Abstract
Histone methylation plays an essential role in regulating chromatin structure and gene expression. Jumonji C (JmjC) domain-containing proteins are generally known as histone demethylases. Circadian clocks regulate a large number of biological processes, and recent studies suggest that chromatin remodeling has evolved as an important mechanism for regulating both plant and mammalian circadian systems. Here, we analyzed a subgroup of JmjC domain-containing proteins and identified Arabidopsis (Arabidopsis thaliana) JMJ30 as a novel clock component involved in controlling the circadian period. Analysis of loss- and gain-of-function mutants of JMJ30 indicates that this evening-expressed gene is a genetic regulator of period length in the Arabidopsis circadian clock. Furthermore, two key components of the central oscillator of plants, transcription factors CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL, bind directly to the JMJ30 promoter to repress its expression, suggesting that JMJ30 regulates the pace of the circadian clock in close association with the central oscillator. JMJ30 represents, to our knowledge, the first JmjC domain-containing protein involved in circadian function, and we envision that this provides a possible molecular connection between chromatin remodeling and the circadian clock.
Circadian rhythms are endogenous biological rhythms with a period of approximately 24 h. The circadian clock, found in organisms ranging from cyanobacteria to mammals, allows organisms to anticipate regular environmental changes thus enhancing evolutionary fitness. The common mechanism of the eukaryotic circadian clock involves multiple interlocked feedback loops. In Arabidopsis (Arabidopsis thaliana), the circadian clock regulates processes such as gene expression, photoperiodic flowering, and leaf movement (McClung, 2006). Several components of the Arabidopsis clock have been identified, although a complete understanding of how the clock generates self-sustaining rhythms is lacking. CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) are morning-expressed MYB transcription factors that are suggested to form a feedback loop in the circadian clock with the evening-expressed pseudoresponse regulator TIMING OF CAB EXPRESSION1 (TOC1; Schaffer et al., 1998; Wang and Tobin, 1998; Strayer et al., 2000). CCA1 and LHY directly repress TOC1 expression by binding to the evening element (EE) in its promoter (Alabadí et al., 2001); TOC1 is in turn believed to activate transcription of CCA1 and LHY through CCA1 HIKING EXPEDITION and other unknown mechanisms (Pruneda-Paz et al., 2009). Other key clock components that interact with the CCA1/LHY/TOC1 loop include GIGANTEA (GI) and PSEUDO RESPONSE REGULATORs (PRRs) 7 and 9 (Farré et al., 2005; Locke et al., 2006).
The methylation status of histones controls chromatin remodeling and gene expression in eukaryotes. Histone modifications have also been implicated in the regulation of the circadian clock in Arabidopsis. For example, the expression of TOC1 is affected by clock-controlled cycles of histone acetylation, although the responsible histone deacetylase(s) is/are not known (Perales and Más, 2007). Jumonji C (JmjC) domain-containing proteins have been shown to be involved in chromatin remodeling, acting as histone demethylases (Tsukada et al., 2006). The name jumonji (which means cruciform in Japanese) was originally derived from a mouse mutation that affected neural tube development and produced a cross-like structure on the neural plate (Takeuchi et al., 1995). The JmjC domain is the catalytic domain, and these proteins catalyze Lys demethylation through an oxidative reaction that requires Fe(II) and α-ketoglutarate as cofactors. The JmjC domain-containing proteins are involved in a broad range of processes, such as neural stem cell differentiation (Jepsen et al., 2007), X-linked mental retardation (Iwase et al., 2007), the posterior development of animals (Lan et al., 2007), and embryonic stem cell self-renewal (Loh et al., 2007).
In Arabidopsis, there are 21 JmjC domain-containing proteins (Lu et al., 2008; Hong et al., 2009), although few have been characterized. EARLY FLOWERING6 (ELF6) and RELATIVE OF ELF6 (REF6) proteins, which contain both JmjC and zinc-finger domains, regulate flowering time in Arabidopsis (Noh et al., 2004). ELF6 represses photoperiodic flowering, whereas REF6 represses FLOWERING LOCUS C (FLC) expression. In addition, they have been shown to modulate gene expression regulated by brassinosteroids by interacting with BRASSINOSTEROID-INSENSITIVE1-EMS-SUPPRESSOR1, a transcription factor that binds to promoters of genes that respond to brassinosteroids (Yu et al., 2008). Two other JmjC proteins, MATERNAL EFFECT EMBRYO ARREST27 and INCREASED EXPRESSION OF BONSAI METHYLATION1 are involved in gametophyte development and repression of cytosine methylation, respectively (Pagnussat et al., 2005). JMJ14, which contains a JmjN and zinc-finger domain in addition to the JmjC domain, prevents early flowering by repressing the expression of FLOWERING LOCUS T (FT), and its homolog TWIN SISTER OF FT, APETALA1, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), and LEAFY (Lu et al., 2010; Yang et al., 2010). More recently, JMJ14 has been shown to be involved in RNA silencing and the maintenance phase of DOMAINS REARRANGED METHYLTRANSFERASE2-mediated RNA-directed DNA methylation (Searle et al., 2010).
In the work presented here, we have identified JMJ30 as, to our knowledge, the first JmjC domain-containing protein involved in circadian function. Since little is known about histone methylation and circadian clock function, our work paves the way to the exploration of a specific pathway linking histone methylation and circadian regulation.
RESULTS
Expression of JMJ30 Oscillates with a Circadian Rhythm
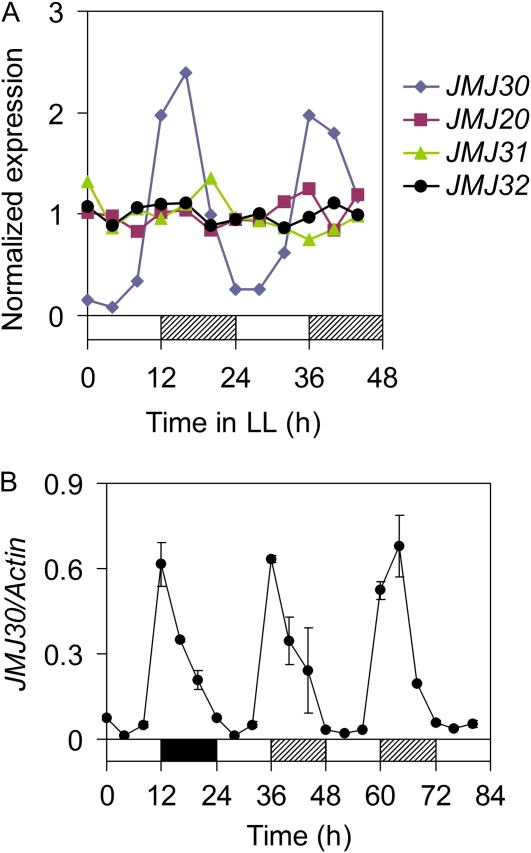
Based on the sequence similarity between JmjC domains, the 21 JmjC domain-containing proteins can be classified into five groups (Hong et al., 2009). JMJ30, encoded by At3g20810, belongs to the JmjC domain-only group that contains four members (Supplemental Fig. S1). JMJ30 is ubiquitously expressed in different tissues (Lu et al., 2008) and has the conserved Fe(II)- and α-ketoglutarate-binding amino acids required for histone demethylation, indicating that JMJ30 might be a functional histone demethylase. In addition, YFP-JMJ30 primarily localizes to the nucleus with a low GFP signal detected in the cytoplasm (Supplemental Fig. S2). Its inclusion in the nucleus further supports our hypothesis that JMJ30 is a histone demethylase in vivo. From the publicly available microarray data, we identified JMJ30 as the only protein of the 21 members that shows a robust circadian rhythm of expression (Mockler et al., 2007; Michael et al., 2008; Fig. 1A). The peak of rhythmic JMJ30 transcript expression occurs around dusk (Zeitgeber time [ZT]-12) under both diurnal and constant white light (LL) conditions (Fig. 1B).
Figure 1.
The JMJ30 transcript level exhibits circadian regulation. A, RNA levels of JmjC domain only containing genes in continuous light (LL) conditions. Data from the DIURNAL Project (http://diurnal.cgrb.oregonstate.edu/) microarray repository were used to compare RNA levels of JMJ30 (At3g20810), JMJ20 (At5g63080), JMJ31 (At5g19840), and JMJ32 (At3g45880). Raw data from experimental set LL12(LDHH) were normalized to the mean of all points to compare relative expression levels among these family members. B, JMJ30 expression oscillates in wild-type Arabidopsis under diurnal and LL conditions. Ten-day-old seedlings were analyzed and qRT-PCR data are presented as the mean of two biological replicates ± sd. Day, night, and subjective night are denoted by white, black, and hatched bars, respectively.
Loss of JMJ30 Function Affects the Free-Running Circadian Period
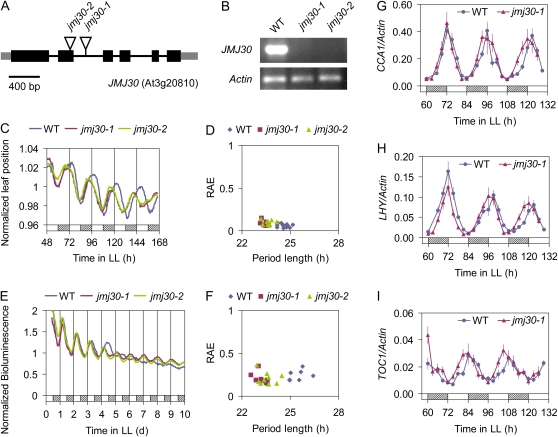
To elucidate the biological roles of JMJ30 in Arabidopsis, we obtained two mutants, jmj30-1 and jmj30-2, in which T-DNA was inserted into the third intron (jmj30-1) or the second exon (jmj30-2) of JMJ30, respectively (Fig. 2A). JMJ30 full-length RNA was not detected in either of these two mutants by real-time (RT)-PCR (Fig. 2B), indicating that they are loss-of-function mutants for JMJ30. Neither of the mutants differed from wild-type Columbia (Col) plants in hypocotyl elongation (data not shown) and flowering time (Supplemental Fig. S3), two physiological responses that can be affected by the circadian clock. To determine directly whether loss of JMJ30 affects the circadian clock, we first examined leaf movement, a well-established circadian response in Arabidopsis (Hicks et al., 1996). Leaf movement rhythms of jmj30 mutant plants had a short period in LL (Fig. 2C; wild-type plants had a period of 24.6 ± 0.3 h, jmj30-1 of 23.4 ± 0.2 h, and jmj30-2 of 23.6 ± 0.3 h). To assess the robustness of the circadian rhythms in individual seedlings, relative amplitude error (RAE) was measured. RAE values can range between 0 (perfect fitted rhythm) and 1 (rhythm not significant). Both of the mutants had RAE values of approximately 0.1, similar to those of the wild type (Fig. 2, C and D).
Figure 2.
The jmj30 mutation shortens the circadian period under LL conditions. A, JMJ30 genomic structure. Exons are represented by black boxes, untranslated regions are represented by gray boxes, and introns are represented by lines. Triangles indicate T-DNA insertions. B, RT-PCR analysis of JMJ30 full-length transcript in wild-type and jmj30 mutants. Ten-day-old seedlings were sampled at ZT-12, and Actin transcript was used as a control. C and D, Assay of circadian leaf movement under LL conditions. C, Normalized positions of primary leaves for wild type (n = 12), jmj30-1 (n = 10), and jmj30-2 (n = 7) are shown. D, Period length and RAE were estimated using fast Fourier transform-nonlinear least-squares analysis. E and F, Assay of CAB2::LUC activity under LL conditions. E, Mean bioluminescence traces of groups of approximately 20 seedlings for wild type (n = 7), jmj30-1 (n = 8), and jmj30-2 (n = 9) are shown. F, Period length and RAE estimates of the CAB2::LUC bioluminescence rhythms shown in E. G to I, qRT-PCR analysis of CCA1 (G), LHY (H), and TOC1 (I) expression in wild-type and jmj30-1 plants under LL conditions. Ten-day-old seedlings that were entrained in a 12L/12D cycle, transferred to LL, and harvested for 3 d at 3-h intervals were analyzed, and the mean of two biological replicates ± sd is shown. Day, night, and subjective night are denoted by white, black, and hatched bars, respectively. All experiments were done at least twice with similar results.
Luciferase (LUC) activity under the control of a circadian promoter is a noninvasive method for monitoring clock function and integrity (Millar et al., 1995). The effect of the JMJ30 mutations on the circadian clock was also analyzed using the circadian reporter CHLOROPHYLL A/B BINDING PROTEIN2::LUC (CAB2::LUC; Millar et al., 1995; Knowles et al., 2008). Consistent with the period shortening observed in leaf movement rhythms, CAB2::LUC oscillations in the jmj30 mutants had shorter periods than the oscillations in wild-type plants (Fig. 2E; wild-type plants had a period of 25.2 ± 0.7 h, jmj30-1 of 23.7 ± 0.5 h, and jmj30-2 of 23.8 ± 0.6 h). The CAB2::LUC rhythms were as robust as the leaf movement rhythms (Fig. 2, E and F).
While jmj30 affects the period of various output rhythms (leaf movement and CAB2::LUC activity), we also tested whether jmj30 affects the expression of central oscillator genes. As shown in Figure 2, G to I, jmj30 mutation shortened the expression period of CCA1, LHY, and TOC1, which function as the central oscillator components (Alabadí et al., 2001). Although jmj30 caused period shortening of central oscillator genes, it has little effect on the amplitude of expression (Fig. 2, G–I). This phenotype is consistent with the finding that the jmj30 mutation affects period length but does not alter the amplitude and robustness of the circadian output rhythms, such as leaf movement rhythms and CAB2::LUC rhythms during free-running conditions (Fig. 2, C–F). Therefore JMJ30 is involved in regulating period length, rather than amplitude and robustness in the Arabidopsis circadian clock.
Constitutive Expression of JMJ30 Affects Flowering Time
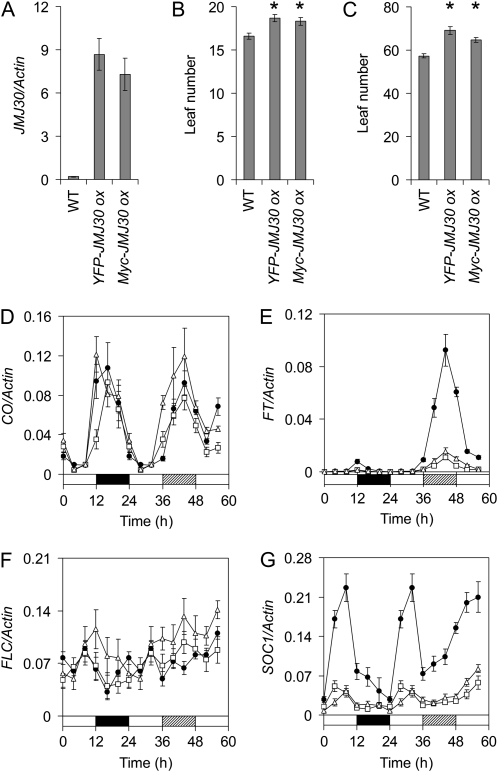
To further investigate the role of JMJ30 in the circadian system, we generated transgenic plants overexpressing the JMJ30 gene. Two homozygous lines with different epitope tags (YFP-JMJ30 ox and Myc-JMJ30 ox) were obtained. Quantitative RT-PCR (qRT-PCR) showed that the expression level of JMJ30 in overexpressing plants is 30 to 50 times higher than that in wild-type plants (Fig. 3A). We further demonstrated that both JMJ30 RNA and protein are constitutively expressed in the overexpressing line under diurnal conditions (Supplemental Fig. S4). In contrast to jmj30, the two lines overexpressing JMJ30 showed delayed flowering (Fig. 3, B and C) in both long-day (LD) and short-day (SD) conditions, although the hypocotyl lengths of JMJ30 ox were similar to that of wild type (data not shown). We therefore examined the expression of CONSTANTS (CO) and FT, two of the key genes in the photoperiodic flowering pathway. Although the rhythmic expression of CO was almost unaffected in JMJ30 ox compared with that of the wild type, expression of FT was greatly reduced in JMJ30 ox plants (Fig. 3, D and E). It is known that FLC directly binds to FT and SOC1 chromatin to repress their expression (Helliwell et al., 2006). We then examined FLC and SOC1 expression, and found that the transcript levels of FLC remained unchanged but the SOC1 transcript levels were strongly reduced in JMJ30 ox relative to wild type (Fig. 3, F and G). This suggests that the delayed-flowering phenotype resulted from down-regulation of FT and SOC1 in the JMJ30 ox plants.
Figure 3.
Constitutive expression of JMJ30 affects flowering time. A, qRT-PCR analysis of JMJ30 expression in wild-type and JMJ30 ox plants. Ten-day-old seedlings were sampled at ZT-12. B and C, Flowering time of wild-type and JMJ30 ox plants under LD (16L/8D; B) and SD (8L/16D; C) conditions. Flowering time was expressed as mean rosette leaf number ± SEM (n = 20–25). Asterisk indicates a significant difference by Student’s two-tail t test (P < 0.01). D to G, qRT-PCR analysis of CO (D), FT (E), FLC (F), and SOC1 (G) expression in wild-type and JMJ30 ox plants under diurnal and LL conditions. Day, night, and subjective night are denoted by white, black, and hatched bars, respectively. All qRT-PCR data are presented as the mean of two biological replicates ± sd. All experiments were done at least twice with similar results.
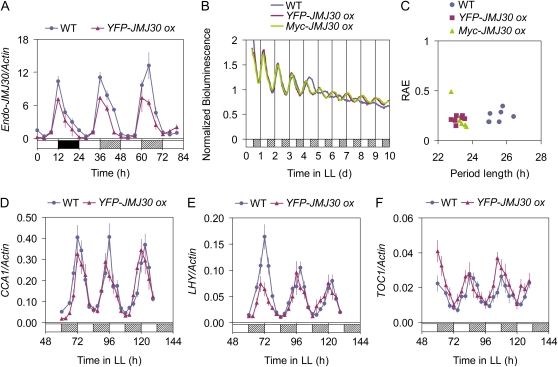
Constitutive Expression of JMJ30 Negatively Regulates Its Own Expression and Affects the Free-Running Circadian Period
Clock components are often controlled by negative feedback regulation. We tested whether JMJ30 regulates its own expression, as other circadian genes do (Schaffer et al., 1998; Wang and Tobin, 1998). Although the endogenous JMJ30 transcript still showed robust oscillations in JMJ30 ox plants, there was a significant reduction in amplitude compared with that in wild-type plants (Fig. 4A). This finding suggests that JMJ30 negatively regulates its own expression. To examine the effect of constitutive expression of JMJ30 on the circadian clock, a CAB2::LUC reporter was introduced into the JMJ30 ox plants. Robust circadian rhythms in CAB2::LUC activity with a short period were observed in JMJ30 ox plants (Fig. 4, B and C; wild-type plants had a period of 25.2 ± 0.7 h, YFP-JMJ30 ox of 23.8 ± 0.5 h, and Myc-JMJ30 ox of 23.9 ± 0.6 h). A similar short-period phenotype was also observed in the expression of central oscillator genes (Fig. 4, D–F). Observation of both mutants and overexpressing plants with short-period phenotypes was also made in characterization of the clock component GI (Mizoguchi et al., 2005). However, the mechanism of this phenomenon is not known. Together, this data shows that JMJ30 is a circadian clock component involved in controlling the circadian period.
Figure 4.
Constitutive expression of JMJ30 affects the circadian period under LL conditions. A, qRT-PCR analysis of endogenous JMJ30 expression in wild-type and JMJ30 ox plants under diurnal and LL conditions. B and C, Assay of CAB2::LUC activity under LL conditions. B, Mean bioluminescence traces of groups of approximately 20 seedlings for wild type (n = 7), YFP-JMJ30 ox (n = 7), and Myc-JMJ30 ox (n = 5) are shown. C, Period length and RAE estimates of the CAB2::LUC bioluminescence rhythms shown in B. D to F, qRT-PCR analysis of CCA1 (D), LHY (E), and TOC1 (F) expression in wild-type and YFP-JMJ30 ox plants under LL conditions. Ten-day-old seedlings that were entrained in a 12L/12D cycle, transferred to LL, and harvested for 3 d at 3-h intervals were analyzed. Day, night, and subjective night are denoted by white, black, and hatched bars, respectively. All qRT-PCR data are presented as the mean of two biological replicates ± sd. All experiments were done at least twice with similar results.
CCA1 and LHY Negatively Regulate JMJ30 Expression
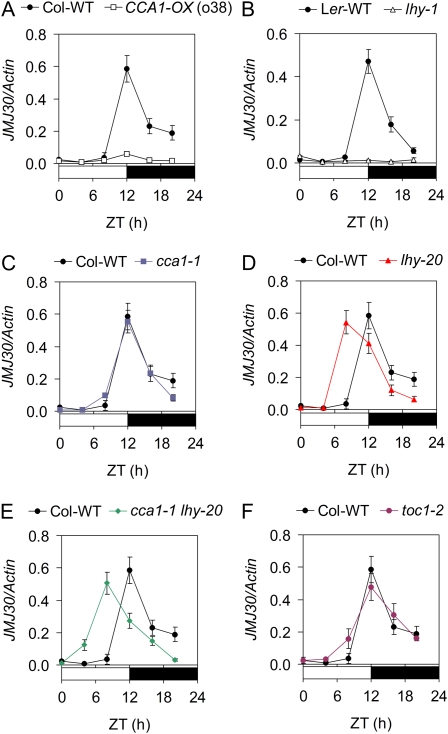
JMJ30 was originally identified among genes represented on a microarray whose expression was repressed by CCA1 and LHY (data not shown), two key components of the central oscillator of plants. To investigate the effect of the transcription factors CCA1 and LHY on the expression of JMJ30, we first measured the expression of JMJ30 in CCA1ox-38 (Wang and Tobin, 1998) and lhy-1 (Schaffer et al., 1998), which overexpress CCA1 and LHY under the strong cauliflower mosaic virus 35S promoter, respectively. The expression of JMJ30 was dramatically reduced in both the CCA1 and LHY overexpressing lines (Fig. 5, A and B), confirming that CCA1 and LHY act negatively on JMJ30 expression. To further evaluate the role of CCA1 and LHY in regulating the expression of JMJ30, we examined the expression of JMJ30 in the cca1-1 (Green and Tobin, 1999), lhy-20 single mutant (Michael et al., 2003), and cca1-1/lhy-20 double mutant (Yakir et al., 2009). The expression profile of JMJ30 in the cca1-1 mutant is indistinguishable from that in the wild-type plants (Fig. 5C). Since the functions of CCA1 and LHY are partially redundant (Mizoguchi et al., 2002), this result suggests that LHY can probably compensate the loss of CCA1 on the regulation of JMJ30. In contrast to cca1-1, the peak of JMJ30 expression in the lhy-20 occurs 4 h earlier than in the wild-type plants (Fig. 5D). This phase shift is not due to the short-period phenotype of the lhy-20 because short-period mutant cca1-1 and toc1-2, another short-period mutant (Strayer et al., 2000), did not display the same effect (Fig. 5F). These results indicate that CCA1 and LHY are not redundant in the regulation of JMJ30 expression and that LHY has a greater effect than CCA1. In the cca1-1/lhy-20 double mutant, JMJ30 levels rise early (comparing the level at ZT-4 in different mutants) and reach a peak at ZT-8 (Fig. 5E), indicating that CCA1 is required to repress JMJ30 expression in the early morning in the absence of LHY. Taken together, the results indicate that CCA1 and LHY have a negative effect on JMJ30 expression, although LHY has a greater effect than CCA1.
Figure 5.
CCA1 and LHY regulate JMJ30 expression. A to F, qRT-PCR analysis of JMJ30 expression in wild type (Col-WT, Landsberg erecta-WT) and CCA1-ox (line o38), LHY-ox (lhy-1), cca1-1, lhy-20, cca1-1 lhy-20, and toc1-2 plants under diurnal conditions. Ten-day-old seedlings were analyzed and the mean of two biological replicates ± sd is shown. Day and night are denoted by white and black bars, respectively. All experiments were done at least twice with similar results.
JMJ30 Is a Direct Target of CCA1 and LHY in Vivo
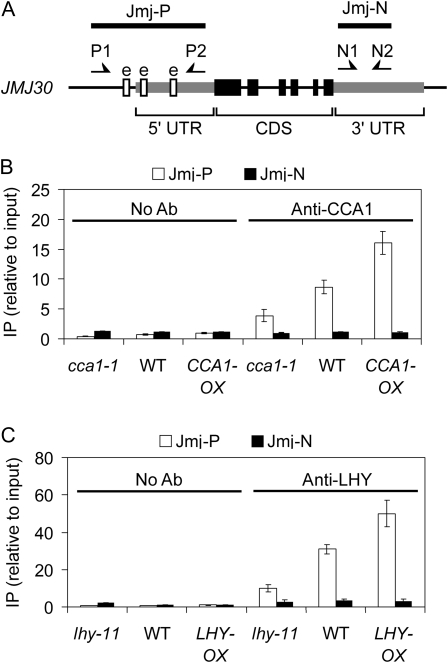
JMJ30 has three EEs in the region −169/−61 upstream of the translational start codon (Fig. 6A). This element has been found in several circadian-regulated genes with evening expression and is known to be bound by CCA1 and LHY (Alabadí et al., 2001; Perales and Más, 2007). To examine whether CCA1 and LHY bind to the EE-containing region of the JMJ30 in vivo, chromatin immunoprecipitation (ChIP) was performed using either an anti-CCA1 antibody or anti-LHY antibody. Figure 6B demonstrated that an anti-CCA1 antibody efficiently immunoprecipitated a fragment of JMJ30 that contains the EEs but not another one located in the 3′ untranslated region. Similar results were observed with the anti-LHY antibody (Fig. 6C), indicating that JMJ30 is a direct target of CCA1 and LHY in vivo.
Figure 6.
JMJ30 is a direct target of CCA1 and LHY. A, Schematic drawing of genomic JMJ30 and the regions examined by ChIP. Exons are represented by black boxes, untranslated regions are represented by gray boxes, and introns are represented by lines. EE elements (e) are represented by white boxes. B and C, Binding of CCA1 and LHY to the JMJ30 promoter in vivo. B, ChIP assays were performed with wild-type (Col), cca1-1, and CCA1-ox (line o38) seedlings collected at ZT-2. C, ChIP assays were performed with wild-type (Landsberg erecta), lhy-11, and LHY-ox (lhy-1) seedlings collected at ZT-2. qRT-PCR was performed on the precipitates by two primer sets. As indicated in A, Jmj-P amplifies the region that contains three EEs and Jmj-N amplifies a region in the 3′ untranslated region. Results were normalized to the input DNA and Actin control. The mean of two biological replicates ± sd is shown. All of these experiments were done at least twice with similar results.
Effects of Pulses of CCA1 and LHY on the Expression of JMJ30
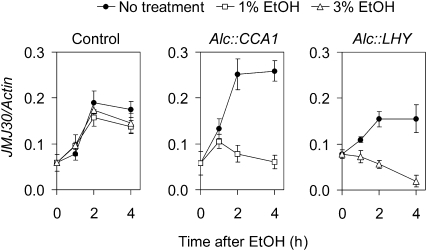
To examine the effect of CCA1 and LHY binding to the JMJ30 promoter, we utilized the ethanol-inducible system (Knowles et al., 2008). The ethanol-inducible system provides a relatively simple way to test the effect of a pulse of virtually any gene on a biological pathway. It has been shown that a pulse of CCA1 or LHY in Alc::CCA1 or Alc::LHY plants can reset the clock and repress the expression of evening-phased genes such as TOC1, LUX, and GI (Knowles et al., 2008). To examine the effect of a pulse of CCA1 or LHY on the expression of JMJ30, ethanol treatment was performed in Alc::CCA1 or Alc::LHY plants (Knowles et al., 2008). A pulse of either CCA1 or LHY at circadian time 8 when the JMJ30 RNA level is increasing resulted in a complete inhibition of the increase within 2 h of the ethanol treatment (Fig. 7). The rapidity of the response further supports the notion that CCA1 and LHY bind directly to the JMJ30 promoter to repress its expression.
Figure 7.
A pulse of CCA1 or LHY expression represses the accumulation of JMJ30 RNA. Control, Alc::CCA1, and Alc::LHY seedlings were given an ethanol pulse at 32 h in LL (circadian time 8). qRT-PCR analysis of JMJ30 expression at the time of induction (0 h), 1, 2, and 4 h after ethanol treatment are represented. The mean of two biological replicates ± sd is shown. All of these experiments were done at least twice with similar results.
DISCUSSION
In this study, we report the identification of a JmjC domain-containing protein JMJ30 involved in the Arabidopsis circadian clock. JMJ30 oscillations peak at dusk (ZT-12) and persist under both diurnal and continuous light conditions (Fig. 1), a hallmark of genes that exhibit circadian expression (McClung, 2006). The levels of endogenous JMJ30 mRNA are reduced in plants overexpressing JMJ30 (Fig. 4A), suggesting that its accumulation is autoregulated at the level of transcription, another hallmark of clock genes involved in the negative feedback loop. Two T-DNA mutants, jmj30-1 and jmj30-2, exhibited shortened periods in rhythms of leaf movement, CAB2::LUC activity, and expression of central oscillator genes (Fig. 2, C–I), indicating that JMJ30 is important for maintaining correct free-running periods of the circadian clock. Interestingly, two overexpressing lines, YFP-JMJ30 ox and Myc-JMJ30 ox, also showed shortened periods in rhythms of CAB2::LUC activity and expression of central clock genes (Fig. 4, B–F). Apparently, this is not due to the cosuppression of JMJ30 in the overexpressing lines because JMJ30 transcript levels in overexpressing plants are much higher than that in wild-type plants (Fig. 3A; Supplemental Fig. S4). Although it is unlikely that the YFP and Myc tags fused to the N-terminal of JMJ30 interrupt the function of the JmjC domain that resides at the C terminus, we cannot rule out the possibility that the addition of the tags cause a dominant negative effect and repress the endogenous JmjC activity. The expression of JMJ30 was dramatically reduced in both the CCA1 and LHY overexpressing lines (Fig. 5, A and B), indicating that CCA1 and LHY negatively regulate JMJ30 expression. ChIP assays conclusively demonstrate that CCA1 and LHY bind to the JMJ30 promoter in vivo (Fig. 6) and the use of the ethanol-inducible system has allowed us to further demonstrate the repression of JMJ30 expression by CCA1 and LHY in real time (Fig. 7).
JMJ30 contains a JmjC domain, which has been implicated in histone demethylation (Lu et al., 2008). To carry out histone demethylation, JmjC domain-containing proteins require the cofactors Fe(II) and α-ketoglutarate. Since JMJ30 contains the two conserved His and one Asp residue required for Fe(II) binding, and a conserved Thr and Lys required for α-ketoglutarate binding, it has the necessary amino acid residues to carry out a histone demethylation reaction (Supplemental Fig. S1). In addition, YFP-JMJ30 localizes primarily to the nucleus (Supplemental Fig. S2), suggesting it may be a functional histone demethylase. An in vitro histone demethylation assay will be a key experiment to determine whether JMJ30 has histone demethylase activity.
The jmj30 mutants and overexpressing plants differ in their flowering phenotypes. Jmj30 mutants do not exhibit any flowering phenotype in either LD or SD conditions (Supplemental Fig. S3), whereas in JMJ30 ox plants, flowering is delayed in LD and SD (Fig. 3, B and C). Since there are 21 JmjC domain-containing proteins in Arabidopsis, it is possible that multiple JmjC domain-containing proteins function redundantly in the control of flowering time. Therefore the loss of JMJ30 may not have a measurable effect on flowering time. In plants constitutively expressing JMJ30, FT and SOC1 are expressed at much lower levels while the expression of CO and FLC are unaffected (Fig. 3, D–G), suggesting that the delayed-flowering phenotype in JMJ30 ox may not be through the photoperiodic flowering pathway. FT expression is enhanced by increased H3K4me3 methylation and repressed by H3K27me3 methylation (Jiang et al., 2008). A recent study showed that JMJ14 is a histone H3 Lys 4 demethylase that represses FT expression to inhibit the floral transition in Arabidopsis (Yang et al., 2010). It is possible that JMJ30 functions as a histone demethylase to repress FT and SOC1 expression in a similar manner to JMJ14.
Circadian clock regulation of the transcriptome is a widespread phenomenon. More than 10% of mammalian transcripts (Duffield et al., 2002) and about 25% of Arabidopsis transcripts oscillate in a circadian manner (Michael et al., 2008). Recent advances in the field have revealed an unexpected link between circadian-regulated gene expression and dynamic changes in chromatin. In Neurospora, rhythmic histone acetylation at the promoter of the central clock gene FREQUENCY was observed (Belden et al., 2007). The CLOCK protein, an essential component of the mammalian circadian system, has been shown to be a histone acetyltransferase (Doi et al., 2006). In Arabidopsis, histone acetylation (H3K9Ac) and dimethylation (H3K4Me2) at the CCA1, LHY, TOC1, and GI promoters positively correlates with their expression (Ni et al., 2009). We have examined the expression of known clock genes in both JMJ30 mutants and overexpressing plants; no significant difference in their expression profile was observed (Supplemental Fig. S5; data not shown). The identification of its target genes will allow elucidation of the function of JMJ30 within the circadian clock and is likely to reveal the importance of histone methylation in circadian clock function.
Taken together, our study shows that Arabidopsis JMJ30 is a novel circadian clock component involved in controlling the circadian period. In addition, CCA1 and LHY directly bind to the JMJ30 promoter to repress its expression, indicating that JMJ30 regulates the pace of the circadian clock in a close association with the central oscillator. JMJ30, a putative histone demethylase, represents, to our knowledge, the first JmjC domain-containing protein involved in circadian function.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana; Col ecotype) was used for all experiments described unless stated otherwise. jmj30-1 (SAIL_811_H12) and jmj30-2 (GK_454C10) are two T-DNA insertion alleles of JMJ30. For JMJ30 overexpression, the full-length coding sequence of JMJ30, including the stop codon, was amplified by specific primers (Supplemental Table S1) and cloned into the pEarleyGate 104 (35S-YFP-JMJ30) and pEarleyGate 203 (35S-Myc-JMJ30) vectors (Earley et al., 2006) using the GATEWAY recombination system (Invitrogen). Seedlings were grown under a 12-h fluorescent light (30 μmol m−2 s−1):12-h dark (12L:12D) photoperiod at a constant temperature of 22°C, unless otherwise stated.
Analysis of Circadian Rhythms
Arabidopsis plants homozygous for jmj30-1, jmj30-2, YFP-JMJ30 ox, Myc-JMJ30 ox, and wild type (Col) were transformed with the CAB2::LUC reporter (Knowles et al., 2008). T2 seedlings from three to six independent transformed lines were entrained for 6 d with 12L/12D cycles before being transferred to LL conditions. Bioluminescence rhythms of groups of approximately 20 seedlings were analyzed as previously described (Knowles et al., 2008). For leaf movement analysis, seedlings were entrained for 10 d under a 12L/12D cycle and then transferred to continuous white light. Seedlings were individually transferred to the wells of upright 24-well tissue culture plates and the positions of the primary leaves were recorded every 20 min for 7 d using a CCD camera (model: LTC 0335) from Bosch. Leaf movement was assessed by measuring the vertical position of the primary leaves using the publicly available software ImageJ. Oscillation properties were analyzed with the BRASS (available from http://www.amillar.org) using the fast Fourier transform-nonlinear least-squares analysis program (Plautz et al., 1997).
Measurement of Flowering Time
Arabidopsis plants were grown on soil under either LD (16-h light/8-h dark) or SD (8-h light/16-h dark) conditions. Flowering time was scored by counting the number of rosette leaves at flowering. Data are presented as mean ± SEM (n = 10–25).
RNA Extraction and qRT-PCR
For all experiments, 1- to 2-week-old seedlings grown on plates were used. For circadian experiments, samples were collected every 3 to 4 h either during the light/dark cycle or in continuous white light. RNA extraction and qRT-PCR were carried out as previously described (Knowles et al., 2008). Actin2 was used as a noncycling reference, and expression levels were normalized to the level of the control. All reactions were performed in triplicate.
ChIP
ChIP was performed as previously described (Ni et al., 2009) using affinity-purified anti-CCA1 antibody (Wang and Tobin, 1998) or affinity-purified anti-LHY antibody (Lu et al., 2009) for immunoprecipitation.
Ethanol Pulse
Ethanol treatment was performed as previously described (Knowles et al., 2008). Control plants contained the regulator construct 35S::AlcR:TNOS. Two-week-old seedlings were treated with 10 min of ethanol vapor of 1% or 3% (v/v) ethanol. Samples were collected at the time of induction (0 h), 1, 2, and 4 h after ethanol treatment.
Tobacco Infiltration and Confocal Microscopy
For transient expression in tobacco (Nicotiana benthamiana), leaves were transfected with 35S::YFP-JMJ30 as described previously (Lu et al., 2009) and analyzed 2 d after infiltration. For stable transgenic Arabidopsis, the leaves of 8-d-old transgenic 35S::YFP-JMJ30 seedlings grown on plates were analyzed. All imaging was done using a Carl Zeiss 510 Meta laser-scanning confocal microscope with a Plan-Apochromat 63×/1.4 oil differential interference contrast objective. YFP was excited with an argon laser at 25% to 50% of its output that was attenuated to 7% to 9% at 514 nm. The emission was sent through a 530 to 600 nm band-pass filter for detection of YFP.
Plant Protein Extracts and Immunoblot Analysis
Proteins were extracted in 1× extraction buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA, 10% glycerol, 1% Triton X-100, 2 mm phenylmethylsulphonyl fluoride, 50 μm MG115, 50 μm MG132, and protease inhibitor cocktail [Roche]). Immunoblotting was performed as described (Lu et al., 2009) with the appropriate primary antibody (anti-GFP [Santa Cruz biotechnology, sc-8334], anti-Actin [MP Biomedicals, Clone C4]).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative data library using the following accession numbers: JMJ30 (At3g20810), JMJ20 (At5g63080), JMJ31 (At5g19840), JMJ32 (At3g45880), CAB2 (At1g29920), FT (At1g65480), CO (At5g15840), FLC (At5g10140), SOC1 (At2g45660), ACT2 (At3g18780), ACT7 (At5g09810), CCA1 (At2g21660), LHY (At1g01060), TOC1 (At5g61380), and PRR3 (At5g60100).
Supplemental Data
Supplemental Figure S1. Alignment of jmjC domains among jmjC domain only containing proteins.
Supplemental Figure S2. YFP-JMJ30 is localized in the nucleus and cytosol.
Supplemental Figure S3. jmj30 mutants have no significant flowering phenotype.
Supplemental Figure S4. Both JMJ30 RNA and protein are constitutively expressed in the overexpression lines.
Supplemental Figure S5. Loss of JMJ30 does not affect the expression of several clock genes.
Supplemental Table S1. List of PCR primer sequences.
Note Added in Proof
Another report of the involvement of the JMJ30 gene (At3g20810) in circadian rhythms in plants (and in animals) was recently published, with the gene designation of JMJD5 (Jones MA, Covington MF, DiTacchio L, Vollmers C, Panda S, Harmer SL [2010] Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci USA 107: 21623–21628).
Supplementary Material
Acknowledgments
We acknowledge the assistance of the Arabidopsis Biological Resource Center, which provided jmj30-1. We thank Angelique Deleris and Steve Jacobsen for the jmj30-2 seeds, Steve Kay for toc1-2 seeds, and Rachael Green for the lhy-20 and cca1-1/lhy-20 seeds.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Belden WJ, Loros JJ, Dunlap JC. (2007) Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell 25: 587–600 [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. (2006) Circadian regulator CLOCK is a histone acetyltransferase. Cell 125: 497–508 [DOI] [PubMed] [Google Scholar]
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. (2002) Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol 12: 551–557 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Green RM, Tobin EM. (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96: 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA. (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Hong EH, Jeong YM, Ryu JY, Amasino RM, Noh B, Noh YS. (2009) Temporal and spatial expression patterns of nine Arabidopsis genes encoding Jumonji C-domain proteins. Mol Cells 27: 481–490 [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. (2007) The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG. (2007) SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450: 415–419 [DOI] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y. (2008) Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles SM, Lu SX, Tobin EM. (2008) Testing time: can ethanol-induced pulses of proposed oscillator components phase shift rhythms in Arabidopsis? J Biol Rhythms 23: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognár L, Gould PD, Fehér B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ. (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. (2007) Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev 21: 2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Liu C, Cao X. (2010) JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res 20: 387–390 [DOI] [PubMed] [Google Scholar]
- Lu F, Li G, Cui X, Liu C, Wang XJ, Cao X. (2008) Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 50: 886–896 [DOI] [PubMed] [Google Scholar]
- Lu SX, Knowles SM, Andronis C, Ong MS, Tobin EM. (2009) CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol 150: 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. (2006) Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR. (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA. (1995) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G. (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, et al. (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17: 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J. (2007) The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol 72: 353–363 [DOI] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS. (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Perales M, Más P. (2007) A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19: 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA. (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Searle IR, Pontes O, Melnyk CW, Smith LM, Baulcombe DC. (2010) JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 24: 986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. (1995) Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev 9: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Yakir E, Hilman D, Kron I, Hassidim M, Melamed-Book N, Green RM. (2009) Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol 150: 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Jiang D, Jiang J, He Y. (2010) A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J 62: 663–673 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Li L, Guo M, Chory J, Yin Y. (2008) Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA 105: 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.