Abstract
Eri1 is a 3′-to-5′ exoribonuclease conserved from fission yeast to humans. Here we show that Eri1 associates with ribosomes and ribosomal RNA (rRNA). Ribosomes from Eri1– deficient mice contain 5.8S rRNA that is aberrantly extended at its 3′ end, and Eri1, but not a catalytically inactive mutant, converts this abnormal 5.8S rRNA to the wild-type form in vitro and in cells. In human and murine cells, Eri1 localizes to the cytoplasm and nucleus, with enrichment in the nucleolus, the site of preribosome biogenesis. RNA binding residues in the Eri1 SAP and linker domains promote stable association with rRNA and thereby facilitate 5.8S rRNA 3′ end processing. Taken together, our findings indicate that Eri1 catalyzes the final trimming step in 5.8S rRNA processing, functionally and spatially connecting this regulator of RNAi with the basal translation machinery.
Eri1 was originally identified in a screen for mutants with enhanced RNA interference (RNAi) in Caenorhabditis elegans neurons1. Also known as Thex1 in some species, Eri1 and its ability to inhibit short interfering RNA (siRNA)-directed gene silencing is evolutionarily conserved from Schizosaccharomyces pombe to humans1–4. Because Eri1 encodes a DEDDh-type exonuclease with RNase activity toward the 3′ ends of RNA in vitro1,4–6, it was proposed that Eri1 counteracts the RNAi pathway by degrading siRNAs7. Alternatively, Eri1 may inhibit RNAi indirectly by sequestering factors involved in siRNA processing into a different protein complex8–10.
Basic amino acid residues in the Eri1 SAP (SAF-A/B, Acinus and PIAS) domain and interdomain linker are necessary for in vitro binding to RNA oligonucleotide mimics of a stem-loop structure at the 3′ end of histone mRNAs6,11. Thus, Eri1 has been implicated in the regulation of both siRNAs and histone mRNAs. However, it remains to be shown which cellular RNAs are physically bound by Eri1 and are processed through its catalytic activity.
The most abundant cellular RNAs are the rRNAs, which form the catalytic core of ribosomes12. The 18S rRNA complexes with 32 ribosomal small subunit proteins (RPS) to form the 40S subunit, and another 47 ribosomal large subunit proteins (RPL) and the 28S, 5.8S and 5S rRNAs assemble into the 60S subunit13.
The 18S, 5.8S and 28S rRNAs derive from a single precursor molecule that is transcribed and processed in the nucleolus. The 28S and 5.8S subunits remain associated with each other throughout ribosome maturation, owing in part to base pairing in a stretch of conserved complementarity at the 3′ end of the mature 5.8S rRNA and the 5′ end of the mature 28S rRNA14–16. The internal transcribed spacer 2 (ITS2) sequence, which divides these complementary strands, is removed by a series of endonucleolytic and exonucleolytic steps that remain incompletely defined. Ribosome biogenesis is tightly coordinated to ensure proper translational control. Inefficient ribosome biogenesis is associated with growth defects in budding yeast17, flies18, mice19,20 and humans21.
Here we report that Eri1-deficient mice show a growth defect and a high rate of neonatal mortality. Eri1 associates with ribosomes and binds to rRNA precursors and 5.8S rRNA in cells. The appearance of a 3′-extended 5.8S rRNA in C. elegans eri-1 mutants indicates that ERI-1 is required for 5.8S rRNA processing10, and our observation of a similar defect in Eri1-deficient mice demonstrates that this is a conserved feature of Eri1 function. Reconstitution experiments in Eri1-deficient cells and in vitro processing reactions with recombinant Eri1 revealed that Eri1 exonuclease activity is necessary and sufficient to catalyze the final step in 5.8S rRNA 3′ end formation.
RESULTS
Growth defects and neonatal mortality in Eri1-deficient mice
To analyze the function of Eri1 in a mammalian system, we generated a conditional knockout mouse model (Supplementary Fig. 1 online). Standard gene-targeting techniques were used to generate mice carrying an allele of Eri1 (hereafter Eri1fl) with loxP sites flanking exon 3, which encodes the C terminus of the SAP domain and part of the catalytic core of the exonuclease domain5. Eri1+/fl mice were bred with CMV-Cre transgenic mice to inactivate Eri1 in the germ line, and Eri1+/− mice were intercrossed to generate Eri1−/− offspring. Eri1 protein was undetectable in Eri1−/− murine embryonic fibroblasts (MEFs) and in T cells from Eri1fl/fl CD4-Cre transgenic animals (Supplementary Fig. 1d).
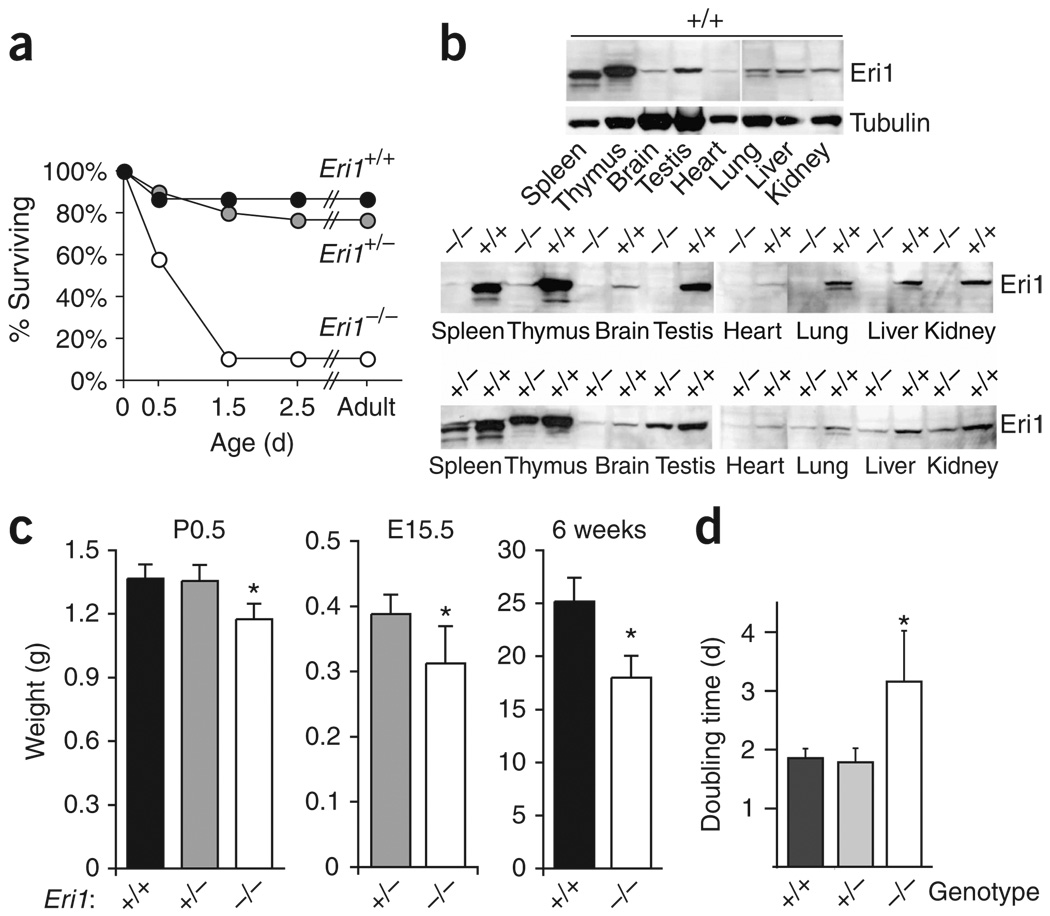
Eri1−/− mice on the C57BL/6 inbred strain background were born at the expected Mendelian frequency, but less than 10% of these animals survived to weaning age (Supplementary Table 1 online). Most Eri1−/− mice died during the first 2 d post partum (Fig. 1a), but standard anatomical and histological analyses of newborn mice did not reveal the cause of neonatal mortality (data not shown). Eri1 was ubiquitously expressed, and it was represented highly in total tissue proteins extracted from the spleen, thymus and testis (Fig. 1b).
Figure 1.
Reduced survival and weight of Eri1-deficient mice. (a) Survival curve of C57BL/6 Eri1−/− mice (open circles), Eri1+/− (gray circles) and wild-type (black circles) littermates. (b) Monoclonal anti-Eri1 immunoblot of NMRI × C57BL/6 F2 Eri1−/−, Eri1+/+ and Eri1+/− mouse tissue extracts. (c) Reduced weight (mean ± s.d.) of C57BL/6 Eri1−/− mice (white), Eri1+/− (gray) and wild-type (black) control littermates on the first morning after birth (P0.5), embryonic day 15.5 (E15.5), and 6 weeks after birth. *, significantly lower weight (one-tailed Student’s t-test, P-values 8 × 10−7 (P0.5), 0.001 (E15.5), 0.0004 (6 weeks)). (d) Reduced growth of primary Eri1−/− MEFs. Mean (± s.d.) doubling times for three independent experiments using two measurements obtained between passages 6 and 9 of fibroblasts derived from a total of six Eri1−/− embryos and controls. *, significantly increased doubling time (ANOVA P = 0.006).
The birth weight of Eri1−/− mice was reduced in comparison with wild-type and heterozygous littermates (Fig. 1c and Supplementary Fig. 1e). Reduced body size was observed as early as embryonic day 15.5 and remained significant in surviving adult mice (Fig. 1c). Neonatal lethality, but not reduced body size, was partially rescued by breeding Eri1−/− mice on outbred strain backgrounds (Supplementary Table 1 and data not shown). Growth defects were also observed for cells cultured in vitro. Using automated microscopy to measure the doubling time of primary MEFs during passages 6 to 9, we found that Eri1-deficient cells grew significantly more slowly (P-value = 0.006) than cells from their wild-type and heterozygous littermates (Fig. 1d).
Eri1 associates with the ribosome
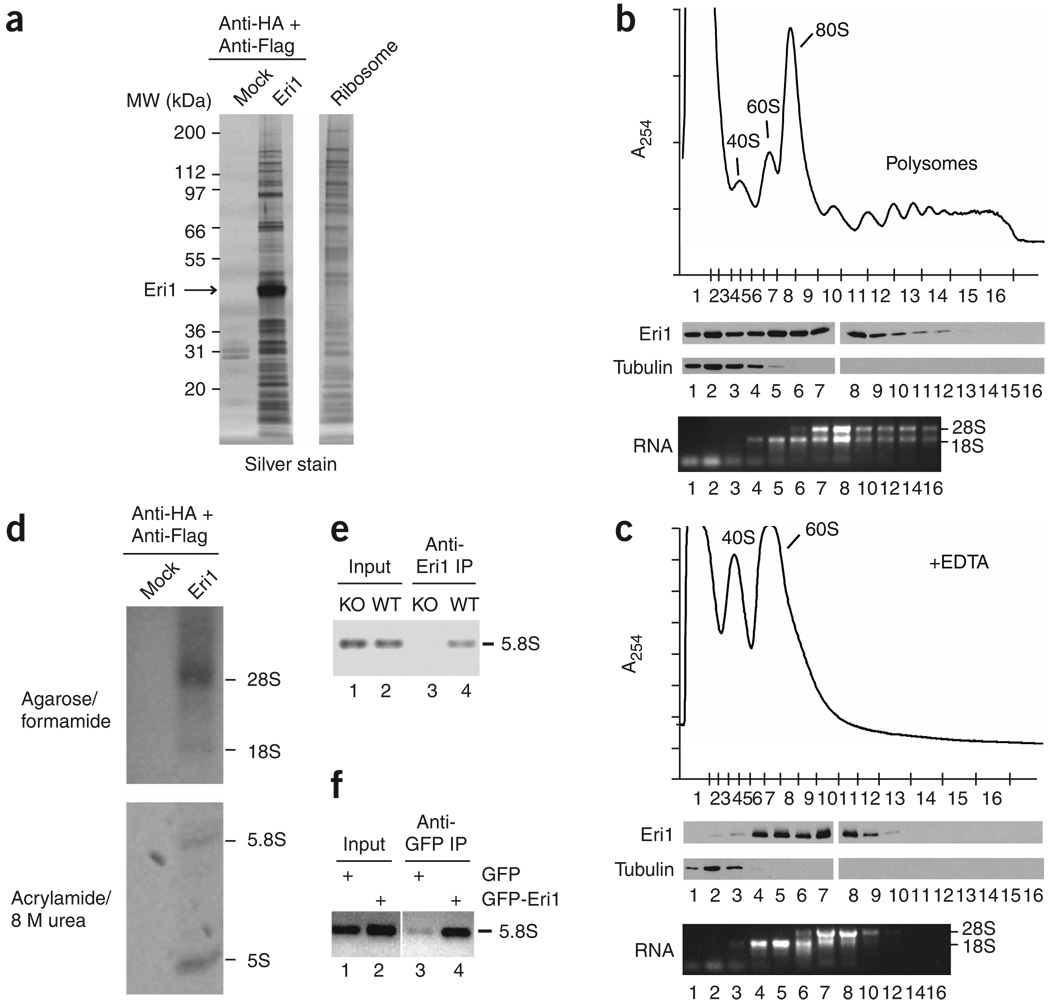
Eri1-interacting proteins were identified by tandem-affinity purification (TAP). Silver staining revealed a pattern of abundant low molecular weight proteins similar to the characteristic protein components of ribosome preparations (Fig. 2a). Complexes purified from nuclear and cytoplasmic extracts of HeLa cells expressing N- or C-terminally epitope-tagged Eri1 seemed similar in composition (Supplementary Fig. 2a online). Liquid chromatography and tandem MS (LC/MS/MS) on a pool of these TAP eluates confirmed the presence of Eri1 and many ribosomal proteins (Supplementary Table 2 online). Ribosomal proteins were not enriched in TAP eluates from untransduced HeLa lysates (Fig. 2a and data not shown).
Figure 2.
Eri1 associates with ribosomes. (a) Silver-stained proteins copurified in Eri1 TAP from cytoplasmic lysates of HeLa cells stably transduced with HA-Flag–Eri1 or untransduced cells (Mock). Ribosomes prepared from untransduced HeLa cells were run on the same gel. MW, molecular weight. (b,c) Sucrose gradient fractionation of 3T3 lysates prepared in the absence (b) or presence (c) of 20 mM EDTA. Above, absorbance profiles at 254 nm (A254) obtained while fractionating sucrose gradients from top to bottom. Approximate positions of collected fractions are indicated on the x axis. Middle, polyclonal anti-Eri1 and anti–α-tubulin immunoblots. Below, RNA from the indicated fractions. Protein fractions were run on two gels per gradient, and immunoblots for all four gels were performed in unison. (d) RNA copurified in Eri1 TAP as in a separated by agarose (above) and polyacrylamide (below) gel electrophoresis. (e,f) RNA immunoprecipitation (RIP) assays for Eri1 binding to 5.8S rRNA. (e) RIP with anti-Eri1 monoclonal antibody in Eri1−/− (KO) and wild-type (WT) MEFs. (f) RIP with anti-GFP polyclonal antibody in HEK 293 cells transduced with LNCX2-GFP or LNCX2-GFP–Eri1.
Endogenous Eri1 from NIH 3T3 cells and overexpressed Flag- and hemagglutinin-tagged Eri1 (Flag-HA–Eri1) from HeLa cells cofractionated with free 40S and 60S ribosomal subunits and assembled 80S ribosomes in sucrose gradients (Fig. 2b,c and data not shown). Eri1 was also present in polysome-containing fractions, although it was less abundant in faster-sedimenting complexes, suggesting some depletion of Eri1 from actively translating ribosomes (Fig. 2b). When ribosomes were dissociated into 40S and 60S subunits by lysing cells in the presence of EDTA, Eri1 cofractionated with both subunits in distinct peaks, indicating that Eri1 can bind to each subunit independently (Fig. 2c and Supplementary Fig. 2).
rRNA also co-immunoprecipitated with Eri1. TAP eluates contained all four major rRNA components (28S, 18S, 5.8S and 5S; Fig. 2d). Under more stringent lysis conditions, 5.8S but not 5S rRNA co-immunoprecipitated with endogenous Eri1 from MEF extracts (Supplementary Fig. 3 online), showing that the interaction between Eri1 and 5.8S rRNA is strong enough to remain intact despite the apparent dissociation of 5S rRNA from the ribosome. Alternatively, Eri1 may reassociate with free 5.8S rRNA in solution.
We confirmed the physical interaction of Eri1 with 5.8S rRNA in intact cells using the RNA immunoprecipitation (RIP) procedure. The RIP assay demonstrated that Eri1 and 5.8S rRNA were associated before cell lysis in MEFs (Fig. 2e) and HEK 293 cells that were transfected with GFP-Eri1 expression constructs (Fig. 2f).
Eri1 is required for proper 5.8S rRNA 3′ end formation
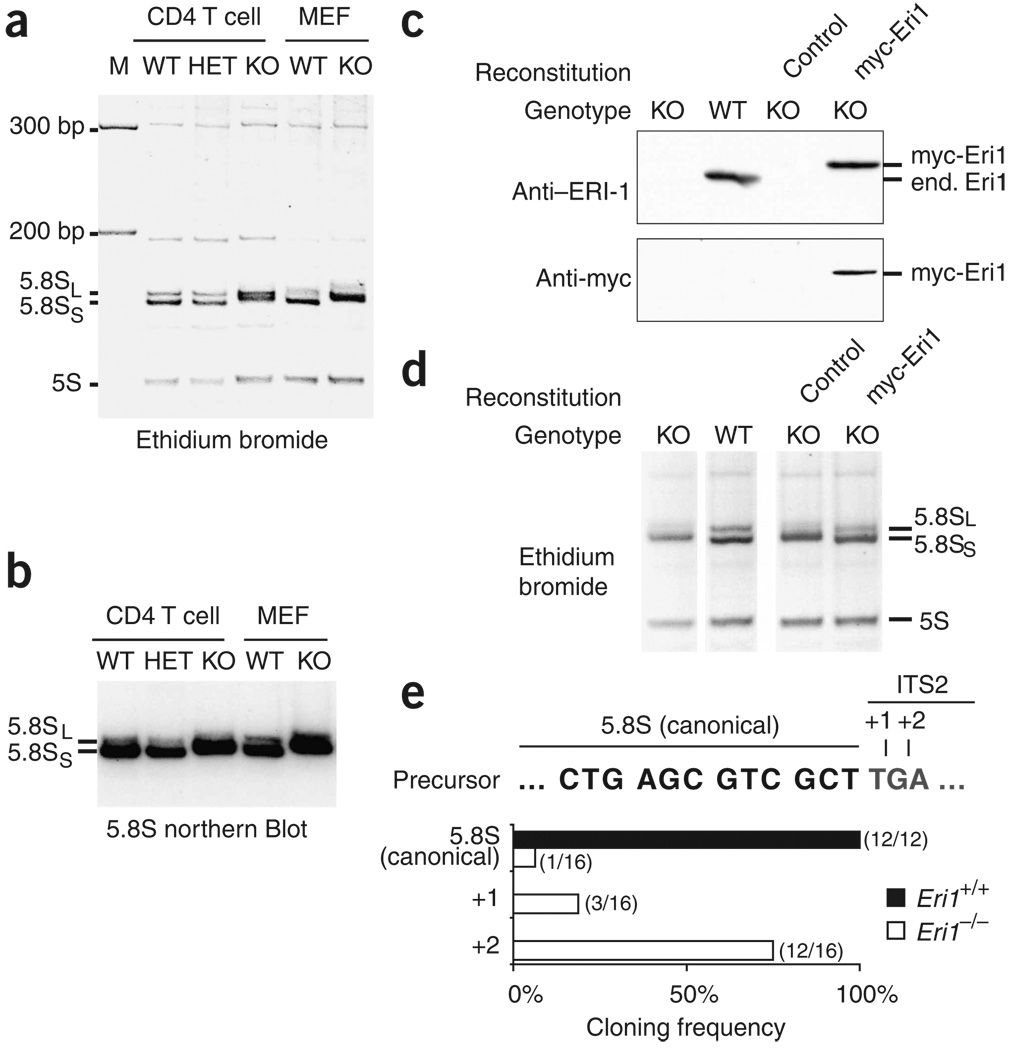
On the basis of the finding that the 5.8S rRNA of C. elegans eri-1 mutants migrates more slowly in polyacrylamide gels10, we looked closely and observed a similar defect in Eri1−/− mouse tissues and cultured cells (Fig. 3a, Supplementary Fig. 4 online and data not shown). Northern blot analysis confirmed that the slow-migrating band was indeed 5.8S rRNA (Fig. 3b). This defect persisted in the outbred mouse background (Supplementary Fig. 4) but could be rescued by reconstituting Eri1-deficient MEFs with myc-tagged Eri1 by retroviral transduction (Fig. 3c,d). Our findings in mice together with the observations by Gabel and Ruvkun in C. elegans and S. pombe demonstrate an evolutionarily conserved requirement for Eri1 in 5.8S rRNA processing.
Figure 3.
Eri1 is required for 5.8S rRNA 3′ end processing. (a,b) RNA from Eri1+/+ CD4-Cre (WT), Eri1+/fl CD4-Cre (HET) and Eri1fl/fl CD4-Cre (KO) primary CD4+ T cells, and Eri1+/+ (WT) and Eri1−/− (KO) MEFs, visualized by ethidium bromide staining (a) and northern blot for 5.8S rRNA (b). (c) Polyclonal anti-Eri1 and monoclonal anti-myc immunoblot analysis of MEFs of the indicated genotypes left untransduced or transduced with KMV (control) or KMV–myc-Eri1. end., endogenous. (d) Ethidium bromide–stained RNA from the same MEF cell populations. All lanes were run on the same gel. (e) Sequences of cloned 5.8S rRNA 3′ ends. Histogram shows the frequency of sequences obtained from Eri1+/+ (black bars; n = 12) and Eri1−/− (white bars; n = 16) that correspond to the predicted 5.8S sequence or extended sequences as shown above. All sequence extensions matched the expected ITS2 sequences (+1, T; +2, TG), except for one +2 clone that contained a TT extension.
During rRNA processing, alternative 5′ endonucleolytic cleavage sites generate the major short (S) and less abundant long (L) forms of the 5.8S rRNA22. Two bands corresponding to these 5.8S isoforms appear in Eri1-deficient cell extracts, and both show a similar retardation in migration compared to the wild-type (Fig. 3a,b,d). Therefore, the apparent molecular weight increase probably reflects changes at the 3′ end. Sequences of cloned wild-type 5.8S rRNA 3′ ends uniformly terminated at the previously reported position. In contrast, the 3′ end of Eri1−/− 5.8S rRNA was variable, and 15 out of 16 clones contained a 1- or 2-nucleotide 3′ extension (Fig. 3e). Thus, Eri1 is crucial to ensure the fidelity of 5.8S rRNA 3′ end maturation.
Eri1 interacts with 5.8S rRNA and its 45S precursor
Overexpressed Eri1 localized to both the cytoplasm and nucleus, and within the nuclear compartment it was enriched in the nucleolus (Supplementary Fig. 5a,b online). Biochemical fractionation showed that endogenous Eri1 was also present in the cytoplasm, nucleus and nucleolus (Supplementary Fig. 5c).
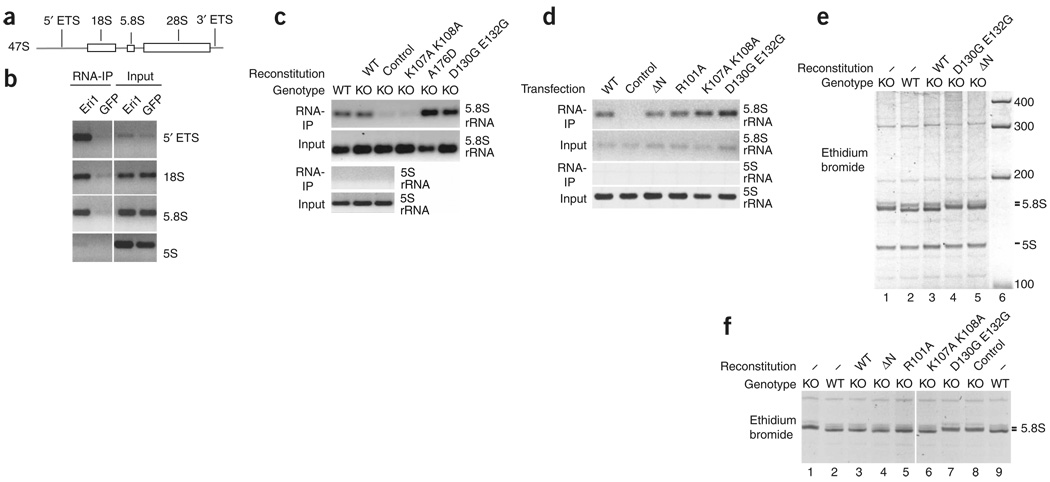
Consistent with its nucleolar localization, Eri1 interacts with early rRNA precursors. Like 5.8S rRNA, sequences from 18S rRNA and the 5′ external transcribed spacer (ETS) specifically co-immunoprecipitated with Eri1 in RIP assays, indicating that Eri1 first interacts with rRNA at least as early as the 45S rRNA precursor stage (Fig. 4a,b). We could not detect a significant interaction of Eri1 with the 3′ ETS, and its interaction with 28S rRNA was detected only at levels close to background (data not shown). These results do not rule out Eri1 interaction with the 47S precursor, but they indicate that Eri1’s interactions are confined to the 5′ part of the 45S and 47S rRNA precursors.
Figure 4.
Eri1 binds directly to rRNA and rRNA precursors, and Eri1 exonuclease activity is required for 5.8S rRNA 3′ end processing. (a) Simplified schematic depiction of the 47S rRNA precursor. ETS, external transcribed spacer. (b) RIP assay performed with anti-GFP polyclonal antibody and HEK 293 cells stably transduced with LNCX2-GFP–Eri1 (Eri1) or LNCX2-GFP (GFP). Primers specific for the 5′ ETS, 18S and 5.8S rRNA were used to identify Eri1 association with the 45S rRNA precursor. (c) RIP assay for Eri1 binding to 5.8S rRNA performed with anti-Eri1 monoclonal antibody and wild-type (WT) and Eri1−/− (KO) MEFs left untransduced or reconstituted by transduction with MSCV encoding myc-Eri1 (WT) and the indicated mutants (Supplementary Fig. 7a). (d) RIP assay for overexpressed GFP-Eri1 binding to 5.8S rRNA using anti-GFP polyclonal antibody and HEK 293 cells transiently transfected with pDEST12.2 expression constructs for GFP-Eri1 (WT), the indicated mutants or GFP-expressing vector (control). (e) RNA from WT and KO MEFs left untransduced (−) or reconstituted with MSCV retroviruses encoding myc-Eri1 (WT) and the indicated mutants. (f) RNA from WT and KO MEFs left untransduced (−) or reconstituted with MSCV retroviruses encoding GFP-Eri1 (WT), the indicated mutants, or GFP alone (control). GFP-expressing cells were FACS sorted for equal GFP fluorescence 1 week before RNA extraction (Supplementary Fig. 7c,d). All lanes were run on the same gel.
Although Eri1 is present during the early stages of rRNA processing, pulse chase assays showed no difference in precursor processing efficiency in immortalized Eri1fl/fl MEFs depleted of Eri1 by adenoviral Cre transduction (Supplementary Fig. 5d,e). These assays were not sensitive enough to resolve the final steps of rRNA processing and did not allow detection of the smaller 5S or 5.8S rRNA species.
We extended our analysis of Eri1 interaction with 5.8S rRNA to include point mutations that have been shown, in the context of human Eri1, to impair histone mRNA binding (ref. 11 and Supplementary Fig. 6d online). RIP assays showed that the catalytically inactive D130G E132G mutant still bound efficiently to the 5.8S rRNA (Fig. 4c and Supplementary Fig. 7a online), whereas the linker region K107A K108A mutant was reproducibly impaired in binding compared to the wild-type protein. We also analyzed a naturally occurring polymorphism in the fourth exon of Eri1 in C57BL/6 mice, which results in an amino acid substitution (A176D) in the catalytic domain of Eri1. The increased negative charge of this amino acid change correlated with a slightly slower migration in SDS-PAGE (Supplementary Figs. 6 and 7a), but it did not affect 5.8S rRNA binding (Fig. 4c) or catalytic activity (data not shown). 5S rRNA did not co-immunoprecipitate with Eri1 in RIP assays, providing further support for the idea that Eri1 has specificity for 5.8S rRNA binding.
We also compared association of 5.8S rRNA with wild-type and mutant forms of Eri1 in HEK 293 cells (Fig. 4d). For these experiments, GFP-tagged Eri1 proteins were overexpressed by transient transfection, and RIP was performed with GFP-specific antibodies that immunoprecipitate all mutants with equal efficiency. In these experiments, sequences in the N-terminal SAP and linker domains were not required for Eri1 interaction with 5.8S rRNA, perhaps because of the high expression levels attained in transfected HEK 293 cells. These findings indicate that the SAP and linker domains have a supportive function in rRNA binding but are not crucial for 5.8S rRNA interaction.
Eri1 catalyzes 5.8S rRNA 3′ end formation
We then asked whether efficient RNA binding or the catalytic activity of Eri1 were required for 5.8S rRNA 3′ end formation. Unlike wild-type Eri1, the myc-tagged catalytically inactive (D130G E132G) mutant and an N-terminal deletion mutant of amino acids 1–106 (ΔN) in Eri1 did not rescue the 5.8S rRNA processing defect of Eri1-deficient fibroblasts (Fig. 4e). Retroviral transduction of Eri1−/− fibroblasts led to detectable expression of all myc-tagged Eri1 constructs at approximately endogenous Eri1 expression levels (Supplementary Fig. 7b, below). To compare different mutant Eri1 proteins at equal expression levels, we used fluorescence-activated cell sorting (FACS) to separate MEF cells transduced with wild-type and mutant forms of GFP-Eri1, obtaining cell populations with the same small range of GFP fluorescence (Supplementary Fig. 7d). The GFP-tagged proteins were expressed at substantially higher levels than the myc-tagged proteins and endogenous Eri1 (Supplementary Fig. 7c and data not shown). Addition of GFP to the N terminus of wild-type Eri1 did not interfere with its ability to process the 3′ end of 5.8S rRNA (Fig. 4f). With these higher (but matched) expression levels, the mutant proteins R101A and K107A K108A, as well as ΔN–Eri1, were able to convert the running behavior of the 5.8S rRNA in Eri1−/− cells to that of the wild-type form (Fig. 4f). Only the catalytically inactive (D130G E132G) mutant remained unable to process the 3′ end of 5.8S rRNA (Fig. 4f).
Thus, only the enzymatic activity of Eri1 is required for 5.8S rRNA 3′ end processing. Our data also support the idea that the SAP domain increases the efficiency of Eri1-mediated 5.8S 3′ end processing, although the RNA binding function of Eri1 is dispensable when the protein is overexpressed at sufficiently high levels.
Finally, we tested whether recombinant Eri1 could trim ribosome-associated 5.8S rRNA in vitro (Fig. 5a). Wild-type Eri1, but not catalytically inactive (D130G E132G) mutant Eri1 efficiently catalyzed conversion of the abnormal 5.8S rRNA of purified Eri1−/− ribosomes to its normal size. This finding provides strong evidence that Eri1 itself is the exoribonuclease responsible for the final step of 5.8S rRNA 3′ end processing and shows that Eri1 can mediate 5.8S processing on fully assembled mature ribosomes.
Figure 5.
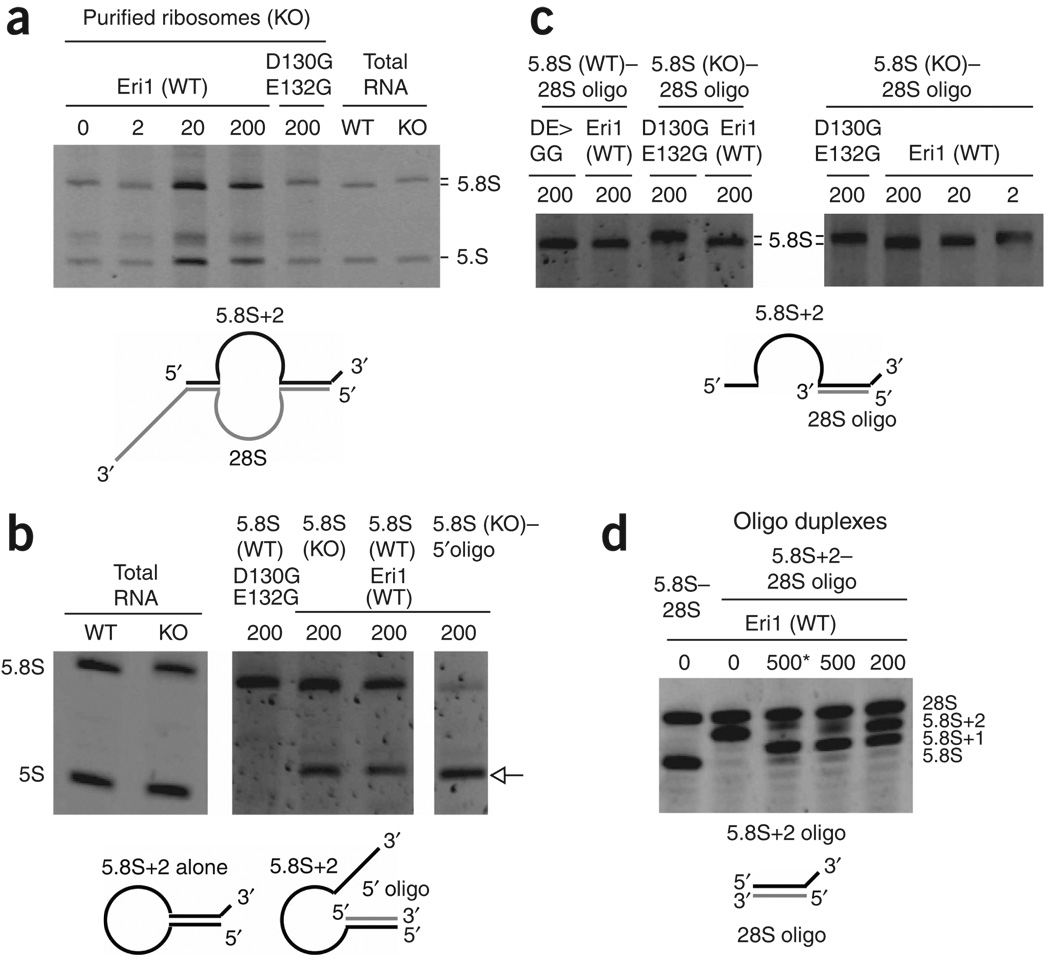
Eri1 catalyzes 3′ end processing of 5.8S rRNA. RNA was subjected to 30 min in vitro processing reactions with the indicated amounts (ng) of recombinant Eri1 or catalytically inactive Eri1 (D130G E132G). Diagrams depict the predicted secondary structure of unprocessed substrates. (a) Ribosomes purified from Eri1−/− liver. Total RNA from wild-type (WT) and Eri1−/− (KO) testis was included to show the migration of the fully processed and 3′-extended 5.8S rRNAs, respectively. (b) Gel-purified WT and KO 5.8S rRNA in isolation, or hybridized with a DNA oligonucleotide complementary to the first 20 nucleotides of the 5.8S rRNA (5′ oligo). All lanes were run on the same gel. The image for total RNA controls was obtained from a shorter exposure. The arrow indicates overprocessed 5.8S rRNA. (c) Gel-purified WT and KO 5.8S rRNA annealed with an RNA oligonucleotide corresponding to the first 22 nucleotides of the 28S rRNA (28S oligo). (d) RNA oligonucleotides corresponding to the last 19 nucleotides of the 5.8S rRNA (5.8S) or the last 21 nucleotides of the typical 2-nucleotide–extended Eri1−/− 5.8S rRNA (5.8S+2), annealed with the 28S oligonucleotide. *, 60 min reaction time.
In the mature ribosome structure, the 3′ end of 5.8S rRNA forms a duplex with the 5′ end of 28S rRNA, leaving a 3′ overhang in Eri1-deficient cells. To determine whether this duplex is important for 5.8S processing, we performed processing reactions with gel-purified Eri1−/− and control 5.8S rRNA. For both substrates, two predominant products emerged: one perhaps slightly shorter than the normal mature 5.8S rRNA, and another approximately 30 nucleotides shorter (Fig. 5b). We proposed that the longer of these two products formed because the 5′ and 3′ ends of the 5.8S, which pair with 28S sequences in the intact ribosome, have substantial complementarity with each other and are capable of forming a duplex that may provide some protection against Eri1-mediated cleavage. To test this possibility, we annealed the purified 5.8S rRNA to a DNA oligonucleotide complementary to the 5′-most 20 nucleotides of the 5.8S. This oligonucleotide effectively sequestered the 5′ end of the 5.8S, exposing the 3′ end and making it vulnerable to degradation (Fig. 5b, right). These experiments demonstrate that the formation of the 5.8S–28S duplex may be sufficient to specify the 3′ end of the 5.8S rRNA by protecting it from overprocessing by Eri1.
To directly test this possibility, we annealed the purified 5.8S to a 28S mimic RNA oligonucleotide equivalent in sequence to the first 22 nucleotides of the 28S (Fig. 5c). The 28S mimic completely protected control 5.8S rRNA from Eri1 cleavage and allowed trimming of the Eri1−/− 5.8S rRNA to the normal length without overprocessing (Fig. 5c, left). Notably, however, processing of the 5.8S rRNA–28S mimic duplex required at least ten times as much recombinant Eri1 as did processing of intact ribosome (compare Fig. 5c, right, and Fig. 5a). Replacing the full-length 5.8S rRNA with an RNA oligonucleotide correponding to the 3′ end of Eri1−/− 5.8S rRNA (5.8S+2) resulted in even less efficient processing, with the expected fully processed product forming in only trace amounts at high concentrations of Eri1 (Fig. 5d). We conclude that the naturally occurring 5.8S–28S duplex is sufficient to specify the final product of Eri1 processing, but efficient Eri1 targeting to this substrate involves interaction with other features of the ribosome, such as more distant regions of double-stranded RNA and possibly ribosomal proteins.
DISCUSSION
Ribosome function in protein synthesis is among the most fundamental of biological processes. Yet our understanding of ribosome biogenesis remains incomplete. In the accompanying manuscript, Gabel and Ruvkun demonstrate that eri-1 is essential for the maturation of the 3′ end of 5.8S rRNA in fission yeast and nematodes10. Together with our findings in mice, these results establish Eri1 as a conserved component of the rRNA processing machinery from fission yeast to mice. Our data further indicate that Eri1 is directly responsible for the final step of 5.8S rRNA 3′ end processing. Biochemical analysis revealed that Eri1 is associated with ribosomes in human and mouse cells: Eri1 co-immunoprecipitated with rRNA, rRNA precursor and ribosomal proteins, and cosedimented with ribosomal subunits and fully assembled ribosomes in sucrose gradients. Reconstitution experiments using Eri1-deficient fibroblasts revealed that the exonuclease activity of Eri1 is required for 5.8S rRNA 3′ end processing. In an in vitro assay of 5.8S rRNA maturation, recombinant Eri1, but not an enzymatically inactive form, was able to access and convert the 3′ end of the 5.8S rRNA of intact Eri1-deficient ribosomes to exactly the same size as mature wild-type 5.8S rRNA. Taken together, our findings strongly support the conclusion that Eri1 catalyzes the removal of unpaired 3′ nucleotides from the end of an already-formed 5.8S–28S rRNA duplex. This characteristic activity of the enzyme is consistent with previous observations that Eri1 activity is enhanced on free 3′ overhangs of siRNA duplexes compared with single-stranded RNA or RNA duplexes1,11.
We found that Eri1 preferentially associates with 5.8S rRNA in solution and in intact cells, even compared with the 5S rRNA, which is also present in the 60S large ribosomal subunit. Previous reports demonstrated that the RNA binding function of the SAP and linker domains of Eri1 are essential for sequence-specific binding to the histone mRNA stem loop and its conserved 3′-terminal ACCCA sequence6,11. In our experiments, the SAP domain and linker sequences were important for stable interaction with the 5.8S rRNA at low or endogenous expression levels. Although they also facilitated 5.8S rRNA 3′ end processing, they were dispensable for binding and processing at high levels of Eri1 protein expression. It is therefore likely that the SAP domain and linker sequences do not engage in a sequence-specific manner with the 5.8S rRNA, but instead stabilize the interaction. We propose that additional protein-protein contacts between Eri1 and ribosomal or other proteins participate in the specific binding and processing of Eri1 RNA targets.
The localization of Eri1 within the nucleolus of murine and human cells and the direct binding of Eri1 to the 45S precursor suggest that Eri1-mediated 5.8S rRNA trimming is temporally and spatially linked with rRNA precursor processing, ITS2 removal and preribosome assembly. However, functional Eri1-GFP fusion proteins seem to be strictly cytoplasmic in C. elegans, and Flag-tagged Eri1 localized only to the cytoplasm in S. pombe1,4,10. Therefore, Eri1-mediated 5.8S rRNA maturation may occur after ribosome assembly and export to the cytoplasm, at least in these species.
A substantial proportion of Eri1 protein was found associated with mature ribosomal subunits and mRNA-bound 80S monosomes. Why does Eri1 remain associated with its substrate after its role in 5.8S end processing is complete? It is tempting to speculate that association with ribosomes may place Eri1 in position to regulate the homeostasis or function of endogenous short RNA species, such as microRNAs (miRNAs), that regulate mRNA translation. Eri1 was relatively depleted from polysome-containing fractions, suggesting that it is preferentially associated with inhibited ribosomal particles. miRNAs have been shown to relocalize targeted mRNAs from polysomes to slower-sedimenting ribonucleoprotein complexes23,24. In addition to enhanced RNAi, increased miRNA expression and defective production of endogenous siRNAs has been observed in eri-1 mutant worms8,9. In fission yeast, the abundance of siRNAs that direct heterochromatin formation is markedly increased in the absence of Eri1 (ref. 3). We have observed modestly enhanced RNAi function in Eri1-deficient mouse cells (ref. 2 and K.M.A., A.D.L., L.C.S. and V.H., unpublished observations), but further research is necessary to characterize the role of Eri1 in endogenous short regulatory RNA pathways in vertebrates.
The budding yeast S. cerevisiae lacks an Eri1 ortholog, and the final step of 5.8S rRNA 3′ end formation instead requires the endonuclease Ngl2 (ref. 25). In addition, a set of exonucleases in the exosome complex act sequentially on the 5.8S 3′ end before Ngl2 action26. Yeast strains lacking the exosome component Rrp6 accumulate 5.8S rRNA with a ~30-nucleotide extended 3′ end that can be isolated from actively translating ribosomes27,28. Similarly, 5.8S rRNA forms that are extended by ~8 nucleotides can be observed in response to the combined inactivation of the Rex1 and Rex2 genes, which encode RNase D family 3′ exonucleases. Neither combined inactivation of Rex1 and Rex2, inactivation of Rrp6 nor inactivation of Ngl2 had a strong impact on the growth of S. cerevisiae at 25 °C25,28,29. However, the evolutionary replacement of Ngl2 endonuclease with an unrelated exonuclease activity from Eri1 in fission yeast underscores the importance of precise 5.8S end formation in eukaryotes.
Eri1-deficient mice showed reduced body size throughout development and into adulthood, and Eri1-deficient MEFs also showed reduced growth in vitro. These phenotypes resemble those of fruitfly Minute mutants, human Diamond-Blackfan anemia patients and the mouse Bst mutant, all of which bear mutations in ribosomal protein genes18,20,21. Compared with other model organisms, such as fruitflies, few viable mutants of ribosomal protein genes have been detected in mice and humans, suggesting that mammals may be less able to compensate for partial loss of function of genes involved in ribosome maturation and function18,20. Consistent with this idea, worms and fission yeast lacking eri-1 are viable, whereas in inbred C57BL/6 mice Eri1 deficiency was associated with perinatal lethality. This bottleneck in viability was largely rescued by breeding the mutation into a mixed strain background, and the large majority of Eri1−/− mice on both backgrounds that survived to 3 d of age subsequently survived into adulthood. Perhaps Eri1 deficiency and impaired 5.8S processing in mice causes a subtle cell type–specific growth defect that would have to be overcome by cell type–specific compensatory mechanisms to ensure survival of the organism. Therefore, the rRNA processing defect common to several Eri1-deficient eukaryotes could be innocuous in worms and fission yeast but manifest in mice as a failure of robust survival. However, the connection between defective rRNA processing and growth and survival defects in Eri1−/− mice remains speculative, and all of the observed phenotypes of Eri1-deficient mice may be dictated by defects in other pathways, such as the homeostasis of endogenous short regulatory RNAs or histone mRNA.
Mice lacking the ATP-dependent DEAD-box RNA helicases Ddx5 or Ddx17 also show defects in 5.8S rRNA production and early lethality19. Ddx5−/− mice do not survive past E11.5, Ddx17 −/− mice die within 2 d following birth with reduced body size and Ddx17 −/− MEF cells show reduced proliferation in vitro. Like Eri1, Ddx5 and Ddx17 perform important functions in 5.8S rRNA biogenesis and short regulatory RNA homeostasis19. Both helicases are found in a complex with the RNase III enzyme Drosha, which is responsible for cropping the hairpins of pre-miRNAs from primary miRNA transcripts30,31. Drosha localizes to the nucleus and also transits into the nucleolus during S phase32. Mouse embryos deficient for either Ddx5 or Ddx17 have reduced levels of 5.8S rRNA and most miRNAs, and this phenotype can be reproduced using siRNAs that target Ddx5, Ddx17 or Drosha in MEFs and HeLa cells19,33. Considering the dual involvement of Eri1, Drosha, Ddx5 and Ddx17 in rRNA processing and the RNAi pathway, one may speculate that, during evolution, factors that were primarily involved in rRNA processing later acquired new functions in the biogenesis and regulation of short regulatory RNA molecules.
METHODS
Mice
Mice were housed in specific pathogen-free barrier facilities and used in accordance with protocols approved by the animal care and use committees of the Immune Disease Institute, Harvard Medical School, and the Helmholtz Center, Munich. We created the Eri1-deficient mice by gene targeting in BRUCE4 C57BL/6 ES cells using standard techniques (Supplementary Fig. 1). Wild-type C57BL/6/J and Actin-FLPe transgenic mice (B6;SJLTg(ACTFLPe)9205Dym/J) were obtained from Jackson Laboratories, CMV-Cre transgenic mice (6.C-Tg(CMV-cre)1Cgn) were provided by K. Rajewsky (Harvard Medical School, Boston), CD4-Cre transgenic mice (TgN(Cd4-Cre)) were provided by C. Wilson (University of Washington, Seattle). ICR and NMRI mice were from Taconic.
Antibodies
We generated monoclonal (anti-Eri1 5G8, rat IgG1) and polyclonal antibodies against Eri1 (anti-Eri1 A28) via immunization of rats or rabbits (Helmholtz Center Munich Monoclonal Antibody Facility and Millbrook), respectively, using full-length Eri1 protein. Recombinant Eri1 was expressed as an N-terminal GST fusion in bacteria, purified on glutathione Sepharose beads and eluted by Prescission protease (GE Healthcare) cleavage. We used Anti–c-myc (9E10), monoclonal anti-GFP (Roche) and anti-tubulin (5-B-1-2 or TU-02, Santa Cruz) in western blots and anti–PES-1 (8E9) for immunofluorescence. For RIP assays, we used polyclonal anti-GFP antibodies (Invitrogen).
Plasmids
pMSCV or pLNCX2 (Clontech) were converted into gateway destination vectors via ligation of the gateway cassette (Invitrogen). Mouse Eri1 cDNA (NM_026067) was PCR amplified from IMAGE clone 5354985 and cloned via SalI/NotI into pENTR11 or pGEX-6P containing either no tag or an N-terminal myc, ECFP, EGFP or GST tag. Recombination reactions from pENTR11 into pDest12.2, pLNCX2 or pMSCVpuro were performed with the LR clonase kit (Invitrogen). Point mutants were generated via site-directed mutagenesis with the Quickchange (Stratagene) kit. Human ERI1 cDNA was amplified from IMAGE clone 5186320 and cloned via XhoI/NotI into pOZ-N and pOZ-C vectors containing N-terminal or C-terminal Flag-HA tags, respectively, and an IRES-CD25 cassette.
Isolation and immortalization of mouse embryonic fibroblasts
MEF cells were isolated by sacrificing pregnant females 13.5 d post coitum and separating the embryos. Fibroblasts of the appropriate genotype were taken into culture and immortalized via ecotropic retroviral infection with SV40 large T antigen using hygromycin selection at 100 µg ml−1 for 2 weeks.
Measurement of cell growth
The Cellscreen (Innovatis) was used according to the manufacturer’s instructions to take pictures of exactly the same cells and their progeny over a culture period of 48 h. Percentages of surface area covered with cells were determined by automated analysis of each picture using the PA adhesion software (Innovatis) and used to calculate cell-doubling time for at least eight replicates of each culture.
Whole-cell protein-extract preparation
Whole-cell extracts for western blot analysis or immunoprecipitation were prepared by lysis of cells or homogenized organs in 0.1% (w/v) NP-40, 150 mM NaCl, 50 mM Tris (pH 7.5), 1 mM EDTA, 1 mM DTT and complete protease inhibitors (Roche). The cells and nuclei were extracted via dounce homogenization using loose and tight pestles and via passage of the lysate through a 26-gauge syringe needle. Insoluble material was removed by centrifugation at 50,000 × g for 10 min. Cell-number equivalents or equal protein content was used for immuoprecipitation or SDS-PAGE and immunoblotting.
Retroviral transduction
MEFs were infected initially with ecotropic retroviruses; infection of HEK 293 cells or the secondary infection of MEF cells were performed with amphotropic retroviruses. For infection, MEFs or HEK 293 cells were seeded in six-well plates (105 cells per well), spin-infected at room temperature for 1 h at 930 × g at a multiplicity of infection (MOI) of ~10–30 with the addition of 5 µg ml−1 polybrene. Cells were grown for 48 h and then selected with 2 µg ml−1 puromycin for 4 d. Cells were sorted after another 6 d of growth on a MoFlo fluorescence-activated cell sorter (Cytomation).
5.8S rRNA cloning and sequencing
5.8S rRNA from Eri1+/+ and Eri1−/− liver was purified by elution from acrylamide-urea gel slices and cloned using an adaptation of a previously described procedure for cloning microRNAs34. Briefly, we used RNA ligase (NEB) in the absence of ATP to attach a preadenylated, oligonucleotide linker (IDT RNA Cloning Linker-1; rAppCTG TAGGCACCATCAAT/3ddC/) to the 3′ end of the 5.8S rRNA. The products of this reaction were eluted from acrylamide-urea gel slices, reverse transcribed with a linker-specific primer (5′-ATTGATGGTGCCTAC-3′), and amplified by PCR with a linker-specific 3′ primer (5′-GCCTTGCCAGCCCGCTCAGATTGATGGTGCCTACAG-3′) and a 5.8S rRNA-specific 5′ primer (5′-ATCGTAGGCACCTGATCATCGACACTTCG-3′). PCR products were cloned in pCR2.1 using a TA cloning kit (Invitrogen) and sequenced using plasmid-specific primers (D. Farber DNA sequencing core facility, Boston).
RNA gel electrophoresis and northern blot
RNA was extracted using Trizol reagent (Invitrogen) and resuspended in RNase free water. We then vacuum dried 1–5 µg RNA and resuspended it in 2× loading buffer (95% (v/v) formamide, 18 mM EDTA, 0.025% (w/v) SDS). Samples were separated on 7–8% (v/v) acrylamide gels containing 8 Murea in 0.5× or 1× Tris-borate with EDTA (TBE). Gels were incubated for 10 min in 1 µg ml−1 ethidium bromide, and images were captured under UV light. RNA was transferred to Highbond N+ membranes (Amersham) with semidry blotting for 45 min at 1.5 mA cm−2 in 1× TBE. The blot was dried, UV cross-linked and incubated for 4 h at 80 °C. Prehybridization was done in Church buffer (0.5 M NaPO4, pH7.2, 7% (w/v) SDS, 1 mM EDTA) for 1 h at 65 °C, and hybridization was performed overnight with 5 pmol end-labeled oligonucleotide 5′-GGC CGC AAG TGC GTT CGA AGT GTC GAT GAT CAA TGT GTC CTG CAA TTC AC-3′. The blot was washed 30 min with 2× SSC–0.1% (w/v) SDS at 65 °C and twice with 0.2× SSC–0.1% (w/v) SDS at 65 °C.
Supplementary Methods
Detailed descriptions of tandem-affinity purification, sucrose gradient fractionation, RIP assays and in vitro exonuclease assays are provided online.
Supplementary Material
ACKNOWLEDGMENTS
We wish to thank W. Hammerschmidt, Helmholtz Center Munich, for the gift of the SV40 large T hygro retrovirus, D. Eick, Helmholtz Center Munich, for the gift of anti–PES-1 antibody and F. Alt, Immune Disease Institute, Boston, for recombinant Cre adenovirus. We thank J. Asara for assistance with MS and data analysis, E. Yanni and C. Gelinas for technical assistance and A. Whynot, T.A. Rapoport and P. Sorger for help and equipment for gradient fractionation. We also thank H. Gabel and G. Ruvkun for communicating unpublished data, the laboratories of D. Eick, M. Meisterernst and I. Jeremias for helpful discussion, and D. Eick and D. Moazed for comments on the manuscript. This work was supported by US National Institutes of Health (NIH) grants AI44432 and AI70788 to A.R., a Cancer Research Institute fellowship and Deutsche Forschungsgemeinschaft grant HE 3359/2 to V.H., and a Burroughs Wellcome Fund Career Award in the Biomedical Sciences to K.M.A. K.M.A. is a Special Fellow of The Leukemia and Lymphoma Society. W.A.P. is a National Defense Science and Engineering Graduate fellow. M.T. is supported by the Harvard Stem Cell Institute and is a Predoctoral Fellow of the Ryan Foundation.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
K.M.A. and V.H. initiated the project, planned and supervised the experiments, performed some experiments, and wrote the manuscript. A.R. provided advice and support and edited the manuscript. W.A.P., N.R. and A.D.L. established tools and assays and performed and analyzed experiments with technical assistance and advice from E.G., C.W., L.C.S., N.P., E.D.L., M.T., J.W.E. and Y.S. The monoclonal anti-Eri1 antibody was generated by E.K.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 2.Buhler M, Mohn F, Stalder L, Muhlemann O. Transcriptional silencing of nonsense codon-containing immunoglobulin minigenes. Mol. Cell. 2005;18:307–317. doi: 10.1016/j.molcel.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 3.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 4.Iida T, Kawaguchi R, Nakayama J. Conserved ribonuclease, Eri1, negatively regulates heterochromatin assembly in fission yeast. Curr. Biol. 2006;16:1459–1464. doi: 10.1016/j.cub.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Y, Patel DJ. Crystallographic structure of the nuclease domain of 3′hExo, a DEDDh family member, bound to rAMP. J. Mol. Biol. 2004;343:305–312. doi: 10.1016/j.jmb.2004.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominski Z, Yang XC, Kaygun H, Dadlez M, Marzluff WF. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- 7.Timmons L. Endogenous inhibitors of RNA interference in Caenorhabditis elegans. Bioessays. 2004;26:715–718. doi: 10.1002/bies.20078. [DOI] [PubMed] [Google Scholar]
- 8.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 9.Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabel HW, Ruvkun G. The exonuclease ERI-1 has a conserved dual role in 5.8S rRNA processing and RNAi. Nat. Struct. Mol. Biol. 2008 April 27; doi: 10.1038/nsmb.1411. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang XC, Purdy M, Marzluff WF, Dominski Z. Characterization of 3′hExo, a 3′ exonuclease specifically interacting with the 3′ end of histone mRNA. J. Biol. Chem. 2006;281:30447–30454. doi: 10.1074/jbc.M602947200. [DOI] [PubMed] [Google Scholar]
- 12.Moore PB, Steitz TA. The involvement of RNA in ribosome function. Nature. 2002;418:229–235. doi: 10.1038/418229a. [DOI] [PubMed] [Google Scholar]
- 13.Lecompte O, Ripp R, Thierry JC, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cote CA, Greer CL, Peculis BA. Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA. 2002;8:786–797. doi: 10.1017/s1355838202023063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph N, Krauskopf E, Vera MI, Michot B. Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res. 1999;27:4533–4540. doi: 10.1093/nar/27.23.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh LC, Lee JC. Structural analysis of the internal transcribed spacer 2 of the precursor ribosomal RNA from Saccharomyces cerevisiae. J. Mol. Biol. 1990;211:699–712. doi: 10.1016/0022-2836(90)90071-S. [DOI] [PubMed] [Google Scholar]
- 17.Chen FW, Ioannou YA. Ribosomal proteins in cell proliferation and apoptosis. Int. Rev. Immunol. 1999;18:429–448. doi: 10.3109/08830189909088492. [DOI] [PubMed] [Google Scholar]
- 18.Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv. Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 20.Oliver ER, Saunders TL, Tarle SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Draptchinskaia N, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 22.Fatica A, Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 23.Pillai RS, et al. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 24.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 25.Faber AW, Van Dijk M, Raue HA, Vos JC. Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′-end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA. 2002;8:1095–1101. doi: 10.1017/s1355838202021027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 27.Allmang C, et al. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 29.van Hoof A, Lennertz P, Parker R. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 31.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 32.Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- 33.Jalal C, Uhlmann-Schiffler H, Stahl H. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res. 2007;35:3590–3601. doi: 10.1093/nar/gkm058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.