Abstract
Double fertilization in flowering plants refers to a process in which two sperm cells, carried by the pollen tube, fertilize both the egg and the central cell after their release into a synergid cell of the female gametophyte. The molecular processes by which the female gametophytic cells express their unique functions during fertilization are not well understood. Genes expressed in egg and synergid cells might be important for multiple stages of the plant reproductive process. Here, we profiled genome-wide gene expression in egg and synergid cells in rice (Oryza sativa), a model monocot, using a nonenzymatic cell isolation technique. We found that the expression profiles of the egg and synergid cells were already specified at the micropylar end of the female gametophyte during the short developmental period that comprises the three consecutive mitotic nuclear divisions after megaspore generation. In addition, we identified a large number of genes expressed in the rice egg and synergid cells and characterized these genes using Gene Ontology analysis. The analysis suggested that epigenetic and posttranscriptional regulatory mechanisms are involved in the specification and/or maintenance of these cells. Comparisons between the rice profiles and reported Arabidopsis (Arabidopsis thaliana) profiles revealed that genes enriched in the egg/synergid cell of rice were distinct from those in Arabidopsis.
The life cycle of plants alternates between a haploid gametophyte stage and a diploid sporophyte stage. The gametophytes are embedded within the sexual organs of the flower. The male gametophytes develop within the anthers (McCormick, 2004), and the female gametophytes develop within the ovule (Yadegari and Drews, 2004; Fig. 1, A–C). In angiosperms, the female gametophyte is formed by meiotic division of a diploid sporophytic cell, followed by mitotic divisions of one or more haploid cells to generate a multicellular gametophyte. In the developing ovule, a megaspore mother cell undergoes meiosis to generate four spores, three of which undergo programmed cell death, leaving only the single spore as the functional megaspore in each ovule. In the most common form, the polygonum type, the megaspore then undergoes three sequential mitotic nuclear divisions to generate the eight nuclei of the mature embryo sac (Maheshwari and Johri, 1950). Subsequent cellularization results in the formation of just seven cells due to nuclear migration and eventual fusion of two nuclei in the large central cell. In the mature embryo sac, the micropylar end of the female gametophyte has an egg cell and two synergid cells, and the chalazal end of the female gametophyte has three antipodal cells (Fig. 1, B and C), which proliferate into a mass of cells before fertilization in rice (Oryza sativa; Dong and Yang, 1989).
Figure 1.
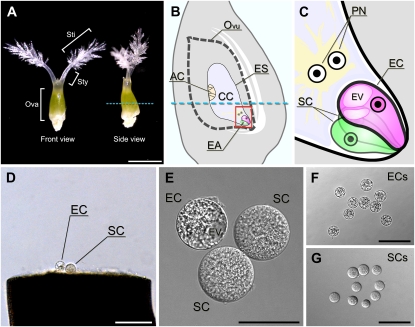
Isolation of rice egg and synergid cells. A, The pistil harvested from a rice flower before anthesis. B, Scheme of the inner structure of the rice ovary. The blue dotted lines in A and B indicate the incision line on the ovary used to isolate the egg and synergid cells. C, An enlarged image of the area enclosed in the square in B. D, The rice egg and synergid cells released from the basal portion of the dissected ovary. E, An isolated egg apparatus collected in a mannitol droplet. F, Isolated egg cells. G, Isolated synergid cells. AC, Antipodal cells; CC, central cell; EA, egg apparatus; EC, egg cell; ES, embryo sac; EV, egg cell vacuole; Ova, ovary; Ovu, ovule; PN, polar nuclei; SC, synergid cell; Sti, stigma; Sty, style. Bars = 1 mm in A, 100 μm in D, F, and G, and 30 μm in E.
Each female gametophyte cell type has specialized roles in reproduction. The egg cell has the potential to receive additional genetic material from the male and to stably transmit it to the next generation. Other cell types in the female gametophyte surrounding the egg cell support the passage of sperm cells into the egg cell. In particular, the synergid cells secrete attractants that guide the pollen tube to the female gametophyte (Higashiyama et al., 2001, 2003; Okuda et al., 2009) and also probably contain factors that control the cessation of pollen tube growth, pollen tube discharge, and gamete fusion (Weterings and Russell, 2004).
The molecular machineries underlying plant reproduction are becoming an area of active research. However, the small number and inaccessibility of these cells have hampered molecular and genome-wide studies; thus, we know little about the molecular basis of cell specification, differentiation, and function in the female gametophyte. The identification of genes expressed in the female gametophyte is essential to understanding how female gametophyte cells become specified and acquire their unique features and functions. Previous research in this field has identified genes that are expressed in specific cells of the female gametophyte in various plant species (Vrinten et al., 1999; Kasahara et al., 2005; Márton et al., 2005; Sprunck et al., 2005; Yang et al., 2006; Steffen et al., 2007; Okuda et al., 2009; Amien et al., 2010; Wang et al., 2010; Wuest et al., 2010). With the exception of Arabidopsis (Arabidopsis thaliana), microarray-based, comprehensive screens for genes exhibiting female gametophyte expression have never been done for plants.
In rice, several methods for isolating egg cells have been reported. Most require a step to degrade the cell wall with catalytic enzymes, such as cellulase (Han et al., 1998; Zhao et al., 2000; Khalequzzaman and Haq, 2005). Egg and synergid cells can also be isolated from unfertilized ovaries by manual manipulation without using enzymes (Zhang et al., 1999; Uchiumi et al., 2006). We collected a large number of rice egg and synergid cells by using manual manipulation instead of enzymes. The quality and quantity of the RNA obtained from the isolated cells were enough to perform transcriptome analysis, and high-quality expression data for rice egg and synergid cells were obtained without cross-contamination. Direct comparisons between the data obtained and the transcriptomes of a variety of diploid tissues showed that the egg and synergid cell transcriptome was distinct. We also identified and characterized a large number of the genes expressed in rice egg and synergid cells. This study provides insights into the roles of genes expressed in the female gametes before, during, and possibly after double fertilization.
RESULTS
Cell Type-Specific Microarrays Using RNA from Isolated Rice Egg and Synergid Cells
Egg and synergid cells can be isolated from unfertilized ovaries by manual manipulation without using enzymes (Zhang et al., 1999; Uchiumi et al., 2006). We chose this nonenzymatic method with some modifications (Takanashi et al., 2010) for the isolation of egg and synergid cells. The sizes of rice egg and synergid cells range between 30 and 50 μm in diameter, which exceeds the size of other cells released from dissected ovaries. Since many vacuoles, ranging in size from 2 to 25 μm, were present in the peripheral region of the egg cells, egg cells can be distinguished from synergid cells by their internal contrast (Fig. 1, D–G). Because an intact egg apparatus composed of a single egg cell and two synergid cells was obtained only rarely, we picked up one or two targeted cell(s) from every ovule. As the synergid cells are more delicate than egg cells, a large number of synergid cells were disrupted during the isolation procedure. We obtained 3,000 egg cells and 1,000 synergid cells from the basal portions of dissected ovaries several days before flowering (Fig. 1; Supplemental Fig. S1) and then extracted the RNA from the isolated cells for microarray analysis. RNA samples derived from egg and synergid cells were of sufficient quality for use in microarray analysis (Supplemental Fig. S2; Supplemental Table S2). We conducted a 44K microarray analysis of egg cell, synergid cell, ovary, and whole plant of japonica rice (cv Nipponbare). In the cell isolation procedure, isolated cells were first washed in mannitol solution and then stored transiently in it (Supplemental Fig. S1). Therefore, we needed to account for any changes in gene expression in the isolated cells caused by the transient storage in mannitol solution. To pay attention to fluctuations in gene expression caused by these washing steps, we prepared two types of ovary RNA: one was derived from ovaries just after cutting the middle portion, and the other was derived from ovaries treated with mannitol after cutting.
Gametic, Nongametic, and Nonreproductive Cell Identities
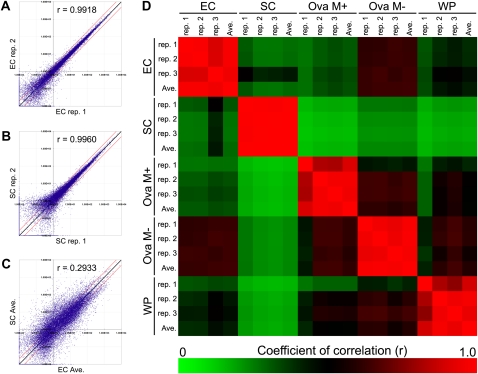
To examine for correlations in gene expression between samples, we created scatterplot diagrams and correlation plots for the microarray data sets. When replicates from the same cell type were compared, the points fell on the 45° identity line (Fig. 2, A and B), showing that the data was highly reproducible for microarray replicates of samples with the same biological origin (egg or synergid cell). In contrast, the points were spread widely when we compared the averaged microarray data set from egg cell samples with that from synergid cell samples (Fig. 2C). This means that the gene expression profiles of egg and synergid cells are very different. Therefore, the cell types are already differentiated with genome-wide transcriptional responses within the egg apparatus at the micropylar end of the female gametophyte during the period comprising the three sequential mitotic nuclear divisions after the creation of the megaspore. These conclusions were supported by correlation plot analysis (Fig. 2D). The expression profiles of the synergid cell were less correlated with the profiles of other samples than were the expression profiles of the egg cell.
Figure 2.
Scatterplot and correlation plot analyses. A to C, Scatterplot analyses of EC replicate 1 versus EC replicate 2 (A), SC replicate 1 versus SC replicate 2 (B), and EC average versus SC average (C). The coefficients of correlation (r) are shown in each plot. Black and red lines indicate the diagonal line y = x and 2-fold ratios of expression level, respectively. D, Correlation plot analysis based on the r values among all samples. The color scale ranges from saturated green (for values of 0, no positive correlation) to saturated red (for values of 1.0, perfect positive correlation). All triplicate data sets had r values of 0.78 or higher. EC, Egg cell; Ova M+/M−, ovary with or without mannitol treatment; SC, synergid cell; WP, whole plant.
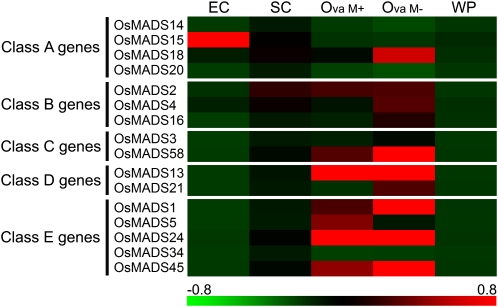
To clearly demonstrate that the egg cell and the synergid cell had already differentiated from the diploid cells that conferred ovule identity, we created a heat map based on the expression patterns of MADS box genes that control floral organ identity (Fig. 3). We confirmed that the levels of the transcripts derived from the three classes of genes (C, D, and E) were higher in both types of ovary samples than in whole plant samples (Fig. 3). Although both egg and synergid cells develop within the ovule, our results show that transcripts from the three classes of genes do not accumulate in these haploid cell samples. In particular, the expression of OsMADS13 was clearly suppressed. OsMADS13 is specifically expressed in the ovule, and its expression is first detected in the ovule primordium, where it persists during further development of the ovule. Inside the ovule, OsMADS13 is expressed in integuments and nucellus tissues (Lopez-Dee et al., 1999). This heat map is consistent with the fact that the egg and synergid cells had already differentiated from the diploid cells that form the ovule and that they had acquired unique gametic and nongametic cell identities, respectively.
Figure 3.
Expression patterns of MADS box genes controlling floral organ identity. Representative gene expression profiles of the MADS box genes controlling floral organ identity are shown. The color scale (representing the average of normalized values) is shown at the bottom. Each column of the matrix represents average expression levels of five samples: egg cell (EC), synergid cell (SC), ovary with mannitol treatment (Ova M+), ovary without mannitol treatment (Ova M−), and whole plant (WP). Class A genes are OsMADS14 (Os03g0752800), OsMADS15 (Os07g0108900), OsMADS18 (Os07g0605200), and OsMADS20 (Os12g0501700); class B genes are OsMADS2 (Os01g0883100), OsMADS4 (Os05g0423400), and OsMADS16 (Os06g0712700); class C genes are OsMADS3 (Os01g0201700) and OsMADS58 (Os05g0203800); class D genes are OsMADS13 (Os12g0207000) and OsMADS21 (Os01g0886200); class E genes are OsMADS1 (Os03g0215400), OsMADS5 (Os06g0162800), OsMADS24 (Os09g0507200), OsMADS34 (Os03g0753100), and OsMADS45 (Os08g0531700).
Interestingly, the heat map of the expression patterns of MADS-box genes showed abundant transcripts for OsMADS15 in egg cell samples. Although the expression of OsMADS15 is developmentally regulated in the differentiating panicle (Furutani et al., 2006), it is possible that OsMADS15 expression is also related to the developmental processes of the egg or embryo.
Genes Enriched in Female Gametophyte Cell Types
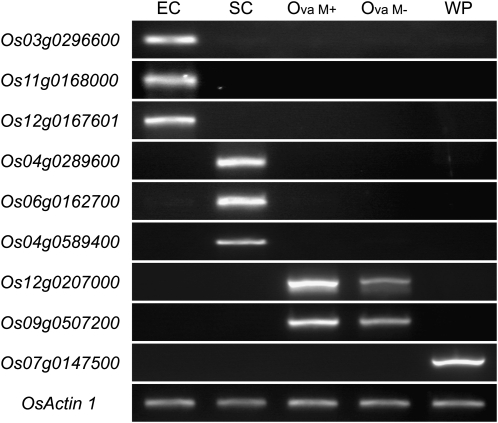
We characterized the gene expression profiles of the egg and synergid cells by focusing on the genes enriched in these cells. Using one-way ANOVA and multiple t test analysis, we identified enriched genes in the egg or synergid cell (see “Materials and Methods”). Based on our criteria, 44 genes (48 probes) and 56 genes (71 probes) were considered as egg cell- and synergid cell-enriched genes, respectively (Supplemental Tables S3 and S4). Among the egg cell-enriched genes (44 genes), we compared the average values of signal intensities between the egg cell and the ovary treated with mannitol and made a list of the top 10 probes by sorting the probes with the EC/Ova M+ value (Table I; these terms are defined in the tables). In a similar way, among the synergid cell-enriched genes (56 genes), we compared the average values of signal intensities between the synergid cell and the ovary treated with mannitol and made a list of the top 10 probes by sorting the probes with the SC/Ova M+ value (Table II). There was a sharp contrast in the expression profiles between the egg cell and the synergid cell, and few genes listed in Tables I and II showed a significant difference in signal intensity between the two types of ovary sample. This suggests that the cell type-dependent accumulation of the transcripts of these listed genes was not caused by artificial mannitol treatment or ovary cutting. We confirmed the expression patterns of several of the listed genes in Tables I and II by reverse transcription (RT)-PCR (Fig. 4). Each surveyed gene showed a cell type-dependent expression pattern that was consistent with the microarray data.
Table I. Ten probes showing genes enriched in the egg cell.
After statistical analysis, 48 probes were considered as egg cell-enriched probes (Supplemental Table S3). The top 10 of 48 probes sorted by log2(EC/M+) are represented here. Values show averages of binary log values of three biological replicates, and values in parentheses are mean P values of the t test with egg cell. EC, Egg cell; Ova M+/M−, ovary with or without mannitol treatment; SC, synergid cell; WP, whole plant.
| Probe Name | Annotation | EC | SC | Ova |
WP | Log2 (EC/M+) | |
| M+ | M− | ||||||
| Os11g0168000|mRNA| AK062599|CDS+3′UTR | Protein of unknown function, DUF1278 family protein (ECA1-like2) | 9.09 | 4.54 (4.74E-03) | −6.64 (5.68E-04) | −4.83 (2.25E-03) | −3.51 (1.48E-03) | 15.73 |
| Os03g0296600|mRNA| AK109176|CDS+3′UTR | Similar to ECA1 protein (ECA1-like1) | 11.21 | 7.42 (9.20E-03) | −3.10 (3.67E-05) | −0.61 (5.37E-04) | −4.39 (2.40E-03) | 14.31 |
| Os11g0187600|mRNA| AK106272|CDS+3′UTR | Similar to heat shock protein 70 | 6.91 | 0.82 (5.11E-03) | −6.17 (1.22E-03) | −5.36 (3.73E-03) | −5.97 (2.66E-03) | 13.08 |
| Os05g0115600|mRNA| AK063589|CDS+3′UTR | Protein of unknown function, DUF674 family protein | 10.99 | 5.69 (4.94E-04) | −1.69 (2.18E-05) | −0.42 (2.14E-06) | −1.99 (1.95E-04) | 12.69 |
| Os06g0228900|COMBINER _EST|Os06g0228900|8 | Hypothetical conserved gene | 10.71 | 5.51 (9.03E-04) | −1.96 (5.83E-04) | −1.70 (1.29E-06) | −2.15 (1.25E-07) | 12.67 |
| Os01g0299700|mRNA| AK108504|CDS+3′UTR | 3′-5′ Exonuclease domain-containing protein | 7.45 | 1.56 (7.50E-04) | −3.75 (1.83E-06) | −1.73 (2.48E-05) | −3.02 (3.90E-06) | 11.20 |
| Os06g0285200|mRNA| AK109660|CDS+3′UTR | Similar to Leafy cotyledon1-like L1L protein | 4.40 | −0.18 (1.53E-03) | −6.64 (6.63E-05) | −6.64 (6.63E-05) | −6.37 (2.13E-04) | 11.05 |
| Os03g0276800|mRNA| AK106371|CDS+3′UTR | Heat shock protein, Hsp70 family protein | 9.64 | 4.59 (1.83E-06) | −1.38 (1.31E-07) | −2.03 (5.88E-05) | −1.81 (2.99E-04) | 11.02 |
| Os11g0157000|COMBINER _EST|CI225501|6 | Hypothetical conserved gene | 4.14 | −6.64 (2.51E-05) | −6.20 (1.63E-03) | −6.37 (4.64E-04) | −6.64 (2.51E-05) | 10.33 |
| Os07g0598400|mRNA| AK072279|CDS+3′UTR | Glycoside hydrolase family 79, N-terminal protein | 7.02 | 1.52 (2.43E-04) | −2.89 (5.07E-04) | −2.80 (3.38E-04) | −3.36 (5.68E-03) | 9.92 |
Table II. Ten probes showing genes enriched in the synergid cell.
After statistical analysis, 71 probes were considered as synergid cell-enriched probes (Supplemental Table S4). The top 10 of 71 probes sorted by log2(SC/M+) are represented here. Values show averages of binary log values of three biological replicates, and values in parentheses are mean P values of the t test with synergid cell. EC, Egg cell; Ova M+/M−, ovary with or without mannitol treatment; SC, synergid cell; WP, whole plant.
| Probe Name | Annotation | EC | SC | Ova |
WP | Log2 (SC/M+) | |
| M+ | M− | ||||||
| Os04g0289600|mRNA| AK111140|CDS+3′UTR | Allergen V5/Tpx-1-related family protein | 6.86 (2.04E-03) | 14.56 | −3.84 (7.64E-06) | −4.19 (4.35E-04) | −6.18 (2.57E-04) | 18.41 |
| Os05g0415700|mRNA| AK102449|CDS+3′UTR | Glycoside hydrolase, family 20 protein | 3.57 (3.10E-06) | 12.56 | −5.21 (5.23E-03) | 2.54 (3.19E-05) | 2.15 (2.87E-04) | 17.77 |
| Os06g0162700|COMBINER| CI446711|x | Similar to MYB98 (myb domain protein 98); DNA-binding/transcription factor | 4.65 (1.39E-04) | 12.10 | −3.26 (1.73E-05) | −2.37 (1.47E-04) | −3.55 (3.42E-03) | 15.36 |
| Os06g0162700|COMBINER| CI435539|6 | Similar to MYB98 (myb domain protein 98); DNA-binding/transcription factor | 2.63 (4.34E-04) | 9.86 | −4.72 (5.31E-03) | −4.66 (4.66E-03) | −6.64 (1.18E-05) | 14.58 |
| Os04g0589400|COMBINER| CI042304|0 | Conserved hypothetical protein | 4.20 (8.36E-03) | 15.41 | 1.73 (1.46E-07) | 1.70 (1.62E-07) | 1.69 (3.76E-05) | 13.68 |
| Os07g0222000|mRNA| AK107870|CDS+3′UTR | Similar to α-amylase/trypsin inhibitor (RBI, RATI) | -2.69 (7.00E-03) | 7.78 | −5.69 (9.56E-04) | −5.99 (3.05E-04) | −6.64 (3.65E-05) | 13.47 |
| Os11g0629100|COMBINER| CI192525|6 | Heavy metal transport/detoxification protein domain-containing protein | 2.57 (4.38E-06) | 6.22 | −5.84 (4.12E-03) | −1.35 (9.82E-05) | −0.64 (3.60E-03) | 12.06 |
| Os04g0289500|COMBINER_EST|Os04g0289500|8 | Allergen V5/Tpx-1-related family proteina | -0.64 (5.02E-03) | 10.56 | −1.25 (1.93E-06) | −1.54 (8.63E-06) | −1.45 (1.54E-06) | 11.82 |
| Os10g0508900|mRNA| AK110739|CDS+3′UTR | Conserved hypothetical protein | 1.20 (4.70E-05) | 11.94 | 0.56 (3.26E-04) | −0.09 (2.03E-05) | 0.68 (7.84E-05) | 11.38 |
| Os11g0529500|COMBINER_EST|CI355192|0 | Polyketide synthase, type III domain-containing protein | 2.67 (1.71E-05) | 11.79 | 0.65 (2.24E-03) | 1.67 (7.03E-04) | 1.81 (2.09E-04) | 11.14 |
A description of the build1 data set. There was no description in the latest data set.
Figure 4.
Confirmatory RT-PCR analysis for genes highly expressed in egg cell and synergid cell. Expression of several genes presented in Tables I and II was tested by RT-PCR of RNA from egg cell, synergid cell, ovary with and without mannitol solution treatment, and whole plant. Os03g0296600, ECA1-like1; Os11g0168000, ECA1-like2; Os12g0167601, ECA1-like3; Os04g0289600, a gene coding allergen V5/Tpx-1-related family protein; Os06g0162700, a gene coding a protein similar to MYB98; Os04g0589400, a gene coding an unknown protein. OsMADS13 (Os12g0207000), OsMADS24 (Os09g0507200), and PSII 10-kD polypeptide (Os07g0147500) were used as negative controls; OsActin1 (Os03g0718100) was used as a positive control. EC, Egg cell; Ova M+/M−, ovary with or without mannitol treatment; SC, synergid cell; WP, whole plant.
In Table I, two of nine genes expressed in egg cells (Os03g0296600, which we named ECA1-like1 gene, and Os11g0168000, which we named ECA1-like2 gene) share significant sequence similarity with egg cell transcripts belonging to the EARLY CULTURE ABUNDANT1 (ECA1) family in barley (Hordeum vulgare; Vrinten et al., 1999), wheat (Triticum aestivum; Sprunck et al., 2005), and Arabidopsis (Steffen et al., 2007; Supplemental Fig. S3). No probes for the third gene of the rice ECA1 gene family (Os12g0167601, which we named ECA1-like3 gene) were present on the microarray chip, but an RT-PCR analysis of the ECA1-like3 gene showed the same expression pattern as the other members (Fig. 4). Two genes (Os11g0187600 and Os03g0276800) encode heat shock protein 70 (HSP70). HSP70 is highly expressed in the egg cells of mouse (Mus musculus; Curci et al., 1987), newt (Pleurodeles waltl; Billoud et al., 1993), maize (Zea mays; Yang et al., 2006), and Arabidopsis (Wuest et al., 2010), and HSP70 is essential for fertilization in bovine (Bos taurus; Matwee et al., 2001) and boar (Sus scrofa domesticus; Spinaci et al., 2005). These results suggest that HSP70 abundance is a common characteristic of animal and plant eggs and that it is required for the fertilization process in both animals and plants. Os06g0285200 encodes a protein that is similar to LEAFY COTYLEDON1-like (L1L). In Arabidopsis, L1L is expressed primarily during seed development and is an essential regulator of embryo development (Kwong et al., 2003). Therefore, it seems likely that L1L is expressed in the rice egg cell before fertilization for swift embryogenesis.
In Table II, we found a gene (Os06g0162700) that had an MYB domain that was highly similar to that of Arabidopsis MYB98. In Arabidopsis, MYB98 controls the development of specific features within the synergid cell during female gametophyte development (Kasahara et al., 2005; Punwani et al., 2007, 2008). Os04g0289600 encodes allergen V5/Tpx-1 related protein, which belongs to a family of Cys-rich secretory proteins called the CAP superfamily (Gibbs et al., 2008).
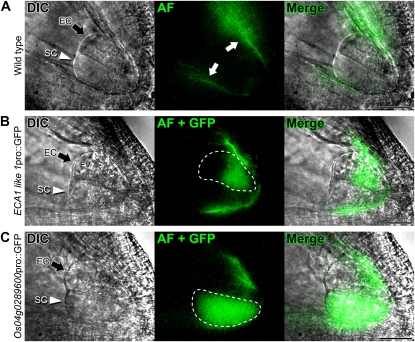
To validate the microarray data, we generated promoter::GFP transgenic rice lines containing promoter regions about 3 kb upstream of the translation start site of the egg cell-expressed ECA1-like1 gene (Os11g0187600) and the synergid cell-expressed allergen V5/Tpx-1-related gene (Os04g0289600). Some site-specific rice ovule autofluorescence was observed at the periphery of the embryo sac, especially at the micropylar end (Fig. 5A, white arrows). In all transgenic rice lines, we observed consistent cell type-dependent GFP expression (Fig. 5, B and C). The chalazal end of the egg cell contains developed vacuoles; therefore, GFP fluorescence driven by the egg cell-dependent promoter was localized to the micropylar end of the egg cell. These results showed that the identified genes were suitable as cell type-specific expression markers.
Figure 5.
Expression analysis of genes expressed in egg and synergid cells from transgenic rice. In each panel, the column on the left shows differential interference contrast (DIC) images, the second column shows autofluorescence (AF) and GFP signals, and the third column shows merged images. A, Autofluorescence observed in the wild-type ovule (white arrows). B, ECA1-like1 pro::GFP expression in the egg cell (enclosed in the white dotted line). C, Os04g0289600 pro::GFP expression in the synergid cell (enclosed in the white dotted line). Black arrows and white arrowheads indicate the egg cell and the synergid cell, respectively. EC, Egg cell; EV, egg cell vacuole; SC, synergid cell. Bars = 30 μm.
Characterization of the Gene Expression Profiles of the Rice Egg and Synergid Cells
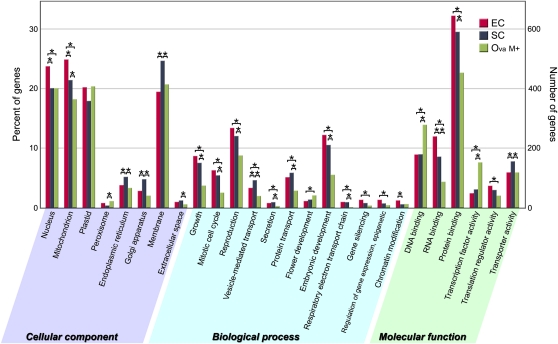
To determine which gene functions are enriched for in the egg or synergid cell, we performed a Gene Ontology (GO) analysis of the 2,000 most highly expressed genes in the egg cell, synergid cell, and ovary after mannitol treatment (Fig. 6). The results showed significant differences in the number of genes categorized by each GO term among these samples. In the egg cell, a high number of genes, whose translated products were estimated to localize to mitochondria and to participate in the respiratory electron transport chain, were expressed. In the synergid cell, a large number of genes involved in secretion were expressed. The fact that genes coding extracellular signaling molecules are expressed preferentially in the synergid cell is a common characteristic of dicots, such as Arabidopsis (Jones-Rhoades et al., 2007), which have a high number of ovules per ovary, and the monocot rice, which has a single ovule per ovary. Furthermore, the results showed that genes involved in transcriptional regulation, such as those involved in DNA binding and transcriptional factor activity, were expressed at a significantly lower level in both the egg and synergid cells than in the ovary. On the other hand, genes involved in gene silencing, regulation of gene expression (epigenetic), and chromatin modification were expressed at significantly higher levels in haploid cell types than in diploid tissues. These results suggest that posttranscriptional regulation or epigenetic control contributes to the establishment of the specification of these haploid cells in the last stages of egg apparatus development and fertilization.
Figure 6.
Gene classification based on GO in the egg cell, synergid cell, and ovary. The 2,000 most highly expressed genes in each sample were classified by GO analysis. In this ontology, cellular component, biological process, and molecular function are treated as independent attributes. Asterisks indicate remarkable relationships, if the P value of the Pearson χ2 test between the gene numbers of the two samples is below the significance level of 0.05. EC, Egg cell; Ova M+, ovary with mannitol treatment; SC, synergid cell.
By focusing on egg cell- and synergid cell-enriched genes, we compared the rice results with those obtained previously (Wuest et al., 2010) from laser-assisted microdissection (LAM)-derived egg and synergid cells from Arabidopsis. We checked whether Arabidopsis homologs of the rice egg/synergid cell-enriched genes existed by searching for candidates with high protein sequence similarities using The Arabidopsis Information Resource (TAIR) BLAST-P. Potential candidates were also screened by the motif pattern-based dendrogram (SALAD database) to rule out the possibility of identifying a different family member as the best Arabidopsis match. We considered candidates that passed both BLAST-P and SALAD database screening as homologs (Supplemental Tables S5 and S6). We excluded genes for which we could not determine corresponding relationships between species due to too many similar genes in the genome.
Initially, we compared Arabidopsis homologs of rice egg cell- or synergid cell-enriched genes (44 or 56 genes) with reported Arabidopsis egg cell- or synergid cell-enriched genes (163 or 144 genes; available in supplemental table S3 of Wuest et al., 2010). This showed that only a few genes were shared by Arabidopsis homologs of rice egg/synergid cell-enriched genes and Arabidopsis egg/synergid cell-enriched genes. The shared genes comprised three egg cell-enriched genes: a gene coding a Cys peptidase active site domain-containing protein (Os04g0599600); a gene coding a ubiquitin-protein ligase/zinc ion-binding protein (Os08g0451900); and a gene coding protein of unknown function belonging to the DUF1278 family (ECA1-like2, Os11g0168000); and two synergid cell-enriched genes: a gene coding a protein similar to MYB98 (Os06g0162700); and a gene coding a glycosyl transferase, family 31 protein (Os09g0452900).
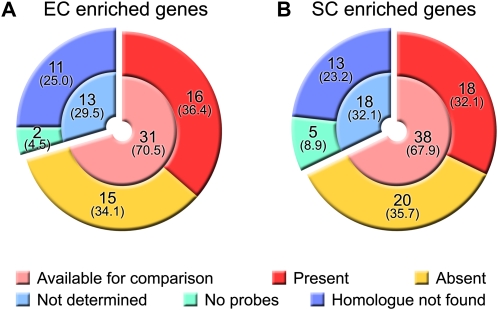
Next, we surveyed a larger number of genes and examined whether Arabidopsis homologs of rice egg cell- or synergid cell-enriched genes were expressed in the Arabidopsis egg or synergid cell by examining a list of Arabidopsis egg cell- or synergid cell-expressed genes (7,171 and 5,628 genes; available in supplemental table S1 of Wuest et al., 2010; Fig. 7). Among the rice egg cell-enriched genes (44 genes), 33 Arabidopsis homologs were found (total of “Available for comparison” and “No probes” in Fig. 7A), and 16 homologs were present in the list of Arabidopsis egg cell-expressed genes, indicating that 15 homologs were not expressed in the Arabidopsis egg cell (Supplemental Table S5). Among the rice synergid cell-enriched genes (56 genes), 43 Arabidopsis homologs were found (Fig. 7B), and 18 homologs were present in the list of Arabidopsis synergid cell-expressed genes, indicating that 20 homologs were not expressed in the Arabidopsis synergid cell (Supplemental Table S6). These results show that a large number of the Arabidopsis homologs of rice egg cell- or synergid cell-enriched genes did not have the same expression pattern in the Arabidopsis egg or synergid cell.
Figure 7.
Comparison of rice and Arabidopsis egg and synergid cell gene expression. We compared the data with reported Arabidopsis data (Wuest et al., 2010). We investigated whether Arabidopsis homologs of the rice egg cell-enriched genes were present in a list of Arabidopsis egg cell-expressed genes (A) and whether Arabidopsis homologs of rice synergid cell-enriched genes were present in a list of Arabidopsis synergid cell-expressed genes (B). Available for comparison means genes fulfilling both of the following conditions: (1) the homolog can be found, and (2) probes for the homolog exist on the microarray chip. No probes means that there were no probes of the homolog on the microarray chip. EC, Egg cell; SC, synergid cell. Values show number of genes, and values in parentheses show their percentages.
From these comparisons, we conclude that some of the homologous genes have conserved functions in rice and Arabidopsis and that nearly half of the homologous genes obtained or lost their roles in the egg or synergid cell through species-specific evolution.
DISCUSSION
Despite previous studies in plant reproduction, the molecular machineries that underlie the development of the female gametophyte and the interactions between the female and male gametophytes have remained elusive. To bring understanding to the molecular basis for these machineries, we performed microarray analysis using pure rice egg and synergid cells. As a result, we found that the cell types at the micropylar end of the female gametophyte had already differentiated with distinct genome-wide expression patterns (Fig. 2C). Our results suggest that these cells are specified rapidly and drastically after the female gametophytic cell fate is conferred by positional information, such as by an auxin gradient. In particular, we found that the gene expression profile of the synergid cell was very different from that of other samples (Fig. 2D). This may be because the synergid cell plays unique roles in many steps of the angiosperm fertilization process, including guiding pollen tube growth toward the female gametophyte. A recent study determined the cell type-specific expression profiles in the female gametophyte of Arabidopsis by using LAM (Wuest et al., 2010). Using their data sets, we calculated the correlation coefficient between gene expression in the Arabidopsis egg and that in the synergid cell. This revealed that the correlation coefficient between the egg and synergid cell was higher in Arabidopsis (r = 0.75) than that in rice (r = 0.29; Fig. 2C). These results indicate that there is no significant difference between the gene expression profiles of the egg cell and that of the synergid cell in Arabidopsis, unlike in rice. This is a major difference between the two studies and might reflect fundamental differences in gene expression of the female gametophyte between Arabidopsis and rice or cross-contamination by neighboring cells, which is difficult to avoid in LAM.
We identified a large number of highly expressed genes in egg and synergid cells as well as genes whose expression profiles were specific to the egg or synergid cell. The cell type-dependent GFP expression in transgenic rice confirmed that our strategy was successful in identifying genes expressed in the egg or synergid cell. Although the genes listed in Tables I and II have no reported function in rice, they might be involved in reproductive events that correlate with their expression patterns in these plants and animals. In particular, we found that the genes included in two gene families, the ECA1 family and the allergen V5/Tpx-1-related protein family, show female gametophyte cell type-dependent expression patterns. The proteins derived from each gene family may have a high degree of functional redundancy. These genes cannot be identified or analyzed using traditional forward-genetic approaches, which rely on the screening of loss-of-function mutants. Our results revealed that allergen V5/Tpx-1-related proteins, which belong to a family of Cys-rich secretory proteins called the CAP superfamily, are abundant in the rice synergid cell. In Torenia fournieri, two secreted, Cys-rich polypeptides in a subgroup of defensin-like proteins are abundantly and predominantly expressed in the synergid cell and act as pollen tube attractants (Okuda et al., 2009). These rice CAP superfamily genes have no known function and therefore may play a role in pollen tube guidance. For the rice MYB98-like gene (Os06g0162700), each of three independent Tos17 insertion lines of this gene exhibits a 50% sterility phenotype (http://tos.nias.affrc.go.jp/), as does the Arabidopsis myb98 mutant (Kasahara et al., 2005). It is possible, therefore, that this gene encodes the rice ortholog of Arabidopsis MYB98. The common features of gametophytic cells in different species suggest that these essential factors for plant reproduction are conserved in angiosperms and could be involved in the molecular mechanisms of not only fertilization but also cell differentiation.
From the GO analysis, we found that a large number of genes related to mitochondrial function were expressed in the rice egg cell. This is consistent with our previous results that the egg cell has a large amount of mitochondrial DNA and that egg cell mitochondria are active in rice (Takanashi et al., 2010). In agreement with a previous report (Wuest et al., 2010), the GO analysis also indicated that not only transcriptional regulation but posttranscriptional regulation and epigenetic regulation contribute to the differentiation of the rice egg and synergid cells. Furthermore, the rice profiles and reported Arabidopsis profiles were compared to screen for genes that exhibit female gametophytic cell type-dependent expression in rice or Arabidopsis. Although the developmental process of the female gametophyte is similar in Arabidopsis and rice (both are of the polygonum type), our comparisons revealed that most of the enriched genes in the egg or synergid cell of rice were distinct from those in Arabidopsis. While differences in the methods of cell isolation and statistical analysis used in this study in rice and those used in the Arabidopsis study (Wuest et al., 2010) may have led to some discrepancies in the data, the results nonetheless suggest that the widely different sets of genes function in the egg/synergid cell between rice and Arabidopsis.
CONCLUSION
To our knowledge, this is the first comprehensive genome-wide transcriptional analysis of female gametophyte cells in monocots. The analysis showed that the gene expression profiles of rice egg and synergid cells are distinct despite their neighboring locations and that complicated machineries exist for the specification of these cells besides the auxin gradient. Our findings enabled us to decipher the differences between individual female gametophyte cell types and to identify new genes expressed in these cells. We are convinced that the transcriptional profiles of female gametophyte cells will help us understand how female gametophyte cells become specified and acquire their unique features and functions. Future research should be directed toward identifying the functions of the egg cell- and synergid cell-enriched genes before, during, and possibly after double fertilization.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type rice plants (Oryza sativa ‘Nipponbare’) were grown in pots under natural conditions. Transgenic rice plants were grown and maintained in a biohazard greenhouse at 30°C (day) and 25°C (night) under natural light conditions.
RNA Samples for Microarrays and RT-PCR
Ovaries were harvested from flowers in stages ranging from heading to just before anthesis (Fig. 1A). The egg and synergid cells were isolated from ovaries by nonenzymatic procedures based on an isolation method for rice egg cells described by Takanashi et al. (2010). Ovary samples were used for RNA extraction steps after cutting and removing the egg and synergid cells from the basal portion of the lower part of the cut ovary. Mannitol treatment consisted of soaking five ovaries at a time in mannitol solution for 8 h. Shoots and roots of 2-week-old rice plants were used as whole plant samples. Total RNA of each cell type and tissue was extracted with a PicoPure RNA isolation kit (Molecular Devices).
Microarray
Microarray experiments were performed as described by Suwabe et al. (2008). Briefly, extracted total RNA was quantified with the Quant-iT RiboGreen RNA reagent and kit (Invitrogen). Three biological replicates were prepared for each sample. A rice 44K oligomicroarray (Agilent Technologies) with 42,000 oligonucleotides based on the Rice Annotation Project was used. Annotation lists available in the Rice Annotation Project database (http://rapdb.dna.affrc.go.jp/, build 5) were used in Tables I and II and Supplemental Tables S3 to S6 (Tanaka et al., 2008). Fluorescent probe labeling using the oligo(dT)-T7 strand-specific amplification method and hybridization were performed in accordance with the manufacturer’s instructions (Agilent Technologies) with slight modifications (The cDNA synthesis reaction time was extended to 6 h [2 h is the manufacturer’s instruction].) The slide images were scanned with a DNA microarray scanner (G2565BA; Agilent Technologies) using the manufacturer’s Feature Extraction software.
Data Analysis
Raw expression data sets were scaled with 75th percentile scaling (per chip) using R software (http://www.r-project.org/). To identify differentially expressed genes, the scaled data were analyzed by one-way ANOVA using oneway.test in the R package (http://cran.r-project.org/). To adjust P values, a Benjamini and Hochberg (1995) correction of the false discovery rate was applied (adjusted P < 0.000001) using R library multtest (http://cran.r-project.org/web/packages/multtest/multtest.pdf), leading to a list of differentially expressed gene sets (3,044 probes; Supplemental Table S1). The scaled data sets of the selected 3,044 probes were transformed to binary log values for further analysis using Microsoft Excel. To identify genes enriched in the egg cell, we performed four patterns of t test (EC versus SC, EC versus Ova M+, EC versus Ova M−, and EC versus WP) and fold change calculations in each pairing using Microsoft Excel. We applied a filter that considers both the fold change (log2 ratio > 3.5) and the P value (P < 0.01) in each pairing, and we considered the genes that fulfilled these criteria among all four of the pairings as egg cell-enriched genes (44 genes in 48 probes; Supplemental Table S3). Using similar methods, 56 genes (71 probes) were considered as synergid cell-enriched genes (Supplemental Table S4).
GO terms for the 2,000 most highly expressed genes in the egg cell, synergid cell, and ovary after mannitol treatment were obtained from the database site AgriGO (http://bioinfo.cau.edu.cn/agriGO/index.php), which was developed on the foundation of EasyGO (Zhou and Su, 2007), and GO terms were analyzed with the Web Gene Ontology Annotation Plotting tool WEGO using default settings (http://wego.genomics.org.cn/cgi-bin/wego/index.pl; Ye et al., 2006). A scaled data set with 75th percentile scaling was used in the scatterplot analysis (Fig. 2, A–C) and correlation plot analysis (Fig. 2D). Z-score-transformed data sets (per chip, using R software) were used in the heat map analysis, and the data were viewed with Java Tree View software (http://jtreeview.sourceforge.net/). Whole microarray data can be found at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE21074) and in Supplemental Table S1.
RT-PCR Analysis
Total RNA (5 ng) from each sample (egg cell, synergid cell, ovary with or without mannitol treatment, and whole plant) was amplified using the SMART mRNA Amplification Kit (Clontech) in accordance with the manufacturer’s instructions. First-strand cDNA was synthesized using 100 ng of each amplified mRNA, 25 μg mL−1 oligo(dT)22 primer, 0.5 mm deoxyribonucleotide triphosphate mix, 1× first-strand buffer, 0.01 m dithiothreitol, 40 units of RNaseOUT (Invitrogen), and 200 units of SuperScript III reverse transcriptase (Invitrogen) in accordance with the manufacturer’s instructions. Each RT-PCR used 1 μL of first-strand cDNA as template in a 20-μL reaction with 0.2 μm of each primer. The primers used for RT-PCR and cycle numbers are listed in Supplemental Table S7.
Analysis of Promoter::GFP Fusions
Genomic sequences of promoter regions about 3 kb upstream of the translation start site of the two egg cell-enriched genes, ECA1-like1 (Os11g0187600) and ECA1-like2 (Os11g0168000), and the two synergid cell-enriched genes, allergen V5/Tpx-1-related gene (Os04g0289600) and Oryzasin1 (Os05g0567100), were amplified by PCR and cloned into pHGWFS7 upstream of the GFP open reading frame using the In-Fusion PCR Cloning System (Clontech). The primers used for plasmid construction are listed in Supplemental Table S7. Rice plants (cv Nipponbare) were transformed as described by Toki et al. (2006). Transgenic plants were selected on a medium containing 50 mg L−1 hygromycin. Hygromycin-resistant plants were transplanted into soil and grown at 30°C (day) and 25°C (night). Isolated ovules of transgenic plants were subjected to vital imaging by using a fluorescence microscope (TE2000-U with 10× [numerical aperture, 0.30] and 40× [numerical aperture, 0.75] objectives; Nikon). GFP signals were visualized with a C-LHG1 HG lamp (Nikon) and GFP(R)-BP filter set. Images were captured by using a cooled CCD camera (Evolution MP; Media Cybernetics). All images were processed using Photoshop 7.0 (Adobe Systems).
Comparisons of the Gene Expression Profiles of Rice and Arabidopsis Egg and Synergid Cells
We compared the egg and synergid cell transcriptome of rice with the genes reported to be expressed in Arabidopsis (Arabidopsis thaliana) egg and synergid cells (Wuest et al., 2010) by examining their homologs. According to Wuest et al. (2010), 163 genes were Arabidopsis egg cell-enriched genes; 5,380 genes were Arabidopsis egg cell-expressed genes; 144 or 3,926 genes were Arabidopsis synergid cell-enriched or -expressed genes. In rice, 44 or 56 genes were rice egg cell- or synergid-cell enriched genes (Supplemental Tables S3 and S4). Homologs of rice egg cell- or synergid cell-enriched genes in Arabidopsis were found by performing amino acid sequence similarity analysis (using the BLASTP program available in TAIR with TAIR9 protein data sets; http://www.arabidopsis.org/Blast/index.jsp) and the motif pattern-based dendrogram (using the SALAD database version 3.0; http://salad.dna.affrc.go.jp/salad/; Mihara et al., 2010). We examined whether the Arabidopsis homologs of rice egg cell- or synergid cell-enriched genes were present in a list of Arabidopsis egg cell- or synergid cell-enriched or -expressed genes (Supplemental Tables S5 and S6).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. How the isolated cells were collected and washed.
Supplemental Figure S2. Assessments of the integrity of the RNA extracted from egg cells and synergid cells.
Supplemental Figure S3. Multiple alignment and phylogenic tree analysis of ECA1 family proteins.
Supplemental Table S1. Microarray data.
Supplemental Table S2. RNA concentrations and integrities in egg and synergid cell samples.
Supplemental Table S3. The probe list of the rice egg cell-enriched genes.
Supplemental Table S4. The probe list of the rice synergid cell-enriched genes.
Supplemental Table S5. Data validation of the genes enriched in the rice egg cell by comparing with the data for genes expressed in the Arabidopsis egg cell.
Supplemental Table S6. Data validation of the genes enriched in the rice synergid cell by comparing with the data for genes expressed in the Arabidopsis synergid cell.
Supplemental Table S7. Primer sequences for RT-PCR and plasmid construction.
Supplementary Material
Acknowledgments
We thank T. Higashiyama and R.D. Kasahara for excellent technical assistance.
References
- Amien S, Kliwer I, Márton ML, Debener T, Geiger D, Becker D, Dresselhaus T. (2010) Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biol 8: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Billoud B, Rodriguez-Martin ML, Berard L, Moreau N, Angelier N, Laine MC. (1993) Constitutive expression of a somatic heat-inducible hsp70 gene during amphibian oogenesis. Development 119: 921–932 [DOI] [PubMed] [Google Scholar]
- Curci A, Bevilacqua A, Mangia F. (1987) Lack of heat-shock response in preovulatory mouse oocytes. Dev Biol 123: 154–160 [DOI] [PubMed] [Google Scholar]
- Dong J, Yang HY. (1989) An ultrastructural study of embryo sac in Oryza sativa L. Acta Bot Sin 31: 81–88 [Google Scholar]
- Furutani I, Sukegawa S, Kyozuka J. (2006) Genome-wide analysis of spatial and temporal gene expression in rice panicle development. Plant J 46: 503–511 [DOI] [PubMed] [Google Scholar]
- Gibbs GM, Roelants K, O’Bryan MK. (2008) The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins. Roles in reproduction, cancer, and immune defense. Endocr Rev 29: 865–897 [DOI] [PubMed] [Google Scholar]
- Han HM, Zhao J, Shi HZ, Yang HY, Zhou C. (1998) Isolation of egg cells and zygotes in Oryza sativa. Acta Bot Sin 40: 186–188 [Google Scholar]
- Higashiyama T, Kuroiwa H, Kuroiwa T. (2003) Pollen-tube guidance: beacons from the female gametophyte. Curr Opin Plant Biol 6: 36–41 [DOI] [PubMed] [Google Scholar]
- Higashiyama T, Yabe S, Sasaki N, Nishimura Y, Miyagishima S, Kuroiwa H, Kuroiwa T. (2001) Pollen tube attraction by the synergid cell. Science 293: 1480–1483 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Borevitz JO, Preuss D. (2007) Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet 3: 1848–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara RD, Portereiko MF, Sandaklie-Nikolova L, Rabiger DS, Drews GN. (2005) MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17: 2981–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalequzzaman M, Haq N. (2005) Isolation and in vitro fusion of egg and sperm cells in Oryza sativa. Plant Physiol Biochem 43: 69–75 [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Dee ZP, Wittich P, Enrico Pè M, Rigola D, Del Buono I, Gorla MS, Kater MM, Colombo L. (1999) OsMADS13, a novel rice MADS-box gene expressed during ovule development. Dev Genet 25: 237–244 [DOI] [PubMed] [Google Scholar]
- Maheshwari P, Johri BM. (1950) Development of the embryo sac, embryo and endosperm in Helixanthera ligustrina (Wall.) Dans. Nature 165: 978–979 [DOI] [PubMed] [Google Scholar]
- Márton ML, Cordts S, Broadhvest J, Dresselhaus T. (2005) Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307: 573–576 [DOI] [PubMed] [Google Scholar]
- Matwee C, Kamaruddin M, Betts DH, Basrur PK, King WA. (2001) The effects of antibodies to heat shock protein 70 in fertilization and embryo development. Mol Hum Reprod 7: 829–837 [DOI] [PubMed] [Google Scholar]
- McCormick S. (2004) Control of male gametophyte development. Plant Cell (Suppl) 16: S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Itoh T, Izawa T. (2010) SALAD database: a motif-based database of protein annotations for plant comparative genomics. Nucleic Acids Res 38: D835–D842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, et al. (2009) Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361 [DOI] [PubMed] [Google Scholar]
- Punwani JA, Rabiger DS, Drews GN. (2007) MYB98 positively regulates a battery of synergid-expressed genes encoding filiform apparatus localized proteins. Plant Cell 19: 2557–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punwani JA, Rabiger DS, Lloyd A, Drews GN. (2008) The MYB98 subcircuit of the synergid gene regulatory network includes genes directly and indirectly regulated by MYB98. Plant J 55: 406–414 [DOI] [PubMed] [Google Scholar]
- Spinaci M, Volpe S, Bernardini C, De Ambrogi M, Tamanini C, Seren E, Galeati G. (2005) Immunolocalization of heat shock protein 70 (Hsp 70) in boar spermatozoa and its role during fertilization. Mol Reprod Dev 72: 534–541 [DOI] [PubMed] [Google Scholar]
- Sprunck S, Baumann U, Edwards K, Langridge P, Dresselhaus T. (2005) The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J 41: 660–672 [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Macfarlane J, Drews GN. (2007) Identification of genes expressed in the Arabidopsis female gametophyte. Plant J 51: 281–292 [DOI] [PubMed] [Google Scholar]
- Suwabe K, Suzuki G, Takahashi H, Shiono K, Endo M, Yano K, Fujita M, Masuko H, Saito H, Fujioka T, et al. (2008) Separated transcriptomes of male gametophyte and tapetum in rice: validity of a laser microdissection (LM) microarray. Plant Cell Physiol 49: 1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanashi H, Ohnishi T, Mogi M, Okamoto T, Arimura S, Tsutsumi N. (2010) Studies of mitochondrial morphology and DNA amount in the rice egg cell. Curr Genet 56: 33–41 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, Numa H, Sakai H, Wu J, Itoh T, Sasaki T, et al. (2008) The rice annotation project database (RAP-DB): 2008 update. Nucleic Acids Res 36: D1028–D1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Komatsu S, Koshiba T, Okamoto T. (2006) Isolation of gametes and central cells from Oryza sativa L. Sex Plant Reprod 19: 37–45 [Google Scholar]
- Vrinten PL, Nakamura T, Kasha KJ. (1999) Characterization of cDNAs expressed in the early stages of microspore embryogenesis in barley (Hordeum vulgare) L. Plant Mol Biol 41: 455–463 [DOI] [PubMed] [Google Scholar]
- Wang D, Zhang CQ, Hearn DJ, Kang IH, Punwani JA, Skaggs MI, Drews GN, Schumaker KS, Yadegari R. (2010) Identification of transcription-factor genes expressed in the Arabidopsis female gametophyte. BMC Plant Biol 10: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings K, Russell SD. (2004) Experimental analysis of the fertilization process. Plant Cell (Suppl) 16: S107–S118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U. (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Yadegari R, Drews GN. (2004) Female gametophyte development. Plant Cell (Suppl) 16: S133–S141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kaur N, Kiriakopolos S, McCormick S. (2006) EST generation and analyses towards identifying female gametophyte-specific genes in Zea mays L. Planta 224: 1004–1014 [DOI] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L, et al. (2006) WEGO: a Web tool for plotting GO annotations. Nucleic Acids Res 34: W293–W297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dong WH, Galli A, Potrykus I. (1999) Regeneration of fertile plants from isolated zygotes of rice (Oryza sativa). Plant Cell Rep 19: 128–132 [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhou C, Yang HY. (2000) Isolation and in vitro culture of zygotes and central cells of Oryza sativa L. Plant Cell Rep 19: 321–326 [DOI] [PubMed] [Google Scholar]
- Zhou X, Su Z. (2007) EasyGO: Gene Ontology-based annotation and functional enrichment analysis tool for agronomical species. BMC Genomics 8: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.