Abstract
Circumsporozoite (CS) protein is a malaria antigen involved in sporozoite invasion of hepatocytes, and thus considered to have good vaccine potential. We evaluated the polymorphism of the Plasmodium vivax CS gene in 24 parasite isolates collected from malaria-endemic areas of Colombia. We sequenced 27 alleles, most of which (25/27) corresponded to the VK247 genotype and the remainder to the VK210 type. All VK247 alleles presented a mutation (Gly → Asn) at position 28 in the N-terminal region, whereas the C-terminal presented three insertions: the ANKKAGDAG, which is common in all VK247 isolates; 12 alleles presented the insertion GAGGQAAGGNAANKKAGDAG; and 5 alleles presented the insertion GGNAGGNA. Both repeat regions were polymorphic in gene sequence and size. Sequences coding for B-, T-CD4+, and T-CD8+ cell epitopes were found to be conserved. This study confirms the high polymorphism of the repeat domain and the highly conserved nature of the flanking regions.
Introduction
Circumsporozoite (CS) protein is an immunodominant antigen abundantly expressed on the sporozoite surface of all Plasmodium species studied to date.1 Humans rendered immune by vaccination with radiation-attenuated malaria sporozoites develop antibodies and lymphocytes that recognize this protein.2
The protein has a typical domain organization among all known Plasmodium species,3,4 with a central region (CR) composed of a tandem repeat sequence that comprises ∼40% of the protein. The CR is flanked by conserved pre-repeat (5′NR) and post-repeat (3′NR) regions.5 These flanking regions contain short, highly conserved sequences denoted as Region I (RI) and RII regions,3 that are the binding domains for glycosaminoglycan heparin sulfate receptors, which are found on the surface of hepatocytes6 and mosquito salivary glands.7 The RI and RII domains appear to play important roles in parasite invasion to host cells, both in the mosquito and the vertebrate host.7,8
The CR of Plasmodium vivax CS protein has been shown to be dimorphic, characterized by tandem repeats of the nonapeptide GDRADGQPA in the VK210 sequence,5 and ANGAGNQPG that corresponds to the VK247 variant sequence.9 Both nonapeptides sequences are repeated ∼20 times in their corresponding proteins; however, there is extensive polymorphism with regard to the number of repeats.10 It has been shown that the prevalent phenotype of the P. vivax parasite in the study sites of Colombia is VK247 (59.7%), whereas VK210 accounts for one-third of the cases 32.8%; the remaining 7.5% corresponds to mixed infection.11
Although previous nucleotide sequence analyses of the flanking regions (3′NR and 5′NR) of P. vivax CS protein have shown high conservation of these regions, recent studies indicated that isolates from Iran, Philippines, China, Brazil, and Korea contained previously undescribed point mutations and an insertion at the beginning of the 3′NR region.10,12–14 Most studies have reported only partial sequence data, thus limiting complete evaluation of the extent of genetic polymorphism found in the gene encoding P. vivax CS protein. Thus, there is a shortage of information on the polymorphism present in relevant immune epitopes, particularly those localized in the flanking regions. We report here a detailed sequence analysis of the full P. vivax CS gene, designed to determine the potential for P. vivax gene polymorphism in such epitopes and their relevance for malaria vaccine development.
Materials and Methods
Origin of blood samples.
Blood samples were collected from 24 subjects attending outpatient clinics in five malaria-endemic areas of Colombia, where transmission of both P. vivax and P. falciparum is unstable. Sites selected for parasite collection were Quibdó (Chocó state), Buenaventura (Valle del Cauca state), Guapi (Cauca state), and Tumaco (Nariño state), located along the Pacific Coastal; and Puerto Asís (Putumayo state), located beyond the Andes Mountains in the Amazonian region (Figure 1). Written informed consent was obtained from volunteers and blood samples were collected before therapy was initiated. Approximately 3 mL of whole blood were collected in EDTA-containing tubes from each individual confirmed to be P. vivax positive by thick smear. Additionally, 1 mL of blood was obtained from an Aotus monkey infected with the reference Sal I strain of P. vivax.
Figure 1.
Map of Colombia depicting the geographical sites where the 24 parasite isolates were collected: Quibdo (Chocó) N = 5; Buenaventura (Valle del Cauca) N = 5; Guapi (Cauca) N = 5; Tumaco (Nariño) N = 5; and Puerto Asís (Putumayo) N = 4.
Parasite DNA extraction and polymerase chain reaction (PCR) amplification of P. vivax CS protein gene.
Parasite genomic DNA was extracted by the salting-out method.15 The DNA samples were coded according to collection site as follows: (Chocó [Ch], Valle del Cauca [Vc], Cauca [Ca], Nariño [Nr], and Putumayo [Pt]) followed by two numerical digits indicating the order of patient arrival. The DNA samples were immediately stored at −20°C and processed by nested PCR to confirm the species-specificity of infection.16
The CS gene was subsequently amplified by PCR using primers CS1 (5′-cagccaaaggctacaagtgtaaac-3′) and CS2 (5′-gggcaagtatttatgtgcatgt-3′). The PCR was performed using the following mixture: 2 μL DNA, 0.125 mM primers, 2.5 U/μL of Platinum Taq DNA Polymerase High Fidelity (Invitrogen Inc., São Paulo, SP, Brazil), and 0.125 mM dNTPs (Invitrogen Corp., São Paulo, Brazil), for a final volume of 20 μL. Reaction cycles comprised an initial denaturation for 5 min at 94°C, then 30 cycles of 1 min at 94°C; followed by cycles of 1 min at 64°C and 1 min at 72°C; and a final 10 min extension step at 72°C. Amplified products (»1,238 bp) were visualized by electrophoresis on 1.2% agarose gels.
Cloning and sequencing.
The PCR products were cloned into the pGEMT Easy Vector (pGEM-T Easy Vector Systems, Promega Corp., Madison, WI) system according to the manufacturer's instructions. Isolation of plasmid DNA was extracted at Minipreparation scale as described previously by Sambrook and others.17
Screening for positive clones (∼1.2 kb gene P. vivax CS protein) was performed by restriction enzyme EcoRI and the digested products were separated on 1.5% agarose gel, stained with ethidium bromide, and visualized under UV. Nucleotide sequences of recombinant clones were determined by the dideoxynucleotide chain termination method with Sequenase and the BigDye terminator sequence kit, version 3.1 (using an ABI PRISM 3100 Avant sequencer, Applied Biosystems, Foster City, CA). The M13F/M13R universal primers, CS1 and CS2 external primers, and PvF2 and PvR2 internal primers were used for sequencing.18
Data analysis.
The DNA from the P. vivax Sal I strain, previously adapted to growth in Aotus monkeys, was produced and used as reference. All sequences of the 5′NR and 3′NR regions were compared with the reference sequence Sal I using Clustal W (European Bioinformatics Institute, Hinxton, Cambridge, UK), with corrections made by visualization. For analysis of the central region, comparisons were made using as references the VK247 databank sequences (M69059) reported previously for the P. vivax Papua New Guinea (PNG) strain phenotype, and the Sal I sequence (VK210).
Sequences were deposited in Genbank with accession nos. GU339059–GU339086. Basic genetic polymorphism parameters were estimated using the DNAsp program.19 The Colombian alleles reported in this work were compared with 14 complete sequences available in GenBank corresponding to the worldwide distribution of P. vivax. This sample of 14 sequences will be further referred to as the global sample.
Evaluation of polymorphism in immunologically relevant protein regions (epitopes) for B-, T-CD4+, and T-CD8+ cells were compared by visual inspection of aligned sequences with sequences of the epitopes previously identified and reported by our group (Table 1).20–22
Table 1.
Specific amino acid sequence of dominant B- and T-cell epitopes of the Plasmodium vivax circumsporozoite (CS) protein
| Peptide | Amino acid sequences | Position | Classification | Reference |
|---|---|---|---|---|
| P6 | HVGQSASRGRGLGENPDDEE | 50–70 | T-Helper | 20 |
| P11 | GDRADGQPA(VK210) | 114–122 | ||
| P15 | GDRAAGQAA (VK210) | 150–158 | ||
| P25 | VRRRVNAANKKPEDLTLNDL | 246–266 | ||
| P8 | GDAKKKKDGKKAEPKNPREN | 71–90 | B-Cell | 21 |
| P11 | GDRADGQPA (VK210) | 96–104 | ||
| P24 | CSVTCGVGVRVRRRVNAANK | 332–351 | ||
| P25 | VRRRVNAANKKPEDLTLNDL ANGAGNQPG (VK247) | 342–361 | ||
| PV1 | YLDKVRATV | 301–309 | T-CD8+ | 22 |
| PV3 | SLGLVILLVL | 365–374 | ||
| PV5 | TLNDLETDV | 341–349 | ||
| PV6 | LLAVSSILL | 6–4 |
Results
Results of the nested PCR for P. vivax and P. falciparum species indicated that all samples corresponded to simple P. vivax infection. The CS genes were successfully amplified from parasite genomic DNA samples obtained from 24 samples of patients and one of Aotus monkeys. All PCR products corresponded to DNA fragments of ∼1.2 Kb.
After cloning, all recombinant plasmids were subjected to enzymatic digestion that resulted in two fragments: the first, a 3.018 bp fragment corresponding to the plasmid vector; and a second fragment of ±1.182 bp corresponding to the CS gene. Digestion of recombinant plasmids from Puerto Asís (Putumayo) presented CS fragments ranging from ±880 to 1.182 bp. All CS fragments were subjected to automated DNA sequencing.
Sequencing data indicated that some parasite isolates presented with more than one CS allele. Three of four parasite isolates from Putumayo showed different sized fragments for the complete CS gene. The Pt04 sample presented two alleles both belonging to the phenotype VK247: Pt04.1 (1.134 bp) (GU339085) and Pt04.5 (840 bp) (GU339086); the Pt02 sample had the following alleles: Pt02.1 (1.164 bp) (GU339082) and Pt02.2 (840 bp) (GU339083); and finally Pt01 isolates showed two alleles: Pt01.3 (894 bp) (GU339080) and Pt01.7 (1.137 bp) (GU339081), both phenotypes VK210.
Overall, P. vivax CS protein was highly conserved when only the non-repetitive regions of the gene were considered; the 5′NR (285 bp) and 3′NR (291 bp) regions together have only an estimated nucleotide diversity of 0.0021 for all available sequences; the 27 from Colombia as reported in this study and 14 from the global sample. The number of synonymous (0.00182) substitutions was higher than the number of non-synonymous substitutions (0.00224) as estimated by the Nei-Gojobori method implemented in DNAsp. The differences were not statistically significant; thus, in contrast with the P. falciparum CS, no evidence of balancing selection was found.23,24
Comparison of the 27 sequences corresponding to the flanking 5′NR and 3′NR regions with the Sal I reference sequence (GU339059) displayed limited genetic polymorphism. The 5′NR region (285 bp) has only two polymorphic sites with an estimated nucleotide diversity (π) of 0.001; 25/27 isolates showed two non-synonymous transitions in the first and second positions of codon 38 AAC → GGC and resulted in an N → G substitution; only the Nr03 and Pt04.1 alleles (GU339072, GU339085) are conserved in this codon. This level of variation is comparable with the one observed in the global sample that has only three polymorphic sites and a π = 0.004. In the 3′NR no polymorphic sites were observed, whereas in the global sample, eight polymorphic sites were observed with a π = 0.005. Both, the Colombian isolates and the global sample, have extensive polymorphism in the form of insertions of 9, 16, and 28 aa in length. The phylogenetic relationships among 40 P. vivax CS protein isolates are shown in Figure 2.
Figure 2.
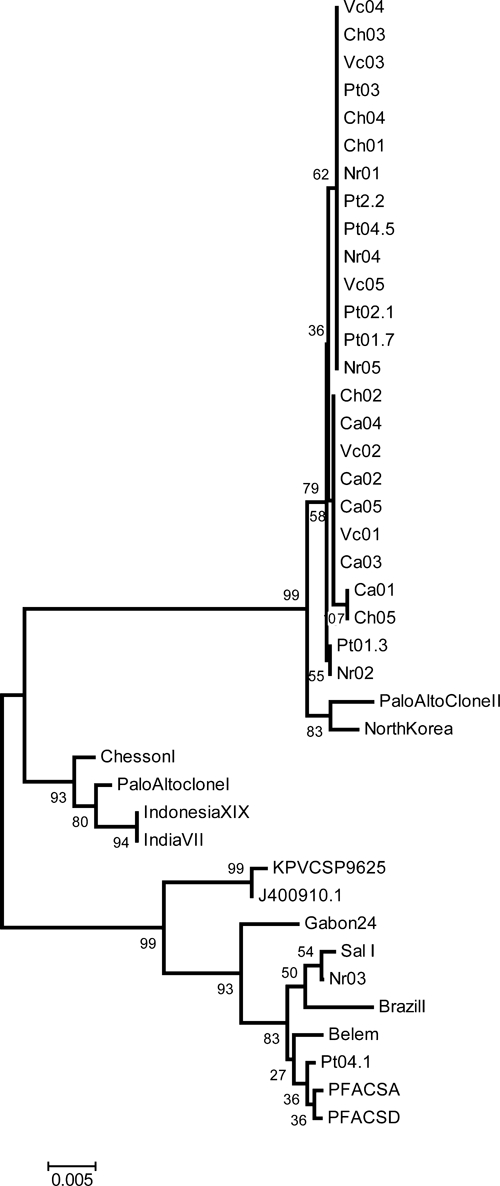
Neighbor-joining tree with a Jukes-Cantor distance. It includes 27 Colombia alleles and a few complete Plasmodium vivax circumsporozoite (CS) protein sequences available in the literature. Clades support was estimated using Bootstrap with 500 pseudo-replications shown as percentages.
In the 3′NR region, a first group of five isolates: Ch02(GU339066), Ch05(GU339066), Ca02(GU339061), Ca04(GU339063), and Ca05(GU339064) had a 16 aa insertion. A second group of eight isolates: Ch03(GU339067), Nr01(GU339070), Nr02(GU339071), Nr05(GU339074), Vc01(GU339075), Vc02(GU339076), Ca01(GU339060), and Ca03(GU339062) had a nine aa insertion; and in the third group of 12 isolates: Ch01(GU339065), Ch04(GU339068), Nr04(GU339073), Vc03(GU339077), Vc04(GU339078), Vc05(GU339079), Pt01.3(GU339080), Pt01.7(GU339081), Pt02.1(GU339082), Pt02.2(GU339083), Pt03(GU339084), and Pt04.5(GU339086) displayed an 18 aa insertion (Figure 3).
Figure 3.

Amino acid sequence alignment of Plasmodium vivax circumsporozoite (CS) amino- and carboxyl-flanking regions of Colombian and of the global sample Sal I reference sequence.
In the CR fragment of the protein, the number of tandem nonapeptide repeats ranged between 8 and 20 among the different 27 alleles. Several synonymous and non-synonymous point mutations were found in the corresponding coding regions without any particular organization (Table 2). Twenty-five (92.6%) of the 27 isolates from the Pacific Coast and from Putumayo state presented with all CS alleles corresponding to the VK247 type, whereas the Nr03 and Pt04.1 isolates (7.4%) displayed alleles corresponding to the common VK210 type. The translated sequence of the VK247 genotype repeats was characterized by the nonapeptide EDGAGDQPG, followed by 4–18 tandem nonapeptide repeats of ANGA(G/D/K)(N/D)QPG, and the terminal nonapeptide ANGAGGQAA. Conversely, on the other hand the Nr03(GU339072) and Pt04.1(GU339085) alleles had 20 nonapeptide repeats of G(D/N)(R/G)A(D/A/G)GQ(P/A)A (Table 2).
Table 2.
Structural organization of the amino acid sequence in the circumsporozoite (CR) of Colombian alleles
| Strains | Central repetitive regions | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
| PNG* | 1 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 |
| Vc01 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Vc02 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 3 | 2 | 2 | 4 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Vc03 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 3 | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – | – | – |
| Vc04 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 4 | 2 | 2 | 4 | 4 | 2 | 4 | 2 | 2 | 5 | – | – | – |
| Vc05 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 4 | 4 | 2 | 2 | 2 | 2 | 5 | – | – | – |
| Ch01 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Ch02 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Ch03 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 2 | 2 | 4 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 |
| Ch04 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 3 | 2 | 2 | 4 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Ch05 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Ca01 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Ca02 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Ca03 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 2 | 2 | 4 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Ca04 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 3 | 2 | 2 | 4 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Ca05 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Nr01 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 3 | 2 | 2 | 4 | 4 | 2 | 4 | 2 | 2 | 7 | 2 | 5 | – |
| Nr02 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 4 | 2 | 2 | 4 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Nr04 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 8 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 5 | – | – | – |
| Nr05 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – |
| Pt01.3 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 5 | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Pt01.7 | 6 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – | – | – | – |
| Pt02.1 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – | – | – |
| Pt02.2 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 5 | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Pt03 | 6 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 5 | – | – | – |
| Pt04.5 | 1 | 2 | 2 | 2 | 2 | 5 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Sal I† | 9 | 9 | 9 | 9 | 10 | 9 | 9 | 9 | 9 | 10 | 10 | 9 | 10 | 9 | 10 | 9 | 10 | 10 | 11 | 11 | |
| Nr03 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 10 | 10 | 10 | 12 | |
| Pt04.1 | 9 | 9 | 9 | 10 | 10 | 9 | 9 | 9 | 9 | 10 | 10 | 9 | 10 | 9 | 10 | 9 | 10 | 10 | 11 | 11 | |
1‡ = EDGAGNQPG 3 = ANGADDQPG 5 = ANGAGGQAA 7 = ANGAGGQPG 9 = GDRADGQPA 11 = GDRAAGQAA
2 = ANGAGNQPG 4 = ANGAGDQPG 6 = EDGAGDQPG 8 = ANGADNQPG 10 = GDRAAGQPA 12 = GNGAGGQAA
Comparison of central repeats of VK210-type Colombian alleles with Sal I reference sequence.
Numbers correspond to repeat type sequence located in repeat position 1–21.
Alignment of sequences coding for defined B, T-CD4+, and T-CD8+ epitopes, described previously,20–22 indicated complete sequence homology to P6, P8, and PV6 epitopes (Figure 3). The central repeat region of the VK210 type found in isolates Pt04.1(GU339085) and Nr03(GU339072) showed homology to the P11 epitope. Only the P15 epitope presented polymorphism in the eighth position (A → P) in the isolate Nr03.
The remaining 25 sequences belonging to the VK247 showed a high degree of homology with variant B-cell epitope ANGAGNQPG. Finally, the 3′NR region of all parasite isolates showed homology in the following epitopes: P25 (T-CD8+), P24, P25 (B-cell); PV1, PV5, and PV3 (T-CD8+) (Figure 3).
Discussion
Our results confirm previous observations that, when only single point mutations are considered, P. vivax CS protein is highly conserved when compared with its homologous gene in P. falciparum. Nevertheless, our results also confirm the existence of extensive polymorphism of the P. vivax CS gene in terms of the number of tandem repeats, particularly in the CR region. The study also confirmed the existence of both VK247 and VK210 CS repeat types in Colombia with a strong predominance of the VK247 type (92.6%), as described previously.11 It is hard to explain the predominance of the VK247 allelic type in Colombia, because in the same regions anti-CS VK210 antibodies are more frequent (68–75%) than those to VK247 (11–20%). We had previously speculated that the VK210 allelic type is more immunogenic than VK247 and that therefore the latter type would be inmunologically selected, however, we do not have formal evidence of this mechanism and studies conducted on P. falciparum CS polymorphism do not appear to confirm this immune selection hypothesis.11,25 A significant size polymorphism was observed for the entire gene that ranged from 840 to 1182 Kb, which depended mainly on the size variability of the CR region. Additionally, the 3′NR region displayed several insertions at the beginning of the sequence.
Size polymorphism was based on the 3′NR variability in number of insertions in the Colombian isolates (NKKAGDA, GAGGQAAGGNAANKKAGDAG, and GGNAGGNA), and in those preceding the CR region. This latter region is usually composed of 19–20 repeat of a nonapeptides G(D/N)(R/G)A(D/A/G)GQ(P/A)A in the case of the VK210 type and ANGA(G/D/K)(N/D)QPG in the VK247; the latter isolates showing variation with 8–20 repeats in this study.
In terms of analyses to determine the immunological potential of this protein for vaccine development, it is interesting that the 5′NR terminal region is well conserved across the isolates analyzed. The gene fragment analyzed here encoded the protein sequence corresponding to 1–95 aa that comprises two T-helper epitopes, P6 (50–70 aa) and P8 (71–90 aa), plus a T-CD8+ cell epitope (PV6, 6–14 aa), which may explain the good immunogenicity described for this fragment.20–22 The non-synonymous substitutions in codon 38 leading to the amino acid mutation (N → G), had been reported previously in isolates from diverse regions of the world.13,26 Whether this substitution has an effect on immunity warrants further investigation; indeed, this epitope exhibits high immunogenicity, both during natural exposure to the parasite and in response to immunization with synthetic peptides.20–22 Our sequences from Colombia contrast to those sequences analyzed by Qari and others14 who identified non-synonymous substitutions that would affect the pre-repeat region, particularly the RI region, in parasites isolated from Brazil and New Guinea. Nevertheless, geographic differences are expected in a parasite with such extensive distribution.
The post-repeat region of the Colombian isolates showed three types of insertions that have already been reported in five different geographically distant regions. The first insertion of 9 aa present in eight isolates had previously been found in Papua New Guinea (M69059, M69060), Brazil (M69062, M69061), and Iran (AY632299, AY632294). A second insertion of 9 aa found in eight parasite isolates had also been seen in isolates from China (U08977, U08978, and U08979) and Korea (M206670, AJ297403, AF164605, AF164603, and AF164603). Finally, 16 aa found in 12 of the Colombian isolates had also been reported for the P30 isolated (AY632258) in Iran.10 The origin of the repeats could be the result of meiotic recombination that occurs during the diploid phase of the malaria life cycle.9
Genetic mechanisms such as slipped-strand mispairing,27 recombination, and gene conversion can be inferred from the formation and separation of such mutations,28 including expansion or reduction of repeat sequences. This is especially true in parasites of the Plasmodium genus, where the immune pressure of the vertebrate host can determine the order and characteristics of sequence repetition.29
The homology observed between 5′NR- and 3′NR-specific regions coding for epitopes recognized by B- and T-lymphocytes (CD4+ and CD8+) cells, both in Colombia and other distant geographical areas, is very encouraging for the design of a P. vivax malaria vaccine for global distribution. However, it is worth noting that the sequences deposited in the Genbank showed substitutions that would alter the sequence of some of the epitopes recognized by Colombian individuals. The G24 isolate (U09737) from Gabon presented a substitution in the epitope PV3 (P → L); the BZLB7–4 isolated (M69062) from Brazil presented a substitution in the P24 (T→A) and P25 epitopes (T →A); and Thay Nyu isolated (M34697) from Thailand presented a substitution in the epitope P15 (Q → P). The limited sequence variation found here for P. vivax is in contrast with the high level of non-synonymous mutations at the sequences encoding the Th2R and Th3R epitopes of the P. falciparum CS in African isolates considered to be the result of host immune selection,30,31 However, it appears that polymorphism, at least in gene sequences coding for T-cell epitopes are unlikely to be selected by immune pressure in the human host.25 The great sequence conservation of this region appears to be similar to that observed for P. falciparum isolates from Southeast Asia and Brazil,32 and together with the great immunogenicity and the presence of functional domains encourage its further development as vaccine subunit (Table 1).20–22
Overall, it is clear that additional comprehensive studies will be required to determine the extent of polymorphism in these and other geographical regions and its immunological impact, if any. Indeed, the influence of parasite polymorphism in the immune response has to be analyzed carefully as the epitopes are defined on the basis of recognition by sera and cells from individuals with diverse major histocompatibility complex (MHC) haplotypes. Therefore, given the diversity of class I and class II haplotypes in any given population, the epitopes recognized in different populations may differ on the basis of the frequency of the MHC haplotypes in endemic areas.
At this point, it is also worth noting that regardless of the growing interest in P. vivax, there have been only 14 complete sequences reported in the literature. Although it is understandable that genetic studies on antigens focus mostly on genes of interest, the malaria research community should consider comprehensive investigations to better understand the polymorphism of potential vaccine candidates. Overall, detailed studies on both the polymorphism of this antigen and the identification of relevant epitopes recognized by individuals from the same endemic areas are required to advance the evaluation of CS as a potential component of an effective malaria vaccine.
ACKNOWLEDGMENTS
We thank Ivan M. Perafán (Cauca), Pilar Pérez (Nariño), Diva Palacios (Chocó), the team from the Programa de Enfermedades Tropicales (PET), and the community of Buenaventura for providing infected samples for this study. We thank the entomology group of the Instituto de Inmunología del Valle (Valle) for their help in obtaining the P. vivax samples we used. We also thank Claudia L. Garcia and Juan F. Delgado for helping with the manuscript preparation.
Footnotes
Financial support: This work was supported by grants from National Institute of Health (NIH), the Instituto Colombiano Francisco José de Caldas para la Ciencia y la Tecnología COLCIENCIAS, and the Colombian Ministry for Social Protection (contract no. 216-2006) and through an International Center of Excellence for Malaria Research NIAID/ICEMR grant no U 19AI089702.
Authors' addresses: Miguel Ángel Hernández Martínez, Myriam Arévalo-Herrera, and Sócrates Herrera, Instituto de Inmunología, Edificio de Microbiología, Facultad de Salud, Universidad del Valle and Centro Internacional de Vacunas, Cali, Colombia, E-mails: miangher@hotmail.com, marevalo@inmuno.org, and sherrera@inmuno.org. Ananías A. Escalante, School of Life Sciences, Arizona State University, Tempe, AZ, E-mail: Ananias.Escalante@asu.edu.
Reprint requests: Sócrates Herrera, Malaria Vaccine and Drug Development Center, Carrera 37 - 2Bis No. 5E - 08, Cali, Colombia, E-mail: sherrera@inmuno.org.
References
- 1.Yoshida N, Nussenzweig RS, Potocnjak P, Nussenzweig V, Aikawa M. Hybridoma produces protective antibodies directed against the sporozoite stage of malaria parasite. Science. 1980;207:71–73. doi: 10.1126/science.6985745. [DOI] [PubMed] [Google Scholar]
- 2.Clyde DF, Most H, McCarthy VC, Vanderberg JP. Immunization of man against sporozite-induced falciparum malaria. Am J Med Sci. 1973;266:169–177. doi: 10.1097/00000441-197309000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Dame JB, Williams JL, McCutchan TF, Weber JL, Wirtz RA, Hockmeyer WT, Maloy WL, Haynes JD, Schneider I, Roberts D, Sanders GR, Reddy EP, Diggs CL, Miller LH. Structure of the gene encoding the immunodominant surface antigen on the sporozoite of the human malaria parasite Plasmodium falciparum. Science. 1984;225:593–599. doi: 10.1126/science.6204383. [DOI] [PubMed] [Google Scholar]
- 4.Escalante A, Barrio E, Ayala FJ. The phylogeny of the Plasmodium species based on the circumsporozoite protein gene. Mol Biol Evol. 1995;12:616–626. doi: 10.1093/oxfordjournals.molbev.a040241. [DOI] [PubMed] [Google Scholar]
- 5.Arnot DE, Barnwell JW, Tam JP, Nussenzweig V, Nussenzweig RS, Enea V. Circumsporozoite protein of Plasmodium vivax: gene cloning and characterization of the immunodominant epitope. Science. 1985;230:815–818. doi: 10.1126/science.2414847. [DOI] [PubMed] [Google Scholar]
- 6.Ancsin JB, Kisilevsky R. A binding site for highly sulfated heparan sulfate is identified in the N terminus of the circumsporozoite protein: significance for malarial sporozoite attachment to hepatocytes. J Biol Chem. 2004;279:21824–21832. doi: 10.1074/jbc.M401979200. [DOI] [PubMed] [Google Scholar]
- 7.Sidjanski SP, Vanderberg JP, Sinnis P. Anopheles stephensi salivary glands bear receptors for region I of the circumsporozoite protein of Plasmodium falciparum. Mol Biochem Parasitol. 1997;90:33–41. doi: 10.1016/s0166-6851(97)00124-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tewari R, Spaccapelo R, Bistoni F, Holder AA, Crisanti A. Function of region I and II adhesive motifs of Plasmodium falciparum circumsporozoite protein in sporozoite motility and infectivity. J Biol Chem. 2002;277:47613–47618. doi: 10.1074/jbc.M208453200. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg R, Wirtz RA, Lanar DE, Sattabongkot J, Hall T, Waters AP, Prasittisuk C. Circumsporozoite protein heterogeneity in the human malaria parasite Plasmodium vivax. Science. 1989;245:973–976. doi: 10.1126/science.2672336. [DOI] [PubMed] [Google Scholar]
- 10.Zakeri S, Abouie Mehrizi A, Djadid ND, Snounou G. Circumsporozoite protein gene diversity among temperate and tropical Plasmodium vivax isolates from Iran. Trop Med Int Health. 2006;11:729–737. doi: 10.1111/j.1365-3156.2006.01613.x. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez JM, Hurtado S, Arévalo-Herrera M, Herrera S. Variants of the Plasmodium vivax circumsporozoite protein (VK210 and VK247) in Colombian isolates. Mem Inst Oswaldo Cruz. 2001;96:709–712. doi: 10.1590/s0074-02762001000500023. [DOI] [PubMed] [Google Scholar]
- 12.Kim T, Kim YJ, Song KJ, Song JW, Cha SH, Kim YK, Shin YK, Suh IB, Lim CS. The molecular characteristics of circumsporozoite protein gene subtypes from Plasmodium vivax isolates in Republic of Korea. Parasitol Res. 2002;88:1051–1054. doi: 10.1007/s00436-002-0699-z. [DOI] [PubMed] [Google Scholar]
- 13.Mann VH, Huang T, Cheng Q, Saul A. Sequence variation in the circumsporozoite protein gene of Plasmodium vivax appears to be regionally biased. Mol Biochem Parasitol. 1994;68:45–52. doi: 10.1016/0166-6851(94)00148-0. [DOI] [PubMed] [Google Scholar]
- 14.Qari SH, Goldman IF, Povoa MM, Oliveira S, Alpers MP, Lal AA. Wide distribution of the variant form of the human malaria parasite Plasmodium vivax. J Biol Chem. 1991;266:16297–16300. [PubMed] [Google Scholar]
- 15.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 18.Kho WG, Park YH, Chung JY, Kim JP, Hong ST, Lee WJ, Kim TS, Lee JS. Two new genotypes of Plasmodium vivax circumsporozoite protein found in the Republic of Korea. Korean J Parasitol. 1999;37:265–270. doi: 10.3347/kjp.1999.37.4.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 20.Herrera S, Escobar P, de Plata C, Avila GI, Corradin G, Herrera MA. Human recognition of T cell epitopes on the Plasmodium vivax circumsporozoite protein. J Immunol. 1992;148:3986–3990. [PubMed] [Google Scholar]
- 21.Arévalo-Herrera M, Roggero MA, Gonzalez JM, Vergara J, Corradin G, Lopez JA, Herrera S. Mapping and comparison of the B-cell epitopes recognized on the Plasmodium vivax circumsporozoite protein by immune Colombians and immunized Aotus monkeys. Ann Trop Med Parasitol. 1998;92:539–551. [PubMed] [Google Scholar]
- 22.Arévalo-Herrera M, Valencia AZ, Vergara J, Bonelo A, Fleischhauer K, Gonzalez JM, Restrepo JC, Lopez JA, Valmori D, Corradin G, Herrera S. Identification of HLA-A2 restricted CD8(+) T-lymphocyte responses to Plasmodium vivax circumsporozoite protein in individuals naturally exposed to malaria. Parasite Immunol. 2002;24:161–169. doi: 10.1046/j.1365-3024.2002.00449.x. [DOI] [PubMed] [Google Scholar]
- 23.Escalante AA, Grebert HM, Isea R, Goldman IF, Basco L, Magris M, Biswas S, Kariuki S, Lal AA. A study of genetic diversity in the gene encoding the circumsporozoite protein (CSP) of Plasmodium falciparum from different transmission areas-XVI. Asembo Bay Cohort Project. Mol Biochem Parasitol. 2002;125:83–90. doi: 10.1016/s0166-6851(02)00216-5. [DOI] [PubMed] [Google Scholar]
- 24.Escalante AA, Lal AA, Ayala FJ. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics. 1998;149:189–202. doi: 10.1093/genetics/149.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumkhaek C, Phra-Ek K, Renia L, Singhasivanon P, Looareesuwan S, Hirunpetcharat C, White NJ, Brockman A, Gruner AC, Lebrun N, Alloueche A, Nosten F, Khusmith S, Snounou G. Are extensive T cell epitope polymorphisms in the Plasmodium falciparum circumsporozoite antigen, a leading sporozoite vaccine candidate, selected by immune pressure? J Immunol. 2005;175:3935–3939. doi: 10.4049/jimmunol.175.6.3935. [DOI] [PubMed] [Google Scholar]
- 26.Arnot DE, Stewart MJ, Barnwell JW. Antigenic diversity in Thai Plasmodium vivax circumsporozoite proteins. Mol Biochem Parasitol. 1990;43:147–149. doi: 10.1016/0166-6851(90)90140-h. [DOI] [PubMed] [Google Scholar]
- 27.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 28.Arnot DE, Barnwell JW, Stewart MJ. Does biased gene conversion influence polymorphism in the circumsporozoite protein-encoding gene of Plasmodium vivax? Proc Natl Acad Sci USA. 1988;85:8102–8106. doi: 10.1073/pnas.85.21.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes AL. The evolution of amino acid repeat arrays in Plasmodium and other organisms. J Mol Evol. 2004;59:528–535. doi: 10.1007/s00239-004-2645-4. [DOI] [PubMed] [Google Scholar]
- 30.Hughes AL. Circumsporozoite protein genes of malaria parasites (Plasmodium spp.): evidence for positive selection on immunogenic regions. Genetics. 1991;127:345–353. doi: 10.1093/genetics/127.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCutchan TF, Good MF, Miller LH. Polymorphism in the circumsporozoite (CS) protein of Plasmodium falciparum. Parasitol Today. 1989;5:143–146. doi: 10.1016/0169-4758(89)90078-1. [DOI] [PubMed] [Google Scholar]
- 32.Shi YP, Alpers MP, Povoa MM, Lal AA. Diversity in the immunodominant determinants of the circumsporozoite protein of Plasmodium falciparum parasites from malaria-endemic regions of Papua New Guinea and Brazil. Am J Trop Med Hyg. 1992;47:844–851. doi: 10.4269/ajtmh.1992.47.844. [DOI] [PubMed] [Google Scholar]


