Abstract
In the fish pathogen Vibrio anguillarum the TonB2 protein is essential for the uptake of the indigenous siderophore anguibactin. Here we describe deletion mutants and alanine replacements affecting the final six amino acids of TonB2. Deletions of more than two amino acids of the TonB2 C-terminus abolished ferric-anguibactin transport, whereas replacement of the last three residues resulted in a protein with wild-type transport properties. We have solved the high-resolution solution structure of the TonB2 C-terminal domain by NMR spectroscopy. The core of this domain (residues 121–206) has an αββαβ structure, whereas residues 76–120 are flexible and extended. This overall folding topology is similar to the Escherichia coli TonB C-terminal domain, albeit with two differences: the β4 strand found at the C-terminus of TonB is absent in TonB2, and loop 3 is extended by 9 Å (0.9 nm) in TonB2. By examining several mutants, we determined that a complete loop 3 is not essential for TonB2 activity. Our results indicate that the β4 strand of E. coli TonB is not required for activity of the TonB system across Gram-negative bacterial species. We have also determined, through NMR chemical-shift-perturbation experiments, that the E. coli TonB binds in vitro to the TonB box from the TonB2-dependent outer membrane transporter FatA; moreover, it can substitute in vivo for TonB2 during ferric-anguibactin transport in V. anguillarum. Unexpectedly, TonB2 did not bind in vitro to the FatA TonB-box region, suggesting that additional factors may be required to promote this interaction. Overall our results indicate that TonB2 is a representative of a different class of TonB proteins.
Keywords: iron transport, nuclear magnetic resonance (NMR), siderophore, TonB, TonB box, Vibrio anguillarum
INTRODUCTION
Iron is an essential micronutrient for nearly all living organisms. However, free iron is not readily available in the vertebrate host or in most natural environments. In the presence of oxygen and at physiological pH, ferric iron is insoluble and therefore the concentration of free iron is estimated to be 10−18 M [1]. Conversely, in the vertebrate host, ferric iron is present at high concentrations, but it is bound to haemoglobin in the red blood cells, whereas in serum it is bound by host proteins such as transferrin and lactoferrin [1]. Bacterial pathogens therefore depend on their ability to scavenge this essential metal from these host proteins, and they have evolved various mechanisms for iron acquisition, solubilization and transport, such as the synthesis of low-molecular-mass siderophores that are able to bind ferric iron with high affinity [2].
Vibrio anguillarum is a Gram-negative bacterium that is responsible for fish epizootics in marine as well as freshwater environments. It causes a highly fatal haemorrhagic septicaemic disease in salmonids and eels. Directly responsible for this high virulence is a 65 kbp pJM1 virulence plasmid that encodes a siderophore-producing and iron-sequestration system that is essential for pathogenicity, i.e. the ferric-anguibactin TonB-dependent transporter (TBDT), FatA, the periplasmic binding protein FatB, the inner-membrane permeases FatD and FatC [3] and some of the genes involved in the biosynthesis of the siderophore anguibactin [4].
Transport of siderophores across the bacterial outer membrane requires the action of the TonB protein [5], which derives its energy from the protonmotive force across the cytoplasmic membrane [6,7]. The TonB protein is anchored in the cytoplasmic membrane, where it forms a complex with the accessory proteins ExbB and ExbD. These two proteins are necessary for the stability of TonB [8] and they work together with TonB to utilize the proton gradient to promote transport through the TBDTs [9]. In V. anguillarum, two different TonB systems, TonB1 and TonB2, have been identified [10]. Although the TonB1 and TonB2 proteins displayed redundant functions in the transport of haem and ferrichrome, only the TonB2 protein is involved in the active transport of the indigenous siderophore anguibactin.
The structures of several different CTD (C-terminal domain) constructs of the E. coli TonB protein have been solved by X-ray crystallography [11-13] and NMR spectroscopy [14]. Constructs consisting of the last 77 or 86 amino acids formed extensively intertwined dimer structures [11,12]. By contrast, a slightly longer construct (92 amino acids), although still forming dimers, resulted in a much smaller protein–protein interface [12]. The solution structure of a longer construct (137 amino acids) was found to be monomeric [14], consistent with analytical-ultracentrifugation studies with similarly sized TonB constructs [12]. Recently the NMR assignments for the backbone of HasB CTD, a TonB-like protein from Serratia marcescens, were reported [15].
When TonB promotes transport through TBDTs, it binds to the N-terminal portion of the cork domain, the so-called ‘TonB-box’ region. The interaction of the E. coli TonB protein with its cognate TBDT has been studied, both with synthetic peptides encompassing their TonB-box-binding regions [14], as well as with the isolated cork domains of E. coli receptors [16]. These studies illustrated that monomeric forms of TonB CTD were capable of binding to the TBDTs, that these complexes had a 1:1 stoichiometry and, on the basis of NMR chemical-shift mapping, that the likely interaction interface was along the β3 strand. Chemical-shift changes were also observed for the β4 strand. These observations were borne out by the crystal structures of two different constructs of E. coli TonB CTD in complex with the intact E. coli ferrichrome receptor FhuA and the vitamin B12 receptor BtuB [17,18], since both demonstrated the formation of 1:1 TonB-TBDT complexes. Most of the interactions between these proteins took place along the β3 strand of TonB in both complexes.
The extreme C-terminal region of TonB has been the subject of close scrutiny. One study described several C-terminally altered E. coli TonB derivatives [19]. A mutant protein in which the last eight amino acids were removed and 13 extra amino acids were added was found to be functional. Another construct, in which the last six amino acids were deleted and the two preceding ones were mutated, was also functional. However, a mutation in which the last eight amino acids were deleted and the preceding seven were altered no longer had TonB-dependent activity. Structurally, the last seven amino acids of TonB correspond to the β4 strand and the turn between the β3 and β4 strands. The terminal 15 amino acids of TonB also include one-half of the β3 strand of the protein. In all these cases, degradation products of these E. coli TonB derivatives were observed. Similar results have also been reported for the TonB protein from Enterobacter aerogenes [20].
In the present work we have characterized the C-terminal domain of V. anguillarum TonB2 protein (residues 106–206) by analysing various deletion and alanine-derivative mutants. Moreover, we have solved the solution structure of the C-terminal folded domain of TonB2 residues 106–206 by NMR spectroscopy.
MATERIALS AND METHODS
Bacterial cultures
The strains and plasmids used in the present work are listed in Tables 1 and 2 respectively. E. coli strains were grown at 37 °C in LB (Luria–Bertani) broth supplemented with antibiotics as appropriate [kanamycin (50 μg/ml), chloramphenicol (30 μg/ml) and ampicillin (100 μg/ml)]. V. anguillarum strains were grown at 25°C in trypticase soy broth and with 1% NaCl supplemented with antibiotics [Cm (10 μg/ml) and rifampicin (100 μg/ml)]. For iron-restricted growth, V. anguillarum was grown in chemically defined CM9 minimal medium [3] supplemented with 10 μM of the iron chelator EDDHA [ethylenediamine-N,N’-bis-(2-hydroxyphenylacetic acid)]. For induction, IPTG (isopropyl β-d-thiogalactoside) was added to a final concentration of 1mM. Chrome Azurol S agar plates were used to detect siderophore production.
Table 1.
Bacterial strains used in the present study
| Organism and strain | Relevant characteristics | Source |
|---|---|---|
| E. coli | ||
| TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZ ΔM15 ΔlacX74 deoR recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| S17-λpir | thi pro hsdR hsdM+ recA RP4-2-Tc::Mu-Km::Tn7 λ-pir | Laboratory stock |
| MM294 | F− endA1 hsdR17 supE44thi-1 λ− harbouring plasmid pRK2013 | Laboratory stock |
| BL21(DE3) | F−ompT[lon] hsdSB (rB − mB−) with DE3, λ prophage carrying the T7 RNA polymerase gene | Invitrogen. |
| DH5α | F−, φ80lacZ ΔM15, endA1, recA1, hsdR17, (rk−mk+), supE44, thi-1, gyrA96, relA1, Δ(lacZYA-argF)U169, λ− | Laboratory stock |
| KP1032 | W3110 tonB::kan | [45] |
| CSL-68 | KP1032/pMMB208 tonBEc | The present study |
| CSL-69 | KP1032/pMMB208 tonB2 V. anguillarum | The present study |
| CSL-70 | KP1032/pMMB208 | The present study |
| CSL-71 | KP1032/pMMB208 exbB2-exbD2-tonB2 | The present study |
| CSL-72 | KP1032/pMMB208 ttpC-exbB2-exbD2-tonB2 | The present study |
| CSL-73 | KP1032/pMMB208 tonB2Va-β4Ec | The present study |
| CSL-74 | KP1032/pMMB208 tonBEcΔβ4 | The present study |
| V. anguillarum | ||
| 775 (pJM1) | Wild-type Pacific Ocean prototype, O1 serotype | Laboratory stock |
| CSL-48 | 775 ΔtonB2 | [24] |
| CSL-52 | 775 ΔtonB2 ΔtonB1 | [24] |
| CSL-54 | CSL-48/pMMB208 wild type tonB2 | The present study |
| CSL-55 | CSL-48/pMMB208 Δ2-C tonB2 | The present study |
| CSL-56 | CSL-48/pMMB208 Δ3-C tonB2 | The present study |
| CSL-57 | CSL-48/pMMB208 Δ4-C tonB2 | The present study |
| CSL-60 | CSL-48/pMMB208 -AAA-C’ tonB2 | The present study |
| CSL-61 | CSL-48/pMMB208 -AAAAAA-C’ tonB2 | The present study |
| CSL-64 | CSL-52/pMMB208 tonBEc | The present study |
| CSL-65 | CSL-52/pMMB208 tonB2Va-β4Ec | The present study |
| CSL-67 | CSL-48/pMMB208 Δ179-195 loop3 | The present study |
| CSL-68 | CSL-48/pMMB208 Δ179-184 loop3 | The present study |
| CSL-69 | CSL-48/pMMB208 Δ185-190 loop3 | The present study |
| CSL-70 | CSL-48/pMMB208 Δ191-195 loop3 | The present study |
Table 2.
Plasmids used in the present study
| Plasmid | Relevant characteristic | Source |
|---|---|---|
| pDM4 | Suicide plasmid R6K origin CmR | [23] |
| pMMB208 |
IncQ, lacIq, Ωcat, ΩPtac, polylinker from M13mp19, CmR |
Laboratory stock |
| pCR2.1® | AmpR KmR | Invitrogen |
| pCR®-BluntII | KmR | Invitrogen |
| pET200 | Expression vector, KmR | Invitrogen |
| pCL22 | pET200 harbouring a partial tonB2 sequence (residues 76–206) |
The present study |
| pCL21 | pET200 harbouring a partial tonB2 sequence (residues 106–206) |
The present study |
Bioassays
The bioassays to determine siderophore utilization in V. anguillarum and E. coli were performed in minimal medium agar supplemented with EDDHA. For E. coli, tryptophan (0.004%) and (0.01%) thiamin were also added. The bacterial cells were included in the agar and, upon solidification, plates were spot-inoculated with the different iron sources and ferric ammonium citrate was used as control.
General molecular-biology procedures
Chromosomal and plasmid DNA purification was performed according to standard protocols [21]. Enzymatic digestions and ligations were carried out according to the manufacturer’s instructions (New England Biolabs Inc., Ipswich, MA, U.S.A.). Total RNA was isolated using the TRI-Reagent® (Ambion, Austin, TX, U.S.A.) following the manufacturer’s specifications.
Reverse-transcriptase assays
Total RNA was extracted as described above and treated with the Turbo DNA-free™ kit (Ambion). Reverse-transcription PCR analysis was performed using the Superscript™ II reverse transcriptase (Invitrogen, Carlsbad, CA, U.S.A.) according to the manufacturer’s instructions.
Constructs and mutant complementation
In-frame deletions of the entire coding sequence of the tonB2 chromosomal gene were generated by SOE-PCR (splicing-by-overlap-extension PCR) [22]. The fragments containing each deletion were cloned in the plasmid pCR2.1®-TOPO (Invitrogen), then subcloned in the suicide vector pDM4 (Table 2). The resulting suicide plasmids were mobilized into V. anguillarum 775 by conjugation using the E. coli MM294 helper strain carrying the pRK2013 vector (Table 2) and the chloramphenicol resistance used for selection purposes. The exconjugants were screened using a sucrose-resistance-based counter-selection as described previously [23]. The tonB2 deletion derivatives were obtained by PCR amplification using the primers FtonB2, delta 2, delta 3 and delta 4 (see Supplementary Table S1 at http://www.BiochemJ.org/bj/418/bj4180049add.htm) and for the alanine derivatives using primers FtonB2, tonB2-AAA and tonB2-6AC’ (Supplementary Table S1). The deletion mutants in the loop 3 region Δ179–195 loop 3, Δ179–184 loop 3, Δ185–190 loop 3, Δ191–195 loop 3 were also constructed using the SOE-PCR method. All the plasmids used in the present study were sequenced at the Core Facility of the Vollum Institute for Advanced Biomedical Research (Portland, OR, U.S.A.), using a 3130xL Genetic Analyzer (Applied Biosystems, Foster City, CA, U.S.A.).
55Fe–anguibactin uptake
The 55Fe–anguibactin complex was prepared using a stock solution of 55FeCl3 in 0.5 M HCl (specific radioactivity 103.46 mCi/mg; PerkinElmer, Boston, MA, U.S.A.). The 55FeCl3 was diluted to a final concentration of 0.8 mM in 0.1 M Mops buffer, pH 7.0, containing 1.2 mM EDDHA. Deferrated anguibactin was prepared as described previously [24] and added to a concentration of 1 mM in 0.1 M Mops, pH 7.0. The V. anguillarum cultures used for this assay were grown in CM9 minimal media with 1 mM IPTG until the exponential phase had been reached [attenuance (D600) 0.5–0.8]. These cultures were then treated for the 55Fe–anguibactin uptake experiment as described previously [24]. Bacterial dilutions were used to calculate the CFU (colony forming units) value. The results were fitted using the GraphPad Prism4 program.
55Fe–ferrichrome uptake
The 55Fe–ferrichrome complex was prepared as indicated by [25] using a 6.7:1 ferrichrome/55FeCl3 molar ratio in Mops buffer (0.1 M pH 7.0). Iron-free ferrichrome was purchased from Sigma–Aldrich. The E. coli cultures used for this assay were grown in M9 supplemented with IPTG until the exponential phase was reached. The cultures were centrifuged (10 000 g for 10 min at 4°C) and resuspended in M9 supplemented with 100 μM 2,2′-dipyridyl and incubated for 2 h to induce iron limitation. The results were fitted using the GraphPad Prism4 program.
Sequence alignments
Sequence alignments were carried out using CLUSTAL W [26] and formatted at the ESPript server (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
SDS/PAGE and Western immunoblotting
V. anguillarum or E. coli cells were cultured in minimal medium until a D600 of 0.4–0.7 was reached. At 20 min after induction with IPTG, 1 ml of culture was harvested by centrifugation. The cellular pellets were resuspended in 0.5 ml of 10% (w/v) trichloroacetic acid and incubated on ice for 1 h [27]. After this incubation period the samples were harvested and the cellular pellets were resuspended in 60 μl of loading buffer [4× sample buffer is 200 mM Tris/HCl, pH 6.8, 8% (w/v) SDS, 0.4% (v/v) glycerol, 1.4 M 2-mercaptoethanol and 0.02% Bromophenol Blue]. After electrophoresis, the proteins were transferred for 4 h at 300 mA into Immobilon™-PQS membranes (0.22 μm pore size; Millipore Corporation, Billerica, MA, U.S.A.) using a Trans-Blot® Electrophoretic Transfer Cell (Bio-Rad Laboratories Inc., Hercules, CA, U.S.A.). The transfer buffer used was 10 mM Caps [3-(cyclohexylamino)propane-1-sulfonic acid]/10% (v/v) methanol, pH 11. After the transfer step, the membranes were blocked at 4°C using 5% (w/v) casein in TBST buffer (20 mM Tris, pH 7.6, 0.3 M NaCl and 0.05% Tween-20). The membranes were washed with TBST buffer and incubated for 1 h with anti-TonB2 polyclonal antibody diluted 1:5000 in TBST/5% (w/v) BSA. After incubation the membranes were washed with TBST buffer and incubated with goat anti-rabbit IgG [H+L; HRP (horseradish peroxidase)-conjugated; Pierce Biotechnology Inc., Rockford, IL, U.S.A.] diluted 1:15 000 in TBST/5% BSA. The membranes were washed with TBST buffer and incubated with the Immobilon™ Western Chemiluminiscent HRP substrate (Millipore). The Western blots were developed using Kodak BioMax MR film.
Cloning, expression and purification of the C-terminal domain of TonB2
The tonB2 gene encoding the CTD of TonB2 residues 106–206 and 76–206 were amplified using as forward primers tonB2–106 and tonB2–76 and as reverse primer tonB2R (Supplementary Table S1), using Taq polymerase (Stratagene, La Jolla, CA, U.S.A.) and subcloned into the pET200 expression vector (Invitrogen), generating pCL21 and pCL22 (Table 2). Isotopically enriched proteins were obtained from cultures grown at 37°C until exponential phase in LB medium, 15N-labelled M9 minimal medium and 15N/13C-labelled M9 minimal medium (Cambridge Isotope Laboratories, Andover, MA, U.S.A.) and protein expression induced by adding 1 mM IPTG. The cultures were incubated for 4–5 h at 37°C and then harvested by centrifugation (10 000 g for 10 min at 4°C). The cellular pellets were resuspended in 10 mM Tris/HCl buffer, pH 7.9, 5 mM imidazole and 500 mM NaCl, containing DNase, RNase, lysozyme and PMSF. The cellular suspensions were lysed by two passes through a French press at 110.4 MPa (16 000 lbf/in2) and the clarified cell lysates purified by nickel affinity chromatography using Fast Flow chelating Sepharose. The eluted protein was buffer-exchanged into enterokinase cleavage buffer (20 mM Tris/HCl, pH 8.0, 100 mM NaCl and 2 mM CaCl2) and was subjected to proteolytic enterokinase cleavage overnight at room temperature (25°C). The buffer was subsequently exchanged to 20 mM Mes, pH 5.5, using low-molecular-mass-cut-off 10 DG gel-filtration columns (Bio-Rad). The sample was further purified using a Resource S cation-exchange column (Pharmacia) using a 0–2 M NaCl gradient. Finally the purified TonB2 CTD was buffer-exchanged into 10 mM NaH2PO4/Na2HPO4 buffer, pH 7.0, and concentrated to ≈0.5 mM for further experiments.
NMR spectroscopy
NMR experiments were performed on Bruker AVANCE 500 and 700 MHz spectrometers equipped with triple resonance inverse cyroprobes or a 600 MHz AVANCE with a digital console and a triple axis gradient probe. Data were processed using NMRPipe [28] and analysed with NMRVIEW [29]. Sequential assignments for HN, N, Cα and Cβ were determined using the automated backbone assignment program MONTE [30], with the data input formatted using the nvAssign module for NMRVIEW [31] using conventional triple-resonance experiments [32-34]. Side-chain assignments were obtained using the H(CCO)NH-TOCSY, C(CCO)NH-TOCSY, HBHA(CO)NH and 3D (three-dimensional) 15N-edited TOCSY experiments. Proton chemical shifts were referenced to DSS (2,2-dimethyl-2-silapentane-5-sulfonate) as 0 p.p.m., and 15N and 13C chemical shifts were referenced indirectly to DSS.
Structure calculation
Distance constraints for TonB2 CTD were derived from a 3D 15N-NOESY-HSQC (15N-NOESY-heteronuclear single quantum correlation) in a 1H2O/2H2O (9:1, v/v) solution and 3D 13C-NOESY-HSQC and 2D 1H/1H NOESY experiments in a 100% 2H2O solution with mixing times ranging from 100 to 150 ms. The distance restraints were calibrated and structures calculated using the CYANA 2.0 program [35,36]. Hydrogen-bond donors were identified from a series of 1H/15N HSQC experiments of previously lyophilized TonB2-CTD dissolved in 2H2O and monitored for 24 h. The corresponding hydrogen-bond acceptors were identified based on the NOE (nuclear Overhauser effect) pattern from the 3D HSQC-NOESY experiments. Dihedral angle restraints were determined using the TALOS program [37].
Interaction with FatA and FhuA TonB-box peptides
Samples of 0.5 mM 15N-labelled E. coli TonB CTD and TonB2 CTD (residues 106–206 and residues 76–206) were titrated with a 1.1:1 ratio of decamer peptides representing the TonB box of FhuA and the proposed TonB box of FatA. Details of the FhuA TonB box peptide have been described previously [15]. The FatA TonB box peptide has the sequence Glu-Ser-Ile-Thr-Val-Tyr-Gly-Glu-Ala, and the FhuA TonB-box peptide has the sequence Glu-Asp-Thr-Ile-Thr-Val-Thr-Ala-Ala-Pro. Both peptides had acetylated N-termini and were synthesized by Anaspec (San Jose, CA, U.S.A.) and purified and analysed by HPLC and MS respectively to a purity of greater than 95%.
RESULTS
Functional analysis of the C-terminal end of the V. anguillarum TonB2 protein
To investigate the functional significance of the C-terminal end of the TonB2 protein, we performed an in vivo analysis of specific tonB2 deletions. For this purpose a V. anguillarum ΔtonB2 null mutant was complemented with a copy of the wild-type gene and a series of C-terminal deletions in which the last two (Δ2-C-tonB2), three (Δ3-C-tonB2) or four (Δ4-C-tonB2) residues were deleted and the resulting strains were assayed for 55Fe–anguibactin transport. Our results indicate that the Δ2-C-tonB2 variant protein has 55Fe–anguibactin transport properties similar to those of the wild-type protein; however, larger deletions resulted in V. anguillarum strains that were no longer able of transporting anguibactin (Figure 1A).
Figure 1. (A) [55Fe]anguibactin uptake kinetics of TonB2 C-terminal deletion mutants and (B) C-terminal alanine derivatives.
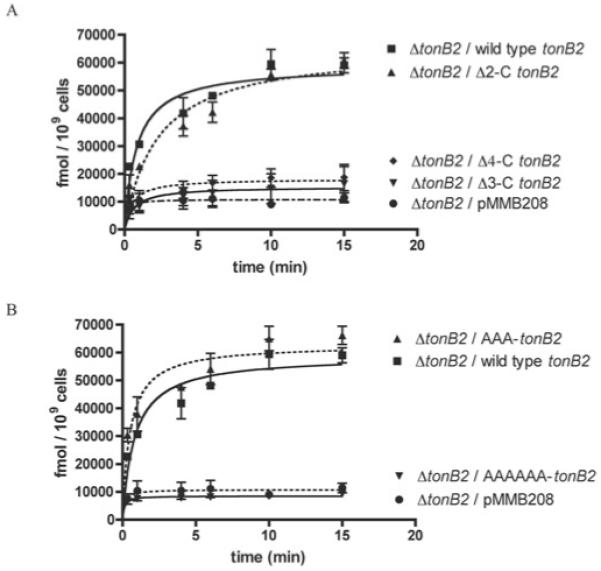
Results are means ± S.D. for at least three independent experiments. The ordinate refers to fmol of [55Fe]anguibactin taken up.
To assess whether the nature of the side chains of the last three amino acids plays a role in function, we constructed a tonB2 variant mutant in which the three C-terminal residues of TonB2, namely Ile-Ala-Lys, were replaced by alanine residues (-AAA-C’ tonB2). This variant protein has similar 55Fe–anguibactin transport rates when compared with the wild-type TonB2, indicating that the specific side chain of residue Ile204 is not critical for the TonB2 function (Figure 1B). Therefore the length of the protein, rather than the specific side chains of the residues at the extreme C-terminus, would seem to be important for the proper functioning of this protein. However, when the last six C-terminal residues, Glu-Phe-Lys-Ile-Ala-Lys, were replaced with a hexa-alanine peptide (-AAAAAA-C’ tonB2), the 55Fe–anguibactin transport through FatA was abolished, underscoring the importance of the nature of residues 201–203 for the action or stability of TonB2 (Figure 1B).
The amounts of the TonB2 mutant or wild-type protein that were present in the cells was determined using published methods [27]. We could only detect the presence of the TonB2 protein in the wild-type tonB2 (lane 2), Δ2-C tonB2 (lane 3) and Δ3-C tonB2 (lane 4) strains, indicating that the Δ4-C tonB2 variant protein is unstable (Figure 2, upper panel). mRNA transcripts were detected for all the mutants (results not shown), indicating that the absence of active TonB2 protein in the Δ4-C tonB2 mutant strain is not due to a lack of transcription, but is more likely caused by instability of the expressed protein.
Figure 2.
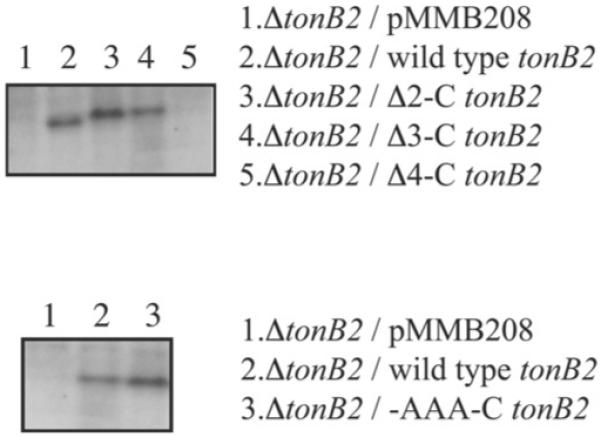
Western blots of the TonB2 C-terminal deletion mutants (upper panel) and the TonB2 C-terminal TonB2 alanine derivative (-AAA-C) (lower panel) using anti-TonB2 specific polyclonal antibodies
Western blots for the -AAA-C’ tonB2 variant demonstrated that this protein is stable (Figure 2, lower panel), whereas the hexaalanine mutant, -AAAAAA-C’ tonB2, was not detected (results not shown). Consistent with the results from the deletion mutants described above, this construct was properly transcribed (results not shown). These results indicate that the C-terminal portion of the V. anguillarum TonB2 protein may be organized in a manner different from that in the same region of E. coli TonB. This observation consequently prompted us to determine the solution structure of the TonB2 protein.
Determination of the structure of the TonB2 CTD
Initial NMR experiments were performed using a TonB2 CTD construct, encompassing residues 76–206, that has a length similar to that of the E. coli TonB CTD 103–239 construct that we previously studied by NMR [14]. Steady-state 15N NOE experiments indicated that, similar to the E. coli construct, a significant portion of the protein was unstructured. To simplify the NMR spectra we used a shorter construct encompassing residues 106–206, which we predicted to encode for the homologous ordered CTD. The 1H/15N HSQC spectrum of this shorter construct overlapped perfectly with that of the longer construct for all the well-dispersed peaks, with all of the missing peaks contained in the region between 7.5 and 8.5 p.p.m., as would be expected for random-coil chemical shifts. Consequently, all further structural studies were performed using the 106–206 construct.
Residues 106–120 of TonB2 had no significant NOEs in the 3D NOESY HSQC spectra, and they have negative steady-state NOE values, indicating that this portion of the protein is relatively flexible and unstructured (results not shown). Our model of residues 121–206 of TonB2 forms a well-defined αββαβ tertiary structure with a backbone RMSD (root mean square deviation) of 0.05 nm (0.5 Å). An overlay of the 20 lowest energy structures calculated for TonB2 (residues 106–206) is shown in Figure 3(A). Figure 3(B) shows a ribbon diagram of TonB2. The structures had only 17 NOE violations greater than 0.01 nm (0.1 Å) and no dihedral violations greater than 5°. Structural statistics and quality checks from PROCHECK NMR [38] and MOLMOL [39] for the refined residues 121–206 are summarized in Table 3 and are typical for a well-resolved high-resolution structure. The overall dimensions of the TonB2-CTD are 4.5 nm × 2.3 nm × 1.8 nm (45 Å × 23 Å × 18 Å). This is slightly longer and narrower the than TonB CTD from E. coli. The secondary structure is organized into a well-structured portion running from residues 121–131, followed by a short α1-helix consisting of residues 132–136, the β1-strand from residues 141 to 148, loop 1 from residues 149 to 151, the β2 strand from residues 152 to 161, loop 2 from residues 162 to 166, the conserved amphipathic α2-helix comprising residues 167–178, loop 3 from residues 179 to 195 and the β3 strand from residues 196 to 201. Together, the three β-strands form a single antiparallel β-sheet. Loop 3 is relatively long and not extremely well defined by the NOE network in comparison with the other loops in TonB2. The amide protons of the N-terminus of the amphipathic α-helix 2 give weak signals in the 1H/15N HSQC, indicating that these are involved in an intermediate exchange process.
Figure 3. Solution structure of TonB2 CTD(106–206) from V. anguillarum.
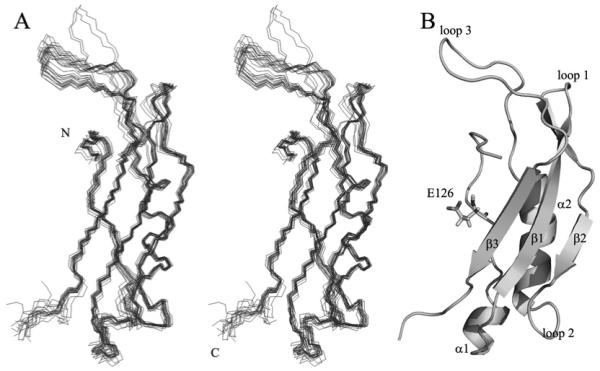
(A) A line stereo overlay of the backbone atoms of the 20 lowest energy structures calculated in the program CYANA. (B) A ribbon diagram of the lowest-energy structure of TonB2 in the same orientation as in (A). The secondary-structure elements are labelled for clarity. The E126 (Glu126) residue from TonB2, which is sequentially analogous to Q160 (Gln126) in E. coli TonB, is indicated.
Table 3.
Summary of the NMR constraints used in the TonB2 CTD structural calculation, as well as the restraint violations and structural statistics for the 20 lowest-energy conformers calculated in CYANA
| Parameter | Value |
|---|---|
| Cross-peaks | |
| With off-diagonal assignment | 2330 |
| With unique assignment | 1795 |
| With short-range assignment, |i−j|<=1 | 1530 |
| With medium-range assignment, 1<|i−j|<5 | 200 |
| With long-range assignment, |i−j|>=5 | 600 |
| Upper distance limits | |
| Total | 1519 |
| Short-range, |i−j|<=1 | 702 |
| Medium-range, 1<|i−j|<5 | 186 |
| Long-range, |i−j|> = 5 | 631 |
| Torsion-angle restraints | |
| H-bond restraints | |
| Distance restraint violations [> 0.01 nm (0.1 Å)] | 17 |
| Angle restraint violations (> 5°) | 0 |
| Average target function value | 2.93 ± 0.22 |
| RMSD (residues 121-202) | |
| Average backbone RMSD to mean [nm (Å)] | 0.049 ± 0.010 (0.49 ± 0.10) |
| Average heavy atom RMSD to mean [nm (Å)] | 0.940 ± 0.008 (0.94 ± 0.08) |
| Ramachandran analysis (residues 19–106) | |
| Residues in most favoured regions | 69.9% |
| Residues in additionally allowed regions | 26.5% |
| Residues in generously allowed regions | 0.3% |
| Residues in disallowed regions | 0.4% |
Comparison of the solution structure of the TonB2 CTD with that the TonB CTD from E. coli
TonB2 CTD from V. anguillarum has two significant differences in tertiary structure when compared with the solution structure of the TonB CTD (residues 103–239) from E. coli. The first major difference is that the loop extending from the α2-helix to the β3-strand is significantly longer in the V. anguillarum protein, extending an extra 0.9 nm (9 Å) into solution. Secondly, the β4-strand that is present in the E. coli protein is totally absent in the V. anguillarum protein (Figure 4, left-hand panel). Gln160 from the E. coli TonB protein, which is in close contact with the TonB box of the TBDTs [18-40], is replaced by a glutamic acid residue in TonB2 (Figure 3B). An overlay of the two proteins comparing residues 153–217 and 226–232 from the E. coli TonB-CTD with residues 119–183 and 196–202 from the V. anguillarum TonB2 reveals that these structures differ by a backbone RMSD of 0.27 nm (2.7 Å). A structure-based sequence alignment of these two TonB proteins clearly shows these structural differences (Figure 4, right-hand panel). Examination of the electrostatics of TonB2-CTD shows that this domain is slightly less positively charged than is the TonB CTD from E. coli (Figure 5), particularly along the face of the protein, where the TonB box from the outer-embrane transporters is known to interact with TonB.
Figure 4. Comparison of TonB2 CTD with TonB CTD from E. coli.
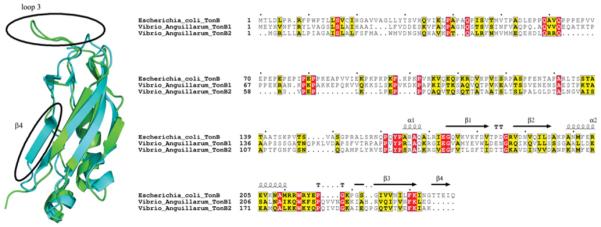
Left-hand panel: ribbon diagram of the backbone atoms of TonB CTD from E. coli (cyan) and TonB2-CTD from V. anguillarum (green) are shown. The largest structural differences are outlined by circles and labelled. Right-hand panel: sequence alignment of TonB proteins from E. coli and V. anguillarum. Sequence alignments were made in Clustal X, and formatting was from the ESPript server (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The secondary structure from PDB ID 1XX3 is diplayed at the top. The lengthened loop 3 between the α2-helix and the β3-strand is apparent in both TonB1 and TonB2 in V. anguillarum, as is the absence of the β4-strand. The gap-opening penalty used to generate this alignment was 3.
Figure 5. Electrostatic plots of TonB proteins.
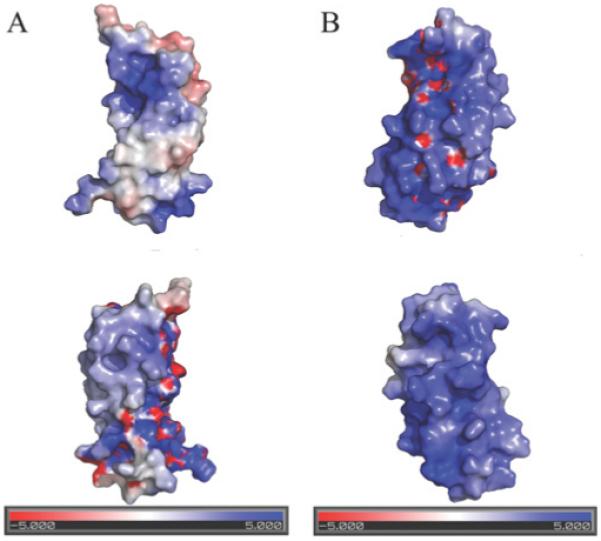
(A) V. anguillarum TonB2 CTD; the top surface diagram is in the same orientation as in Figure 4, and the bottom is rotated 180° around the y-axis. (B) E. coli TonB CTD; the top surface diagram is in the same orientation as in (A), and the bottom is rotated 180° around the y-axis. The electrostatic potential was calculated using the APBS (http://apbs.sourceforge.net/) plugin for Pymol (http://pymol.sourceforge.net/).
TonB-CTD from E. coli can bind to the TonB box from the TonB2-dependent transporter FatA
TonB2-CTD is slightly less basic overall than TonB-CTD from E. coli, the theoretical pI values for TonB2-CTD and TonB CTD being 9.52 and 10.41 respectively. As the TonB box of many receptors contains primarily hydrophobic and acidic residues, we wondered whether the composition of the electrostatic surface might be important in the recruitment of the TonB-box. We have shown previously that the TonB2 protein from V. anguillarum cannot complement the E. coli KP1032 tonB mutant in vivo, even in the presence of the accessory proteins ExbB2 and ExbD2 and the recently found TtpC [41] protein from V. anguillarum (Figure 6, upper panel; [10]). Since we were able to demonstrate that the isolated E. coli TonB CTD can bind to TonB box peptides of native receptors [14], we examined the ability of V. anguillarum TonB2 CTD and E. coli TonB CTD to bind the TonB box of the V. anguillarum ferric-anguibactin FatA TBDT.
Figure 6. [55Fe]Ferrichrome-uptake kinetics after 10 min of incubation in the E. coli KP1032 tonB strain (upper panel) and Western blot using anti-TonB2 specific polyclonal antibodies (lower panel).
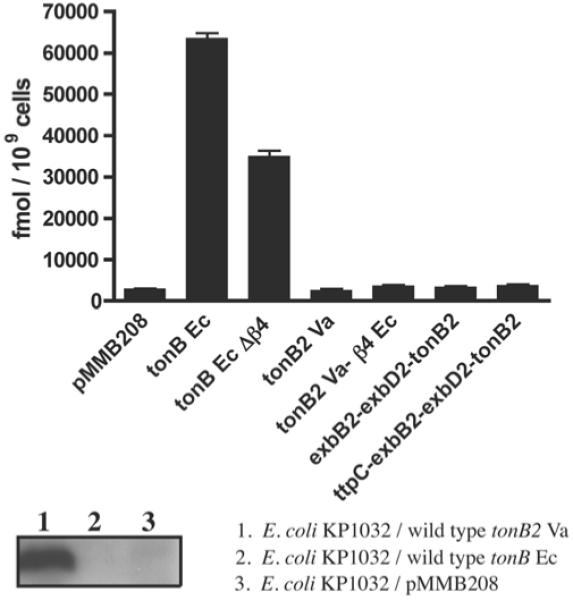
In the upper panel, results are means ± S.D. for three independent experiments.
Synthetic peptides corresponding to the E. coli FhuA and the V. anguillarum FatA TBDTs were added to the CTD constructs of both TonB and TonB2, yielding strikingly different results. Although similar chemical-shift changes were noted for E. coli TonB CTD binding to both the FhuA and FatA TonB boxes (Figure 7), essentially no chemical-shift changes are observed for both TonB2 constructs with the FhuA TonB box (results not shown). Moreover, only very minor shifts were observed with the FatA TonB box (Supplementary Figure S1 at http://www.BiochemJ.org/bj/418/bj4180049add.htm). In order to test whether the TonB-box peptide of FatA included any residues N-terminal to our original decamer peptide, we also tested the binding of a FatA TonB-box peptide, including two extra amino acids at the N-terminus from the native FatA receptor. However, no binding was detected for this peptide either.
Figure 7. Titration of E. coli TonB CTD with FhuA and FatA peptides.
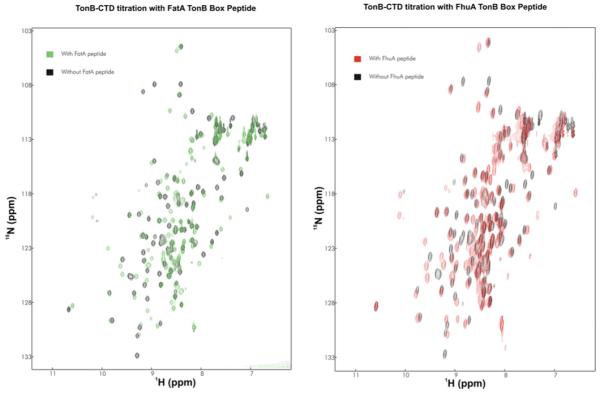
Left-hand panel: 1H/15N-HSQC spectrum of E. coli TonB CTD with (green) and without (black) the FatA TonB box peptide. Right-hand panel: 1H/15N-HSQC spectrum of E. coli TonB CTD with (red) and without (black) the FhuA TonB box.
The E. coli TonB protein can substitute for TonB2 function in V. anguillarum ferric-anguibactin transport
In order to determine whether the in vitro binding of the E. coli TonB CTD to the FatA TonB box could translate into in vivo functionality, we decided to verify whether the TonB protein from E. coli could complement a double V. anguillarum ΔtonB1 ΔtonB2 mutant in anguibactin transport. The rationale for using this double ΔtonB1 ΔtonB2 non-polar mutant is that, a priori, we did not know whether E. coli TonB could use the V. anguillarum ExbB1D1 or ExbB2D2 complexes. Hence, using this genetic background we avoid the titration of any of these complexes. [55Fe]Anguibactin transport studies in V. anguillarum demonstrate that the E. coli TonB protein is indeed capable of substituting for TonB2 in the transport of anguibactin through FatA (Figure 8). Interestingly, neither enterobactin nor the catechol-type siderophore vanchrobactin, a recently characterized siderophore from a V. anguillarum O2 serotype strain [42], which are normally transported via the TonB2 system, were utilized as iron sources (results not shown), suggesting that TonB might not be able to recognize their specific TBDTs.
Figure 8. [55Fe]Anguibactin-uptake kinetics of the TonB2–β4Ec chimaera and the E. coli TonB (tonBEc) in V. anguillarum.
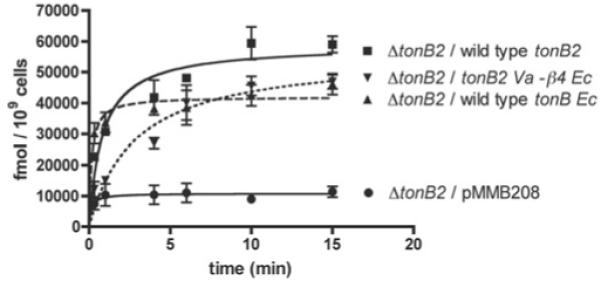
Results are means ± S.D. for at least three independent experiments.
A TonB2 chimaera with an extended C-terminus was unable to complement the function of the E. coli TonB
The TonB proteins from E. coli and V. anguillarum share 36% identity and 55% similarity over their entire length. However, TonB2 is 38 amino acid residues shorter than TonB and lacks the extra β4 strand at the extreme C-terminus. As mentioned above, TonB2 is unable to complement the function of the E. coli TonB even in the presence of its accessory inner membrane proteins (Figure 6, upper panel). We also determined that this lack of function was not due to TonB2 protein instability in the E. coli cells (Figure 6, lower panel). To further identify whether the lack of complementation was due to a shorter C-terminus, we designed a protein chimaera in which the β4 strand from the E. coli TonB protein (β4Ec) was fused to the V. anguillarum TonB2 protein (TonB2Va). The results indicate that this TonB2Va–β4Ec chimaera is still able to energize the transport of [55Fe]anguibactin at levels similar to those of the wild-type TonB2 in V. anguillarum (Figure 8). Moreover, it was also able to utilize enterobactin and vanchrobactin as iron sources, as assessed by bioassays (results not shown). On the other hand, similar to the wild-type TonB2 protein, the TonB2Va–β4Ec protein chimaera could not substitute for the E. coli TonB in ferrichrome (Figure 6, upper panel) or enterobactin transport (results not shown).
A specific region of the loop 3 from the V. anguillarum TonB2 is essential for anguibactin transport
As mentioned above, one of the differences observed between the E. coli and the V. anguillarum TonB2 protein was the presence of a longer loop extending from the α2 helix to the β3 strand of TonB2. To evaluate the functionality of this loop, we constructed the four mutants depicted in Figure 9(A). The results indicate that the Δ185–190-TonB2 and Δ191–195-TonB2 variant proteins displayed lower [55Fe]anguibactin transport rates compared with the wild-type TonB2; however, deletion of the complete loop 3 (Δ179–195-TonB2) resulted in a lack of transport (Figure 9B). Interestingly, the Δ179–184-TonB2 variant protein also showed a lack of [55Fe]anguibactin transport, suggesting that this region of the loop is essential for the proper function of the protein. Reverse-transcription PCR analysis of RNA isolated from these strains showed that the constructs were properly transcribed (results not shown). Moreover, by Western-blot analysis we were able to detect the four variant proteins (Figure 9C), indicating that the lack of transport detected in two of them was not due to a lack of protein expression.
Figure 9. (A) Partial sequence of the wild-type TonB2 from amino acid residues 168–206, (B) uptake kinetics and (C) Western blots.
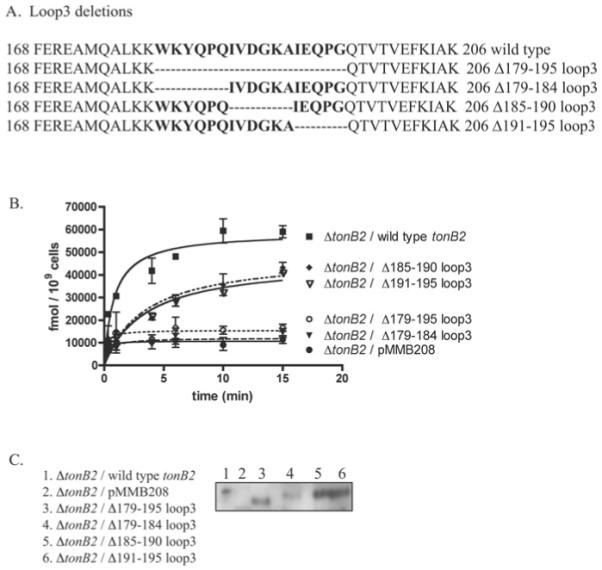
Residues within loop 3 are in bold. The remaining residues of loop 3 after each deletion are depicted. (B) [55Fe]Anguibactin-uptake kinetics of mutants in loop 3. Results are means ± S.D. for at least three independent experiments. (C) Western Blots of loop-3 deletion derivatives.
DISCUSSION
Our results obtained with both deletion and alanine-derivative mutants of the V. anguillarum TonB2 protein demonstrate that, without a fully intact β3-strand, TonB2 is non-functional, most likely due to increased susceptibility to degradation in vivo. This instability is consistent with the previously reported results in other bacteria such as E. coli [19,20]. This indicates that a complete three-stranded β-sheet is essential for both TonB2 and TonB stability. Bioinformatics analysis of the known genomic TonB sequences demonstrate that virtually all of them contain at least a three-stranded β-sheet TonB core and that most commonly the protein ends without a fourth β-strand [43]. These in silico findings are supported by two recently determined structures, the V. anguillarum TonB2 CTD reported here, as well as the preliminary HasB secondary structure from Serratia marcescens [15].
The absence of the β4 strand in the V. anguillarum TonB2 CTD has implications with regard to the role of dimerization of the TonB protein. In all the crystal structures of the E. coli TonB CTD in which there is a protein dimer interface, the β4 strand is crucial for a large portion of the interaction [11-13]. In vivo studies with mutants in which this part of the protein was deleted indicated that the remaining protein (TonBEc Δβ4) showed a reduced ability to energize the TBDT as compared with the full-length TonB (Figure 6, upper panel; [19]), suggesting that this region, although not essential, is important for its function. In the case of the crystal structure of the C-terminal 92 residues of E. coli TonB [13], the dimer interface consists of 13.19 nm2 (1320 Å2) of buried surface area per monomer; however, in the V. anguillarum TonB2 CTD, assuming a similar type of dimeric interface, there is a maximum of 4.24 nm2 (425 Å2) of surface area available for potential dimerization. This is less than the 5.99 nm2 (600 Å2) that has been suggested as the minimum interface required for the formation of protein–protein dimers [44]. Since TonB2 lacks sufficient surface to make these types of inter-strand interactions, it is unlikely that this particular dimer form of TonB can play a role in the mechanism of TonB2 binding to the TBDTs. In the case of the intertwined dimer, a significant portion of the dimeric interface is provided by an extension of the β1-strand, which would be possible in the V. anguillarum TonB2 protein, but the dimeric interface would nonetheless be greatly diminished without the β4-strand extension (Figure 10). Our results do not exclude the possibility of different forms of TonB dimerization and higher-order oligomerization in TonB-dependent energy-transduction mechanism, as has been suggested by previous works [45,46].
Figure 10. (A) Crystal structure of TonB-92 (92-residue C-terminal fragment) from E. coli, redrawn from the co-ordinates given by Koedding et al. [13].
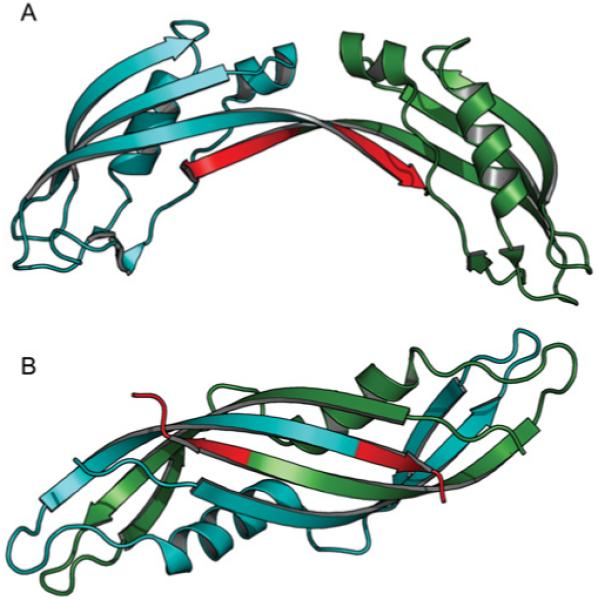
Subunit A is coloured blue and subunit B is coloured dark green. Residues 235–239 are highlighted in red to emphasize which residues are absent in TonB2 from V. anguillarum. (B) Crystal structure of an intertwined dimer form of TonB-CTD from E. coli [11,12]. The colour scheme is the same as in (A).
In the present study we have also demonstrated that the addition of the β4 strand from E. coli TonB to the C-terminus of V. anguillarum TonB2 did not affect the function of the protein. Moreover, this chimaera was able to energize the TBDT FatA during ferricanguibactin transport, indicating that the extended C-terminus did not alter the recognition of the Vibrio TBDT TonB boxes. On the other hand, an extended C-terminus in TonB2 was not sufficient to allow the protein to act as a functional energy transducer in E. coli. The absence of complementation of the V. anguillarum TonB2 and its derivative in E. coli might be due to the inability of these proteins to recognize the E. coli TBDT, since even in the presence of the accessory inner-membrane proteins ExbB2, ExbD2 and TtpC, the TonB2 protein is not functional in E. coli.
One of the unanswered questions in the TonB-mediated iron-transport field is how the TonB protein recognizes the various TBDTs to promote transport of a variety of substrates through the outer membrane. E. coli has only one single TonB protein, and all of the TBDTs have recognizable and similar TonB boxes. However, many bacteria, including V. anguillarum, encode two or more TonB proteins in their genome [43]. Often these TonB proteins do not have fully overlapping functions with respect to the TBDTs in which they can facilitate transport. Although the precise molecular determinants of this specificity are not clear, it is presumably defined by the structure of the C-terminus of the particular TonB protein and the structure and/or sequence of the corresponding TonB box. Our in vivo as well as the in vitro results indicate that (an)other factor(s) might also be involved in the molecular recognition process in this particular form of TonB.
The fact that the TonB CTD from E. coli appears to be able to bind to both the FhuA and the FatA TonB box, which has a fairly different amino acid sequence compared with most of the native E. coli TonB receptors, was surprising. In V. cholerae the −1 residue preceding the TonB box has an important effect on the ability of E. coli TBDTs to be recognized by the V. cholerae TonB1 protein [47]. Specifically, the presence of an aromatic residue in this position is essential for protein interaction. With regard to the V. anguillarum TonB boxes, only three TBDTs have been characterized so far: FatA, the TBDT for anguibactin, (TonB-box Asp-Glu-Ser-Ile-Thr-Val-Tyr-Gly-Gln; the −1 residue is indicated in bold), FvtA, the TBDT for vanchrobactin and enterobactin [48] (TonB-box Gln-Asp-Thr-Val-Val-Val-Val-Gly-Glu), and HuvA [49], the TBDT for haem (TonB-box Phe-Asp-Glu-Val-Val-Val-Ser-Ala-Thr). In V. anguillarum TonB2 energizes these three TBDTs, even though TonB1 can also act on the HuvA receptor. Considering these TonB-box sequences, it seems that unlike the situation in the V. cholerae TonB1, an aromatic residue at position −1 is not essential for the recognition of TonB2 towards the TBDT. Given the different electrostatic surface observed with the V. anguillarum TonB2 as compared with the E. coli TonB and the absence of a β4 strand along the presumed binding site for the TonB boxes, more studies focused on the differences in binding to various TonB-boxes are needed.
What is the purpose of the extra-long loop that joins the α2-helix to the β3 strand in the V. anguillarum TonB2? It is clear that this extra-long loop is present in the majority of known sequenced TonB proteins [43]. Moreover, this loop is proximal to the region of the protein thought to interact with the periplasmic binding protein FhuD in the case of E. coli TonB [50]. The 55Fe-anguibactin transport kinetic studies performed with various mutants within this loop revealed that its complete presence is not essential for the TonB2 function. Interestingly, the first six amino acid residues at the beginning portion of loop 3 (Figure 10A), which is more conserved with respect to the E. coli loop 3, is important for TonB2 function, as a deletion of these residues resulted in a lack of anguibactin transport. It is worth mentioning that this TonB2 variant protein lacks two of the aromatic residues (Trp179 and Tyr181) that are well conserved in the E. coli and V. anguillarum TonB proteins and that were ascribed as important for TonB in vivo oligomerization and function in E. coli [45]. Therefore it seems possible that they could contribute to the oligomerization of TonB2 as well; however, we cannot rule out the possibility that this region of the loop could also play a role in the interaction of TonB2 with periplasmic binding proteins. Experiments to test our hypotheses are currently being performed.
TonB proteins are critical to the viability and pathogenicity of a number of clinically important micro-organisms. Most of the structural and biophysical work that has been reported for TonB proteins in the past has focused on the E. coli protein. However, it appears that the structure of V. anguillarum TonB2 has two obvious differences when compared with E. coli TonB. On the basis of the absence and elongation of several structural features and a differing electrostatic profile, TonB2 from V. anguillarum likely represents a different structural and functional class of TonB protein distinct from that of TonB from E. coli. Our understanding of these different types of TonB proteins is critical for effective structure-based drug-design programs, and organism-specific TonB-targeting antimicrobial compounds may be a possibility in the future.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Kathleen Postle (Department of Biochemistry and Molecular Biology, Pennsylavania State University, University Park, PA, U.S.A.) for providing the KP1032 strain used in the present study. We are indebted to Dr Deane McIntyre (Bio-NMR Centre, University of Calgary, Calgary, AB, Canada) for the upkeep of the NMR spectrometers.
FUNDING
This work was supported by grants [nos. AI19018, GM64600] from the National Institutes of Health to J.H.C. Operating support was also obtained from the Canadian Institutes of Health Research to H.J.V. R.S.P. was the recipient of Studentship awards from the Natural Sciences and Engineering Research Council of Canada and the Alberta Heritage Foundation for Medical Research. H.J.V. is the recipient of a Scientist award from the Alberta Heritage Foundation for Medical Research.
Abbreviations used
- CFU
colony forming units
- CTD
C-terminal domain
- 3D
three-dimensional
- DSS
2,2-dimethyl-2-silapentane-5-sulfonate
- EDDHA
ethylenediamine-N,N-’bis-(2-hydroxyphenylacetic acid)
- HRP
horseradish peroxidase
- HSQC
heteronuclear single quantum correlation
- IPTG
isopropyl β-d-thiogalactoside
- LB
Luria–Bertani
- NOE
nuclear Overhauser effect
- RMSD
root mean square deviation
- TBDT
TonB-dependent transporter
- SOE-PCR
splicing-by-overlap-extension PCR
Footnotes
The co-ordinates for the ensemble of structures can be found in the Protein Data Bank (http://www.rcsb.org) (entry code 2k9k), and the NMR restraints are deposited at the BioMagResBank (http://www.bmrb.wisc.edu) (entry code 15988).
REFERENCES
- 1.Krewulak KD, Vogel HJ. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta. 2008;1778:1781–1804. doi: 10.1016/j.bbamem.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 2.Braun V, Braun M. Iron transport and signaling in Escherichia coli. FEBS Lett. 2002;529:778–785. doi: 10.1016/s0014-5793(02)03185-x. [DOI] [PubMed] [Google Scholar]
- 3.Crosa JH. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron-sequestering system. Nature. 1980;284:566–568. doi: 10.1038/284566a0. [DOI] [PubMed] [Google Scholar]
- 4.Alice AF, Lopez CS, Crosa JH. Plasmid- and chromosome-encoded redundant and specific functions are involved in biosynthesis of the siderophore anguibactin in Vibrio anguillarum 775: a case of chance and necessity? J. Bacteriol. 2005;187:2209–2214. doi: 10.1128/JB.187.6.2209-2214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadner RJ. Vitamin B12 transport in Escherichia coli: energy coupling between membranes. Mol. Microbiol. 1990;4:2027–2033. doi: 10.1111/j.1365-2958.1990.tb00562.x. [DOI] [PubMed] [Google Scholar]
- 6.Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol. 1993;175:3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen RA, Thomas MG, Postle K. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 1999;31:1809–1824. doi: 10.1046/j.1365-2958.1999.01317.x. [DOI] [PubMed] [Google Scholar]
- 8.Ahmer BM, Thomas MG, Larsen RA, Postle K. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J. Bacteriol. 1995;177:4742–4747. doi: 10.1128/jb.177.16.4742-4747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Held KG, Postle K. ExbB and ExbD do not function independently in TonB-dependent energy transduction. J. Bacteriol. 2002;184:5170–5173. doi: 10.1128/JB.184.18.5170-5173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stork M, Di Lorenzo M, Mourino S, Osorio CR, Lemos ML, Crosa JH. Two tonB systems function in iron transport in Vibrio anguillarum, but only one is essential for virulence. Infect. Immun. 2004;72:7326–7329. doi: 10.1128/IAI.72.12.7326-7329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang C, Mooser A, Pluckthun A, Wlodawer A. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem. 2001;276:27535–27540. doi: 10.1074/jbc.M102778200. [DOI] [PubMed] [Google Scholar]
- 12.Koedding J, Howard P, Kaufmann L, Polzer P, Lustig A, Welte W. Dimerization of TonB is not essential for its binding to the outer membrane siderophore receptor FhuA of Escherichia coli. J. Biol. Chem. 2004;279:9978–9986. doi: 10.1074/jbc.M311720200. [DOI] [PubMed] [Google Scholar]
- 13.Koedding J, Killig F, Polzer P, Howard SP, Diederichs K, Welte W. Crystal structure of a 92-residue C-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments. J. Biol. Chem. 2005;280:3022–3028. doi: 10.1074/jbc.M411155200. [DOI] [PubMed] [Google Scholar]
- 14.Peacock RS, Weljie AM, Peter Howard S, Price FD, Vogel HJ. The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J. Mol. Biol. 2005;345:1185–1197. doi: 10.1016/j.jmb.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 15.Lefevre J, Simenel C, Delepelaire P, Delepierre M, Izadi-Pruneyre N. 1H, 13C and 15N resonance assignments of the C-terminal domain of HasB, a specific TonB like protein, from Serratia marcescens. Biomol. NMR Assign. 2007;1:197–199. doi: 10.1007/s12104-007-9055-7. [DOI] [PubMed] [Google Scholar]
- 16.Peacock RS, Andrushchenko VV, Demcoe AR, Gehmlich M, Lu LS, Herrero AG, Vogel HJ. Characterization of TonB interactions with the FepA cork domain and FecA N-terminal signaling domain. Biometals. 2006;19:127–142. doi: 10.1007/s10534-005-5420-0. [DOI] [PubMed] [Google Scholar]
- 17.Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 18.Shultis DD, Purdy MD, Banchs CN, Wiener MC. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 19.Anton M, Heller KJ. Functional analysis of a C-terminally altered TonB protein of Escherichia coli. Gene. 1991;105:23–29. doi: 10.1016/0378-1119(91)90509-a. [DOI] [PubMed] [Google Scholar]
- 20.Bruske AK, Heller KJ. Molecular characterization of the Enterobacter aerogenes tonB gene: identification of a novel type of tonB box suppressor mutant. J. Bacteriol. 1993;175:6158–6168. doi: 10.1128/jb.175.19.6158-6168.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook B, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY; Cold Spring Harbor Laboratory Press: 2001. [Google Scholar]
- 22.Senanayake SD, Brian DA. Precise large deletions by the PCR-based overlap extension method. Mol. Biotechnol. 1995;4:13–15. doi: 10.1007/BF02907467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milton DL, O’Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez CS, Alice AF, Chakraborty R, Crosa JH. Identification of amino acid residues required for ferric-anguibactin transport in the outer-membrane receptor FatA of Vibrio anguillarum. Microbiology. 2007;153:570–584. doi: 10.1099/mic.0.2006/001735-0. [DOI] [PubMed] [Google Scholar]
- 25.Larsen RA, Postle K. Conserved residues Ser16 and His20 and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J. Biol. Chem. 2001;276:8111–8117. doi: 10.1074/jbc.M007479200. [DOI] [PubMed] [Google Scholar]
- 26.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaskula JC, Letain TE, Roof SK, Skare JT, Postle K. Role of the TonB amino terminus in energy transduction between membranes. J. Bacteriol. 1994;176:2326–2338. doi: 10.1128/jb.176.8.2326-2338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 29.Johnson B, Blevins R. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 30.Hitchens T, Lukin J, Zhan Y, McCallum S, Rule G. MONTE: An automated Monte Carlo based approach to nuclear magnetic resonance assignment of proteins. J. Biomol. NMR. 2003;25:1–9. doi: 10.1023/a:1021975923026. [DOI] [PubMed] [Google Scholar]
- 31.Kirby NI, DeRose EF, London RE, Mueller GA. NvAssign: protein NMR spectral assignment with NMRView. Bioinformatics. 2004;20:1201–1203. doi: 10.1093/bioinformatics/bth064. [DOI] [PubMed] [Google Scholar]
- 32.Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31-kDa protein. J. Magn. Reson. 1992;96:432–440. [Google Scholar]
- 33.Kay LE, Xu GY, Yamazaki T. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O Saturation. J. Magn. Reson. 1994;109:129–133. [Google Scholar]
- 34.Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance three-dimensional nmr experiments with improved sensitivity. J. Magn. Reson. 1994;103:203–216. [Google Scholar]
- 35.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 36.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 37.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 39.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 40.Cadieux N, Kadner RJ. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10673–10678. doi: 10.1073/pnas.96.19.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stork M, Otto BR, Crosa JH. A novel protein, TtpC, is a required component of the TonB2 complex for specific iron transport in the pathogens Vibrio anguillarum and Vibrio cholerae. J. Bacteriol. 2007;189:1803–1815. doi: 10.1128/JB.00451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soengas RG, Anta C, Espada A, Paz V, Ares IR, Balado M, Rodriguez J, Lemos ML, Jimenez C. Structural characterization of vanchrobactin, a new catechol siderophore produced by the fish pathogen Vibrio anguillarum serotype O2. Tetrahedron Lett. 2006;48:3021–3024. [Google Scholar]
- 43.Chu BC, Peacock RS, Vogel HJ. Bioinformatic analysis of the TonB protein family. Biometals. 2007;20:467–483. doi: 10.1007/s10534-006-9049-4. [DOI] [PubMed] [Google Scholar]
- 44.Janin J, Miller S, Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol. 1988;204:155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- 45.Ghosh J, Postle K. Disulphide trapping of an in vivo energy-dependent conformation of Escherichia coli TonB protein. Mol. Microbiol. 2005;55:276–288. doi: 10.1111/j.1365-2958.2004.04384.x. [DOI] [PubMed] [Google Scholar]
- 46.Sauter A, Howard SP, Braun V. In vivo evidence for TonB dimerization. J. Bacteriol. 2003;185:5747–5754. doi: 10.1128/JB.185.19.5747-5754.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mey AR, Payne SM. Analysis of residues determining specificity of Vibrio cholerae TonB1 for its receptors. J. Bacteriol. 2003;185:1195–1207. doi: 10.1128/JB.185.4.1195-1207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naka H, Lopez CS, Crosa JH. Reactivation of the vanchrobactin siderophore system of Vibrio anguillarum by removal of a chromosomal insertion sequence originated in plasmid pJM1 encoding the anguibactin siderophore system. Environ. Microbiol. 2007;10:265–277. doi: 10.1111/j.1462-2920.2007.01450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazoy R, Osorio CR, Toranzo AE, Lemos ML. Isolation of mutants of Vibrio anguillarum defective in haem utilisation and cloning of huvA, a gene coding for an outer membrane protein involved in the use of haem as iron source. Arch. Microbiol. 2003;179:329–338. doi: 10.1007/s00203-003-0529-4. [DOI] [PubMed] [Google Scholar]
- 50.Carter DM, Miousse IR, Gagnon JN, Martinez E, Clements A, Lee J, Hancock MA, Gagnon H, Pawelek PD, Coulton JW. Interactions between TonB from Escherichia coli and the periplasmic protein FhuD. J. Biol. Chem. 2006;281:35413–35424. doi: 10.1074/jbc.M607611200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


