Abstract
Brassica napus is an allotetraploid (AACC) formed from the fusion of two diploid progenitors, Brassica rapa (AA) and Brassica oleracea (CC). Polyploidy and genome-wide rearrangement during the evolution process have resulted in genes that are present as multiple homologs in the B. napus genome. In this study, three B. napus homologous genes encoding endoplasmic reticulum-bound sn-glycerol-3-phosphate acyltransferase 4 (GPAT4) were identified and characterized. Although the three GPAT4 homologs share a high sequence similarity, they exhibit different expression patterns and altered epigenetic features. Heterologous expression in yeast further revealed that the three BnGPAT4 homologs encoded functional GPAT enzymes but with different levels of polypeptide accumulation. Complementation of the Arabidopsis (Arabidopsis thaliana) gpat4 gpat8 double mutant line with individual BnGPAT4 homologs suggested their physiological roles in cuticle formation. Analysis of gpat4 RNA interference lines of B. napus revealed that the BnGPAT4 deficiency resulted in reduced cutin content and altered stomatal structures in leaves. Our results revealed that the BnGPAT4 homologs have evolved into functionally divergent forms and play important roles in cutin synthesis and stomatal development.
Polyploidy has long been considered a prominent evolutionary force for flowering plants (Udall and Wendel, 2006). Over 95% of the lineages of angiosperms have undergone at least one event of polyploidization over their evolutionary time, suggesting that most of the existing flowering plants evolved from ancient polyploids (Bennett and Leitch, 1997; Chen, 2007; Gaeta et al., 2007). Polyploidy, along with genomic segmental duplications, could benefit plants by increasing overall gene expression levels and cell sizes and providing sources for novel variants and genome “buffering” of deleterious mutations (Udall and Wendel, 2006). Genes duplicated by such events could undergo three primary evolutionary fates over the long term (Wendel, 2000; Blanc and Wolfe, 2004; Lukens et al., 2004; Whittle and Krochko, 2009): (1) pseudogenization (loss or silencing), whereby duplicated genes with redundant functions accumulate deleterious mutations and are eventually lost without detrimental effects on plant fitness; (2) neofunctionalization, whereby some redundant genes develop new adaptive functions by positive Darwinian selection; and (3) subfunctionalization, a process in which the ancestral gene functions become subdivided among the duplicated genes.
Brassica napus (AACC; n = 19) is an allotetraploid oilseed crop that evolved from the hybridization of two diploid progenitors, Brassica rapa (AA; n = 10) and Brassica oleracea (CC; n = 9) during human cultivation (over 10,000 years ago; U, 1935; Cheung et al., 2009). The Brassica species are closely related to the model plant Arabidopsis (Arabidopsis thaliana), all of which belong to the same tribe (Brassiceae) and share a common recent ancestry (20 million years ago; Yang et al., 1999). Comparative mapping studies of the genomic microstructures of B. oleracea, B. rapa, Brassica nigra, B. napus, and Arabidopsis revealed extensive triplications in the genomes of the diploid Brassica progenitors and strongly suggested that the extant Brassica diploid species evolved from a common hexaploid ancestor (O’Neill and Bancroft, 2000; Rana et al., 2004; Park et al., 2005). A previous study also showed that the majority of the Arabidopsis conserved genomic regions could be mapped to six conserved segments within the allotetraploid genome of B. napus (Parkin et al., 2005). There are a few exceptions, however, where fewer or more copies of certain segments have been detected in the genome of B. napus. This could be caused by multiple rounds of duplication (either segmental or the result of polyploidy) along with genome-wide rearrangements and segmental deletions during the evolution process (Cheung et al., 2009). Thus, the complex genome structure of the diploid Brassica progenitors, together with the extensive genome rearrangements after speciation, have led to genes being represented as multiple homologs in the allotetraploid B. napus.
Although there are numerous studies comparing the genomic structures of the Brassica species and Arabidopsis, little is known about molecular and functional variances among homologous genes arising from polyploidy and genomic segmental duplications in B. napus. In part, this is due to experimental challenges in distinguishing highly identical transcripts and polypeptides, which are inherited from two genomically similar and evolutionarily related progenitors. Such information, however, is fundamentally important for a better understanding of the complex mechanisms involved in variant biological pathways in the Brassica polyploid species. Additionally, in the genetic engineering of plants with polyploid backgrounds, knowledge of the transcriptional and functional behavior of individual homologs is essential to avoid pleiotropic effects. For example, in Triticum aestivum (bread wheat), three WLHS1 homoeologous genes (originating from A, B, and D genomes, respectively) are associated with different effects on flowering time (Shitsukawa et al., 2007). Such an understanding would allow for the manipulation of specific genes relevant to the targeted metabolic process without compromising overall plant fitness.
The sn-glycerol-3-phosphate acyltransferases (GPATs; EC 2.3.1.15) are involved in catalyzing the initial step in the assembly of glycerolipids (Zheng et al., 2003). In Arabidopsis, 10 genes have been identified as encoding GPAT enzymes located in various subcellular compartments, such as plastids (ATS1), mitochondria (AtGPAT1–AtGPAT3), and endoplasmic reticulum (ER; AtGPAT4–AtGPAT9; Zheng et al., 2003; Xu et al., 2006; Gidda et al., 2009). Recent studies in Arabidopsis have shown that several members in the ER-bound GPAT family are involved in lipid polyester synthesis (i.e. cutin and suberin; Beisson et al., 2007; Li et al., 2007). In this study, we identified and characterized three B. napus GPAT4 homologs. Phylogenetic analysis of the genomic DNA sequences of the GPAT4 genes from B. napus, B. rapa, and B. oleracea strongly suggested that two of the BnGPAT4 homologs originated from the C genome and the third originated from the A genome. Heterologous expression in yeast revealed that all three BnGPAT4 homologs encoded functional GPAT enzymes with different levels of accumulation. The gene expression, epigenetic variations, and phenotypic rescue of the gpat4 gpat8 Arabidopsis double mutant indicated that the three BnGPAT4 genes have evolved through functional divergence and are involved in lipid polyester synthesis. In addition, analysis of the gpat4 RNA interference (RNAi) lines of B. napus revealed changes in overall cutin load and stomatal structure of the rosette leaves.
RESULTS
Identification of GPAT4 Homologs in B. napus
The cDNA sequence of Arabidopsis GPAT4 (At1g01610) was used as a reference to query the B. napus EST database in GenBank (National Center for Biotechnology Information) for UniGene entries (Pontius et al., 2003). Overall, 43 UniGene entries were identified (UniGene accession no. Bna.684). After analyzing their alignment, the 43 UniGene sequences were condensed into three groups, which were denoted as three BnGPAT4 homologs.
In B. napus, the A and C genomes of the diploid progenitors became homoeologous subgenomes. Due to this complex genome background, the three identified BnGPAT4 genes may represent paralogs (i.e. duplicated genes originating from the same subgenome) and homoeologs (i.e. orthologous genes originating from different subgenomes coexisting in the newly synthesized allotetraploid genome). As the homology between the subtypes is complex, the GPAT4 genes from B. napus were referred to as homologs, according to the recommendation from Fitch (2000).
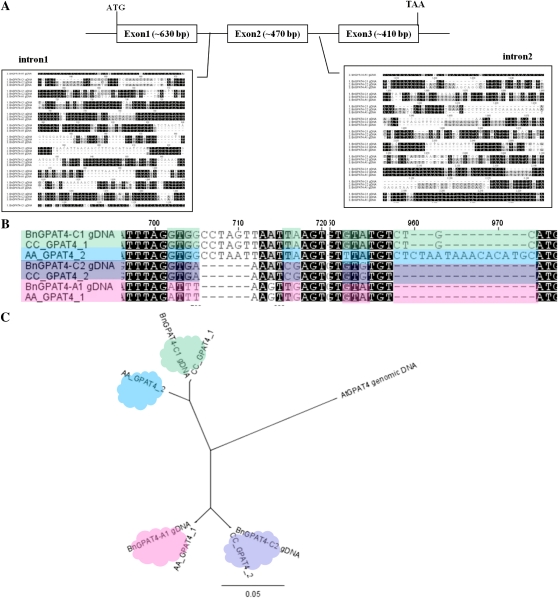
The full-length cDNAs and genomic DNA sequences of the three BnGPAT4 homologs were cloned by PCR using homolog-specific primer pairs, which were designed based on single-nucleotide polymorphisms in the sequences. The cDNA sequences of the three BnGPAT4 homologs and AtGPAT4 shared over 90% similarity. The deduced amino acid sequences shared even greater sequence similarity (over 94%; Table I). The major sequence differences between the three homologs were found in the two intron regions as shown in Figure 1A.
Table I. Sequence identity of the genomic DNA, cDNA, and protein of the three BnGPAT4s and AtGPAT4.
| GPAT4s | BnGPAT4-C2 |
BnGPAT4-A1 |
AtGPAT4 |
||||||
| Genomic DNA | cDNA | Protein | Genomic DNA | cDNA | Protein | Genomic DNA | cDNA | Protein | |
| % | |||||||||
| BnGPAT4-C1 | 87 | 93 | 97 | 87 | 94 | 97 | 82 | 90 | 96 |
| BnGPAT4-C2 | 96 | 97 | 99 | 81 | 90 | 95 | |||
| BnGPAT4-A1 | 81 | 90 | 94 | ||||||
Figure 1.
Genome structures of the BnGPAT4 homologs and sequence analysis of the GPAT4 genes from B. napus and its diploid progenitors, B. rapa and B. oleracea. A, The major sequence differences among the homologs are in the intron regions, as shown in the two blocks. B, Partial genomic DNA sequence alignment of the GPAT4 genes. Sequences labeled with the same color indicate higher sequence similarity. C, Phylogenetic tree of the genomic sequences of the GPAT4 genes. AA_GPAT4, GPAT4 genes isolated from B. rapa; CC_GPAT4, GPAT4 genes isolated from B. oleracea. The phylogenetic tree was built using PHYML (Guindon and Gascuel, 2003).
Genome Origins of the Three BnGPAT4 Homologs
B. napus is an allotetraploid (AACC) formed from hybridization of two diploid progenitors, B. rapa (AA) and B. oleracea (CC; U, 1935). To study the genome origins of the three BnGPAT4 homologs, the genomic DNA sequences of GPAT4 genes from B. rapa and B. oleracea were isolated and compared with the BnGPAT4 homologs. Using primer pairs designed from the BnGPAT4 sequences, two GPAT4 genes were isolated from B. rapa and two were isolated from B. oleracea. A comparison of the genomic DNA sequences, particularly in the intron regions, revealed that two of the BnGPAT4 homologs exhibited highest sequence similarity with two B. oleracea GPAT4 genes (CC_GPAT4_1 and CC_GPAT4_2) and were thus named BnGPAT4-C1 and BnGPAT4-C2. The third BnGPAT4 homolog exhibited highest sequence similarity with one of the GPAT4 genes isolated from B. rapa (AA_GPAT4_1) and was thus named BnGPAT4-A1 (Fig. 1B). Phylogenetic analysis of the genomic DNA sequences further confirmed the homology among GPAT4 genes from the three Brassica species and Arabidopsis (Fig. 1C). Additionally, based on the genome origin and sequence similarity level, BnGPAT4-C1 is predicted to be paralogous to BnGPAT4-C2 and BnGPAT4-C2 is homoeologous to BnGPAT4-A1.
Notably, the second GPAT4 gene cloned from B. rapa (AA_GPAT4_2) did not have a counterpart within the three BnGPAT4 homologs. To examine whether a fourth, unidentified BnGPAT4 gene homologous to AA_GPAT4_2 exists in B. napus, PCR was performed using B. napus genomic DNA as a template and AA_GPAT4_2-specific primers. No amplicon was generated from the PCR using B. napus genomic DNA as a template; however, the target amplicon of approximately 420 bp was obtained using B. rapa genomic DNA as a template (Supplemental Fig. S1). This 420-bp amplicon was further sequenced and confirmed to be the target region of the AA_GPAT4_2 gene. This result confirmed that AA_GPAT4_2 did not have a counterpart copy in the B. napus genome.
BnGPAT4 Homologs Encode Functional GPAT Enzymes with Different Levels of Polypeptide Accumulation
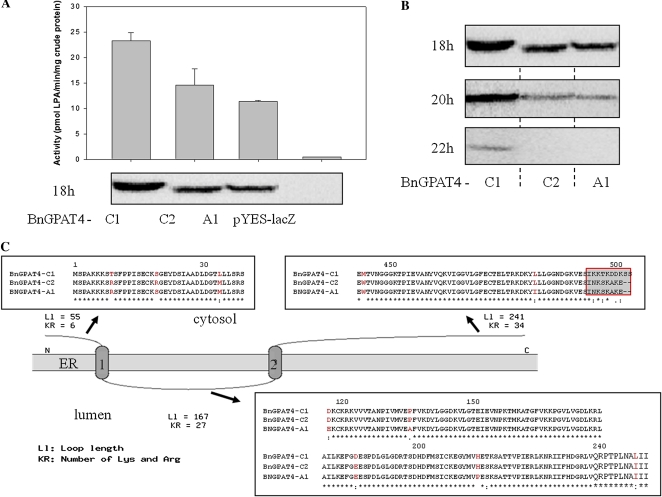
The cDNAs of each of the three BnGPAT4 homologs were cloned in frame (without a stop codon) with the V5 epitope and polyhistidine tag of the yeast expression vector, pYES2.1/V5-His-TOPO (Invitrogen). A yeast mutant strain (gat1Δ) deficient in the major ER-bound GPAT activity (Zheng and Zou, 2001) was used for Gal-induced expression of the BnGPAT4 recombinant proteins. The gat1Δ yeast strain has a very low GPAT activity background and has been commonly used as a test strain for putative GPATs cloned from a number of species (Zheng and Zou, 2001, Zheng et al., 2003). In Arabidopsis, GPAT4 was confirmed to be an ER-bound enzyme (Zheng et al., 2003). Thus, we assayed GPAT activity in microsomal fractions of gat1Δ yeast expressing BnGPAT4 cDNAs or a null vector.
As shown in Figure 2A, all three BnGPAT4 proteins exhibited GPAT enzyme activity after 18 h of Gal induction, but the levels of activity varied. Results from a western blot of the same protein samples indicated that the higher enzyme activity of BnGPAT4-C1 was likely due to a higher abundance of the BnGPAT4-C1 protein in the transformed yeast. We further investigated the protein accumulation levels upon longer Gal induction time (up to 22 h). The abundance of all BnGPAT4 proteins decreased with the increasing time of induction (Fig. 2B). In addition, the abundance of BnGPAT4-C1 was always higher than BnGPAT4-A1 and BnGPAT4-C2 in the yeast expression system.
Figure 2.
The three BnGPAT4 homologs encode functional GPAT enzymes but with different activities. A, The activities of the BnGPAT4 enzymes and the corresponding protein amounts detected by western blot after 18 h of Gal induction in the gat1Δ yeast expression system. B, The abundance of the BnGPAT4 polypeptides detected by western blot after 18, 20, and 22 h of Gal induction in the gat1Δ yeast expression system. C, Predicted protein topology of the three BnGPAT4 enzymes and partial amino acid sequence alignment. Conserved residues are denoted by asterisks, strongly similar amino acids by colons, weakly similar amino acids by periods, and the different amino acids among the three BnGPAT4 proteins are shown in red. [See online article for color version of this figure.]
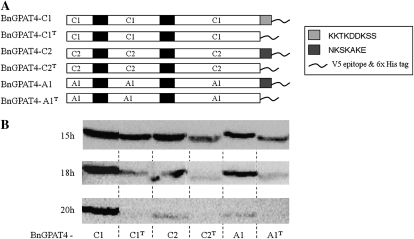
The polypeptide sequences of the three forms of BnGPAT4 share up to 99% similarity (Table I). Protein topology prediction (TopPred; Claros and von Heijne, 1994) indicated that all three forms of BnGPAT4 proteins possessed two ER transmembrane domains at the same positions (Fig. 2C). The major difference among the amino acid sequences of the three forms of BnGPAT4 is in the final five to seven amino acid residues at the C termini (Fig. 2C). BnGPAT4-C2 and BnGPAT4-A1 have the same C-terminal sequences but are different from BnGPAT4-C1, and both BnGPAT4-C2 and BnGPAT4-A1 exhibited lower protein abundance than BnGPAT4-C1. To test whether the different C termini are important to the BnGPAT4 protein abundance in yeast, we made deletions of the C termini of all three BnGPAT4 proteins (Fig. 3). Western blots were used to compare the quantity of the BnGPAT4 proteins in yeast after 15 to 20 h of induction. As shown in Figure 3, all BnGPAT4 proteins with truncated C termini exhibited much lower abundance compared with the unmodified proteins. In addition, we also exchanged the C termini between the BnGPAT4 proteins (Supplemental Fig. S2). Western-blot results suggested that the exchange of C termini had no effect on protein abundance (Supplemental Fig. S2). Collectively, these results indicated that the C termini of BnGPAT4 proteins may play a role in regulating the level of polypeptide accumulation (through synthesis and/or degradation) of the BnGPAT4 enzymes; however, the sequence differences in the C termini do not appear to be related to the differences in protein abundance.
Figure 3.
The C termini of the BnGPAT4s are important for controlling the level of polypeptide accumulation. A, The structures of native and C-termini-deleted BnGPAT4 proteins. Black bars represent the putative transmembrane domains. The light gray bar (KKTKDDKSS) represents the C terminus of BnGPAT4-C1. The dark gray bars (NKSKAKE) represent the C termini of BnGPAT4-C2 and BnGPAT4-A1. B, Western-blot analysis of the protein abundances of the native and C-termini-deleted BnGPAT4 proteins in transformed gat1Δ yeast with different Gal induction times.
The Three BnGPAT4 Homologs Exhibit Distinct Expression Patterns
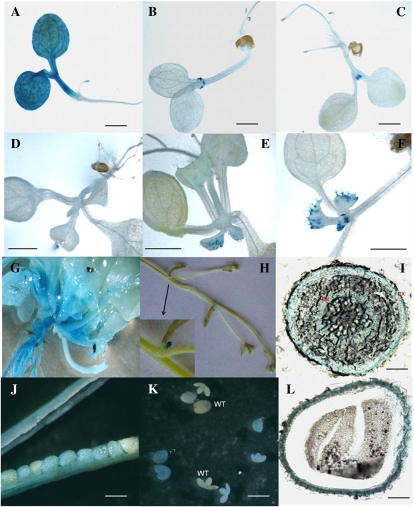
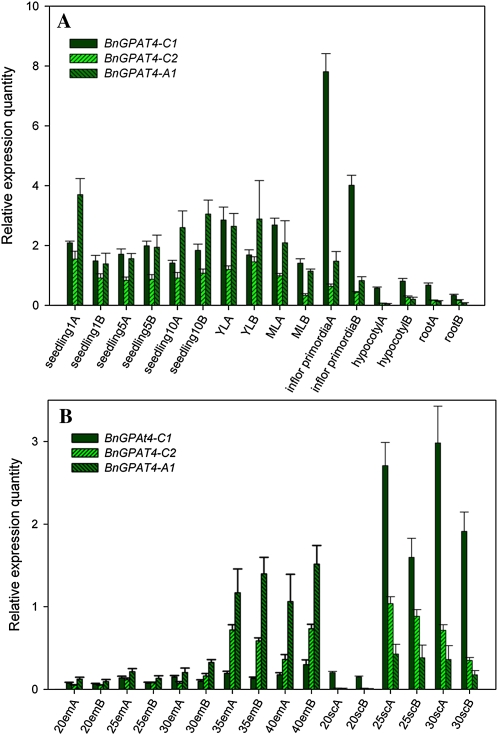
The promoter regions of the three BnGPAT4 homologs have a much lower sequence similarity compared with their protein-coding sequences (Supplemental Fig. S3). To study the gene expression patterns of the three BnGPAT4 homologs, GUS assays (using promoter-GUS fusion constructs) and TaqMan quantitative real-time reverse transcription (qRT)-PCR were performed. Fragments consisting of over 600 bp upstream of the first ATGs of the three BnGPAT4 homologs were used to drive the expression of the GUS reporter gene in transgenic Arabidopsis. Three to five independent transgenic lines for each construct were analyzed for GUS activities. The results from histochemical staining revealed that the promoters of different homologs directed GUS expression in various plant tissues and organs (Fig. 4). The differences in GUS expression patterns between different BnGPAT4 promoters were most distinct in young seedlings (Fig. 4, A–F); at older stages, the expression patterns were more similar between all three promoters.
Figure 4.
Histochemical analysis of the GUS activities directed by individual BnGPAT4 promoters in transgenic Arabidopsis. Similar GUS activities were observed in at least three independent transgenic lines for each construct. Only one transgenic line is shown in each panel. A to C, Five-day-old seedlings. GUS activities were driven by the three promoters of BnGPAT4-C1, BnGPAT4-C2, and BnGPAT4-A1, respectively. Bars = 0.5 mm. D to F, Ten-day-old seedlings. GUS activities were driven by the three promoters of BnGPAT4-C1, BnGPAT4-C2, and BnGPAT4-A1, respectively. Bars = 5 mm. G to L, Images represent similar GUS activities observed with all three BnGPAT4 promoters. G, GUS activity driven by the BnGPAT4-C2 promoter. H to L, GUS activities driven by the BnGPAT4-A1 promoter. G, A 20-d-old whole plant. H, Inflorescence stem. The inset shows an enlarged image of the inflorescence primordia. I, Cross-section of the main root. EN, Endodermis; EX, exodermis. Bar = 0.3 mm. J, Middle- to late-stage seeds in a silique. Bar = 0.5 mm. K, Dissected embryos and seed coats. WT, Wild type. Bar = 1 mm. L, Cross-section of a whole seed. Bar = 0.1 mm.
Considering that other regulatory factors and elements could also be involved in regulating gene expression in vivo, and the limitations of the promoter-GUS assay (such as the lack of accurate quantification and dosage effects of the transgene), we further performed TaqMan qRT-PCR to quantify the transcript levels of the individual BnGPAT4 homologs in wild-type B. napus. Based on the preliminary results from the GUS assay, we tested several tissues and organs from B. napus, including tissues where the GUS assays indicated that the BnGPAT4 homologs might be highly expressed. The qRT-PCR results (Fig. 5) were generally consistent with the earlier observations in the GUS assay. In general, the BnGPAT4 genes had higher expression levels in vegetative tissues than in seeds. In particular, BnGPAT4-C1 was most highly expressed in inflorescence primordia (Fig. 5A). During embryo and seed coat development, BnGPAT4-C1 and BnGPAT4-A1 had quite distinct expression patterns. In the developing embryos, all BnGPAT4 homologs exhibited increasing transcript abundance as the developing embryo matured, and BnGPAT4-A1 was expressed at a much higher level than the other two homologs. In the developing seed coat, the transcript abundance of all BnGPAT4 homologs was very low at the 20-d after pollination (DAP) stage and increased substantially after the 25-DAP stage. Notably, BnGPAT4-C1 was expressed at a very high level in the seed coat when compared with the other homologs; in contrast, its expression level was the lowest in embryos. We were only able to investigate the transcript abundance in seed coats excised from the developing seeds at 20, 25, and 30 DAP, since no RNA could be extracted from the samples at later developmental stages. This is due to the fact that as the seed matures, most of the seed coat cells undergo cell death and become compressed cell layers impregnated with brown pigments (Supplemental Fig. S4; Wan et al., 2002; Haughn and Chaudhury, 2005).
Figure 5.
Gene expression patterns of the three BnGPAT4 homologs. A, Expression in vegetative tissues and organs. B, Expression in embryos and seed coats. Seedling 1, 5, and 10 refer to 1-, 5-, and 10-d-old seedlings, respectively; ML, mature leaves; YL, young leaves; 20em to 40em, 20- to 40-DAP embryos; 20sc to 30sc, seed coats excised from 20- to 30-DAP seeds. A and B after the sample names refer to the two biological replicates. Three technical replicates were performed for each sample. [See online article for color version of this figure.]
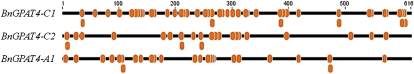
Epigenetic Variation among the BnGPAT4 Homologs
Cytosine DNA methylation has been recognized as an important mechanism for the regulation of gene expression and has been found to be a common epigenetic feature in most eukaryotes (Zilberman et al., 2007). Recent work in allotetraploid Arabidopsis suecica, allohexaploid T. aestivum, and resynthesized allotetraploid B. napus has shown that DNA methylation may play a crucial role in establishing homolog expression patterns in allopolyploid plants (Chen, 2007; Gaeta et al., 2007; Shitsukawa et al., 2007). In contrast to animals, in which methylation occurs primarily at CG dinucleotides, cytosine DNA methylations in plants occur at sites containing CG, CNG (where N is any nucleotide), and CHH (an asymmetric site, where H is A, C, or T; Chan et al., 2005). Another important feature of plant cytosine DNA methylation is that a large number of genes (approximately 33.3%) are methylated in their coding regions (Zhang et al., 2006). Since the coding regions of BnGPAT4 homologs share high sequence similarity (93%–97%), it was of interest to investigate the methylation patterns within the coding regions of the three homologs. We investigated the methylation status of the first exon region (approximately 600 bp) of each BnGPAT4 homolog, using sodium bisulfite-treated genomic DNA isolated from a mixed collection of plant tissues and organs as a PCR template. Twelve clones of each PCR amplicon were sequenced. The ratios of the methylated cytosines (presented as the number of clones having methylated cytosines/the number of total clones) were indicated by percentages. As shown in Figure 6, all the locations of methylated cytosines were labeled with orange dots, and only those with methylation ratios over 17% are listed in Table II. In general, the three homologs had very different methylation patterns. Notably, BnGPAT4-C2 had more methylated cytosines with ratios over 17% than the other two homologs. This may be related to the lower gene expression levels observed for the BnGPAT4-C2 homolog (Fig. 5). These very different methylation patterns again indicated that the homologous genes have evolved and are regulated differentially at the molecular level.
Figure 6.
Cytosine DNA methylation patterns of the first exon regions of the three BnGPAT4 homologs. All methylated cytosine locations are indicated by orange dots. [See online article for color version of this figure.]
Table II. Summary of the methylated cytosines with methylation ratios over 17% in the first exon regions of the three BnGPAT4 homologs.
|
BnGPAT4-C1 |
BnGPAT4-C2 |
BnGPAT4-A1 |
||||||
| Location | Methylation Ratio | Sequence | Location | Methylation Ratio | Sequence | Location | Methylation Ratio | Sequence |
| % | % | % | ||||||
| 138 | 17 | CGC | 7 | 17 | CCG | 234 | 17 | CAT |
| 141 | 25 | CAT | 24 | 17 | CAG | 264 | 17 | CGG |
| 213 | 17 | CTT | 178 | 17 | CTT | 419 | 25 | CGT |
| 257 | 17 | CCT | 211 | 17 | CTC | 472 | 33 | CCC |
| 282 | 17 | CAT | 246 | 17 | CAT | 473 | 33 | CCA |
| 342 | 17 | CAG | 270 | 17 | CAA | |||
| 417 | 25 | CGT | 300 | 17 | CGC | |||
| 542 | 17 | CCA | 311 | 33 | CAC | |||
| 546 | 17 | CTT | 326 | 17 | CGG | |||
| 509 | 17 | CTG | ||||||
| 572 | 17 | CGG | ||||||
Phenotypic Rescue of Arabidopsis gpat4 gpat8 Double Mutant Lines
In Arabidopsis, GPAT4 and GPAT8 are closely related (greater than 80% sequence similarity between the cDNAs) and display functional redundancy in cutin synthesis; double T-DNA insertional mutations at both genes result in severe defects in cutin synthesis and increased cuticle permeability (Li et al., 2007). A BLAST search of the full-length cDNA sequence of AtGPAT8 against the B. napus EST database showed that all the positive hits (i.e. those with the highest scores, sequence similarities greater than 80%) were from the sequences of BnGPAT4 homologs. Thus, it is likely that AtGPAT8 emerged as a duplicated copy of AtGPAT4 during Arabidopsis evolution after the divergence of Arabidopsis and the Brassica species.
To determine whether the BnGPAT4 homologs also function in epidermal cell lipid polymer biosynthesis, we expressed the three BnGPAT4 cDNAs under the control of their native promoters in the Arabidopsis gpat4 gpat8 double T-DNA lines (the schematic diagram of the binary vector is presented in Supplemental Fig. S5). At least three independent T2 transgenic lines for each construct were analyzed. In order to determine if the BnGPAT4 homologs were able to rescue the defective cuticle phenotype of Arabidopsis gpat4 gpat8 mutant plants, we examined the cuticle permeability of the transgenic Arabidopsis plants using a toluidine blue test. The toluidine blue test was developed by Tanaka et al. (2004) and has been extensively used for the rapid visualization of cuticle defects of plant tissues. Varying degrees of phenotypic rescue were observed in the T2 plants of gpat4 gpat8 lines transformed with BnGPAT4 homologs (Fig. 7). Segregation analysis indicated that the transgenic lines that had phenotypes closest to being fully restored had more than one copy of the transgene; however, none of the transgenic lines displayed a complete restoration of the defective cuticle phenotype. These results suggest that BnGPAT4 homologs are also involved in cutin synthesis. The fact that expression in individual BnGPAT4 homologs could not fully replace the function of AtGPAT4 and AtGPAT8 in Arabidopsis also indicated that the BnGPAT4 homologs have evolved through functional divergence.
Figure 7.
Toluidine blue test of the cuticle permeability of gpat4 gpat8 Arabidopsis transformed with individual BnGPAT4 homologs. A, Plants before toluidine blue treatment. Bar = 3 cm. B to F, Plants after toluidine blue treatment. WT, Wild type. Bars = 1 cm.
B. napus gpat4 RNAi Lines Exhibit Alterations in Cuticle Load and Stomatal Structure Resulting in Increased Water Loss
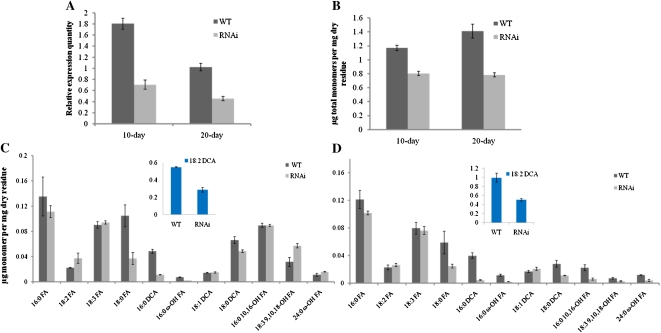
To further confirm the involvement of BnGPAT4 homologs in cutin biosynthesis and explore other physiological functions of the BnGPAT4 genes in B. napus, an RNAi approach targeting the expression of BnGPAT4 homologs was used. Due to the high probability of functional overlap among the three homologs, silencing one particular homolog may not give a detectable phenotype; therefore, an RNAi construct (Supplemental Fig. S6) was designed to silence all three BnGPAT4 homologs. The overall transcript abundance of the BnGPAT4 homologs in the RNAi lines was determined using SYBR-Green qRT-PCR (Fig. 8A). The overall expression levels of the BnGPAT4 homologs were decreased by approximately 40% in both young and mature rosette leaves of the transgenic plants. To confirm that the RNAi silencing is specific to GPAT4, we also analyzed the expression level of GPAT6 in the RNAi lines, because GPAT6 shares the highest cDNA sequence similarity (approximately 60%) to GPAT4 in the GPAT gene family. As shown in Supplemental Figure S7, the transcriptional levels of the GPAT6 gene were unchanged in both wild-type and gpat4 RNAi lines.
Figure 8.
Silencing of GPAT4 homologs in B. napus results in decreased cutin monomer load. A, Overall expression levels of BnGPAT4 homologs in the rosette leaves of wild-type (WT) and gpat4 RNAi B. napus lines. Compared with the wild type, the transcription levels of GPAT4 were 43.3% and 35.8% lower in the rosette leaves at 10 and 20 d post emergence of the RNAi lines. n = 3. B, Total amount of monomers per mg of dry residues. The rosette leaves of RNAi lines had 31.2% and 44.5% reductions in total cutin monomer load in comparison with the wild-type lines at 10 and 20 d post emergence. n = 3. C and D, Cutin monomer profile of the rosette leaves at 10 (C) and 20 (D) d post emergence. FA, Fatty acid. n = 3. [See online article for color version of this figure.]
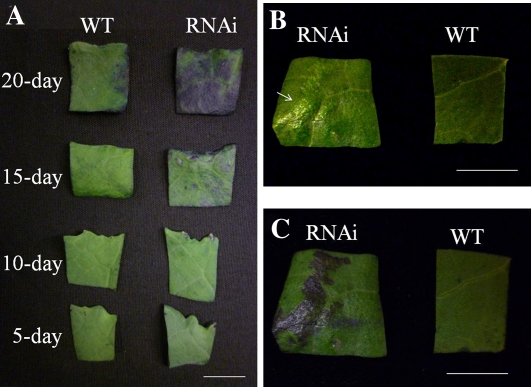
Young rosette leaves (5 and 10 d after emerging) of the gpat4 RNAi lines did not exhibit visible morphological differences when compared with the wild-type lines (Fig. 9A); however, in older leaves (approximately 15 d post emergence), several small protruding areas with a different texture (referred as glossy texture) appeared on the adaxial leaf surface of RNAi lines (Fig. 9B), which exhibited increased cuticle permeability in the toludidine blue test (Fig. 9C). By the 20-d stage, increased cuticle permeability was observed over most of the leaf surface. Analysis of cutin monomers extracted from 10- and 20-d-old leaves showed that the overall content of cutin aliphatic monomers was decreased by 31.2% and 44.5% in the 10- and 20-d-old leaves of the gpat4 RNAi lines, respectively (Fig. 8B). Interestingly, the cutin monomer compositions were quite different between 10- and 20-d-old leaves (Fig. 8, C and D), except for the fact that the dominant cutin monomer was 18:2 dicarboxylic acid (18:2 DCA) at both stages. In general, at both stages, the monomer species exhibiting the greatest reductions were 18:2 DCA (approximately 50%), 16:0 DCA (approximately 80%), and 18:0 fatty acid (approximately 58%). Notably, even though there was a significant decrease (31.2%) in the overall level of cutin monomers in 10-d-old rosette leaves, we did not observe any significant cuticle permeability defect using the toluidine blue test (Fig. 9A). One explanation could be that the younger staged leaves had sufficient cutin monomers per unit of leaf surface to maintain regular cuticle permeability; while at an older stage, when the leaf size was much larger, the amount of cutin monomers per leaf surface unit was too low to maintain a normal level of cuticle permeability.
Figure 9.
Cuticle permeability of B. napus gpat4 rosette leaves at different stages with the toluidine blue test. A, Comparison of the rosette leaves of wide-type and gpat4 lines after treatment with the toluidine blue solution. B, The rosette leaves at 15 d post emergence without toluidine blue treatment. The glossy area is indicated by an arrow. C, The same leaf samples in B were treated with toluidine blue. WT, Wild type. Bars = 1 cm.
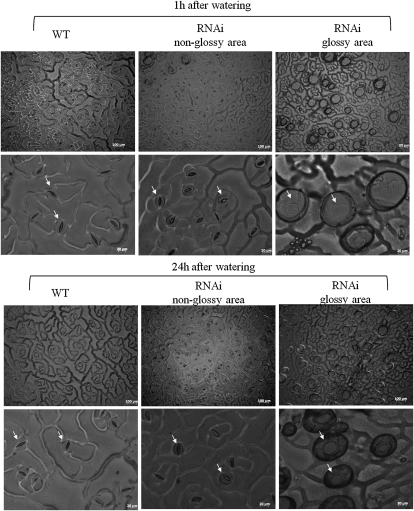
We also observed that the gpat4 B. napus lines exhibited increased water loss during the daytime (Supplemental Fig. S8). To further investigate the surface structure of gpat4 RNAi lines exhibiting the defective cuticle phenotype, surface casts of the attached leaves were prepared at 1 and 24 h after watering to observe the pavement cells. We observed that the stomata were widely open in the glossy areas of the rosette leaves of the RNAi plants (Fig. 10). In addition, the stomata opened much wider 24 h after watering as compared with 1 h after watering, and the surrounding guard cells were stretched much thinner than normal, which could possibly explain the observed increase of water loss in the RNAi lines. Our results suggested that an intact cuticle layer is essential for the guard cells to function properly and for maintaining a normal stomatal structure.
Figure 10.
Comparison of stomatal structures between wild-type (WT) and gpat4 B. napus lines. The bubbles covering the widely opened stomata in the glossy area were likely caused by a high rate of water vapor release when the leaf surface was sealed. The same phenotype was observed in four RNAi lines.
DISCUSSION
Genome Origins and Evolutionary Fates of BnGPAT4 Homologs
In this study, we identified three GPAT4 homologs in B. napus and were able to determine the genome origin of each homolog based on its sequence identity with GPAT4 genes from B. rapa and B. oleracea. Two BnGPAT4 homologs showed greatest sequence similarity with GPAT4 sequences from B. oleracea and therefore are believed to originate from the C genome, while only one A genome-derived BnGPAT4 homolog (showing similarity to GPAT4 from B. rapa) was identified. The recent sequencing of B. rapa (http://brassicadb.org:8081/brad/index.php) uncovered two GPAT4 UniGene sequences (accession nos. Bra033249 and Bra032643), which were highly similar (more than 99%) to the two GPAT4 genes we isolated from B. rapa. Given the complex genetic background and incomplete genome sequence information of B. napus, we are unable to completely exclude the possible existence of other as yet unidentified BnGPAT4 homologs (that may exist as pseudogenes or functional genes). Genome sequencing of B. napus and B. oleracea is still ongoing; thus, our identification of homologous GPAT4 genes in B. napus is limited by the incomplete genome/EST sequence database. Nevertheless, our results here represent, to our knowledge, the most up-to-date information on expressed and functional BnGPAT4 homologs.
The evolutionary consequences of genes after polyploidy and genomic segmental duplications include loss or silencing, maintaining the ancestral function, and functional divergence, which includes neofunctionalization and subfunctionalization (Wendel, 2000; Whittle and Krochko, 2009). Neofunctionalization could result in the acquisition of new catalytic/structural functions or a new expression domain; while subfunctionalization could result in the expression domain or the catalytic/structural functions of an ancestral gene being divided and shared by the existing duplicated genes (Force et al., 1999; Flagel and Wendel, 2009; Liu and Adams, 2010). In the case of BnGPAT4 homologs, because the expression domain of the ancestral gene is unclear, the different expression patterns of the BnGPAT4 homologs could be the result of either subfunctionalization or neofunctionalization.
In addition, the different tissue/organ-specific expression patterns indicated that the BnGPAT4 homologs are developmentally regulated. For example, in developing seeds, BnGPAT4-A1 was expressed dominantly in maturing embryos but was low in developing seed coats; in contrast, BnGPAT4-C1 was expressed at low levels in the embryo but at high levels in the seed coats (Fig. 5B). A previous study with allotetraploid cotton (Gossypium hirsutum) also discovered such developmental regulation (Adams et al., 2003). In that study, several pairs of homologous genes exhibited developmentally regulated reciprocal silencing, whereby one homolog was expressed in certain organs but was low in others and its counterpart was expressed in a complementary way. This developmental regulation was predicted to be a frequent consequence of polyploidy (Adams et al., 2003). The possible mechanisms regulating the expression of homologous genes could include changes in DNA sequence, epigenetic modifications, cis- and trans-acting effects, and RNA-mediated pathways (Chen, 2007). These mechanisms are also believed to be important in facilitating the adaptation of polyploid crops during the evolution and domestication processes (Chen, 2007).
Functional Divergence between the Paralogous and Homoeologous BnGPAT4 Genes
Within the three BnGPAT4 homologs, BnGPAT4-C2 is paralogous to BnGPAT4-C1 and homoeologous to BnGPAT4-A1. The gene structures of the paralogous BnGPAT4-C1 and BnGPAT4-C2 have diverged in several key ways, such as the promoter sequences, intron sequences, and DNA methylation patterns. These different features may ultimately contribute to the different expression patterns between BnGPAT4-C1 and BnGPAT4-C2. A striking difference between their expression patterns is that BnGPAT4-C1 is expressed at a higher level than BnGPAT4-C2 in almost all surveyed plant tissues/organs but not in the developing embryos (Fig. 5). It is clear that their expression patterns are regulated differently according to the developmental stages.
BnGPAT4-A1, originating from the A genome, is considered as a homoeolog of BnGPAT4-C2. Different from BnGPAT4-C1, BnGPAT4-A1 shares a higher level of similarity with BnGPAT4-C2 in the promoter and intron sequences (Fig. 1; Table I; Supplemental Fig. S3). In the promoter-GUS expression assay, the GUS activities directed by the promoters of BnGPAT4-A1 and BnGPAT4-C2 are more similar in comparison with BnGPAT4-C1, particularly at the seedling stage (Fig. 4, A–F). Nevertheless, qRT-PCR results indicated that the expression patterns of BnGPAT4-A1 and BnGPAT4-C2 are quite different. These results suggested that in addition to the promoter, other regulatory factors associated with the individual BnGPAT4 homologs have evolved divergently in order to regulate the different gene expression patterns of the BnGPAT4 homologous genes.
The BnGPAT4 Homologs Encode Functional GPAT Enzymes
In this study, the three BnGPAT4 homologs were functionally expressed in yeast. All three proteins exhibited GPAT enzyme activities, but with different levels of accumulation in the ER membrane of transformed yeast. We demonstrated that the last five to seven amino acids at the C termini of the BnGPAT4 proteins play an important regulatory role for protein accumulation (Fig. 3). The sequence difference between the C termini, however, was not related to their differential accumulation in yeast (Supplemental Fig. S2). Protein abundance is determined by the opposing processes of synthesis and degradation; it remains unclear which of these processes account for the observed differences in BnGPAT4 protein accumulation in transformed yeast.
One possible mechanism behind our observations could be that yeast has different codon usage efficiencies for the three BnGPAT4 homologs, which affects the synthesis rate of the encoded proteins. Another possible mechanism could be differential degradation mediated by the other different amino acids within the protein sequences (Fig. 2C). A previous comparative study of the ER-bound fatty acid desaturases (Fad3) from tung (Vernicia fordii) and B. napus indicated that the BnFad3 contained a degradation signal in its N terminus (approximately 60 amino acid residues), which was responsible for its higher protein turnover rate than the tung Fad3 (O’Quin et al., 2010). In the case of BnGPAT4, there are very minor differences at the N termini (Fig. 2C), which are less likely to cause different protein half-lives. It also remains to be determined whether the observations on different levels of polypeptide accumulation for the BnGPAT4s produced in the yeast expression system reflect the in planta situation.
BnGPAT4 May Be Involved in Suberin Biosynthesis in Root and Seed Coat
Cutin and suberin are two types of insoluble fatty acid- and glycerol-based lipid polymers that are deposited in the cell walls of different plant tissues. They are different from each other based on their monomer compositions and deposition locations. In general, suberin deposits in the seed coat and root, whereas cutin is found mostly at the cutical layer that covers the aerial epidermis (Beisson et al., 2007). An earlier study in Arabidopsis revealed that GPAT5, another member of the ER-bound GPAT family, was involved in suberin synthesis in seed coat and root (Beisson et al., 2007). In this study, we found that all three BnGPAT4 homologs were expressed (at different levels) in seed coat during the period from 20 to 30 DAP (Fig. 5), which corresponds to the timing of suberin deposition in the seed coat (Molina et al., 2008). In addition, the promoter-GUS assay also indicated that the GUS gene was expressed in the periderm and endodermis parts of the root (Fig. 4I), where suberin is normally deposited to strengthen the cell wall and contributes to the control of water movement (Enstone et al., 2003). Therefore, we suggest that the BnGPAT4 homologs (particularly BnGPAT4-C1, which exhibited the highest expression levels) are very likely to be involved in suberin synthesis.
The Cuticle Layer Is Closely Related to Stomatal Development and Structure
The stomata are tiny pores in leaves that regulate the exchange of gas for photosynthesis and the release of water vapor by transpiration. The guard cells surrounding the stomata control the opening and closing of the stomata via turgor-related shape changes (Kappen and Haeger, 1991). A number of environmental and internal factors, such as humidity, light, temperature, CO2, hormones, and ion content, interact with the guard cells through a series of metabolic processes to control the stomatal apparatus (Zeiger, 1983).
As the first line of contact with the environment, the cuticle is mainly composed of wax, cutin, and polysaccharides (Bird and Gray, 2003). In addition to its function as a protective barrier, the cuticle has been shown to be important for stomatal development. It has been shown that defective cuticular wax biosynthesis affects stomatal development with altered stomatal densities in Arabidopsis (Zeiger and Stebbins, 1972; Gray et al., 2000; Chen et al., 2003). It has been proposed that environmental effects on stomatal development are significantly affected by changes in cuticular wax (Bird and Gray, 2003). In comparison with wax, cutin is another important lipid-derived component deposited in the cuticle layer. In this study, we observed “glossy” textured areas on the leaf surface of gpat4 RNAi lines, which were caused by the lack of cuticle deposition (as indicated by the toluidine blue test). Instead of the altered stomatal densities seen in other studies, we observed that the stomata were widely open in the glossy areas of the mature rosette leaves (Fig. 10). It is obvious that a cutin-defective cuticle surface has led to abnormal stomatal morphology. Li et al. (2007) observed defective cuticular ledges on the guard cells in the Arabidopsis gpat4 gpat8 line. A previous study on Vicia faba leaves showed that stomatal opening could be caused by the reduction in epidermal turgor when the leaf was perturbed by a stream of dry air (Mott and Franks, 2001). In the case of B. napus gpat4, it is possible that the defective cuticle layer altered the microenvironmental condition (i.e. lower humidity) for the epidermis, changing the turgor of the epidermal cells surrounding the guard cells and in turn causing the stomata to open. Nevertheless, given the complexity of the mechanisms involved in regulating the stomatal pores (Schroeder et al., 2001), a more comprehensive interpretation of how the defective cuticle layer affects stomatal aperture requires further study.
CONCLUSION
We have characterized three GPAT4 homologous genes in the allotetraploid B. napus, focusing primarily on their genomic origins and functional divergence. All three BnGPAT4 genes were confirmed to encode functional GPAT enzymes but with different levels of polypeptide accumulation when expressed in yeast. The gene expression patterns, epigenetic variations, and phenotypic rescue of the gpat4 gpat8 Arabidopsis double mutant indicated that the three BnGPAT4 genes have evolved through functional divergence. Suppression of GPAT4 expression in B. napus resulted in reduced cutin deposition and alterations in stomatal structure in leaves, which revealed an important role of the cuticle in stomatal function.
MATERIALS AND METHODS
Plant Materials
Brassica napus double haploid line (DH12075) plants were grown in a greenhouse under a 16-h-day/8-h-night cycle at 23°C. Arabidopsis (Arabidopsis thaliana ecotype Columbia) plants were grown in a growth chamber at 23°C under a photoperiod of 18 h. Seeds of homozygous double T-DNA mutant Arabidopsis gpat4 gpat8 plants were obtained from Dr. Yonghua Li-Beisson (CNRS).
Cloning of GPAT4 Genes from B. napus
The Arabidopsis GPAT4 (At1g01610) cDNA sequence was used to query the B. napus EST database in GenBank (National Center for Biotechnology Information) using the megablast program. By analyzing the alignment of the positive hits, we identified three putative GPAT4 homologs in B. napus. Based on single-nucleotide polymorphisms in the cDNA sequences, homolog-specific primer pairs (Supplemental Table S1) were designed to amplify the full-length cDNA and genomic DNA sequences. For cDNA amplification, total RNA prepared from young B. napus seedlings was used in RT-PCR. The promoter regions of the BnGPAT4 homologous genes were cloned using the Universal Genome Walker kit (Clontech Laboratories) according to the manufacturer’s instructions. The corresponding primer pairs are listed in Supplemental Table S1. All the generated PCR amplicons were subcloned into the pCR4-TOPO vector (Invitrogen) and sequenced.
Heterologous Expression in Yeast and in Vitro GPAT Enzyme Assay
The cDNAs of three BnGPAT4 homologs were subcloned into the yeast expression vector pYES2.1/V5-His-TOPO (Invitrogen) and sequenced to confirm the PCR fidelity. The PCR primers used are listed in Supplemental Table S1. The plasmids were then transformed into yeast (Saccharomyces cerevisiae) GPAT mutant strain gat1Δ (Zheng et al., 2003). Gal-induced expression of the BnGPAT4 cDNAs in yeast was performed according to the manual of the pYES2.1/V5-His-TOPO TA expression kit (Invitrogen). LacZ-transformed gat1Δ yeast strain was used as a negative control. Yeast homogenates were prepared with glass beads in a lysate buffer as described previously (Zheng and Zou, 2001). The crude homogenates were first centrifuged at 2,500g at 4°C for 10 min to pellet the cell debris; the supernatant was further centrifuged at 100,000g at 4°C for 1 h to pellet the microsomal fractions, which were then resuspended in lysate buffer for enzyme assay. Protein concentration was determined using the Bradford assay (Bio-Rad) with bovine serum albumin as a standard.
The GPAT enzyme assay was performed at 30°C for 10 min in a 50-μL reaction mixture containing 40 mm HEPES, 5 mm EDTA, 1 mm dithiothreitol, 2.5 mg mL−1 bovine serum albumin, 100 μm [14C (U)]glycerol-3-phosphate (30 Ci mol−1; GE Healthcare), 20 μm oleoyl-CoA (18:1-CoA), and 40 μg of yeast crude microsomal protein. The reaction was quenched with 2 mL of chloroform:methanol (1:2, v/v) and 1 mL of 1 m KCl in 0.2 m H3PO4. The lower organic phase was extracted, dried under nitrogen, resuspended in 70 μL of chloroform, and applied to a precoated thin-layer chromatography plate (layer: 0.25 mm, SIL G-25; DC-Fertigplatten), which was subsequently developed in a solvent system containing chloroform:methanol:acetic acid:5% aqueous sodium bisulfite (75:30:9:3, v/v). The radiolabeled final products (14C-labeled glycerolipids) on the thin-layer chromatography plates were scraped and subjected to scintillation counting.
Western Blotting
The microsomal fractions (40 μg of protein) from yeast expressing BnGPAT4 or LacZ were separated by SDS-PAGE and electrotransferred onto polyvinylidene difluoride membranes following the manufacturer’s instructions (Amersham). The membranes were then incubated in a blocking buffer (GE Healthcare) for 1 h and followed by incubation with horseradish peroxidase-conjugated anti-His antibodies (Invitrogen) at 1:10,000 dilution for 1 h. Horseradish peroxidase activity was visualized by chemiluminescence using the ECL Advance Kit (GE Healthcare).
Promoter-GUS Fusion Analysis
The upstream sequences of the first ATG in BnGPAT4 homoeologs (615 bp for BnGPAT4-C1, 640 bp for BnGPAT4-C2, 656 bp for BnGPAT4-A) were cloned as a HindIII-XbaI fragment into the binary vector pBI121, in which the original GUS reporter gene was replaced with a GUS-eGFP bifunctional reporter gene fusion (Thilmony et al., 2006). The sequences of the primers used for amplification of the promoter sequences are listed in Supplemental Table S1. The three constructs (BnGPAT4-C1Pro:GUS-eGFP, BnGPAT4-C2Pro:GUS-eGFP, BnGPAT4-A1Pro:GUS-eGFP) were introduced into wild-type Arabidopsis plants by Agrobacterium tumefaciens-mediated floral dip (Clough and Bent, 1998). The T2 and T3 progeny from several individual transgenic plants were analyzed for GUS gene expression. Fresh tissues were immersed, vacuum infiltrated, and incubated at 37°C overnight in a staining solution (10 mm sodium phosphate, pH 7.0, 10 mm EDTA, 0.5–2 mm potassium ferricyanide/potassium ferrocyanide, 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, and 0.1% Triton X-100). Stained tissues were soaked in ethanol:acetic acid (3:1, v/v) to clear the chlorophyll and fixed in a mixture of ethanol:acetic acid:37% formaldehyde:water (50:5:27:18, v/v). Images of whole-mount tissues were taken by a Wild M8 dissecting microscope and Zeiss Scope A1 microscope with digital cameras.
TaqMan qRT-PCR
Total RNA was extracted from different B. napus samples at various developmental stages using the RNeasy Plant Mini Kit (Qiagen), except for extraction of RNA from seed coat and root samples, where the cetyl-trimethyl-ammonium bromide method (Gambino et al., 2008) and TRIzol reagent (Invitrogen) were used. First-strand cDNA synthesis was performed in a 20-μL reaction mixture with 1 μg of DNase (Qiagen)-digested total RNA using the QuantiTect reverse transcription kit (Qiagen) according to the manufacturer’s instructions. The 20-μL final cDNA product was then diluted to 500 μL, and 8.5 μL was used in each qRT-PCR. Real-time PCR was performed on a Fast Optical 96-Well Reaction Plate on the ABI PRISM 7900 HT Real-Time PCR System (both from Applied Biosystems). The 25-μL reactions contained 1× TaqMan Universal PCR Master Mix (Applied Biosystems), 360 nm primers, 320 nm probes, and 8.5 μL of cDNA. The following standard thermal profile was used: 50°C for 2 min, 95°C for 5 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Three technical replicate reactions were performed with each cDNA sample and individual primer pairs. According to the recommendation of Chen et al. (2010), three internal reference genes, UBC21, TIP41, and ACT7, were used to normalize the qRT-PCR data. The analysis of the qRT-PCR data (including the calculation of se) was performed according to the geNORM manual (Vandesompele et al., 2002). The sequences of the primers and probes used in this study are listed in Supplemental Table S1.
SYBR-Green qRT-PCR
SYBR-Green qRT-PCR was used to quantify the overall transcription level of the three BnGPAT4 homologs in gpat4 RNAi B. napus lines. The reactions were performed as described by Chen et al. (2010) with minor modifications. In brief, the 10-μL reaction contained 1× SYBR-Green Master Mix (Molecular Biology Facility, University of Alberta), 450 nm primers, and 2.5 μL of 50× diluted cDNA sample (the cDNA samples were prepared as described above under “TaqMan qRT-PCR”). The following standard thermal profile was used: 95°C for 2 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. Amplicon dissociation (melting) curves were recorded thereafter by using the following cycle: 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s, with a ramp rate of 2%. Four technical replicate reactions were performed with each cDNA sample and individual primer pairs.
Genomic DNA Cytosine Methylation Analysis
Genomic DNA was isolated from a mixture of different B. napus vegetative samples, including seedlings, leaves, stems, inflorescence primordia, and flowers. The EpiTect Bisulfite Kit (Qiagen) was used for bisulfite conversion and cleanup of DNA according to the manufacturer’s handbook. For PCR amplification of the bisulfite-converted genomic DNA, three pairs of homolog-specific primers were designed for each homolog to give three amplicons that would cover the 625 bp of target sequence. The sequences of the primers are listed in Supplemental Table S1. The PCR products were then gel purified and subcloned into the pCR4-TOPO TA cloning vectors (Invitrogen). For each PCR amplicon, 12 clones were sequenced.
Constructs for the Expression of Different BnGPAT4 Homologs in the Arabidopsis gpat4 gpat8 Double Mutant Line
The full-length cDNA and promoter of individual BnGPAT4 homologs together with a NOS terminator were subcloned into the binary vector pGreenII0229 (Supplemental Fig. S5; Hellens et al., 2000). The sequences of the associated primers are listed in Supplemental Table S1. The new plasmids, which were then named pGreenII0229- BnGPAT4-C1pro:cDNA, BnGPAT4-C2pro:cDNA, and BnGPAT4-A1pro:cDNA, were transformed into Arabidopsis gpat4 gpat8 double T-DNA mutant lines (provided by Dr. Yonghua Li-Beisson) via the floral dip method as described earlier.
Preparation of the RNAi Construct for Silencing GPAT4 Homologs in B. napus
Two identical fragments of approximately 250 bp, which were amplified from the coding region of BnGPAT4, were subcloned into pKannibal (Wesley et al., 2001) at two orientations of sense (at the sites of XhoI and KpnI) and antisense (at the sites of HindIII and XbaI). The XhoI-XbaI cassette, which was then excised from the resulting pKannibal construct, together with a double cauliflower mosaic virus 35S promoter (with SacI and XhoI at the two ends) were subcloned into a binary plasmid RD400 (kindly provided by Dr. Elzbieta Mietkiewska) at the sites of SacI and XbaI (Supplemental Fig. S6). The sequences of the primers used here are listed in Supplemental Table S1.
Generating gpat4 RNAi B. napus Lines
B. napus plants (DH12075) were transformed as described by Bondaruk et al. (2007). Benzyladenine (4.5 mg L−1) and naphthalene acetic acid (0.1 mg L−1) were used to induce shoot and root formation from transformed callus. The resulting transgenic plants were transferred to soil once roots were established. Genomic DNA PCR and real-time PCR were then performed to confirm the presence of the transgene cassette.
Toluidine Blue Test
The toluidine blue test was used according to the method described by Tanaka et al. (2004). Plant tissues were incubated in an aqueous solution of 0.05% (w/v) toluidine blue for 2 min and then rinsed with water.
Preparation of Surface Casts of Live Leaves and Microscopy
The surface casts were prepared by painting the adaxial surface of attached leaves with clear fingernail polish. After 10 min, the cast was pulled from the leaf surface using a piece of scotch tape (Brewer, 1992). Light microscopy was performed with a Zeiss Imager M1 microscope coupled to a Zeiss AxioCam HRm camera.
Analysis of Cutin Monomers
The method for analyzing cutin monomers was adapted from Bonaventure et al. (2004) with slight modifications. Briefly, delipidated residues from 300 mg of fresh leaves were treated with sodium methoxide for depolymerization and methanolysis. The products of methanolysis were derivatized with pyridine and N,O-bis(trimethylsilyl)-trifluoroacetamide to prepare trimethylsilyl derivatives. The final monomer derivatives were dissolved in 200 μL of heptanes:toluene (1:1, v/v) and then analyzed by gas chromatography-mass spectrometry. Gas chromatography-mass spectrometry analysis was performed using an Agilent 6890N gas chromatograph with an Agilent 5975 Inert Mass Selective Detector. Chromatographic separation was achieved using an HP-5MS capillary column (30 m × 0.25 mm × 0.25 μm; Agilent Technologies) with a constant helium flow rate of 1 mL min−1 and with temperature programmed from 140°C to 300°C at 3°C min−1. The inlet was operated in split mode (10:1 split ratio, 1-μL injection) at 310°C. For the mass spectra condition, the solvent delay was 4 min, ionization energy was 70 eV, and data were acquired in scan mode with a range from 35 to 500 atomic mass units.
Gene sequence data of Arabidopsis (AtGPAT4 and AtGPAT8) from this article can be found in the GenBank data libraries under accession numbers NM_100043 and NM_116264, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PCR amplification of the fourth putative BnGPAT4 homolog.
Supplemental Figure S2. The C termini of the BnGPAT4s are important for controlling protein metabolism.
Supplemental Figure S3. Sequence alignment of the promoter regions of the three BnGPAT4 homologs.
Supplemental Figure S4. Developing B. napus seeds at the stages from 20 to 40 DAP.
Supplemental Figure S5. Schematic diagram of the constructs used for the expression of different BnGPAT4 homologs in Arabidopsis gpat4 gpat8 double mutant lines.
Supplemental Figure S6. Schematic diagram of the construct used for RNAi silencing BnGPAT4 homologs in B. napus.
Supplemental Figure S7. Comparison of the BnGPAT6 expression levels in the rosette leaves of wild-type and RNAi B. napus lines.
Supplemental Figure S8. Water loss of excised rosette leaves recoded over 60 min measured as a percentage of the initial weight.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Elzbieta Mietkiewska for providing the RNAi vector and for her generous suggestions on the RNAi construct design; Drs. Yonghua Li-Beisson, Fred Beisson, and John Ohlrogge for kindly providing the gpat4 gpat8 Arabidopsis seeds; Dr. Enrico Scarpella for his help in the GUS staining assay; Dr. Wei Deng for suggestions and discussions in the preparation of the leaf surface cast; Drs. Disa Brownfield, Rodrigo Siloto, and Shi Xiao for suggestions in revising the manuscript; Dr. Roger Thilmony for providing the GUS-eGFP fusion protein; and Ginette Séguin-Swartz and Gerhard Rakow for providing B. napus DH12075. We are grateful to CSIRO Australia for providing the pKANNIBAL vector.
References
- Adams KL, Cronn R, Percifield R, Wendel JF. (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100: 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. (2007) The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19: 351–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. (1997) Nuclear DNA amounts in angiosperms: 583 new estimates. Ann Bot (Lond) 80: 169–196 [Google Scholar]
- Bird SM, Gray JE. (2003) Signals from the cuticle affect epidermal cell differentiation. New Phytol 157: 9–23 [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G, Beisson F, Ohlrogge J, Pollard M. (2004) Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J 40: 920–930 [DOI] [PubMed] [Google Scholar]
- Bondaruk M, Johnson S, Degafu A, Boora P, Bilodeau P, Morris J, Wiehler W, Foroud N, Weselake R, Shah S. (2007) Expression of a cDNA encoding palmitoyl-acyl carrier protein desaturase from cat’s claw (Doxantha unguis-cati L.) in Arabidopsis thaliana and Brassica napus leads to accumulation of unusual unsaturated fatty acids and increased stearic acid content in the seed oil. Plant Breed 126: 186–194 [Google Scholar]
- Brewer CA. (1992) Responses by stomata on leaves to microenvironmental conditions. Test Stud Laboratory Teach 13: 67–75 [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE. (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Chen X, Goodwin SM, Boroff VL, Liu X, Jenks MA. (2003) Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Truksa M, Shah S, Weselake RJ. (2010) A survey of quantitative real-time polymerase chain reaction internal reference genes for expression studies in Brassica napus. Anal Biochem 405: 138–140 [DOI] [PubMed] [Google Scholar]
- Chen ZJ. (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung F, Trick M, Drou N, Lim YP, Park JY, Kwon SJ, Kim JA, Scott R, Pires JC, Paterson AH, et al. (2009) Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell 21: 1912–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, von Heijne G. (1994) TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci 10: 685–686 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Enstone DE, Peterson CA, Ma F. (2003) Root endodermis and exodermis: structure, function, and responses to the environment. J Plant Growth Regul 21: 335–351 [Google Scholar]
- Fitch WM. (2000) Homology: a personal view on some of the problems. Trends Genet 16: 227–231 [DOI] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. (2009) Gene duplication and evolutionary novelty in plants. New Phytol 183: 557–564 [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. (2007) Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambino G, Perrone I, Gribaudo I. (2008) A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem Anal 19: 520–525 [DOI] [PubMed] [Google Scholar]
- Gidda SK, Shockey JM, Rothstein SJ, Dyer JM, Mullen RT. (2009) Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol Biochem 47: 867–879 [DOI] [PubMed] [Google Scholar]
- Gray JE, Holroyd GH, van der Lee FM, Bahrami AR, Sijmons PC, Woodward FI, Schuch W, Hetherington AM. (2000) The HIC signalling pathway links CO2 perception to stomatal development. Nature 408: 713–716 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A. (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Kappen L, Haeger S. (1991) Stomatal responses of Tradescantia albiflora to changing air humidity in light and in darkness. J Exp Bot 42: 979–986 [Google Scholar]
- Li YH, Beisson F, Koo AJK, Molina I, Pollard M, Ohlrogge J. (2007) Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA 104: 18339–18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Adams KL. (2010) Dramatic change in function and expression pattern of a gene duplicated by polyploidy created a paternal effect gene in the Brassicaceae. Mol Biol Evol 27: 2817–2828 [DOI] [PubMed] [Google Scholar]
- Lukens L, Quijada PA, Udall J, Pires JC, Schranz ME, Osborn T. (2004) Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol J Linn Soc Lond 82: 675–688 [Google Scholar]
- Molina I, Ohlrogge JB, Pollard M. (2008) Deposition and localization of lipid polyester in developing seeds of Brassica napus and Arabidopsis thaliana. Plant J 53: 437–449 [DOI] [PubMed] [Google Scholar]
- Mott KA, Franks PJ. (2001) The role of epidermal turgor in stomatal interactions following a local perturbation in humidity. Plant Cell Environ 24: 657–662 [Google Scholar]
- O’Neill CM, Bancroft I. (2000) Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J 23: 233–243 [DOI] [PubMed] [Google Scholar]
- O’Quin JB, Bourassa L, Zhang D, Shockey JM, Gidda SK, Fosnot S, Chapman KD, Mullen RT, Dyer JM. (2010) Temperature-sensitive post-translational regulation of plant omega-3 fatty-acid desaturases is mediated by the endoplasmic reticulum-associated degradation pathway. J Biol Chem 285: 21781–21796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Koo DH, Hong CP, Lee SJ, Jeon JW, Lee SH, Yun PY, Park BS, Kim HR, Bang JW, et al. (2005) Physical mapping and microsynteny of Brassica rapa ssp. pekinensis genome corresponding to a 222 kbp gene-rich region of Arabidopsis chromosome 4 and partially duplicated on chromosome 5. Mol Genet Genomics 274: 579–588 [DOI] [PubMed] [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ. (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontius JU, Wagner L, Schuler GD. (2003) UniGene: a unified view of the transcriptome. McEntyre J, Ostell J, , The NCBI Handbook. National Center for Biotechnology Information, Bethesda, MD, http://www.ncbi.nlm.nih.gov/books/NBK21101/ [Google Scholar]
- Rana D, van den Boogaart T, O’Neill CM, Hynes L, Bent E, Macpherson L, Park JY, Lim YP, Bancroft I. (2004) Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J 40: 725–733 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Shitsukawa N, Tahira C, Kassai K, Hirabayashi C, Shimizu T, Takumi S, Mochida K, Kawaura K, Ogihara Y, Murai K. (2007) Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 19: 1723–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y. (2004) A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J 37: 139–146 [DOI] [PubMed] [Google Scholar]
- Thilmony R, Guttman M, Chiniquy D, Blechl A. (2006) pGPro1, a novel binary vector for monocot promoter characterization. Plant Mol Biol Rep 24: 57–69 [Google Scholar]
- U N (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7: 389–452 [Google Scholar]
- Udall JA, Wendel JF. (2006) Polyploidy and crop improvement. Crop Sci (Suppl) 46: S3–S14 [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: H0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Xia Q, Qiu X, Selvaraj G. (2002) Early stages of seed development in Brassica napus: a seed coat-specific cysteine proteinase associated with programmed cell death of the inner integument. Plant J 30: 1–10 [DOI] [PubMed] [Google Scholar]
- Wendel JF. (2000) Genome evolution in polyploids. Plant Mol Biol 42: 225–249 [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Whittle CA, Krochko JE. (2009) Transcript profiling provides evidence of functional divergence and expression networks among ribosomal protein gene paralogs in Brassica napus. Plant Cell 21: 2203–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Yu B, Cornish AJ, Froehlich JE, Benning C. (2006) Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants deficient in acyl-ACP glycerol-3-phosphate acyltransferase. Plant J 47: 296–309 [DOI] [PubMed] [Google Scholar]
- Yang YW, Lai KN, Tai PY, Li WH. (1999) Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J Mol Evol 48: 597–604 [DOI] [PubMed] [Google Scholar]
- Zeiger E. (1983) The biology of stomatal guard cells. Annu Rev Plant Physiol 34: 441–475 [Google Scholar]
- Zeiger E, Stebbins L. (1972) Developmental genetics in barley: a mutant for stomatal development. Am J Bot 59: 143–148 [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Xia Q, Dauk M, Shen W, Selvaraj G, Zou J. (2003) Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15: 1872–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Zou J. (2001) The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J Biol Chem 276: 41710–41716 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.