minimum of five kelch repeats. Therefore, they match the prerequisites to form a closed propeller structure with stabilized interactions between the first and last blades. We found, however, that the vast majority of FBKs in plants contain less than three kelch repeats (Fig. 2C). It is questionable, therefore, whether functional β-propellers can be formed in such proteins. A survey of the Pfam database for kelch motif-containing proteins revealed a total of approximately 400 different protein architectures (irrespective of the species background and the presence of other functional domains). Approximately 65% of the proteins underlying these architectures contain only one or two kelch repeats. Since it is unlikely that this majority of kelch domains is nonfunctional, several scenarios are possible. (1) It is conceivable that FBK proteins with fewer kelch repeats dimerize to achieve a full set of propeller blades. (2) Or they operate via a completely different mechanism to interact with target proteins. (3) Alternatively, a significant number of these genes may be nonfunctional and represent the remains of once functional FBKs. (4) Lastly, because of poor sequence conservation of the kelch motif, the missing repeats are present but not recognized by Pfam. In agreement with the latter notion, we detected conserved C-terminal residues that resemble the kelch motif structure. Furthermore, other motif recognition algorithms/databases such as Interpro recognize a complete kelch β-propeller motif (IPR015915) in the majority of the FBKs. Hence, we assume that most plant FBKs do indeed form functional β-propeller-like tertiary structures. Such conserved C-terminal residues indicate gradual degeneration of the kelch consensus sequence toward the C terminus. In contrast to earlier studies from Drosophila (Xue and Cooley, 1993; Bork and Doolittle, 1994), we found that kelch motifs at given positions within the kelch repeat domain between proteins are more similar to each other than different kelch motifs within a protein (Supplemental Fig. S8). In agreement with this, we derived specific consensus sequences for each repeat position (Supplemental Fig. S1), and FBK proteins in the all-species tree generally clustered according to their number of kelch repeats (Fig. 2A). This may simply reflect the progressive degeneration of the C-terminal kelch repeat primary sequence or, alternatively, may indicate that each blade position in the β-propeller may require specific functional residues.
The All-Species Tree Identifies Superstable, Stable, and Unstable Genes
The all-species tree contains valuable information on several levels. It reveals the phylogenetic architecture of FBKs between species and identifies clades with genes conserved across species as well as lineage-specific clades. In a similar approach, Thomas (2006) performed a detailed characterization of F-box proteins in three Caenorhabditis species. The author divided the genes into stable and unstable categories. It was hypothesized that stable genes most likely became devoted to specific endogenous substrates long ago, while unstable genes continued to evolve by birth-death evolution and are primarily involved in recognizing foreign proteins and targeting them for degradation (Thomas, 2006). Accordingly, he found strong evidence for positive selection in the C-terminal substrate-binding domains of unstable genes, while those of stable genes seemed to be under purifying selection. In this study, we adopted similar gene categories and identified similar signatures of natural selection acting on FBKs in At. While superstable FBKs had very low Ka/Ks values, indicating purifying selective pressures, Ka/Ks ratios of unstable FBKs were significantly higher (Fig. 5A). Sliding window analyses showed that numerous regions in the unstable genes seemed to be under strong positive selection (Fig. 5C). While in most cases the F-box domain is rather conserved, it is the substrate-recruiting kelch domain that seems to be positively selected for. Hence, a picture emerges in which kelch repeats evolve in a manner that supports the constant development of novel substrate specificities. Furthermore, as predicted, we found unstable FBKs from At to be strongly clustered with respect to their chromosomal localization.
If this large pool of unstable FBKs in At is indeed used to recruit targets for specialized environmental responses with potential adaptive importance, it is clear why to date only a tiny fraction of members of the plant F-box gene superfamily have been identified in forward genetic screens, which are usually designed to identify components of more general mechanisms of development or physiology. Consequently, the few biologically characterized FBKs are, as expected, ancient genes with conserved functions. AFR, ZTL, FKF1, and LKP2 are all members of phylogenetic clades with orthologs in several species (Supplemental Fig. S4) and perform functions in essential physiological processes such as the regulation of light responses and circadian rhythms. And, not surprisingly, all but one of the 38 F-box genes functionally characterized to date in At have orthologs in one or several other of the species analyzed in this study (data not shown). The only exception is SUPPRESSOR OF NIM1-1 (SON1; Kim and Delaney, 2002). The substrate/target protein of SON1 is not known. But remarkably, SON1 plays a role in pathogen response and therefore perfectly fits Thomas’ (2006) category of unstable genes with a possible function in the recognition of foreign proteins. While unstable genes are dominating in At, we identified significantly fewer or no unstable FBKs at all in the other species analyzed in this study. Following the initial hypothesis, the vast majority of FBKs in these species most likely perform conserved functions. In agreement with this, more than 50% of Vv, Pt, Os, Sb, and Sm FBKs fall into the ancient or superstable category (Fig. 2B). Therefore, at least for FBKs, the emerging paradigm of rapidly evolving gene families organized in tandem repeats cannot be easily transferred from At to other plant species.
Functional Redundancy and Subfunctionalization
The total number of FBKs among the plant genomes assessed in this study was highest for At, which made this species a suitable model to study possible functional consequences of gene family expansion. Although Gagné et al. (2002) argued that direct sequence alignments between members of the same clade suggested that most of the At F-box proteins do not have obvious functional paralogs, molecular characterization of numerous F-box proteins has meanwhile demonstrated the opposite. At least partial functional redundancy could be shown for the auxin receptors (Dharmasiri et al., 2005), the ethylene signaling components EIN3-BINDING F-BOX PROTEIN1/2 (An et al., 2010) and EIN2-TARGETING PROTEIN1/2 (Qiao et al., 2009), VIER F-BOX PROTEINE1 to VIER F-BOX PROTEINE4, which are involved in root development (Schwager et al., 2007), and also for the FBKs ZTL, FKF1, and LKP2 (Baudry et al., 2010). Hence, although we have no functional information on the vast majority of FBKs, rapid gene family expansion suggests scenarios wherein natural selection favors additional copies either for increased dosage or an increased arsenal of molecular weaponry via subfunctionalization (Demuth and Hahn, 2009). Our expression analysis of AtFBKs supports the latter and clearly shows differences in transcriptional regulation within phylogenetic subclades. We found similar patterns for subclades with both genetically unlinked members and those that are organized in tandem repeats (Fig. 6, B and C). Hence, the transcriptional divergence seems to be independent of the genetic mechanism that led to the increase in copy numbers. Similar transcriptional and posttranscriptional diversification (by microRNAs) could be shown by GUS-reporter assays for the auxin receptors, members of the F-box LRR subfamily (Dharmasiri et al., 2005; Parry et al., 2009), suggesting that differential expression indeed contributes to functional diversification. Furthermore, although unstable genes had significantly lower mean expression values across a large number of tissues (Fig. 6A), this is most likely due to the specialization of spatiotemporal expression patterns. While the higher expressed stable genes had significant expression values in many tissues, the expression of unstable genes was specific for few selected tissues and/or developmental stages (Supplemental Data Set S1), again arguing that duplicates here subfunctionalized.
On the other hand, our molecular characterization of a selected subclade containing seven FBKs revealed conservation of protein features such as subcellular localization and ASK-binding patterns. This indicates that the family members are generally able to integrate into the same SCF complexes and act in the same cellular compartments. Therefore, we hypothesize that after copy number expansion, two genetic mechanisms mainly contributed to potential subfunctionalization of FBK family members in At: (1) different transcriptional regulation, and (2) positive selection acting primarily on the kelch domain of unstable FBKs, likely resulting in modified substrate specificities.
CONCLUSION
F-box proteins with C-terminal kelch repeats are classic multidomain proteins. While the F-box connects the protein via a restricted set of ASK adaptor proteins to the rest of the SCF complex, the C-terminal domains most likely recruit target proteins destined for proteasomal degradation. The diversity of potential substrates is mirrored by the number of different interaction domains C terminal to the F-box with kelch repeats being widespread in land plant genomes. Although there is hardly any experimental evidence addressing the function of the vast majority of these genes, their patterns of evolution are strongly suggestive. All species analyzed in this study contained numerous stable and superstable FBK genes, which are under purifying selection and potentially perform conserved functions in land plant development and physiology. Depending on the species background, several clades dramatically expanded in a lineage-specific manner. These clades contain genes that most likely evolved (or still evolve) to perform functions specific for the respective lineage. They may contribute to adaptational processes, as signatures of positive selection suggest.
Evolutionary and phylogenetic analyses of F-box protein subfamilies with other C-terminal domains (e.g. LRR, F-box associated domain) are desired to enable direct comparisons with our findings for the FBK subfamily. We hypothesize that detailed analyses of additional subfamilies with a focus on the evolutionary categories (stable/unstable) and selective pressures may reveal subfamily-specific patterns. Certain subfamilies with characteristic protein-protein interaction domains may be more capable to generate novel functions that could confer selective advantages and therefore drive adaptational processes. Naturally, subfamilies with large portions of unstable genes would be candidates for this category. Other subfamilies may contain predominantly stable or superstable genes and perform primarily conserved functions across species. However, the assignment of specific subfamilies to either category may be species dependent, as this study shows for F-box proteins with C-terminal kelch repeat domains. Additional insight will be gathered by molecular population genetic analyses primarily in At that will become feasible in the near future through the 1,001 Genomes Project (Weigel and Mott, 2009). Lastly, this analysis should provide a good basis to select promising candidates for reverse genetic characterization of FBKs.
MATERIALS AND METHODS
Identification of FBKs in Different Land Plant Genomes
The most recent annotated version of cDNAs and proteins from each of the genomes was downloaded from the respective genome sequence sites (status March 2009: Arabidopsis thaliana from The Arabidopsis Information Resource [ftp://ftp.arabidopsis.org/home/tair, arabi_cds_v.090704]; Populus trichocarpa [version 1.1], Physcomitrella patens [version 1.1], and Selaginella moellendorffii [filtered model 3] from the Joint Genome Institute [http://www.jgi.doe.gov/]; Vitis vinifera [version 1] from Genoscope [http://www.cns.fr/externe/GenomeBrowser/Vitis/]; Oryza sativa [Osjaponica_cds300503] from The Rice Genome Annotation Project [ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs]; Sorghum bicolor [version 1.4] from the Munich Information Center for Protein Sequences [http://mips.helmholtz-muenchen.de/plant/sorghum]). Published FBK protein sequences (Gagné et al., 2002; Yang et al., 2008; Xu et al., 2009) were used as first queries for FBKs using BLASTP and TBLASTN (Altschul et al., 1997). Identified full-length cDNAs were translated in the correct frame. Putative FBK sequences were aligned using ClustalX (Thompson et al., 2002). Alignments were verified manually, and a consensus sequence was created for each of the motifs of interest with the help of the Weblogo software package (http://weblogo.berkeley.edu/logo.cgi). Consensus sequences were then used in a BLASTP search to identify more FBKs from the downloaded genomes. This alignment was used to generate an HMM model using the program hmmbuild from the HMMER program suite (Eddy, 1998). The HMM model was further improved by calculating HMM parameters with the hmmcalibrate package (Eddy, 1998). Using hmmsearch, the HMM model was applied in a search against the most recent protein annotations from each plant. To confirm the presence of both F-box and kelch domains in the obtained sequences (e < 3.8), we further compared the results from hmmsearch and the Pfam databases (Sonnhammer et al., 1997) with the hmmpfam package. Our domains of interest are annotated in Pfam as PF00646 (F-box), PF01344 (kelch domain 1), PF07646 (kelch domain 2), PF04300 (FBA_1), and PF08268 (FBA_3). We did not use the domain DUF1668 (PF07893), classified as a member of the kelch family in Pfam, because this domain was not detected in our searches.
Construction of F-Box and Kelch Consensus Sequences
To construct consensus sequences of the F-box and kelch 1 repeat, complete protein sequences of FBKs of all seven plant species were aligned using ClustalW (Thompson et al., 2002) and corrected manually. To create consensus sequences of the kelch 2, kelch 3, kelch 4, and kelch 5 repeats, only FBKs were aligned that actually include the required number of kelch repeats. Gaps occurring in the alignment were deleted if more than 75% of the aligned sequences contained a gap in the same position. Positions of F-box and kelch domains were predicted according to the matches in Pfam (Sonnhammer et al., 1997).
Alignments of Kelch Repeats and Phylogenetic Reconstruction
With the hmmalign package from the HMMER suite, alignments for the phylogenetic reconstruction were created by applying the HMM-calibrated model obtained previously. All 407 FBKs and the outgroup protein sequences aligned with hmmalign were used to generate a final tree constructed with the NJ algorithm implemented in PHYLIP (bootstrap = 100, seqboot; Felsenstein, 1989). To confirm the robustness of the NJ tree, we built trees using at least one representative sequence of each of the defined clades in the NJ tree and built phylogenies using two additional methods: maximum likelihood (PHYLIP; bootstrap = 100, amino acid substitution model, Jones-Taylor-Thornton matrix) and MrBayes 3.1.2 (Huelsenbeck and Ronquist, 2001) with the following parameters: ngen = 1 × 106, aamodel = mixed. These trees were then compared using the Shimodaira and Hasegawa (1999) and one-sided Kishino and Hasegawa (1989) tests, which calculate the likelihood of each tree, using maximum likelihood distances and the Jones-Taylor-Thornton amino acid substitution model. P values were obtained by a χ2 test (Strimmer and Rambaut, 2002).
Comparison of Kelch Repeats within and between Proteins
Genetic distances among different kelch repeats within the same protein and among different proteins were estimated using protdist from the PHYLIP suite (Felsenstein, 1989). A permutation test was performed to estimate whether the similarity of kelch repeats within a protein is significantly lower than between proteins. As test statistic, the difference of the means and 1 million permutations was used. Test statistic d was defined as follows: d = mean(simKwP) – mean(simKbP), with simKwP as similarities of kelch repeats within a protein and simKbP as similarities of kelch repeats between different proteins (increasing similarity is denoted by decreasing simKxP values). A value of d > 0 means a lower similarity of repeats within proteins in contrast to repeats between proteins. Subsequently, the P value was calculated as the relative frequency of all random values greater than or equal to our measured value. Calculations were performed with R version 2.10.0 (Suzuki and Shimodaira, 2006).
Estimation of the Maximum Number of Gained and Lost FBKs
To determine the degrees of gene family expansion in the analyzed plant lineages, we divided the phylogeny into ancestral clades (those containing at least one representative of the lower land plants, monocots, and eudicots), recent clades (monocot specific, eudicot specific, or lower land plant specific), and species-specific clades. Nodes basal to the split among lineages denote the MRCA and are labeled as N1 to N5. In Figure 3, N0 was added as eukaryotic MRCA on basis of FBKs identified in Supplemental Table S2.
Divergence Levels and Sliding Window Analysis
For analysis of Ka/Ks ratios and sliding window plots, orthologous A. thaliana and Arabidopsis lyrata protein sequences were identified in Supplemental Figure S7. A. thaliana-A. lyrata gene pairs were considered orthologs when they clearly formed a single subclade and possible A. lyrata paralogs were more distantly related. Homologous protein sequences were aligned using ClustalW (Thompson et al., 2002). Codon alignments generated with PAL2NAL (Suyama et al., 2006) were used to compute divergence levels (Ka/Ks ratios) with DnaSP 5.0 (Librado and Rozas, 2009) using the sliding window option (window size, 150 bp; step size, 9 bp).
Correlation Analysis of Expression Data
To create the dendrogram for the cluster analysis of the expression data, the R package pvclust (Suzuki and Shimodaira, 2006) was used. The expression data of the A. thaliana FBKs were extracted from the AtGenExpress extended tissue series (Schmid et al., 2005). Only 81 of 103 A. thaliana FBKs were represented on the ATH1 microarray. In pvclust, a hierarchical clustering was performed using the Pearson correlation as a similarity measurement (dist = 1 – cor[x,y]) between the expression of the genes and the UPGMA method as cluster distance function. To calculate the stability of the dendrogram, a bootstrapping with 1,000 repeats was performed.
Measurement of Relative Transcript Levels by Quantitative Reverse Transcription-PCR in A. thaliana
Shoots were harvested from 7-d-old seedlings and roots from 10-d-old seedlings. Ecotype Columbia-0 seedlings were cultivated on sterile A. thaliana solution agar medium (Lincoln et al., 1990). After 2 d of stratification, the seedlings were cultivated under long-day conditions (16 h of light/8 h of dark) at 20°C. Additionally, 5-week-old Columbia-0 plants were cultivated in growth chambers under long-day conditions at 20°C, and cauline leaves, rosette leaves, open flowers, flower buds, stems, and siliques were harvested for RNA isolation. RNA isolation, cDNA synthesis, and quantitative reverse transcription-PCR were performed according to Delker et al. (2010). Gene-specific primer sequences can be found in Supplemental Table S6.
Subcellular Localization
To examine the subcellular localization of A. thaliana FBKs, 35S::GFP-FBK fusion constructs were created using the Gateway cloning system (Invitrogen) according to the manufacturer’s protocols. pDONR221 was used as entry vector and pGWB6 (Nakagawa et al., 2007) as destination vector. Using Agrobacterium tumefaciens strain GV3101, the fusion constructs were transiently transformed into leaves of 6-week-old Nicotiana benthamiana plants grown in the greenhouse at 20°C and long-day conditions. GFP fluorescence was detected by confocal laser scanning microscopy (LSM510 META; Zeiss).
Yeast Two-Hybrid Assays
Initially, the ASK and FBK genes were cloned into pDONR221 vectors using the Gateway cloning system (Invitrogen) according to the manufacturer’s protocols. The inserts were then recombined into “gatewayized” pGADT7, resulting in the expression of a GAL4 activation domain (AD) fusion protein. ASK constructs were cloned into pGBST7, resulting in the expression of a GAL4 DNA-binding domain (BD) fusion protein. Fusion constructs were transformed into Saccharomyces cerevisiae haploid strains AH109 (MATa; Invitrogen; James et al., 1996) and Y187 (MATα; Invitrogen; Harper et al., 1993). Following yeast mating, a dilution series of the diploid yeast cell suspension was grown at 30°C for 3 d on nonselective (−Leu, −Trp) and strong selective (−Leu, −Trp, −His, −Ade) media. To exclude autoactivation, all FBK and ASK constructs were cloned into pGADT7 and plated on selective (−Leu, −Trp) media. As a negative control, we performed interaction studies with human LaminC, which neither forms complexes nor interacts with most other proteins (Bartel et al., 1993; Ye and Worman, 1995).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic view of a plant F-box kelch protein.
Supplemental Figure S2. Alignment of F-box associated domains and individual kelch repeats from A. thaliana proteins.
Supplemental Figure S3. Rooted NJ trees including FBKs and FBAs of V. vinifera, P. trichocarpa, O. sativa, S. bicolor, and S. moellendorffii.
Supplemental Figure S4. NJ tree generated using full-length FBK protein sequences of A. thaliana, P. trichocarpa, V. vinifera, O. sativa, S. bicolor, S. moellendorffii, and P. patens.
Supplemental Figure S5. Representative phylogenetic trees.
Supplemental Figure S6. Protein sequence alignment of 43 representative FBKs.
Supplemental Figure S7. NJ tree of A. thaliana and A. lyrata FBKs.
Supplemental Figure S8. Density plot of the permutation test.
Supplemental Figure S9. Relative transcript level of closely related FBKs.
Supplemental Table S1. Identifiers of F-box kelch proteins in A. thaliana, P. trichocarpa, V. vinifera, O. sativa, S. bicolor, S. moellendorffii, and P. patens.
Supplemental Table S2. Number of F-box kelch proteins in nonplant model species.
Supplemental Table S3. Identifiers of F-box proteins with F-box associated domains in A. thaliana, P. trichocarpa, V. vinifera, O. sativa, S. bicolor, S. moellendorffii, and P. patens.
Supplemental Table S4. Comparison of three tree topologies obtained with NJ, maximum likelihood, and Bayesian algorithms.
Supplemental Table S5. Absolute numbers of unstable, stable, ancient, and superstable FBKs in plant genomes.
Supplemental Table S6. Sequences of quantitative reverse transcription-PCR primers.
Supplemental Data Set S1. Tissue-specific expression data of A. thaliana FBKs extracted from AtGenExpress_Plus extended tissue series.
Supplementary Material
Acknowledgments
We thank the members of the Quint laboratory, Doug Grubb, and two anonymous reviewers for helpful comments on the manuscript and Thomas Lahaye for yeast strains and vectors. Furthermore, we are grateful to Mariana Mondragon-Palomino for her phylogenetic expertise, to Anja Raschke and Nina Dombrowski for technical support, and to Wolfgang Knogge and Steffen Neumann.
References
- Adams J, Kelso R, Cooley L. (2000) The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol 10: 17–24 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade MA, González-Guzmán M, Serrano R, Rodríguez PL. (2001) A combination of the F-box motif and kelch repeats defines a large Arabidopsis family of F-box proteins. Plant Mol Biol 46: 603–614 [DOI] [PubMed] [Google Scholar]
- Bartel P, Chien CT, Sternglanz R, Fields S. (1993) Elimination of false positives that arise in using the two-hybrid system. Biotechniques 14: 920–924 [PubMed] [Google Scholar]
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, et al. (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, Walker JM, Gagné JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD. (2007) The Arabidopsis EIN3 binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Book AJ, Smalle J, Lee KH, Yang P, Walker JM, Casper S, Holmes JH, Russo LA, Buzzinotti ZW, Jenik PD, Vierstra RD. (2009) The RPN5 subunit of the 26s proteasome is essential for gametogenesis, sporophyte development, and complex assembly in Arabidopsis. Plant Cell 21: 460–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Doolittle RF. (1994) Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol 236: 1277–1282 [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong JK, Paterson AH. (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 5: 739–751 [DOI] [PubMed] [Google Scholar]
- Chae E, Tan QK, Hill TA, Irish VF. (2008) An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342 [DOI] [PubMed] [Google Scholar]
- Delker C, Pöschl Y, Raschke A, Ullrich K, Ettingshausen S, Hauptmann V, Grosse I, Quint M. (2010) Natural variation of transcriptional auxin response networks in Arabidopsis thaliana. Plant Cell 22: 2184–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarsy E, Fankhauser C. (2009) Higher plants use LOV to perceive blue light. Curr Opin Plant Biol 12: 69–74 [DOI] [PubMed] [Google Scholar]
- Demuth JP, Hahn MW. (2009) The life and death of gene families. Bioessays 31: 29–39 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Wang L, Zhang Y, Zhang Y, Deng X, Xue Y. (2006) An auxin-inducible F-box protein CEGENDUO negatively regulates auxin-mediated lateral root formation in Arabidopsis. Plant Mol Biol 60: 599–615 [DOI] [PubMed] [Google Scholar]
- Du ZY, Xiao S, Chen QF, Chye ML. (2010) Depletion of the membrane-associated acyl-coenzyme A-binding protein ACBP1 enhances the ability of cold acclimation in Arabidopsis. Plant Physiol 152: 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1989) PHYLIP: Phylogeny Inference Package (version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Fukamatsu Y, Mitsui S, Yasuhara M, Tokioka Y, Ihara N, Fujita S, Kiyosue T. (2005) Identification of LOV KELCH PROTEIN2 (LKP2)-interacting factors that can recruit LKP2 to nuclear bodies. Plant Cell Physiol 46: 1340–1349 [DOI] [PubMed] [Google Scholar]
- Fülöp V, Jones DT. (1999) Beta propellers: structural rigidity and functional diversity. Curr Opin Struct Biol 9: 715–721 [DOI] [PubMed] [Google Scholar]
- Gagné JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza ZH, Rompa U, Peters JL, Bhatt AM, Wagstaff C, Stead AD, Roberts JA. (2007) Hawaiian skirt: an F-box gene that regulates organ fusion and growth in Arabidopsis. Plant Physiol 144: 1370–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M, Su N, Zheng J, Huai J, Wu G, Zhao J, He J, Tang D, Yang S, Wang G. (2009) An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J 60: 757–770 [DOI] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C. (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313: 903–919 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Han L, Mason M, Risseeuw EP, Crosby WL, Somers DE. (2004) Formation of an SCF(ZTL) complex is required for proper regulation of circadian timing. Plant J 40: 291–301 [DOI] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH. (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon FG, Kay SA. (2003) The F box protein AFR is a positive regulator of phytochrome A-mediated light signaling. Curr Biol 13: 2091–2096 [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816 [DOI] [PubMed] [Google Scholar]
- Hermand D. (2006) F-box proteins: more than baits for the SCF? Cell Div 1: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. (2007) F-box proteins in rice: genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143: 1467–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaso-Friedmann L, Peterson DS, Gonzalez DS, Evans DL. (2002) The antigen receptor (NCCRP-1) on catfish and zebrafish nonspecific cytotoxic cells belongs to a new gene family characterized by an F-box-associated domain. J Mol Evol 54: 386–395 [DOI] [PubMed] [Google Scholar]
- Jawad Z, Paoli M. (2002) Novel sequences propel familiar folds. Structure 10: 447–454 [DOI] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. (2004) Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 18: 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. (1997) The origin and early evolution of plants on land. Nature 389: 33–39 [Google Scholar]
- Kim HS, Delaney TP. (2002) Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 14: 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim OK, Jung JH, Park CM. (2010) An Arabidopsis F-box protein regulates tapetum degeneration and pollen maturation during anther development. Planta 232: 353–366 [DOI] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE. (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Pagano M. (2000) The F-box protein family. Genome Biol 1: REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino H, Hasegawa M. (1989) Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol 29: 170–179 [DOI] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. (2006) F-box proteins everywhere. Curr Opin Plant Biol 9: 631–638 [DOI] [PubMed] [Google Scholar]
- Leung KC, Li HY, Mishra G, Chye ML. (2004) ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with kelch motifs that bind oleoyl-CoA. Plant Mol Biol 55: 297–309 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang D, Hannink M, Beamer LJ. (2004) Crystal structure of the kelch domain of human Keap1. J Biol Chem 279: 54750–54758 [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucotelli E, Belloni S, Marone D, De Leonardis A, Guerra D, Di Fonzo N, Cattivelli L, Mastrangelo A. (2006) The e3 ubiquitin ligase gene family in plants: regulation by degradation. Curr Genomics 7: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-García S, Vert G, Yin Y, Caño-Delgado A, Cheong H, Chory J. (2004) Nuclear protein phosphatases with kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Ou CY, Pi H, Chien CT. (2003) Control of protein degradation by E3 ubiquitin ligases in Drosophila eye development. Trends Genet 19: 382–389 [DOI] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. (2009) Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA 106: 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prag S, Adams JC. (2003) Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics 4: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR. (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL. (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J 34: 753–767 [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. (2000) Comparative genomics of the eukaryotes. Science 287: 2204–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schwager KM, Calderon-Villalobos LIA, Dohmann EMN, Willige BC, Knierer S, Nill C, Schwechheimer C. (2007) Characterization of the VIER F-BOX PROTEINE genes from Arabidopsis reveals their importance for plant growth and development. Plant Cell 19: 1163–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16: 1114–1116 [Google Scholar]
- Skaar JR, D’Angiolella V, Pagan JK, Pagano M. (2009a) SnapShot: F box proteins II. Cell 137: 1358. [DOI] [PubMed] [Google Scholar]
- Skaar JR, Pagan JK, Pagano M. (2009b) SnapShot: F box proteins I. Cell 137: 1160. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Durbin R. (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28: 405–420 [DOI] [PubMed] [Google Scholar]
- Sterck L, Rombauts S, Jansson S, Sterky F, Rouzé P, Van de Peer Y. (2005) EST data suggest that poplar is an ancient polyploid. New Phytol 167: 165–170 [DOI] [PubMed] [Google Scholar]
- Stone SL, Callis J. (2007) Ubiquitin ligases mediate growth and development by promoting protein death. Curr Opin Plant Biol 10: 624–632 [DOI] [PubMed] [Google Scholar]
- Strimmer K, Rambaut A. (2002) Inferring confidence sets of possibly misspecified gene trees. Proc Biol Sci 269: 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Shi L, Li W, Yu W, Liang J, Zhang H, Yang X, Wang Y, Li R, Yao X, et al. (2009) JFK, a kelch domain-containing F-box protein, links the SCF complex to p53 regulation. Proc Natl Acad Sci USA 106: 10195–10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34: W609–W612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzui N, Nakamura S, Fujiwara T, Hayashi H, Yoneyama T. (2006) A putative acyl-CoA-binding protein is a major phloem sap protein in rice (Oryza sativa L.). J Exp Bot 57: 2571–2576 [DOI] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Kuroda H, Kuromori T, Hirayama T, Seki M, Shinozaki K, Shimada H, Matsui M. (2004) Expression and interaction analysis of Arabidopsis Skp1-related genes. Plant Cell Physiol 45: 83–91 [DOI] [PubMed] [Google Scholar]
- Thomas JH. (2006) Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res 16: 1017–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics August: chapter 2, unit 2.3 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wang L, Dong L, Zhang Y, Zhang Y, Wu W, Deng X, Xue Y. (2004) Genome-wide analysis of S-locus F-box-like genes in Arabidopsis thaliana. Plant Mol Biol 56: 929–945 [DOI] [PubMed] [Google Scholar]
- Weigel D, Mott R. (2009) The 1001 genomes project for Arabidopsis thaliana. Genome Biol 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Ma H, Nei M, Kong H. (2009) Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci USA 106: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. (2002) The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue F, Cooley L. (1993) Kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72: 681–693 [DOI] [PubMed] [Google Scholar]
- Yang X, Kalluri UC, Jawdy S, Gunter LE, Yin T, Tschaplinski TJ, Weston DJ, Ranjan P, Tuskan GA. (2008) The F-box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant Physiol 148: 1189–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Worman HJ. (1995) Protein-protein interactions between human nuclear lamins expressed in yeast. Exp Cell Res 219: 292–298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.