Abstract
Cellulose from plant biomass is the largest renewable energy resource of carbon fixed from the atmosphere, which can be converted into fermentable sugars for production into ethanol. However, the cellulose present as lignocellulosic biomass is embedded in a hemicellulose and lignin matrix from which it needs to be extracted for efficient processing. Here, we show that expression of an Arabidopsis (Arabidopsis thaliana) transcription factor, SHINE (SHN), in rice (Oryza sativa), a model for the grasses, causes a 34% increase in cellulose and a 45% reduction in lignin content. The rice AtSHN lines also exhibit an altered lignin composition correlated with improved digestibility, with no compromise in plant strength and performance. Using a detailed systems-level analysis of global gene expression in rice, we reveal the SHN regulatory network coordinating down-regulation of lignin biosynthesis and up-regulation of cellulose and other cell wall biosynthesis pathway genes. The results thus support the development of nonfood crops and crop wastes with increased cellulose and low lignin with good agronomic performance that could improve the economic viability of lignocellulosic crop utilization for biofuels.
Crop residues are a vast resource of lignocellulose feedstock available for conversion to biofuels, and their utilization does not compete with food supplies, unlike grain-based feedstocks (Haigler et al., 2001). Rice (Oryza sativa) straw itself constitutes half the crop waste worldwide, which is either burnt or wasted (Sticklen, 2006). Nonfood perennial grasses such as switchgrass (Panicum virgatum) and miscanthus (Miscanthus giganteus) as well as fast-growing woody crops make up the bulk of lignocellulosic resources. In either case, plant lignocellulosic cell walls are quite resistant to digestion of the complex polysaccharides (cellulose) into simple sugars before fermentation due to the presence of heavily cross-linked lignin. Therefore, methods to lower lignin and improve the availability and levels of cellulose are important to make the conversion into biofuels economically feasible.
Cellulose is the most abundant biopolymer on earth, comprising 25% to 50% of plant biomass with an estimated 100 billion tons synthesized annually as a result of photosynthesis (Haigler et al., 2001; Sticklen, 2006). Cellulose is made up of Glc units and is synthesized at the plasma membrane by the cellulose synthase (CESA) complex, comprising multiple CESA proteins that belong to multigene families in plants (Somerville, 2006). Long-chain cellulose polymers are organized into microfibrils that make up the core content of plant cell walls, contributing to the strength, structure, and development of plants (Sticklen, 2006). Hemicelluloses are polysaccharides in plant cell walls that include xyloglucans, xylans, mannans and glucomannans, and β-(1→3,1→4)-glucans and are synthesized by glycosyltransferases located in the Golgi membranes. The most important biological role of hemicelluloses is their contribution to strengthening the cell wall by interaction with cellulose and, in some cell walls, with lignin (Scheller and Ulvskov, 2010). Despite its importance, the details regarding the synthesis of hemicelluloses remain very elusive, and very little is known about the regulation of the cellulose biosynthesis pathway.
Lignin, the second most abundant polymer, is a complex composed of guaiacyl (G), syringyl (S), and p-hydroxylphenyl (H) phenylpropanoid units (Supplemental Fig. S1), contributing to lignin heterogeneity (Boerjan et al., 2003). Angiosperm dicot lignin is primarily composed of G and S units, and monocot lignin is a mixture of G, S, and H units (Supplemental Fig. S1). Among these, the G lignins (found characteristically in abundance in softwoods of gymnosperms like pines) are more resistant to chemical degradation, making the composition of lignin (the relative ratio of G to S units), along with its quantity, crucial for the digestibility of crops for conversion into biofuels and cellulosic products. The monolignol biosynthetic genes, therefore, have been used in engineering lignin content and composition in several plants (Vanholme et al., 2008). Many of these studies were first reported in nonfeedstock model dicot plants such as tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana; Zhou et al., 2009), and the expectation is that similar approaches can be applied to cellulosic feedstock crops, but very few detailed engineering studies have been reported in the grasses, which are a major lignocellulosic resource.
In the grasses, the maize (Zea mays) and sorghum (Sorghum bicolor) brown midrib mutations (Li et al., 2008) show alterations in lignin content and digestibility; the maize brittle stalk2 (bk2) and rice brittle culm1 (bc1) mutations of a similar gene have a brittle phenotype due to reduction in cellulose and cell wall composition with no compensatory changes in lignin (Li et al., 2003b); and the rice flexible culm1 mutant has reduced lignin, H, and G residues (Li et al., 2009). However, significant reductions in lignin or digestibility in the monocot crops, including the brown midrib and other mutants, are also accompanied by reductions in plant growth, biomass, stalk strength, or pathogen resistance (Li et al., 2008).
Several transcription factors (TFs) have also been shown to affect cellulose and lignin content and composition (Mele et al., 2003; Kubo et al., 2005; Zhong et al., 2006; Zhong and Ye, 2009). Transcriptional regulation is achieved by top-level NAC TFs SECONDARY WALL-ASSOCIATED NAC DOMAIN1 (SND1), NAC SECONDARY WALL THICKENING PROMOTING1/2 (NST1/2), and VASCULAR-RELATED NAC-DOMAIN6/7 (VND6/7) that activate a nexus of intermediate TFs, mostly MYBs. These intermediate TFs in turn activate low-level MYB TFs that bind to and activate target cell wall biosynthetic genes, presumably achieving high levels of specificity. Such a multilayered regulatory network that affects multiple target genes offers the cell a robust mechanism to achieve coherent changes in the flux through the pathways. Additionally, the network also points to the possibility of the existence of multiple knobs and switches that can be tuned to execute specific regulation of different cell wall pathways in order to optimize secondary cell wall composition. Specifically, cell walls with increased cellulose content in combination with reduced lignin for enhanced sugar and ethanol would yield valuable cellulosic feedstock (Jakob et al., 2009).
The Arabidopsis SHINE/WAX INDUCER (SHN/WIN) clade of three genes, belonging to the AP2/ERF TF family, was previously shown to be involved in wax/cutin lipid regulation and drought tolerance in Arabidopsis (Aharoni et al., 2004; Broun et al., 2004; Kannangara et al., 2007). A homolog, NUDUM, has also been found in barley (Hordeum vulgare), which is responsible for adhesion of the hulls covering the barley seed (probably mediated by a lipid layer), an important trait in the domestication process (Taketa et al., 2008). Following our findings in Arabidopsis, the SHN2 gene (Aharoni et al., 2004) under the control of the cauliflower mosaic virus (CaMV) 35S promoter was transformed into rice and shown to confer drought resistance and enhanced water use efficiency with a slight increase in cuticular wax (Karaba, 2007). In this study, we describe our discovery of a novel function of the SHN gene as a key regulator of lignin and cellulose/cell wall biosynthesis pathways, coordinating the down-regulation of lignin biosynthesis and the up-regulation of cellulose and other cell wall biosynthesis pathway genes.
RESULTS
Expression of the Arabidopsis SHN Gene in Rice Causes Coordinate Regulation of Cell Wall Biosynthetic Genes
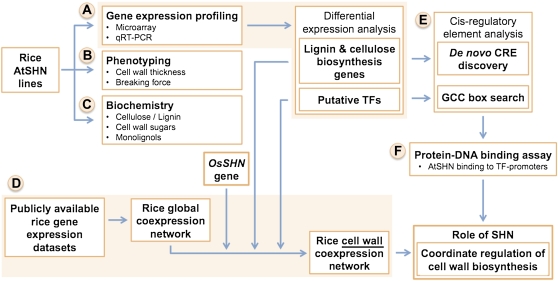
Rice genotypes expressing the Arabidopsis SHN2 gene (Karaba, 2007), hereafter called rice AtSHN lines, were analyzed for their molecular and biological phenotypes. The overall strategy used in this study is depicted in Figure 1 and presented in detail below. Gene expression analysis of the rice AtSHN lines using Affymetrix GeneChips revealed coordinate regulation of cell wall biosynthesis genes (as annotated in rice from Yokoyama and Nishitani, 2004), with a distinctive up-regulation of cellulose and other cell wall biosynthesis genes and down-regulation of lignin biosynthesis genes (Supplemental Table S1). Since we observed that the expression of homologous members of large gene families were being altered in the SHN lines, we sought to reliably quantify the expression levels of individual genes in these multigene families (e.g. 4-coumarate-CoA ligase [4CL], cinnamyl alcohol dehydrogenase [CAD], and CESA). Therefore, gene expression was quantified based on a reannotation of the rice Affymetrix GeneChip probe sets (described in “Materials and Methods”) to distinguish between members of gene families to the extent that was possible with the available probes. This reannotation can distinguish 35,161 rice genes and thus reliably characterize the differential expression of many cell wall pathway genes.
Figure 1.
Integrated systems analysis workflow for elucidation of the role of SHN in the regulation of cell wall biosynthesis in rice. A, Gene expression profiling of rice AtSHN lines was used to characterize differentially expressed genes (compared with the wild type). B and C, Phenotyping and biochemical analysis were used to discern the status of cell wall biosynthesis in rice AtSHN lines. D, Coexpression analysis was used to establish the transcriptional network of cell wall-related genes, identify the relevance of the rice SHN (OsSHN) gene to this network, and independently validate the gene expression profile (from A). E, Cis-regulatory element analysis was used to predict potential promoter regions that could be involved in gene regulation by SHN and other putative cell wall TFs. F, Protein-DNA binding assay was used to evaluate AtSHN interaction with promoter regions of predicted target genes. Our observations from this integrative computational and experimental approach were used to reveal the role of SHN as a key regulator of cell wall biosynthesis in rice. [See online article for color version of this figure.]
In addition, several TFs were observed to be regulated by SHN in the microarray experiment (Supplemental Table S1). These included rice homologs of several NAC and MYB transcriptional activators of the cell wall biosynthetic pathways uncovered in Arabidopsis and other plant species (Zhong and Ye, 2009).
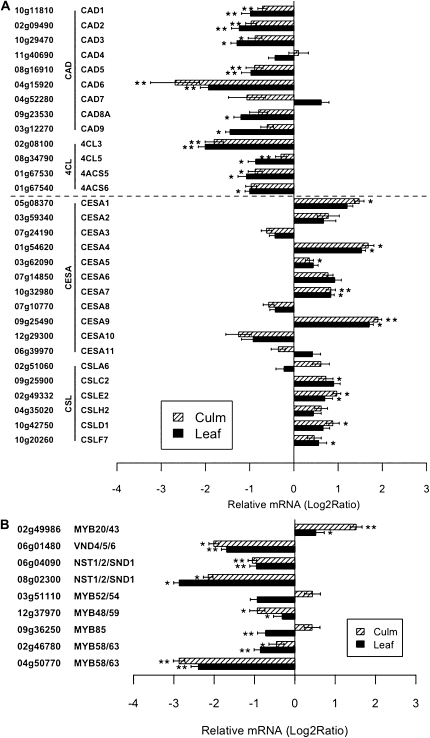
To verify SHN regulation of the monolignol and cell wall biosynthesis pathways as observed in the microarray, we carried out rigorous quantitative real-time (qRT)-PCR experiments to determine the abundance and tissue specificity of biosynthetic genes in two different tissue types, leaf and culm, in SHN and wild-type plants. We found that transcript levels of seven CADs and four 4CLs were significantly repressed in one of the tissues (Fig. 2A, top), confirming their regulation in leaf and culm by SHN. Two genes, CAD4 (Os11g40690) and CAD7 (Os04g52280), were either not responsive or slightly induced in the culm and leaf tissue, respectively. Pathway-wide repression of lignin biosynthesis, including the 4CL and CAD genes that catalyze specific branches of lignin biosynthesis leading to different lignin monomers, suggested possible alterations in lignin composition along with overall reduction in response to SHN expression.
Figure 2.
Expression analysis of cell wall biosynthetic genes and their putative transcriptional regulators assessed through qRT-PCR. A, Relative expression of lignin and cellulose biosynthesis genes in SHN leaf and culm compared with that in the wild type. B, Relative expression of putative cell wall TF genes in SHN leaf and culm compared with that in the wild type. Data are expressed as mean relative transcript levels in SHN lines compared with the wild type (log2 ratio) in each tissue type (leaf and culm). Error bars represent se (n = 3; three wild-type and three SHN lines). Asterisks indicate levels of significance of differential expression (t test; * P ≤ 0.05, ** P ≤ 0.01).
Expression of a battery of cellulose and other cell wall biosynthesis genes, on the other hand, was induced in SHN leaf and culm compared with the wild type (Fig. 2A, bottom; Supplemental Fig. S2). The varying levels of expression of individual members of cell wall (cellulose or hemicellulose) biosynthetic gene families in culm and leaf point to selective expression in specific tissues or organs. Nevertheless, five out of 11 CESA genes in rice are significantly up-regulated by SHN in culm tissue (Fig. 2A, bottom). Induction of three out of the five CESA genes is consistent across leaf and culm. Together, these findings suggest a role of SHN in inducing the expression of certain cellulose and other cell wall biosynthetic genes in rice.
In addition, seven putative rice cell wall biosynthesis TFs, three NAC genes and four MYB genes, were confirmed to be down-regulated in the leaf tissue (Fig. 2B). Six of these TFs were also repressed in culm. One TF, MYB20/43 (Os02g49986), however, was up-regulated in both tissue types (Fig. 2B). These modes of differential gene expression of TFs may account for the coordinate expression of their biosynthetic targets in new spatial or temporal patterns as required to generate the functional integrity of the pathway. Taken altogether, these data indicate that AtSHN is a key regulator of monolignol and other cell wall biosynthesis genes. These gene expression results urged us to test for associated phenotypic and biochemical changes.
Rice AtSHN Lines Have Altered Cell Walls in Mechanical Tissues
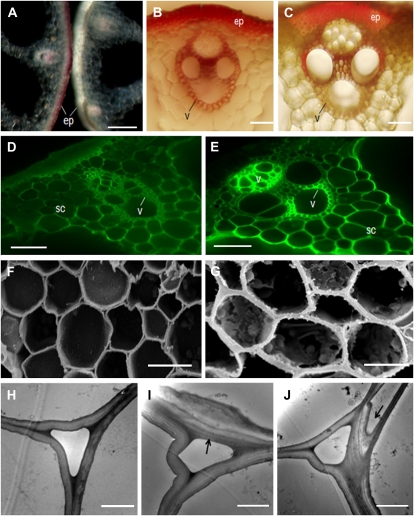
To assay for changes in lignin and cellulose, transverse sections of the culms of wild-type and rice AtSHN lines were histochemically stained with phloroglucinol and calcofluor solutions. Phloroglucinol stain reacts with coniferaldehyde groups in lignin, and the color intensity approximately reflects the total lignin content, while cellulose and other cell wall components show fluorescence staining with calcofluor. The color differences in mechanical tissues, especially in the subepidermal and vascular bundles, between wild-type (Fig. 3, A [left], B, and D) and SHN (Fig. 3, A [right], C, and E) plants were evident, indicating an apparent decrease in lignin quantity and an increased level of cellulose in SHN plants.
Figure 3.
Phenotypic characterization of rice AtSHN lines. A to C, Phloroglucinol staining for lignin in culm (stem) sections of rice AtSHN (right) and wild-type (left) lines; sections were visualized by light microscopy (A, 5× magnification; B and C, 20× magnification). Lignin in epidermal (ep) and vascular (v) tissue is significantly reduced in SHN lines. D and E, Calcofluor staining of wild-type (D) and SHN (E) culm sections, showing increased cellulose in cell walls of sclerenchyma (sc) and vascular bundles (v) in SHN lines. F and G, Scanning electron micrographs of transverse sections of wild-type (F) and SHN (G) culms. H to J, Transmission electron micrographs of wild-type (H) and SHN (I and J) parenchyma cell walls. Arrows show the thickened and folded walls. Bars = 200 μm in A, 40 μm in B and C, 50 μm in D and E, 10 μm in F and G, and 4 μm in H to J.
Electron microscopy analysis demonstrated that the rice AtSHN lines also showed heavily thickened sclerenchyma cell walls (Fig. 3G) and a thickened folded cell wall structure (Fig. 3, I and J) compared with that of the wild type (Fig. 3, F and H). These analyses together suggest a thickening of secondary walls due to the deposition of cellulose in place of lignin in SHN lines. Further quantitative analysis showed that the wall thickness of sclerenchyma and bundle sheath fiber cells was increased by approximately 45% and 48%, respectively, in the SHN overexpressors compared with the wild type (Table I).
Table I. Quantitative comparison of the wall thickness of sclerenchyma and bundle sheath cells in the culms of wild-type and SHN lines.
Wall thickness was measured from transmission electron micrographs, and the data are presented as means ± se of 20 cells (t test; ** P ≤ 0.01).
| Line | Wall Thickness |
|
| Sclerenchyma | Bundle Sheath | |
| μm | ||
| Wild type | 1.42 ± 0.10 | 0.76 ± 0.07 |
| SHN_10 | 2.02 ± 0.12** | 1.09 ± 0.06** |
| SHN_15 | 2.12 ± 0.11** | 1.13 ± 0.09** |
| SHN_18 | 2.03 ± 0.11** | 1.14 ± 0.09** |
Since the rice AtSHN plants are morphologically indistinguishable from wild-type plants, we tested if the reduced lignin had an effect on its physical properties. To quantitatively compare the strength of the tissues, we determined the breaking forces of rice AtSHN and wild-type plants (Supplemental Table S2), which showed that the force required to break the leaves and culms of rice AtSHN lines was almost identical to that of the wild type.
Rice AtSHN Lines Have Enhanced Cellulose and Reduced Lignin
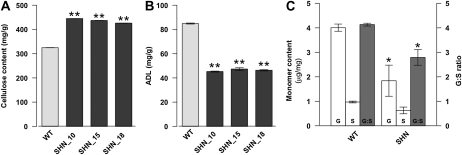
Expression analysis and cellular phenotypes implied that the cellulose/cell wall biochemical composition in the rice AtSHN lines may be altered. Therefore, we analyzed the major secondary cell wall constituents like cellulose and lignin contents in rice AtSHN and wild-type plants. Chemical analysis of rice culms showed a significant 34% increase in cellulose content (Fig. 4A) and a 45% reduction in lignin (Fig. 4B) in the rice AtSHN lines compared with the wild type. Furthermore, analysis of neutral cell wall-associated carbohydrates (Ara, Fuc, Gal, Glc, Man, Rha, and Xyl) showed that the amount of Glc and Xyl, the main sugars in cellulose and hemicellulosic polysaccharides (Li et al., 2003b), respectively, were significantly increased in the SHN lines (Table II). For the above analyses, we used equal amounts of dry weight, because the cell wall preparation, calculated as the ratio of dry weight to fresh weight, was not significantly different between wild-type and SHN lines. Hence, we suggest that the lower amount of lignin is mass balanced by hemicelluloses and/or cellulose, without change in total dry weight. Taken together, these results support the hypothesis that SHN causes a decrease in lignin and a compensatory increase in hemicellulose and cellulose.
Figure 4.
Cell wall composition analysis of rice AtSHN culms. A, Cellulose content measured in rice AtSHN lines and the wild type (WT). B, Lignin content estimated as the percentage of acid detergent lignin in terms of dry matter. C, Lignin monomer ratio G:S assayed by GC-MS analysis of rice AtSHN lines and the wild type. Data represent means ± se (n = 6 for each genotype in A, n = 3 [each replicate being a pool of two plants] for each genotype in B, and n = 3 [with replicates corresponding to independent wild-type or SHN lines] in C). Asterisks indicate levels of significance compared with the wild type (t test; * P ≤ 0.05, ** P ≤ 0.01).
Table II. Comparison of the contents of cell wall sugars between rice wild-type and SHN culms.
The sugar contents of cell walls of wild-type and SHN lines are given as means ± se (mg g−1) of three independent assays. Asterisks indicate levels of significance compared with the wild type (t test; ** P ≤ 0.01).
| Sugar | Wild Type | SHN_10 | SHN_15 | SHN_18 |
| Rha | 0.45 ± 0.38 | 0.60 ± 0.23 | 0.57 ± 0.21 | 0.51 ± 0.28 |
| Fuc | 0.26 ± 0.04 | 0.19 ± 0.06 | 0.15 ± 0.02 | 0.18 ± 0.02 |
| Ara | 7.64 ± 0.78 | 9.71 ± 0.47 | 9.52 ± 0.27 | 9.94 ± 0.60 |
| Xyl | 33.71 ± 0.72 | 45.32 ± 0.85** | 46.87 ± 0.56** | 46.95 ± 0.66** |
| Man | 0.95 ± 0.05 | 0.79 ± 0.10 | 0.80 ± 0.05 | 0.80 ± 0.09 |
| Gal | 1.55 ± 0.53 | 1.88 ± 0.44 | 1.79 ± 0.42 | 1.72 ± 0.49 |
| Glc | 208.34 ± 2.28 | 256.71 ± 2.37** | 254.45 ± 3.05** | 253.73 ± 2.65** |
The digestibility of lignocellulosic feedstock for feed and biofuel is dependent, in addition to lignin content, on the composition, manifested as the ratio of G and S monomers (Vanholme et al., 2008). Since enzymes involved in specific parallel branches of lignin biosynthesis were altered in expression, we conceived an effect of SHN expression on the composition of lignin in addition to its content. Gas chromatography-mass spectrometry (GC-MS) analysis (Lu and Ralph, 1997) revealed that the rice AtSHN lines, compared with the wild type, have a 54% reduction in G content and consequent reduction in G-S ratio to two-thirds (Fig. 4C). The S content is not significantly changed, possibly due to a selective reduction in activity of the 4CL, CCR, and CAD genes involved in G synthesis, as observed in the gene expression analysis (Fig. 2; Supplemental Table S1). Alternatively, AtSHN might cause equal inhibition of the individual steps, but under such circumstances, the branch leading to coniferyl alcohol might be more efficient at channeling limited precursors than the branch leading to sinapyl alcohol. Selective inhibition in monolignol biosynthesis has been observed previously in tobacco by expressing o-methyl transferase (Atanassova et al., 1995) and Antirrhinum MYB genes (Tamagnone et al., 1998).
Coexpression Network Underlying the Regulation of Lignin and Cellulose Biosynthesis
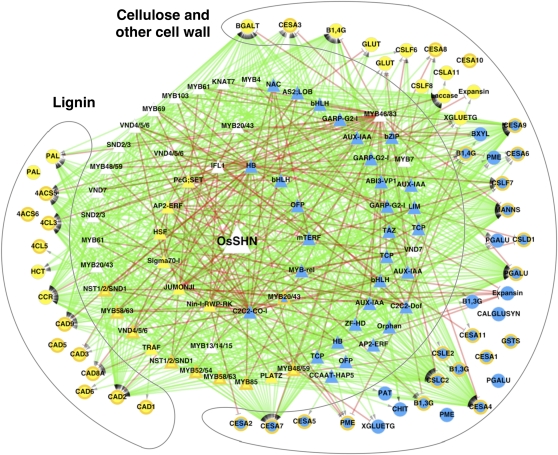
In order to evaluate if the rice SHN gene Os06g40150 (OsSHN), homolog of Arabidopsis SHN2, also has an intrinsic association with the cell wall pathways, an extensive analysis of coexpression in rice was undertaken. A global coexpression network of rice genes was constructed based on public gene expression data sets in rice. Raw Affymetrix rice expression profiles pertaining to “response to environmental conditions” were collected from the Gene Expression Omnibus (GEO; Barrett et al., 2009) and ArrayExpress (Parkinson et al., 2009). Gene expression values across the diverse conditions were extracted using the custom gene-centric probe set definitions, and pairwise gene-gene correlations were calculated to create the global network (see “Materials and Methods”). From this global network, we explored the connectivity between (1) AtSHN-regulated lignin and other cell wall-related genes, (2) TFs associated with these pathways and other TF genes differentially expressed in the AtSHN microarray, and (3) the OsSHN gene. In addition to finding that the biosynthesis genes were intimately connected to the TFs, including the NACs and MYBs, remarkably, the OsSHN gene was also found to be connected to these TFs, together forming a dense coexpression network (Fig. 5).
Figure 5.
Coexpression network analysis and model of cell wall synthesis in rice. The coexpression network connects TFs (triangles) to pathway gene targets (circles) through directed edges and TFs to each other through undirected edges. Positive and negative correlation edges are colored green and red, respectively. Nodes are labeled with the gene name/family and colored based on the direction of regulation in response to SHN expression (microarray data supplemented by qRT-PCR) with blue, yellow, and white for up-regulation, down-regulation, and no regulation, respectively. Genes tested for differential expression using qRT-PCR are denoted with thick orange borders. The outer rings of TFs are all positively coexpressed with each other; hence, all the green edges connecting them to each other have been removed for clarity. See Supplemental Table S1 for identifiers, names, and annotations of all the genes in the network. This network is provided in Supplemental File S1 and can be imported into Cytoscape.
We then cross-referenced this network with our independent SHN expression profile of rice genes to gauge their correspondence. Overlaying the differential expression of the genes (from microarray and qRT-PCR) onto the nodes of the network showed that almost all the gene regulation observed due to the expression of Arabidopsis SHN in rice was explained by the signs of the rice network coexpression edges: positively correlated gene pairs were regulated in the same direction (both up-regulated or both down-regulated), and negatively correlated gene pairs were regulated in opposite directions. OsSHN was directly connected to the homolog of VND6 through a negative edge, supporting the down-regulation of the VND6 homolog in the expression study. Here, for simplicity in transferring functional information between species, the rice TF genes are referred to by the names of Arabidopsis homologs (Zhong and Ye, 2009). VND6 is well connected to several other NAC (NST1/2/SND1) and MYB (MYB46/83, MYB52/54, MYB85, and MYB58/63) TFs and to the lignin biosynthetic genes through positive edges, vindicating the down-regulation of this homologous transcriptional machinery by AtSHN leading to the repression of the lignin biosynthetic pathway. As expected, this large-scale repression also causes the down-regulation of several other cell wall pathway genes. On the other hand, among the putative cell wall TFs known from Arabidopsis, OsSHN is also directly positively connected to the MYB20/43 homolog, an interaction again supporting the up-regulation of this gene in response to AtSHN expression. This gene, in turn, is positively correlated with cellulose/cell wall genes that are also found to be up-regulated. Moreover, OsSHN is positively correlated with several other TFs (up-regulated by SHN expression), which are positively correlated with many cell wall genes found to be up-regulated in the expression studies, including seven CESA genes and four CSL genes (Fig. 2A) up-regulated by SHN expression.
AtSHN Directly Binds to the Promoters of NAC and MYB Genes
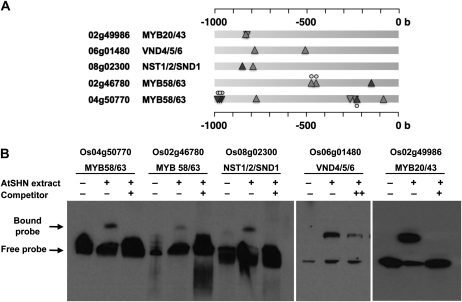
While independently seeking evidence for transcriptional regulation of NAC and MYB TF genes by SHN, as observed in the coexpression analysis, the promoter regions of these genes were found to contain GCC box motifs ([AG]CCGNC) known to be bound by AP2-ERF TFs (Ohme-Takagi and Shinshi, 1995; Fig. 6A), suggesting that AtSHN could regulate these TFs by direct binding. Hence, based on the coexpression network (Fig. 5), confirmed gene expression changes (using qRT-PCR; Fig. 2B), and promoter analysis (Fig. 6A), the NAC switches VND6 (Os06g01480) and SND1/NST1/2 (Os08g02300, Os06g04090) and three downstream MYBs, MYB20/43 (Os02g49986) and MYB58/63 (Os04g50770, Os02g46780), were predicted to be direct targets of AtSHN. All these TFs have been shown to have roles in the regulation of cell wall biosynthesis in Arabidopsis (Zhong and Ye, 2009). These homologous TFs were hence hypothesized to be the regulators of cell wall biosynthesis in rice and mediators of the coordinate regulation of the biosynthetic pathways by SHN.
Figure 6.
Gel retardation assay of AtSHN binding to the promoter sequence of putative secondary cell wall TF genes. A, Locations of the GCC box motif ([AG]CCGNC) in the 1-kb upstream sequences of the SHN-regulated TF genes. Upright and inverted triangles represent + and – strands, respectively. Dark triangles signify the presence of the GCC core (GCCGCC). Small circles above or below a triangle represent another overlapping GCC box motif in the + or – strand, respectively. B, The locations of DNA fragments used for electrophoretic mobility shift assay are selected based on the abundance of GCC box motifs shown in A. Each of the end-labeled domains was competed with 100-fold excess of the competing unlabeled domain, whereas ++ was 60-fold. + and − indicate the presence and absence of the respective component in the binding reaction. The labeled “free probe” and DNA-protein complex “bound probe” positions are indicated, showing competition with an unlabeled fragment for each promoter domain.
To examine this hypothesis, we used a recombinant 6His-AtSHN fusion protein for protein-DNA binding studies. Gel mobility shift assays showed binding between affinity-purified recombinant AtSHN protein and promoter regions of putative rice secondary wall TFs MYB58/63, NST1/2/SND1, VND4/5/6, and MYB20/43 (Fig. 6B), the specificity shown by competition with an excess of unlabeled oligonucleotides of putative binding sites (Fig. 6A). The recombinant AtSHN protein bound to the MYB and the NAC promoters with almost same affinity, where merely 10 times excess of cold probes could significantly reduce the extent of binding. Promoter analysis and binding assay, along with the observed gene expression changes, thus suggested that AtSHN could regulate these NAC and MYB TFs, the effect of which cascades into the biosynthetic pathways.
DISCUSSION
Cellulose is the most abundant biopolymer and a major renewable energy resource, which can be processed into bioethanol as liquid fuel. In spite of its significance, very little is known about the regulation of cellulose biosynthesis and its coregulation with the synthesis of other cell wall biopolymers. We present a systems-level analysis of the complex regulation of cellulose and lignin biosynthesis, showing an unprecedented significant increase (one-third) in cellulose and a decrease in lignin using the grass/crop model rice. These results offer novel ways for engineering nonfood grasses and crop wastes for the production of lignocellulosic feedstocks that can be efficiently processed into biofuels. Expression of SHN leads to coordinate regulation of multiple steps in the pathways for monolignol and cellulose biosynthesis. Repression of the lignin pathway was exhibited, as a moderate reduction in expression levels of many enzymes spread across the pathway rather than a drastic reduction of a few enzymes (Supplemental Fig. S3). The moderate reduction in terms of the absolute expression levels indicates that gene expression is not completely shut off but allows a background flux through the pathway. Moreover, repression of 4CL and CAD genes that catalyze specific branches of lignin biosynthesis, leading to different lignin monomers, suggests possible alterations in lignin composition along with overall reduction in response to SHN expression.
Differential expression of genes was observed in the microarray (Supplemental Table S1) and was confirmed using qRT-PCR analysis using gene-specific primers of multigene family members at two different tissues of the plant (Fig. 2). SHN overexpressors had significantly repressed transcript levels of lignification genes such as CAD and 4CL family members (Fig. 2A, top) and induced levels of cell wall and CESA family genes (Fig. 2A, bottom; Supplemental Fig. S2) compared with the wild type. It is noteworthy that OsCAD2, which is significantly repressed in SHN lines, is the rice ortholog of the maize CAD that maps to the bm1 locus, knockout of which causes an 80% reduction in lignin (Tobias and Chow, 2005). Such large decreases in lignin content, either due to abolished CAD activity or transcriptional down-regulation, as observed in SHN plants, point to possibilities whereby grasses can be made more digestible. Likewise, in support of our hypothesis that SHN up-regulates hemicellulose and cellulose biosynthesis, several CESA-like glycosyltransferases are up-regulated (Fig. 2A, bottom) that are most likely involved in making hemicelluloses (Scheller and Ulvskov, 2010). In addition, three rice CESA genes (OsCesA4, OsCesA7, and OsCesA9) known to be involved in the synthesis of cellulose in the secondary cell walls (Tanaka et al., 2003) and responsible for the overall strength of the plant are up-regulated in SHN lines in both leaf and culm (Fig. 2A, bottom). While expression changes estimated by microarrays and qRT-PCR were concordant for several genes, more genes, including CESA genes, whose changes were not statistically significant (indicating either noisy or no change) using microarray were detected by qRT-PCR as being differentially expressed.
Several other lines of evidence support the role of the SHN gene in coordinate regulation of cell wall synthesis pathways. First, the involvement of AtSHN in reduced lignin and increased cellulose levels correlate well with the regulation of NAC and MYB TFs (Fig. 2B) that control the activity of various branches of lignin and cellulose biosynthesis and the downstream regulation of the biosynthetic genes (Fig. 2; Supplemental Figs. S2 and S3). Second, light microscopy analysis using phloroglucinol (showing total lignin autofluorescence) provided good resolution of the reduced accumulation patterns of lignin in SHN lines, particularly at subepidermal and vascular bundles (Fig. 3C). Likewise, the antisense down-regulation of 10 individual enzymes in the monolignol pathway generated in a series of otherwise isogenic alfalfa (Medicago sativa) lines showed variation in lignin content and composition (Nakashima et al., 2008), such variation also being evident at the level of individual cell wall layers (Vanholme et al., 2010). Third, the rice AtSHN phenotypic changes for lignin content are very similar to the effects reported for the down-regulation of PAL, 4CL, CAD, and target TF genes in Arabidopsis and other plants (Vanholme et al., 2008). Finally, extensive biochemical analyses revealed evident qualitative differences in the lignin and cellulose levels of the SHN plants when compared with wild-type plants (Fig. 4).
The change in cellulose and lignin did not adversely affect the rice AtSHN lines, as they displayed normal plant phenotypes, maturity, and seed yield under greenhouse conditions (data not shown). In addition, the strength of the stem/culm of rice AtSHN lines was unaltered. The tensile or bending strength of grass tissue, such as that of maize, has been shown to correlate with the cellulose content, whereas lignin is thought to play a role in resistance to compression (Dhugga, 2007). The increase in cellulose in secondary walls of SHN lines also probably offsets any reduction in mechanical strength due to reduced lignin. Such a compensatory increase in cellulose with reduced lignin has also been observed due to the inhibition of a 4CL gene in aspen (Populus spp.) trees that leads to better growth (Hu et al., 1999). Similarly, plants expressing both antisense 4CL and sense CALd5H have been found to have 52% less lignin and 30% more cellulose (Li et al., 2003a). Maize brown midrib mutants have also been shown to harbor reduced lignin and increased hemicellulose (with no change in cellulose content; Vermerris et al., 2010). On the other hand, the rice bc1 mutant and maize bk2 mutant, which have reduced cellulose content, have been shown to contain more lignin (Li et al., 2003b; Ching et al., 2006; Sindhu et al., 2007). These studies, along with our study presented here, provide evidence for complex interdependent regulation of the different cell wall pathways. However, we provide novel evidence for a TF (SHN) coordinating such a compensatory regulatory mechanism.
Coexpression network analysis was performed as an independent route to validating the function of SHN (Fig. 5). Using a large gene expression compendium in rice, correlations between the expression profiles of all the cell wall regulatory and biosynthetic genes along with OsSHN were calculated. Coexpression network analysis has found use in dissecting transcriptional regulation in Arabidopsis (Usadel et al., 2009) and rice (Wang et al., 2009; Fu and Xue, 2010) to gain biological insights into general and case-specific regulation of gene expression. This approach has also been used to identify novel members of biochemical processes, including cellulose synthesis (Persson et al., 2005). In this study, we have used the rice coexpression network as a predictive tool to first assess the expected associations between TFs and their targets, as in the case of NAC and MYB TFs and their putative lignin and other cell wall biosynthetic targets. Second, it was used to discover novel genes that might have a role in a pathway/process of interest using guilt by association, as in the case of OsSHN. And finally, in conjunction with an independent gene expression data set (here the SHN microarray), it was used to demarcate positive and negative interactions and to propose a regulatory model for genes of interest.
This analysis shows that OsSHN, the rice homolog of AtSHN, has a strong association with the cell wall regulatory and biosynthetic machinery in rice. The nature of the association between the cell wall TFs and the biosynthetic genes is strongly positive, as expected. The intriguing finding is the differential association of OsSHN with the TFs, via a negative connection to VND6 and a positive connection to MYB20/43 (and several other TFs), which are positively correlated with lignin and cellulose/other cell wall biosynthetic genes, respectively. In addition, most of the coexpression associations in the rice network agree with, and hence corroborate, the expression changes of the TFs and biosynthetic genes in response to AtSHN expression. Finally, AtSHN expressed in rice functions in the context of rice genes, established by the coexpression network, to perform its functions, a context in which OsSHN is expected to function. Here, it is relevant to note that the OsSHN gene is not regulated upon the expression of AtSHN in rice; hence, all the observed differential expression is solely due to AtSHN. Based on this analysis, we hypothesized that OsSHN has a native association with cell wall regulatory and biosynthetic pathways, with an ability to coordinately regulate the lignin and cellulose pathways by shutting down the main switches (NACs) and intervening by directly regulating downstream MYBs: repressing MYBs specific to lignin biosynthesis (in addition to release of NAC activation) and activating MYBs and other TFs specific to cellulose/other cell wall biosynthesis genes.
To further pursue this hypothesis, we were interested in finding if there were any clues for potential SHN regulation of the NAC and MYB TFs. We performed de novo motif discovery on the upstream regions of all the TFs in the network. However, the analysis showed no significant motifs, which we reasoned to be because SHN could bind to and regulate just a few major TFs that could then regulate other TFs and biosynthetic genes. Therefore, we restricted ourselves to the few TF candidates that showed confirmed gene expression changes and were homologous to Arabidopsis TFs associated with the secondary cell wall biosynthetic pathways. We then searched the upstream regions of these TF genes for the presence of a GCC box motif, a putative binding site of AP2-ERF TFs (Ohme-Takagi and Shinshi, 1995). The identification of several GCC box motifs in these sequences (Fig. 6A) led us to postulate that SHN could directly bind to and regulate these TFs. We then experimentally confirmed SHN binding to these promoter regions using mobility shift assay (Fig. 6B).
With evidence for SHN regulating the putative NAC and MYB TFs, we gleaned support for MYB-mediated transcriptional regulation of the cell wall pathway genes in rice to that in other species. To this end, we performed de novo sequence motif discovery on the promoter regions of the lignin and other cell wall genes. This analysis led to the identification of a rice AC element, CACCA[ACG]NC[AC] (Supplemental Fig. S3), that is similar to the ACII element, CACCAACCC, known to mediate vascular tissue-specific expression of the cell wall biosynthetic genes in other plant species, with evidence for MYB TFs binding to this element (Zhong and Ye, 2009). This suggested that a similar transcriptional machinery of MYB TFs (downstream of NACs) could exist in rice and that SHN could regulate genes involved in lignin and cell wall biosynthesis, possibly by regulating these TFs.
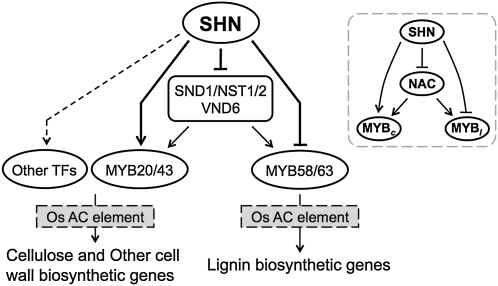
Several TFs with both activator and repressor functions have been well characterized in plants (González-Lamothe et al., 2008; Ikeda et al., 2009) and animals (Adkins et al., 2006; Zheng et al., 2007). The hypothetical model in Figure 7 summarizes our findings on the transcriptional regulation of cell wall biosynthesis in rice involving a plausible bifunctional activity of SHN. SHN represses the NAC TFs, SND1/NST1/2 and VND6, which are known to be the main switches of cell wall biosynthesis. But SHN also directly represses the MYB TFs MYB58/63 and directly activates MYB20/43. Due to these three activities, SHN bypasses the main switches (NACs) and selectively up- and down-regulates downstream TFs (MYBs) specific to cellulose (and other cell wall) and lignin biosynthetic genes, respectively. As depicted in the inset of Figure 7, an interesting feature of this mechanism is its resemblance, in architecture, to a coupled system of feed-forward loops (Mangan and Alon, 2003), one incoherent (type 4; left) and the other coherent (type 2; right); the relevance of this feature in a dynamic sense remains to be determined. We also believe that TFs other than those tested here could be involved in mediating SHN up-regulation of cellulose and other cell wall genes, candidates for which have been identified as those up-regulated in response to SHN expression and positively correlated with the up-regulated cell wall genes.
Figure 7.
Hypothetical model of transcriptional regulation of cell wall biosynthesis in rice. Dashed arrows are hypothesized interactions based on the coexpression network and gene expression changes. Thick arrows emanating from SHN represent the confirmed interaction of SHN to the upstream regions of the TFs. In the inset, NAC represents the NAC main switches and MYBc and MYBl represent downstream MYB TFs hypothesized to be specific to cellulose/other cell wall genes and lignin genes, respectively.
Since several insights about cell wall biosynthesis have been gleaned in Arabidopsis, it is also important to put our findings in rice in that context and deliberate the probable role of AtSHN in Arabidopsis. To gain some understanding about AtSHN function in relation to secondary cell wall biosynthesis, we reanalyzed the gene expression profiles of Arabidopsis AtSHN (WIN1) overexpression lines compared with wild-type controls from Broun et al. (2004). Since the design lacks replication, we quantified the relative expression levels of approximately 390 (present in the 8 K Affymetrix Arabidopsis genome array) cell wall-related genes (out of approximately 930 total in the genome) in a “strong” WIN1 overexpressor compared with the wild type (Supplemental Table S3). We observed here that quite a few lignin genes (including PAL2, PAL3, and 4CL3) and cellulose/hemicellulose biosynthetic pathway genes (including CESA2, CSLB01, CSLB02, CSLB03, CSLB04, CSLC12, and CSLG3) were up-regulated. On the other hand, two cellulose (CESA1 and CSLA03) and two lignin (CCR and CCoA-OMT) biosynthetic genes were down-regulated. Moreover, along with the biosynthetic genes, the NAC TF NST1 (a functional paralog of SND1) is also up-regulated. Based on the above observations, overexpression of AtSHN in Arabidopsis seems to cause a nominal up-regulation (along with a certain amount of down-regulation) of both lignin and cellulose/hemicellulose biosynthesis and, therefore, probably does not cause any large absolute or relative change in the amounts of lignin and cellulose. In support of this notion, simple sugar measurements made by Kannangara et al. (2007) and our observations of Arabidopsis cross-sections (data not shown) for lignin quantification, respectively, indicate that there are no significant changes in either cellulose or lignin in Arabidopsis SHN lines compared with the wild type.
Transcriptional regulation has been found to exhibit high evolutionary plasticity (Wagner and Lynch, 2008), and several orthologous TFs have been reported to have marginally or drastically diverged in function (Studer and Robinson-Rechavi, 2009; Lavoie et al., 2010). Here, we propose that AtSHN does have an association with secondary cell wall biosynthesis in Arabidopsis as far as transcriptional regulation of the regulatory and biosynthetic genes is concerned. This includes AtSHN’s ability to both up- and down-regulate the concerned genes. Therefore, an overall similar association with secondary cell wall regulation and biosynthesis is conserved across Arabidopsis and rice, with different details: we show that AtSHN in rice causes an inverse regulation of the pathways by differently regulating various TFs of the lignin and cellulose biosynthetic pathways.
Furthermore, to explore the expression pattern of OsSHN in different rice organs and tissues, we used the rice eFP browser (Winter et al., 2007) to identify that the gene is expressed in the inflorescence stages P3, P4, P5, and P6, with the strongest expression at the P5 stage. Using the rice expression atlas (Jiao et al., 2009) resource, we observed that OsSHN is expressed in the coleoptile (0 h) and fresh whole leaf and to a lesser extent in epiblast (12 h) and seedling blade (Supplemental Table S4). These cell types represent young growing tissue where lower lignin deposition is expected, consistent with SHN’s proposed role in up-regulating cellulose synthesis and down-regulating lignin synthesis pathways in rice.
The SHN master regulator orchestrates coordinated regulation of the cellulose and lignin pathways to provide enhanced cellulose and decreased lignin deposition. The two other functions ascribed to SHN/WIN are in cuticle (cuticular wax and cutin) formation (Aharoni et al., 2004; Broun et al., 2004; Kannangara et al., 2007). Most strikingly, all these processes could have evolved in organismal organization simultaneously when land plants emerged, to give them a protective cover as well as strength to remain erect and transport water upward. Coordination of these processes was probably maintained by the master regulatory functions of the SHN gene family. Altogether, SHN regulates the accumulation of cellulose, lignin, and cutin, the top three plant biomass polymers, and can help in improving plant feedstock for these components. The activity of SHN in the grass model rice shows that it has a potential role in engineering the cellulose-lignin composition and content in grasses or other suitable biomass producers. The SHN master regulator can also be used as a tool to express in plants of interest and unravel the regulatory pathway in cellulose and lignin biosynthesis. The Arabidopsis-rice homologous TFs and biosynthetic genes described here, and the genetic model of their interaction, can serve as a dicot-monocot conserved model to understand the coordinate regulation of cellulose, hemicellulose, and lignin biosynthesis in a number of other plant species.
MATERIALS AND METHODS
Vector Construction and Generation of Transgenic Rice Plants
An Arabidopsis (Arabidopsis thaliana) AtSHN gene overexpression construct was made by assembling fragments of the CaMV 35S promoter (Pietrzak et al., 1986), the coding region of Arabidopsis SHN2 (At5g11190), and the CaMV 35S terminator (Pietrzak et al., 1986) into the binary vector pMOG22 (Zeneca-Mogen), which contains a chimeric CaMV 35S-hygromycin phosphotransferase-tNos for selection during transformation. Agrobacterium tumefaciens-mediated transformation of rice (Oryza sativa subsp. japonica ‘Nipponbare’) was done as described (Greco et al., 2001).
Plant Genotyping and Statistical Analysis
Out of 15 transgenic rice AtSHN lines generated (Trijatmiko, 2005), T3 progeny of different transgenic SHN lines segregating for single inserts were tested for stable expression pattern using qRT-PCR and genotyped for the presence of the AtSHN transgene locus. Based on the results of qRT-PCR amplification of AtSHN, it was evident that three lines showed significant and stable expression, and these were used for further analysis described here.
Fifty progeny of three independent lines expressing AtSHN were grown in controlled growth chambers. For all analyses, six plants were used for each of the three transgenic lines and the wild type. The data in the experiments of qRT-PCR, cell wall composition analysis, breaking-force measurements, and measurement of cell wall thickness were analyzed using Student’s t test. All t tests performed were two sided.
Histochemical Staining and Microscopy
Histochemical localization of the lignin and cellulose was done using phloroglucinol and calcofluor staining as described (Li et al., 2003b) using approximately 20-μm-thick hand-cut sections from rice culms. The stained sections were examined and photographed with a light microscope (Nikon Eclipse E600) for lignin (phloroglucinol) and with an inverted fluorescence microscope (Nikon TE 2000-U) for cellulose (calcofluor) staining. For scanning electron microscopy, rice culm tissues were excised and fixed in 2.5% (v/v) glutaraldehyde. The fixed samples were washed twice in 0.1 m sodium cacodylate buffer for 15 min each, postfixed in 1% OsO4 for 1 h, dehydrated through an ethanol gradient, and infiltrated. Samples were critical point dried, sputter coated with gold in an E-100 ion sputter, and viewed with a scanning electron microscope (Carl Zeiss EV040). For transmission electron microscopy, ultrathin sections were made using an ultramicrotome (MT-X; RMC), and the sections were thoroughly stained with aqueous 2% uranyl acetate for 10 min followed by lead citrate for 2 min. The sections were viewed with a JEM-1010 electron microscope (JEOL) operating at 60 kV. The wall thickness of sclerenchyma and bundle sheath fiber cells was measured using the ImageJ program (Rasband, 1997).
Measurement of Breaking Force
The breaking force of rice culms and leaves, defined as the force required to break the segment, was measured using a force-testing device (Kokubo et al., 1989) assembled in-house with parts obtained from Instron (www.instron.com) and Fisher Scientific (www.fishersci.com). The first internodes of culms and flag leaves were used for immediate fresh tissue measurements.
Cell Wall Composition Analysis
Rice culms from wild-type and SHN lines were used for acid detergent lignin (ADL) analysis (Van Soest, 1967) and cellulose content as described (Scott and Melvin, 1953; Updegraff, 1969). For ADL analysis, samples were dried at 55°C for 48 h and ground to pass a 1-mm screen in a cyclone mill. After that, 0.5 ± 0.05 g of air-dried sample was directly placed into the filter bags for acid detergent fiber determinations (ANKOM Technology) using a fiber analyzer and followed by ADL analysis by treating with 72% sulfuric acid for 3 h. The data from wild-type and SHN lines were determined in replicates through gravimetric analysis and are calculated as percentage on a dry matter basis. For cellulose, the dried powder was hydrolyzed with acetic-nitric reagent for 1 h at room temperature and centrifuged at 8,000 rpm for 5 min. The resulting pellet was extracted twice with acetone and dried under vacuum at 45°C. The resulting precipitate was resuspended in 67% (v/v) H2SO4 for 1 h at room temperature, and cellulose content was determined at A625 on an appropriate dilution of the sulfuric acid solution by the anthrone method (Scott and Melvin, 1953). Cell wall monosaccharide analysis was carried out essentially as described (Albersheim et al., 1967; Foster et al., 2010). Briefly, dried cell wall material from culms of both wild-type and SHN lines was treated with 2 m trifluoroacetic acid for 90 min at 121°C, derivatizing the resulting solubilized monosaccharide to their alditol acetates (Albersheim et al., 1967), and analyzed on an GC-MS apparatus equipped with an SP-2380 column (30 mm × 0.25 mm × 0.25 μm; Supelco). The peaks are identified by mass profiles and/or retention times of standards. Monosaccharides are quantified based on standard curves.
Lignin Monomer Analysis by Derivatization Followed by Reductive Cleavage
Derivatization followed by reductive cleavage is a method for lignin compositional analysis that produces analyzable monomers and dimers by cleaving α- and β-ethers in lignins (Lu and Ralph, 1997, 1998). Dry plant samples (approximately 20 mg) were suspended in acetyl bromide:acetic acid (20%, v/v) in a 10-mL round-bottom flask and gently stirred at 50°C for 3.5 h. Solid residues were filtered off, and the filtrate was evaporated by a rotary evaporator at 50°C under reduced pressure. After evaporation, the residue was dissolved in 2.5 mL of dioxane:acetic acid:water (5:4:1, v/v/v), and zinc dust (50 mg) was added to the solution as it was stirred. Stirring was maintained for 30 min. Before GC-MS analysis, a solid-phase extraction was applied for the collection of monomers while dimers and oligomers were removed. Thus, crude acetylated derivatization followed by reductive cleavage products in 50 to 100 μL of dichloromethane were loaded onto a preconditioned SPE column (3 mL of silica; normal phase) and eluted with 12 mL of cyclohexane:ethyl acetate (5:1, v/v). The eluent was concentrated to about 0.1 mL for GC-MS analysis.
Gene Expression Analysis
Total RNA was isolated from the rice leaf and culm tissue of wild-type and SHN lines using the RNeasy plant kit (Qiagen), and RNA quantity/quality were measured by the Agilent 2100 Bioanalyzer (Agilent Technolgies). For each sample, 4 μg of total RNA was used to generate first-strand cDNA with a T7-Oligo(dT) primer. Following second-strand synthesis, in vitro transcription was performed using the GeneChip IVT labeling kit. The preparation and processing of labeled and fragmented complementary RNA targets, as well as hybridization to rice Affymetrix GeneChips, washing, staining, and scanning, were carried out according to the manufacturer’s instructions (http://www.affymetrix.com). The RNA samples used for the microarray experiments were also used to synthesize cDNA templates for qRT-PCR analysis. The comparative threshold cycle (Ct) method of quantitation was used with the actin gene as a reference (Ambavaram and Pereira, 2010). The relative fold change for each of the selected genes was detected from both the wild-type and SHN transgenic lines. Three independent biological replicates of each sample and two technical replicates of each biological replicate were used for expression analysis. For each sample, 1 μg of total RNA from one of the biological replicates was converted into cDNA using oligo(dT) 15-mer (Promega) and SuperScript III reverse transcriptase (Invitrogen Life Technologies). This cDNA was diluted to 250 μL in sterile water. Validation experiments were performed on five to six log dilutions of each of the target genes together with the actin reference to determine if the amplification efficiencies were equal. Triplicate qRT-PCRs were performed using iQSYBR Green Supermix in a Bio-Rad iQ5 thermocycler (Bio-Rad). Melting curve analysis, by applying increasing temperature from 55°C to 95°C (0.5°C 10 s−1), and gel electrophoresis of the final product confirmed single amplicons. Negative control reactions using untranscribed RNA were also run to confirm the absence of genomic DNA. To determine relative fold differences for each sample in each experiment, the Ct value for each gene was normalized to the Ct value for actin and was calculated relative to a calibrator using the equation 2−ΔΔCt (Livak and Schmittgen, 2001).
Reannotation of Rice GeneChip Probe-Gene Mapping
A high-quality custom chip definition file (CDF) was built for the rice GeneChip array by uniquely mapping 442,810 probe sequences (http://www.affymetrix.com/analysis/downloads/data/) to 35,161 rice gene-based probe sets in the following manner: (1) probes that have perfect sequence identity with a single target gene were selected; (2) probes mapping to reverse complements of genes were annotated separately as antisense probes (not used in the above counts); and finally (3) probes were grouped into probe sets, each corresponding to a single gene, and probe sets with at least three probes were retained (greater than 98% of probe sets have five or more probes). Note that these stringent criteria used to construct the CDF make it possible to reliably measure expression values of members of multigene families (free from cross-hybridization between paralogs showing high sequence similarity) and to get around “one gene to multiple probe sets” ambiguities. Moreover, improving probe-gene mapping also considerably aids in better quantification of coexpression by filtering out false-positive correlations (Casneuf et al., 2007). This new custom CDF is available from the National Center for Biotechnology Information (NCBI) with the GEO accession number GPL11322.
Analysis of Differential Gene Expression
Raw data from the SHN overexpression experiment were background corrected, normalized, and summarized according to the custom CDF using robust multichip average (RMA; Ihaka and Gentleman, 1996; Irizarry et al., 2003; Gentleman et al., 2004), followed by nonspecific filtering of genes that do not have enough variation (interquartile range across samples less than median interquartile range) to allow reliable detection of differential expression. A linear model was then used to detect differential expression of the remaining genes (Smyth, 2004). The P values from the moderated t tests were converted to q values to correct for multiple hypothesis testing (Storey and Tibshirani, 2003), and genes with q < 0.1 were declared as differentially expressed in response to AtSHN expression. WIN1 overepxression data (Broun et al., 2004) were obtained from NCBI GEO (accession no. GSE1071), and raw data were similarly preprocessed using RMA based on a custom CDF for the Arabidopsis genome array obtained from http://brainarray.mbni.med.umich.edu/Brainarray/ (Dai et al., 2005).
Curation of Lignin and Cellulose Biosynthetic Genes and Putative Regulators
Rice genes involved in cell wall biosynthesis and regulation were identified as homologs of Arabidopsis cell wall-related genes (Remm et al., 2001; Yokoyama and Nishitani, 2004; Zhong and Ye, 2009) and from direct annotations (rice genome annotation database; Ouyang et al., 2007). A further 153 TFs that were regulated in the SHN expression microarray (see below) were added to the list of putative regulators. The OsSHN gene (Os06g40150) was determined as the rice homolog of AtSHN (At5g11190) based on BLASTP (Altschul et al., 1990).
Coexpression Network Analysis
Twenty-nine publicly available Affymetrix rice GeneChip gene expression data sets (414 samples; 150 groups after gathering biological replicate samples into single groups) were collected from NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/; Barrett et al., 2009) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/; Parkinson et al., 2009), and the largest subset of experiments (10 data sets, 129 samples, 45 groups) with a similar biological context corresponding to studies of response to some environmental condition was used for coexpression analysis (Supplemental Table S5).
Raw data were background corrected, normalized, and summarized according to the custom CDF using justRMA (Irizarry et al., 2003), and expression values were averaged across replicates. Pearson correlations were first calculated between every pair of genes (Huttenhower et al., 2008; see “Note” below), which were then Fisher z transformed (David, 1949) and standardized to get coexpression scores (zcs) with a N(0,1) distribution. This formulation was robust and highly interpretable as deviations from the expected value and even by level of significance, where |zcs| values greater than 1.645, 1.96, and 2.58 correspond to 10%, 5%, and 1% extremes of the distribution of zcs scores.
TFs of interest were connected to each other when they had a strong correlation (|zcs| > 1.645). TFs were connected to pathway genes based on a more rigorous procedure. For each set of “pathway” genes, “lignin,” and “other cell wall,” a pathway correlation matrix was first created taking the pathway genes along the columns and the pathway genes plus the putative TFs along the rows. Here, cell (i,j) contained the coexpression score zcs(i,j) between genes i and j. This was the same as the adjacency matrix of the pathway genes, only with extra rows of TFs. Thus, each row i contained the vector of zcss of gene i to all the pathway genes, measuring its “association” with the entire pathway. Pearson correlations were then calculated between rows, and TF-pathway gene pairs with absolute correlation greater than 0.8 were selected. This correlation measures how well the genes agree with the regulatory program of the pathway. TF-TF edges were left undirected, while TF-pathway gene edges were directed from TF to a putative target. Among the pathway genes, only those regulated by SHN (from the expression studies) were included in the final network. The network was visualized using Cytoscape version 2.7.0 (Shannon et al., 2003) and is available as Supplemental File S1, which can be imported into Cytoscape.
Note: There are popular methods to derive regulatory networks from gene expression data based on the calculation of mutual information (MI), for example, ARACNE (Basso et al., 2005) and CLR (Faith et al., 2007), that perform better than simple correlation-based methods. But these methods require very large amounts of gene expression data to contain expression of the genes across a large dynamic range to calculate MI reliably. Hence, with relatively less data in rice, especially when considering those similar in biological context, using MI-based methods would not be possible. Then, for measuring simple correlations, the Spearman rank correlation metric is a good choice. However, given that we have carefully chosen data sets similar in biological context, identical in experimental platform, and resolved ambiguity in hybridization using a redefinition of probe-gene mapping in the array, we sought to use a metric more sensitive to the actual expression values, like the Pearson correlation coefficient, rather than one that works on the relative ranks of the values, like the Spearman rank correlation coefficient.
Promoter Analysis
For promoter analysis in rice, FIRE (Elemento et al., 2007) was used to discover motifs specific to the cell wall pathway genes by comparing the motif content of 1-kb upstream sequences of these genes with that of the rest of the genome. Briefly, FIRE seeks to discover motifs whose patterns of presence/absence across all considered regulatory regions (motif profile) are most informative about the expression of the corresponding genes (expression profile). The sole reported motif, CACCA[ACG]NC[AC], was compared with known cis-elements in public databases (Higo et al., 1999; Crooks et al., 2004; Mahony and Benos, 2007) to identify it as one similar to the Arabidopsis AC II element. This de novo approach was taken since cis-regulatory motifs could diverge quickly across species, making them hard to find simply by searching. A Perl script was used to search for GCC box motifs ([AG]CCGNC) in the 1-kb upstream sequences of SHN-regulated TFs.
Expression of AtSHN in Escherichia coli, Protein Purification, and Electrophoretic Mobility Shift Assays
The AtSHN coding region was PCR amplified using proofreading DNA polymerase with a gene-specific forward primer carrying the NdeI restriction site (5′-GGAATTCCATATGGTACATTCGAGGAAGTTCC-3′) and the reverse primer with the EcoRI restriction site (5′-GGCCGAATTCTCAACTCCAATTCAGCAAC-3′). The amplified PCR product was gel purified, digested with NdeI and EcoRI, and cloned into the corresponding sites of pET28a(+) with an N-terminal His tag fusion. The sequence confirmed that the construct was transformed into the E. coli strain BL-21 (DE3) and used for the induction and purification of the recombinant protein. The recombinant AtSHN protein from induced cells was purified using nickel-chelate agarose affinity chromatography (Qiagen), and the identity of the purified protein was confirmed by western blotting (data not shown) using the His tag antibody.
The binding reaction and electrophoretic mobility shift assays were carried out using a standard protocol according to the manufacturer’s instructions (LightShift Chemiluminescent EMSA Kit). Specific sets of primers (Supplemental Table S6) were designed for 100- to 250-bp flanking regions of each putative SHN-responsive promoter sequence of the OsMYB and OsNAC genes, and the promoter sequences were PCR amplified using Nipponbare rice genomic DNA as a template. The amplified promoter fragments were biotin labeled at the 3′ end using the Biotin 3′ End DNA Labeling Kit (Pierce). The binding reaction was carried out in a buffer containing 10 mm Tris, pH 7.5, 50 mm KCl, 1 mm dithiothreitol, 2.5% glycerol, 5 mm MgCl, 0.05% Nonidet P-40, and 50 ng μL−1 poly(dI-dC). For competition analysis, the binding reaction was incubated for 10 min on ice before adding 10- or 100-fold excess of unlabeled competitor DNA, and the reaction mixture was further incubated for 20 min at room temperature before loading onto a 5% native polyacrylamide gel. The resolved DNA-protein complexes were electroblotted onto nylon membranes and subsequently detected using the chemiluminescence detection kit.
The rice genome annotation or the locus identifier numbers for the genes investigated in this study are as follows: OsSHN (Os06g40150), OsCAD1 (Os10g11810), OsCAD2 (Os02g09490), OsCAD3 (Os10g29470), OsCAD4 (Os11g40690), OsCAD5 (Os08g16910), OsCAD6 (Os04g15920), OsCAD7 (Os04g52280), OsCAD8A (Os09g23530), OsCAD9 (Os03g12270), Os4CL3 (Os02g08100), Os4CL5 (O08g34790), Os4ACS5 (Os01g67530), Os4ACS6 (Os01g67540), polygalacturonase (Os03g03350), mannosidase (Os11g32260), β-1,3-glucanase (Os01g64170), β-1,4-glucanase (Os02g50040), glucosyltransferase (Os08g23780), CESA1 (Os05g08370), CESA2 (Os03g59340), CESA3 (Os07g24190), CESA4 (Os01g54620), CESA5 (Os03g62090), CESA6 (Os07g14850), CESA7 (Os10g32980), CESA8 (Os07g10770), CESA9 (Os09g25490), CESA10 (Os12g29300), CESA11 (Os06g39970), OsCSLA6 (Os02g51060), OsCSLC2 (Os09g25900), OsCSLE2 (Os02g49332), OsCSLH2 (Os04g35020), OsCSLD1 (Os10g42750), OsCSLF7 (Os10g20260), MYB20/43 (Os02g49986), MYB52/54 (Os03g51110), MYB48/59 (Os12g37970), MYB85 (Os09g36250), MYB58/63 (Os02g46780), MYB58/63 (Os04g50770), VND4/5/6 (Os06g01480), NST1/2/SND1 (Os06g04090), and NST1/2/SND1 (Os08g02300). The gene expression data reported here are available from the NCBI GEO database with the accession number GSE26092.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Overview of the monolignol biosynthesis pathway.
Supplemental Figure S2. qRT-PCR expression analysis of cell wall-related genes in SHN leaf and culm.
Supplemental Figure S3. Gene expression and regulation of lignin biosynthetic pathway genes.
Supplemental Table S1. Gene expression data for SHN-regulated lignin and other cell wall biosynthetic genes and putative TFs.
Supplemental Table S2. Breaking force of rice wild-type and AtSHN lines.
Supplemental Table S3. Differential expression levels of cell wall genes in WIN1 overexpression lines (Broun et al., 2004).
Supplemental Table S4. Expression level of Os06g40150 (OsSHN) in various developmental stages obtained from RiceAtlas (http://bioinformatics.med.yale.edu/riceatlas/).
Supplemental Table S5. Rice microarray data sets used for coexpression analysis.
Supplemental Table S6. The primer sequences used to amplify the promoter regions of the MYB and NAC TFs.
Supplemental File S1. The rice cell wall coexpression network presented in Figure 5 provided as a Cytoscape-readable (.cys) file for further exploration.
Supplementary Material
Acknowledgments
We give special thanks to Dr. Sang-Wook Park (Virginia Bioinformatics Institute) for help with the recombinant protein expression. We thank Dr. John Ralph (U.S. Department of Agriculture-Agricultural Research Service) for the gift of lignin monomer standards and Dr. Aarati Karaba and Dr. Shital Dixit (Wageningen University and Research Centre) for sharing results on the agronomic performance of the rice genotypes used. We also thank Dr. Utlwang Batlang (Virginia Bioinformatics Institute) for help in growing plants, Dr. John Fike (Crop and Soil Environmental Sciences) for help with lignin analysis, and Kathy Lowe (Morphology Laboratory) for help in the use of the microscope facility. We are grateful to Dr. Olivier Elemento (Weill Medical College of Cornell University), Dr. Manhong Dai (University of Michigan), and Dr. Curtis Huttenhower (Harvard School of Public Health), respectively, for discussions on using FIRE, building the custom CDF, and using Sleipnir.
References
- Adkins NL, Hagerman TA, Georgel P. (2006) GAGA protein: a multi-faceted transcription factor. Biochem Cell Biol 84: 559–567 [DOI] [PubMed] [Google Scholar]
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albersheim P, Nevins DJ, English PD, Karr A. (1967) A method for the analysis of sugars in plant cell-wall polysaccharides by gas-liquid chromatography. Carbohydr Res 5: 340–345 [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ambavaram MM, Pereira A. (2010) Setting up reverse transcription quantitative-PCR experiments. Methods Mol Biol 678: 45–54 [DOI] [PubMed] [Google Scholar]
- Atanassova R, Favet N, Martz F, Chabbert B, Tollier MT, Monties B, Fritig B, Legrand M. (1995) Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J 8: 465–477 [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. (2009) NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 37: D885–D890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. (2005) Reverse engineering of regulatory networks in human B cells. Nat Genet 37: 382–390 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 101: 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casneuf T, Van de Peer Y, Huber W. (2007) In situ analysis of cross-hybridisation on microarrays and the inference of expression correlation. BMC Bioinformatics 8: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching A, Dhugga KS, Appenzeller L, Meeley R, Bourett TM, Howard RJ, Rafalski A. (2006) Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 224: 1174–1184 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. (2005) Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David FN. (1949) The moments of the Z and F distributions. Biometrika 36: 394–403 [PubMed] [Google Scholar]
- Dhugga KS. (2007) Maize biomass yield and composition for biofuels. Crop Sci 47: 2211–2227 [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. (2007) A universal framework for regulatory element discovery across all genomes and data types. Mol Cell 28: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. (2007) Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol 5: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M. (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: carbohydrates. J Vis Exp 37: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu FF, Xue HW. (2010) Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol 154: 927–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe R, Boyle P, Dulude A, Roy V, Lezin-Doumbou C, Kaur GS, Bouarab K, Després C, Brisson N. (2008) The transcriptional activator Pti4 is required for the recruitment of a repressosome nucleated by repressor SEBF at the potato PR-10a gene. Plant Cell 20: 3136–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco R, Ouwerkerk PB, Sallaud C, Kohli A, Colombo L, Puigdomènech P, Guiderdoni E, Christou P, Hoge JH, Pereira A. (2001) Transposon insertional mutagenesis in rice. Plant Physiol 125: 1175–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP. (2001) Carbon partitioning to cellulose synthesis. Plant Mol Biol 47: 29–51 [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai CJ, Chiang VL. (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17: 808–812 [DOI] [PubMed] [Google Scholar]
- Huttenhower C, Schroeder M, Chikina MD, Troyanskaya OG. (2008) The Sleipnir library for computational functional genomics. Bioinformatics 24: 1559–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. (1996) R: a language for data analysis and graphics. J Comput Graph Statist 5: 299–314 [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. (2009) Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21: 3493–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jakob K, Zhou FS, Paterson A. (2009) Genetic improvement of C4 grasses as cellulosic biofuel feedstocks. In Vitro Cell Dev Biol Plant 45: 291–305 [Google Scholar]
- Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, Ceserani T, Chen M, Ma L, Holford M, et al. (2009) A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet 41: 258–263 [DOI] [PubMed] [Google Scholar]
- Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, Höfte H, Pauly M, Riechmann JL, Broun P. (2007) The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaba A. (2007) Improvement of water use efficiency in rice and tomato using Arabidopsis wax biosynthetic genes and transcription factors. PhD thesis Wageningen University, Wageningen, The Netherlands [Google Scholar]
- Kokubo A, Kuraishi S, Sakurai N. (1989) Culm strength of barley: correlation among maximum bending stress, cell wall dimensions, and cellulose content. Plant Physiol 91: 876–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Hogues H, Mallick J, Sellam A, Nantel A, Whiteway M. (2010) Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol 8: e1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhou Y, Cheng X, Sun J, Marita JM, Ralph J, Chiang VL. (2003a) Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc Natl Acad Sci USA 100: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Weng JK, Chapple C. (2008) Improvement of biomass through lignin modification. Plant J 54: 569–581 [DOI] [PubMed] [Google Scholar]
- Li X, Yang Y, Yao J, Chen G, Li X, Zhang Q, Wu C. (2009) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69: 685–697 [DOI] [PubMed] [Google Scholar]
- Li Y, Qian Q, Zhou Y, Yan M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, et al. (2003b) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15: 2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu FC, Ralph J. (1997) Derivatization followed by reductive cleavage (DFRC method), a new method for lignin analysis: protocol for analysis of DFRC monomers. J Agric Food Chem 45: 2590–2592 [Google Scholar]
- Lu FC, Ralph J. (1998) The DFRC method for lignin analysis. 2. Monomers from isolated lignins. J Agric Food Chem 46: 547–552 [DOI] [PubMed] [Google Scholar]
- Mahony S, Benos PV. (2007) STAMP: a Web tool for exploring DNA-binding motif similarities. Nucleic Acids Res 35: W253–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U. (2003) Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA 100: 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele G, Ori N, Sato Y, Hake S. (2003) The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev 17: 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima J, Chen F, Jackson L, Shadle G, Dixon RA. (2008) Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): effects on lignin composition in specific cell types. New Phytol 179: 738–750 [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35: D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. (2009) ArrayExpress update: from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res 37: D868–D872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzak M, Shillito RD, Hohn T, Potrykus I. (1986) Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res 14: 5857–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. (1997) ImageJ. National Institutes of Health, Bethesda, MD [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL. (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Scott TA, Melvin EH. (1953) Determination of dextran with anthrone. Anal Chem 25: 1656–1661 [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu A, Langewisch T, Olek A, Multani DS, McCann MC, Vermerris W, Carpita NC, Johal G. (2007) Maize Brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol 145: 1444–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3 [DOI] [PubMed] [Google Scholar]
- Somerville C. (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Sticklen M. (2006) Plant genetic engineering to improve biomass characteristics for biofuels. Curr Opin Biotechnol 17: 315–319 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer RA, Robinson-Rechavi M. (2009) How confident can we be that orthologs are similar, but paralogs differ? Trends Genet 25: 210–216 [DOI] [PubMed] [Google Scholar]
- Taketa S, Amano S, Tsujino Y, Sato T, Saisho D, Kakeda K, Nomura M, Suzuki T, Matsumoto T, Sato K, et al. (2008) Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc Natl Acad Sci USA 105: 4062–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C. (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H. (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol 133: 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias CM, Chow EK. (2005) Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta 220: 678–688 [DOI] [PubMed] [Google Scholar]
- Trijatmiko KR. (2005) Comparative analysis of drought resistance genes in Arabidopsis and rice. PhD thesis Wageningen University, Wageningen, The Netherlands [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Usadel B, Obayashi T, Mutwil M, Giorgi FM, Bassel GW, Tanimoto M, Chow A, Steinhauser D, Persson S, Provart NJ. (2009) Coexpression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ 32: 1633–1651 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Ralph J, Boerjan W. (2008) Lignin engineering. Curr Opin Plant Biol 11: 278–285 [DOI] [PubMed] [Google Scholar]
- Van Soest PJ. (1967) Development of a comprehensive system of feed analysis and its application to forages. J Anim Sci 26: 119–128 [Google Scholar]
- Vermerris W, Sherman DM, McIntyre LM. (2010) Phenotypic plasticity in cell walls of maize brown midrib mutants is limited by lignin composition. J Exp Bot 61: 2479–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Lynch VJ. (2008) The gene regulatory logic of transcription factor evolution. Trends Ecol Evol 23: 377–385 [DOI] [PubMed] [Google Scholar]
- Wang X, Haberer G, Mayer KF. (2009) Discovery of cis-elements between sorghum and rice using co-expression and evolutionary conservation. BMC Genomics 10: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. (2004) Genomic basis for cell-wall diversity in plants: a comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol 45: 1111–1121 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. (2007) Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445: 936–940 [DOI] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye ZH. (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18: 3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. (2009) Transcriptional regulation of lignin biosynthesis. Plant Signal Behav 4: 1028–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH. (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.