Abstract
Objective
The objective of the study is to report 2 new genotypic forms of protease-sensitive prionopathy (PSPr), a novel prion disease described in 2008, in 11 subjects all homozygous for valine at codon 129 of the prion protein (PrP) gene (129VV). The 2 new PSPr forms affect individuals who are either homozygous for methionine (129MM) or heterozygous for methionine/valine (129MV).
Methods
Fifteen affected subjects with 129MM, 129MV, and 129VV underwent comparative evaluation at the National Prion Disease Pathology Surveillance Center for clinical, histopathologic, immunohistochemical, genotypical, and PrP characteristics.
Results
Disease duration (between 22 and 45 months) was significantly different in the 129VV and 129MV subjects. Most other phenotypic features along with the PrP electrophoretic profile were similar but distinguishable in the 3 129 genotypes. A major difference laid in the sensitivity to protease digestion of the disease-associated PrP, which was high in 129VV but much lower, or altogether lacking, in 129MV and 129MM. This difference prompted the substitution of the original designation with “variably protease-sensitive prionopathy” (VPSPr). None of the subjects had mutations in the PrP gene coding region.
Interpretation
Because all 3 129 genotypes are involved, and are associated with distinguishable phenotypes, VPSPr becomes the second sporadic prion protein disease with this feature after Creutzfeldt-Jakob disease, originally reported in 1920. However, the characteristics of the abnormal prion protein suggest that VPSPr is different from typical prion diseases, and perhaps more akin to subtypes of Gerstmann-Sträussler-Scheinker disease.
Human prion diseases are prominently heterogeneous. In sporadic Creutzfeldt-Jakob disease (sCJD), the most prevalent prion disease, heterogeneity is largely predicated on the common methionine (M)/valine (V) polymorphism at codon 129 of the prion protein (PrP) gene and the disease-associated PrP (PrPDis) that are distinguished in types 1 and 2 based on the electrophoretic mobility of their protease-resistant regions.1
However, despite this remarkable heterogeneity, all well-established sporadic prion diseases (here operationally defined as nonacquired prion diseases free of mutations in the PrP gene coding region) have been shown to share the same basic pathogenetic mechanism; PrPDis interacts with the normal or cellular PrP and converts it into PrPDis, triggering an autocatalytic process that leads to the accumulation of PrPDis and ultimately to the clinical disease.2
In 2008, we described 11 cases affected by a new disease involving PrP; we named this disease protease-sensitive prionopathy (PSPr).3 Subsequently, 2 additional cases of PSPr have been independently reported.4,5 PSPr differed from known sporadic prion diseases in the clinical presentation, in the histopathologic and immunohistochemical features, and in the basic characteristics of the PrPDis. Furthermore, all 11 cases had the 129VV genotype and no mutation in the PrP gene open reading frame (ORF).
We now report 15 additional cases, all of which bear features of the PSPr as originally reported. However, the new cases also include, in addition to new 129VV subjects, individuals who are 129MV heterozygous and 129MM homozygous. Although the affected subjects belonging to the 3 genotypes share several important characteristics, they also display basic variations that allow the 3 corresponding phenotypes to be distinguished. Therefore, the new cases show that the disease originally described as PSPr, like sCJD, affects all 3 129 genotypes and to some extent mimics the 129-related phenotypic heterogeneity of sCJD, although the PSPr characteristics underline basic differences from sCJD and similarities with Gerstmann-Sträussler-Scheinker disease (GSS), a rare phenotype, which to date has been reported as exclusively associated with PrP gene mutations. In view of the increased protease-resistance of the PrPDis associated with the new 129 genotypes compared to that of the 129VV cases, we propose to revise the original PSPr label to VPSPr or “variably protease-sensitive prionopathy.” Parts of these findings have been presented previously.6–9
Subjects and Methods
Subjects
A total of 15 affected subjects, including 3 129MM, 6 129MV, and 6 previously unreported 129VV, were examined. Thirteen affected subjects were referred to the National Prion Disease Pathology Surveillance Center (NPDPSC) (Cleveland, OH) between 2002 and 2010. All cases were symptomatic except 1 of the 129MM subjects, who died suddenly of heart problems while participating in a dementia study as a negative control, underwent autopsy, and was referred to the NPDPSC because it was noted to have spongiform degeneration (SD) on histological examination. One 129MM subject was received by Dr Fabrizio Tagliavini10 (National Neurological Institute, Instituto Nazionale Neurologico Carlo Besta, Milan, Italy), and 1 129MV subject was received by Dr Piero Parchi (Department of Neurological Sciences, University of Bologna, Dipartimento di Scienze Neurologiche, Universitá di Bologna, Bologna, Italy). All the subjects including those serving as positive control as indicated were examined at autopsy following analyses of fixed and frozen tissues. Consent was obtained for using tissues for research, including genetic analyses.
Tissue Processing
Fixed and frozen brain tissues were processed as previously described; a different procedure was followed for the case received from Dr Tagliavini.3,10–12
Histopathology and Immunohistochemistry
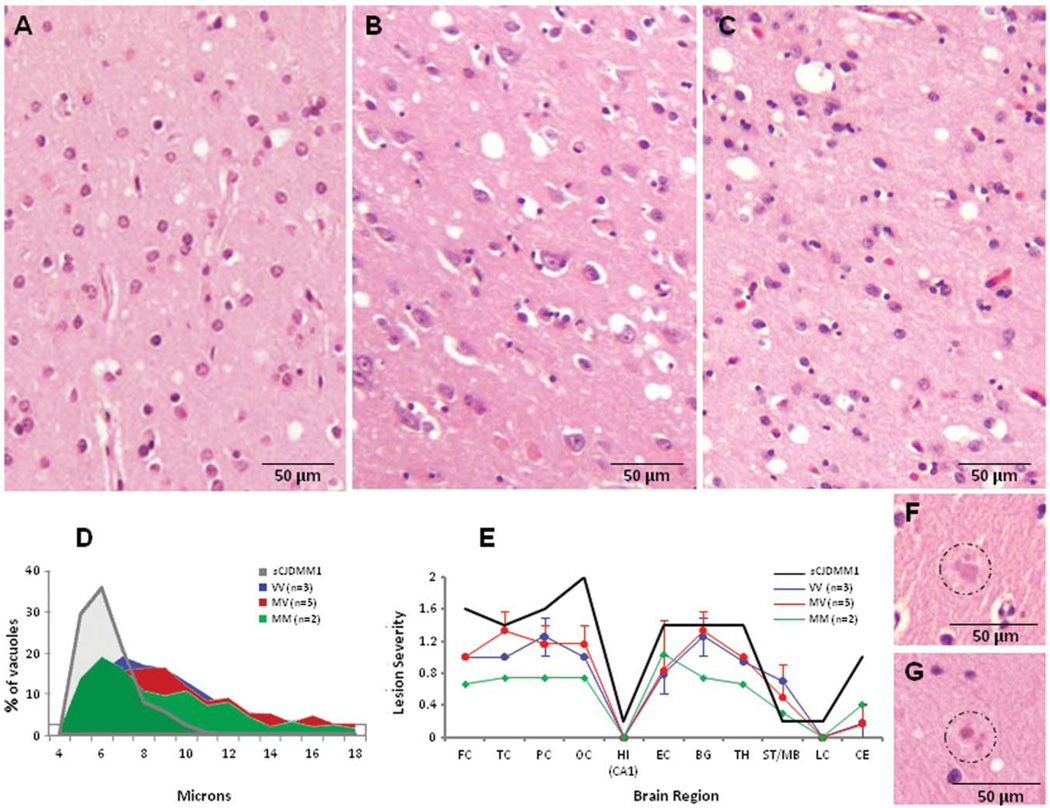
Samples obtained from up to 18 brain regions were processed according to previously described procedures.3,12 Lesion profiles were constructed using semiquantitative evaluation of SD and astrogliosis in 11 brain regions from 10 subjects, including the 3 129 genotypes. SD and astrogliosis were scored (Fig 1), and the scores from each of the brain regions were summed for each subject separately; values were averaged, their standard deviations determined, and they were plotted according to the brain region.3,12 Vacuoles with >4µm diameter were measured individually on random photomicrographs of frontal neocortex (10/subject, ×180) using Spotsoftware version 4.6 after calibration (Diagnostic Instruments, Sterling Heights, MI).3
FIGURE 1.
Histopathology with vacuole size and lesion profiles in the cases belonging to the 3 129 genotypes of variably protease-sensitive prionopathy (VPSPr). The spongiform degeneration is qualitatively similar in all 3 129 genotypes (A, 129VV; B, 129MV; C, 129MM). (D) As originally shown in the 129VV cases,3 the spongiform degeneration is made of a significant percentage of relatively large and midsize vacuoles on average significantly larger than those of common sporadic Creutzfeldt-Jakob disease (sCJD) subtypes (diameters: VPSPr [combined] 9.3 ± 3.4µ vs sCJDMM1 5.8 ± 1.2µ; p < 0.0001 [Student t test]), resulting in an elongated vacuole size distribution in the vacuole size histogram. (E) The lesion profiles are very similar in the 3 129 genotypes, but show less severe lesions in the 129MM genotype than in the 129VV and 129MV genotypes. FC, TC, PC, and OC = frontal, temporal, parietal, and occipital cortices; HI = CA1 of hippocampus; EC = entorhinal cortex; BG = basal ganglia; TH = thalamus (medial-dorsal nucleus); ST/MB = striatum/midbrain; LC = pons (locus coeruleus); CE = cerebellar cortex. The vertical bars refer to standard deviations. Spongiform degeneration was scored on a 0 to 4 scale (0, not detectable; 1, mild; 2, moderate; 3, severe; and 4, confluent), and astrogliosis on a 0 to 3 scale (0, not detectable; 1, mild; 2, moderate, and 3, severe). (F, G) Homogeneous micro deposits with the appearance of plaques were observed in the molecular layer of the cerebellum in some cases associated with the 129VV (F) and 129MV (G) genotypes, but not in the 3 129MM cases. (A–C, F, G) Hematoxylin & eosin. M = methionine; V = valine.
Sections from the frontal and occipital neocortices, hippocampus, basal ganglia, thalamus, cerebellar hemisphere, and midbrain were processed for PrP immunohistochemistry with the monoclonal antibody (mAb) 3F4 or 1E4 (Cell Sciences, Canton, MA).11–17 Selected brain regions were also immunostained with the mAbs 4G8 to amyloid β or PHF1 to the tau protein.3
Molecular Genetics
The entire PrP ORF was amplified by polymerase chain reaction using genomic DNA (extracted from unfixed brain tissue or blood) and the primers 42F (CATAACTTAGGGTCACATTTGTCC) and 45R (CCAGATTAACCAATGGTTATTTGC); sequencing was done directly or after cloning into plasmid pSTBlue 1 (Novagen, Madison, WI) by automated sequencing.13
Prion Protein Characterization
REAGENTS AND ANTIBODIES
Phenylmethylsulfonyl fluoride was purchased from Sigma Chemical Co. (St. Louis, MO). Peptide N-glycosidase F (PNGase F) was purchased from New England Biolabs (Beverly, MA) and used following the manufacturers protocol. Reagents for enhanced chemiluminescence (ECL Plus) were from Amersham Pharmacia Biotech (Piscataway, NJ). Antibodies to various sequences of human PrP included anti-C, a rabbit antiserum (220–231), 3F4, a mouse mAb (106–110), and 1E4, a mAb (97–108).3,14–17
BRAIN HOMOGENIZATION
The 10% (weight/volume) brain homogenates were prepared in 9 volumes of lysis buffer (100mM Tris, 150 mM NaCl, 0.5% Nonidet P40, 0.5% deoxycholate, 5mM ethylenediaminetetraacetic acid, pH 8.0) on ice using pestles with Eppendorf tubes driven by a cordless motor as previously described.14 When required, brain homogenates were centrifuged at 1,000 × g for 10 minutes at 4°C to collect supernatant.
IMMUNOBLOT ANALYSIS
Samples were resolved on 15% Tris-HCl Criterion precast gels (Bio-Rad Laboratories, Hercules, CA) for gel electrophoresis and Western blotting as described previously.15 The proteins on the gels were transferred to Immobilon-P membrane polyvinylidene fluoride (Millipore, Billerica, MA) for 2 hours at 70V. For probing PrP, the membranes were incubated for 2 hours at room temperature with anti-PrP antibodies. Following incubation with horseradish peroxidase-conjugated sheep antimouse immunoglobulin G (IgG) or donkey antirabbit IgG at 1:3,000, the PrP bands were visualized on Kodak film (Eastman Kodak, Rochester, NY) by ECL Plus as described by the manufacturer.
Results
Clinical Features
The cases, grouped according to the 129 genotype, demographics, and clinical data and tests, along with the previous 129VV cases, are summarized in the Table.
TABLE.
Summary of Cases
| Codon 129 | Distribution, % (No.) |
Onset, yr ± SD (range) |
Duration, mo ± SD (range) |
Main Neurological Signs |
Diagnostic Tests |
Family History of Dementia |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Psychiatric,a % |
Aphasia, % |
Parkinsonism, % |
Ataxia, % |
Myoclonus,b % | Frontal Type Dementia |
14.3.3, Positive/ Total or % |
EEG, Typical/ Total or % |
MRI,c Typical/ Total or % |
|||||
| VPSPr | |||||||||||||
| MM | 12 (3)d | 64 & 78e | 41 & 50 | 0 | 50 | 100 | 50 | 100 | 0 | 1/2 | 1/2 | 0/2 | 1/2 |
| MV | 23 (6) | 72 ± 7f(65–81) | 45 ± 24g(7–72) | 83 | 17 | 67 | 67 | 40 | 50 | 0/2 | 0/2 | 0/6 | 0/5 |
| VV | 65 (17)h | 65 ± 8e,f (48–77) | 23 ± 17g(10–60) | 71 | 47 | 30 | 71 | 5 | 60 | 1/7 | 0/14 | 1/17 | 7/13 |
| Total % distribution | 100 | 67 ± 10 (48–81) | 30 ± 21 (7–72) | 68 | 48 | 48 | 64 | 20 | 52 | 2/12 | 1/18 | 1/25 | 8/20 |
| sCJDi | |||||||||||||
| MM1 | 68 | 65 (42–91) | 4(1–18) | 28 | 23 | 7 | 33 | 97 | NA | 95% | 80% | 75% | — |
| MV1 | 2 | 62 (51–72) | 12 | 25 | 0 | 75 | 100 | NA | 71% | ||||
| VV1 | 1 | 37 (19–55) | 21(10–49) | 0 | 33 | 33 | 0 | 67 | NA | 100% | 0% | 100% | — |
| MM2 | 2 | 64 (49–77) | 16 (9–36) | 0 | 33 | 33 | 17 | 67 | NA | 75% | 0% | 43% | — |
| MV2 | 9 | 60 (41–81) | 17 (5–72) | 34 | 11 | 22 | 81 | 77 | NA | 80% | 7% | 86% | — |
| VV2 | 16 | 6 (3–18) | 19 | 0 | 6 | 100 | 66 | NA | 7% | 70% | — | ||
| Totals | 98j | 63 | 6 | 26 | 18 | 8 | 49 | 87 | NA | 89% | 56% | 72% | |
Psychiatric symptoms include depression, psychosis, and personality/behavioral changes.
Generally appeared late in the disease.
Twenty-one of 25 VPSPr patients showed significant cerebral atrophy.
One of the 3 129MM subjects who died of accidental causes before onset of clinical disease has been excluded.
p < 0.025;
p < 0.045;
p < 0.03 (statistical analysis by GraphPad [La Jolla, CA] Prisma 5 software).
Including the 11 129VV cases previously published. 3
Data adapted from Gambetti et al,1 Parchi et al,12 and Zou et al17; cases with co-occurrence of disease-associated prion protein type 1 and 2 have been omitted.14,18,19
Does not include sporadic fatal insomnia (2%).20
SD = standard deviation; EEG = electroencephalogram; MRI = magnetic resonance imaging; VPSPr = variably protease-sensitive prionopathy; M = methionine; V = valine; sCJD = sporadic Creutzfeldt-Jakob disease.
In 129VV cases, the presentation was characterized by 1 or more components of a triad comprising psychiatric signs, in the form of behavior and mood changes, speech deficit, and cognitive impairment. Behavior and mood changes, expressed as disinhibition, euphoria, and impulsivity or loss of interest and apathy, were the most frequent (80% of the cases). Language deficits, observed in half of the cases, were characterized by anomic or semantic aphasia, or by dysarthria. Cognitive impairment, mostly of the frontal lobe type, was present at onset in 50% of the cases, alone or together with the behavioral changes and language deficits.
In the 129MV subtype, psychiatric signs were often associated with parkinsonism, followed by ataxia and myoclonus, whereas aphasia was rare; in these cases, the mean age at onset (72 years) and duration (45 months) of the disease were the most advanced and the longest, respectively, of the 3 subtypes and the duration was significantly different from those of the 129VV genotype (p < 0.017).
Both symptomatic 129MM subjects (1 died apparently before clinical onset of disease) presented with Parkinsonism and ataxia followed by progressive diffuse cognitive impairment and myoclonus; aphasia was reported in 1 case, but neither showed psychiatric symptoms.
As for the diagnostic tests, just 1 VV case showed signal changes consistent with CJD on magnetic resonance imaging and electroencephalography; all the other cases revealed various degrees of brain atrophy and diffuse slowing of cerebral electrical activity.
Familial occurrence of dementia was reported in about 50% (7/14 available family histories) of the 129VV cases (1 case in the present series), and in 1 129MM, but not in the 129MV genotype.
Histopathology
The hallmark common to all 129 genotypes was the presence of moderate SD comprising vacuoles in the major cerebral regions, which were relatively larger than those observed in sCJDMM1 but overall smaller than those of sCJDMM2 (see Fig 1A–E and Fig 1A–C of Gambetti et al3). Occasionally, the molecular layer of the cerebellum contained small homogeneous formations, with the appearance of microplaques in the 129VV and 129MV cases (see Fig 1F, G). On average, all these lesions were more severe in the 129VV and 129MV than in the 129MM cases (see Fig 1E).
The pattern of PrP immunostaining was slightly different in the 3 genotypes.
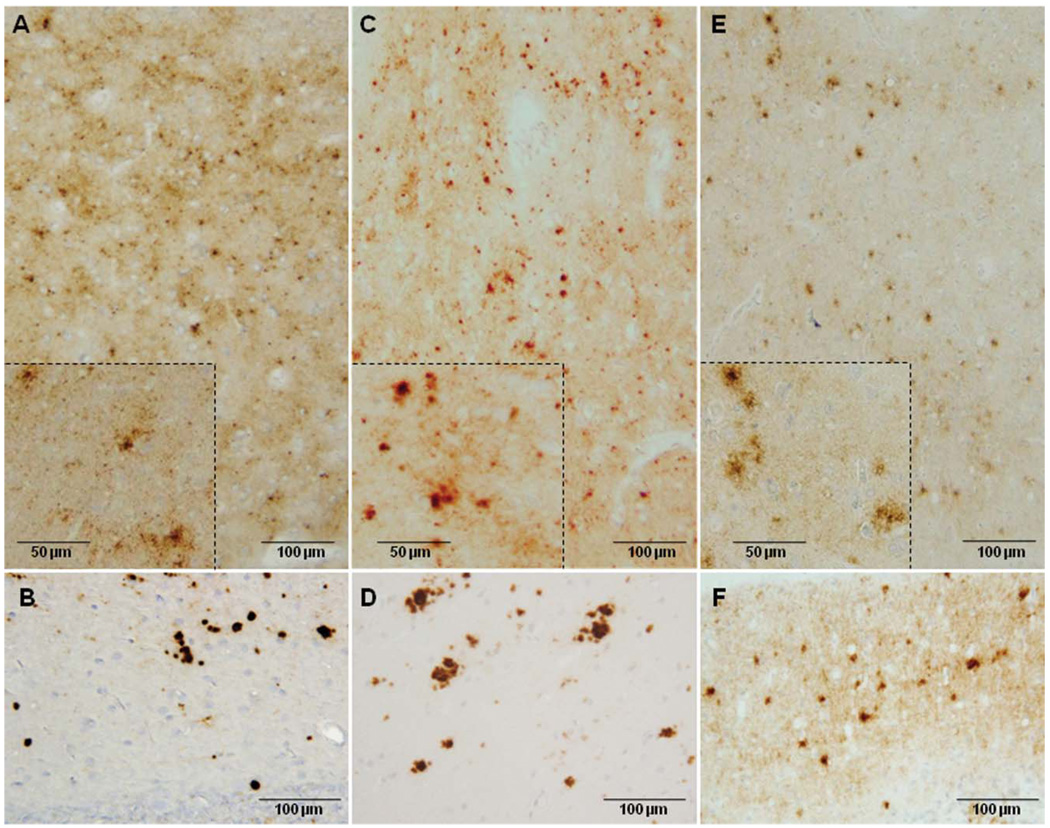
In the 129VV cases, the PrP immunostaining, as originally described, was targetlike in the cerebrum (Fig 2A) and dotlike in the cerebellar molecular layer (see Fig 2B).
FIGURE 2.
Prion protein immunohistochemistry in the 3 variably protease-sensitive prionopathy 129 genotypes. The cerebral cortex (A, C, E) and cerebellar molecular layer (B, D, F) best exemplify the predominant immunostaining patterns. (A) pattern in the 129VV genotype is often targetlike, with a larger stained granule or clusters of granules surrounded by smaller granules in a focal or more diffuse background of punctate or synaptic staining (inset: higher magnification of the same cortical region). (B) The molecular layer of the cerebellum shows relatively large granules that are often compact and intensely stained. (C) In the 129MV genotype, the targetlike pattern is generally less obvious, as large granules are more often isolated; focal or larger areas of synaptic staining are also present (inset: as above). (D) In the cerebellum, the granules are fewer, more loose, and have a plaquelike appearance. (E) The 129MM genotype often shows a plaquelike immunostaining pattern (inset: as above). (F) The cerebellum shows small formations. Immunostaining was done with monoclonal antibody 3F4. M methionine; V = valine.
In the 129MV genotype, the targetlike pattern was less recognizable (see Fig 2C). The cerebellar molecular layer showed a more plaquelike immunostaining pattern (see Fig 2D).
In the 129MM subjects, the predominant immunostaining pattern was plaquelike (see Fig 2E). The cerebellum occasionally showed small plaquelike formations (see Fig 2F).
With mAb 1E4, the patterns of PrP immunostaining were similar to those revealed by 3F4 (data not shown).
Various degrees of amyloid β immunostaining, apparently age-correlated, were also observed (data not shown).
Characterization of PrPDis in the 3 129 Genotypes
PRPDIS ELECTROPHORETIC PROFILE AND PROTEINASE K RESISTANCE
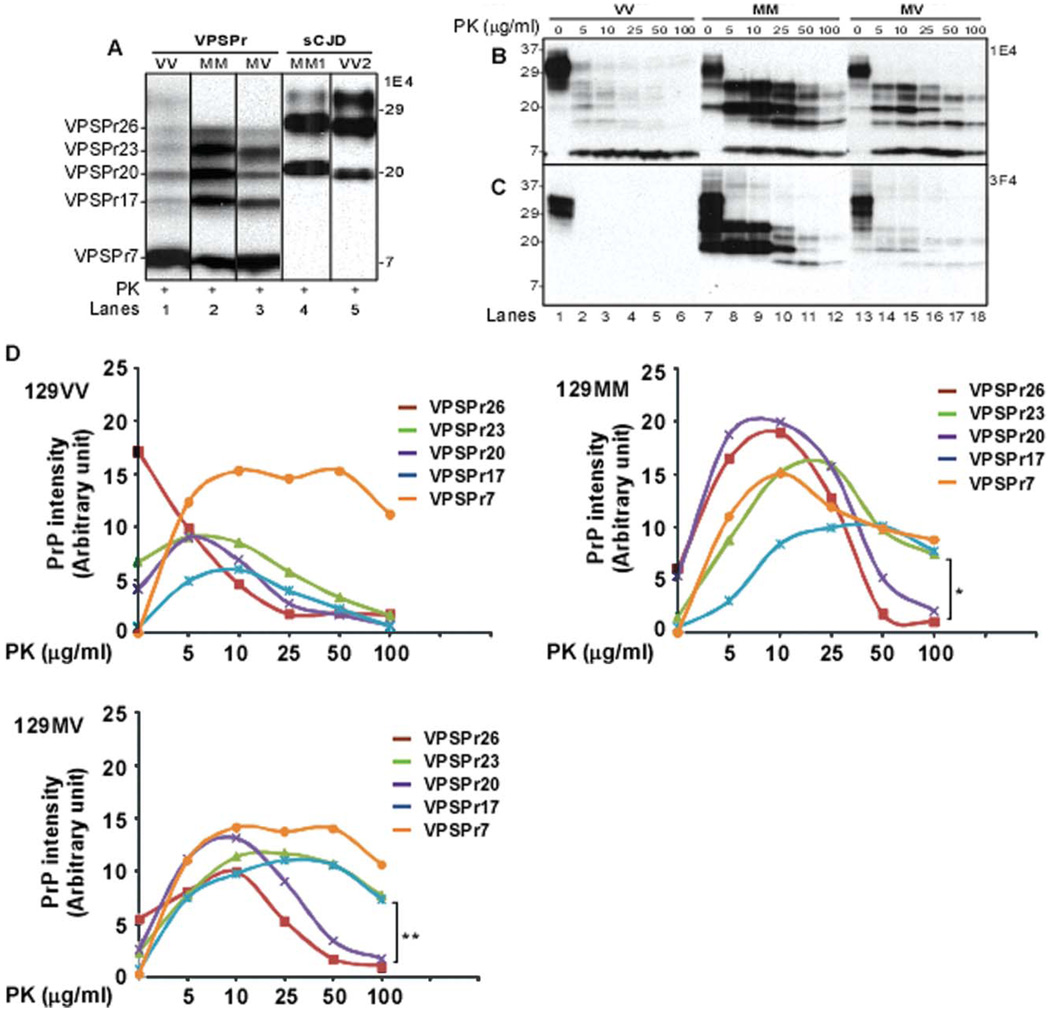
The ladderlike electrophoretic profile of the proteinase K (PK)-resistant PrPDis fragments, the distinctive feature of the PSPr 129VV cases, was shared by all the affected subjects belonging to the 129MM and 129MV genotypes, although, due to the higher PK resistance, the representation of most PrPDis fragments was greater in 129MM and 129MV than in 129VV (Fig 3A).3 The ladderlike profile demonstrated with 1E4 consisted of 5 major bands migrating at approximately 26kDa, 23kDa, 20kDa, 17kDa, and 7kDa (hereafter identified as VPSPr26, VPSPr23, VPSPr20, VPSPr17, and VPSPr7) (see Fig 3A). In contrast, PrPDis types 1 and 2 from sCJD formed the classical pattern of 3 bands migrating at 32/30kDa, 28/26kDa, and 21/19kDa (see Fig 3A).
FIGURE 3.
Electrophoretic profiles and proteinase K (PK) titration of PK-resistant disease-associated prion protein (PrPDis) from variably protease-sensitive prionopathy (VPSPr) associated with the 129VV, 129MM, or 129MV genotype. (A) The Western blots of the total brain homogenates (BHs) treated with 25µg/ml of PK and probed with the monoclonal antibody 1E4 reveal 5 PrP bands migrating approximately to 26kDa, 23kDa, 20kDa, 17kDa, and 7kDa, forming a ladderlike pattern in all 3 (129VV, 129MM, and 129MV) genotypes of VPSPr (VPSPr26, VPSPr23, VPSPr20, VPSPr17, and VPSPr7) (lanes 1–3). The faint band that migrates at approximately 30kDa in VPSPr-129VV (lane 1) likely represents the incomplete PK digestion of the normal diglycosylated, N-terminus truncated PrP fragment or associated monoglycosylated full-length PrP. In contrast, BHs from sporadic Creutzfeldt-Jakob disease (sCJD) associated with the 129MM genotype and the PrPDis type 1 (sCJDMM1) or sCJDVV2 (sCJD with the 129VV genotype and PrPDis type 2) show the typical 3 PK-resistant PrP fragments of type 1 and 2 migrating between 31kDa and 19kDa (lanes 4 and 5). (B, C) PK titration of PrPDis. Brain homogenates from 129VV, 129MM, and 129MV genotypes were treated with PK at various concentrations between 0 and 100µg/ml. (B) Probed with 1E4. (C) Probed with 3F4. (D) PK titration with quantitative analysis of the individual VPSPr fragments. The curves represent the relative amounts of the individual VPSPr fragments at increasing PK concentrations (0–100µg/ml) after probing the immunoblots with 1E4 in each of the 3 129 genotypes. The relative representations of the bands corresponding to the VPSPr fragments were determined by densitometry and expressed as averages of 129VV (n = 3), 129MM (n = 2), and 129MV cases (n = 3). Comparative analysis of the curves from each of the 3 129 genotypes confirms the PK sensitivity of all the fragments in 129VV cases, with the exception of VPSPr7, which is resistant to PK in all 3 genotypes. The remaining 4 fragments follow similar patterns in both the 129MM and 129MV genotypes; VPSPr26 and VPSPr20 form rapidly but are digested at PK concentrations >10µg/ml; VPSPr23 and VPSPr17 are resistant up to 100µg/ml of PK (*p < 0.02; **p < 0.005). M = methionine; V = valine.
PrPDis preparations from the 3 genotypes were probed with mAb 1E4 or 3F4 after treatment with various amounts of PK. When PrPDis fragments were considered all together, both 1E4 and 3F4 confirmed the relatively high PK resistance of PrPDis in the 129MM cases, intermediate in the 129MV cases, and low or entirely lacking in the 129VV cases (see Fig 3). However, both mAbs also showed the heterogeneous resistance of the individual fragments to PK, which was confirmed with PK titration experiments. These analyses showed that the VPSPr7 fragment was highly PK-resistant in all 3 genotypes. In contrast, the other 4 fragments appeared to follow 2 distinct patterns, which were similar and involved pairs of the same fragments in both 129MV and 129MM; VPSPr26 and VPSPr20 increased and decreased rapidly in amount peaking at 10µg/ml of PK and generated fairly narrow bell-shaped curves. In contrast, both PSPr23 and PSPr17 increased at a lower rate, peaked between 25 and 50µg/ml of PK, and remained relatively well represented even at 100µg/ml of PK. The representations of the 2 pairs of fragments were significantly different at 100µ/ml PK concentration in both 129 genotypes (129MM, p < 0.02; 129MV, p < 0.005). As expected, the PK resistance of the 129VV fragments was much lower, except for VPSPr7. Combined, the immunoblots and quantitative analyses argue that VPSPr23 and VPSPr17 have the strongest resistance to PK and likely form secondarily from VPSPr26 and VPSPr20 following treatment with high PK concentrations. It has to be noted, however, that the PK sensitivity of the 129VV preparations was in part related to the mAb used. When probing with 1E4 instead of 3F4, all fragments present in the VPSPr-129MM and −129MV preparations were also detectable in the preparations from the 129VV genotype, even if they displayed different ratios. Therefore, the PK treatment might not only break down the PrPDis associated with the 129VV genotypic form, but also generate fragments relatively undetectable by the mAb 3F4. Alternatively, PrPDis associated with VPSPr-129VV might have a low immunoreactivity with 3F4, even without PK treatment.
IDENTIFICATION OF THE PRPDIS CORE FRAGMENTS AND THEIR COMPARISON WITH THOSE OF THE GSS VARIANT LINKED TO THE A117V MUTATION
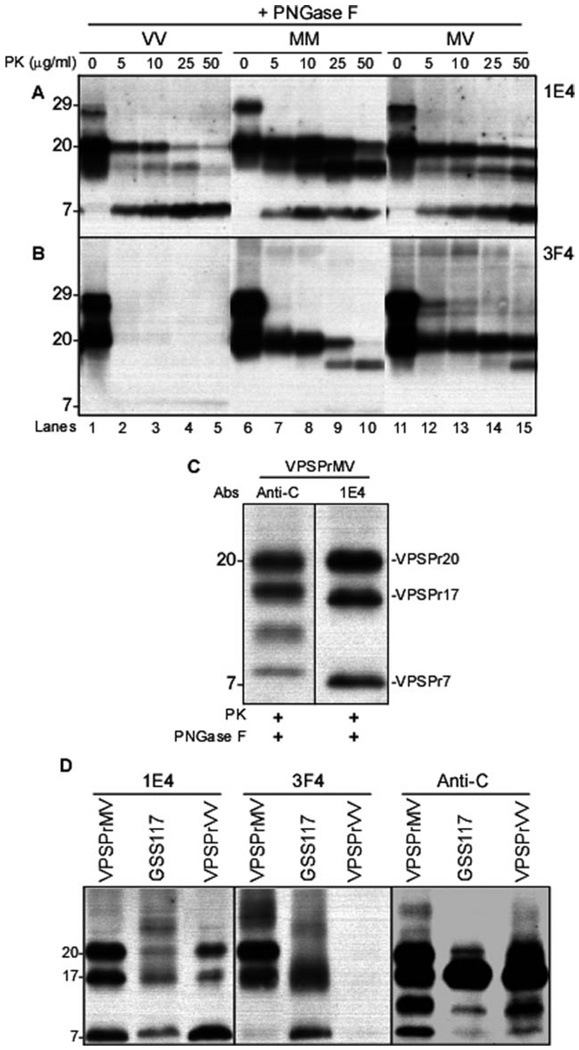
Various amounts of VPSPr20, VPSPr17, and VPSPr7 were demonstrated by 1E4 in all 3 129 genotypes after deglycosylation and up to 50µg/ml of PK treatment (Fig 4A). Because the deglycosylation eliminated VPSPr26 and VPSPr23, these 2 fragments likely are the glycosylated isoforms of VPSPr20 and VPSPr17, respectively. This would explain the shared level of PK resistance of these 2 fragments (see Fig 3D). Of notice, the same PK-resistant fragments were well represented also without deglycosylation, suggesting that VPSPr20 and VPSPr17 are present as both glycosylated and unglycosylated isoforms (see Fig 3A, B). With 3F4, only VPSPr20 and VPSPr17 were detectable in the 129MM and 129MV cases, whereas again the 129VV genotype showed no PK-resistant PrP (see Fig 4B). The combined and individual resistance of the deglycosylated fragments to PK was comparable to that of the glycosylated isoforms.
FIGURE 4.
Proteinase K (PK)-resistant core fragments of variably protease-sensitive prionopathy (VPSPr) and their comparison with the disease-associated prion protein (PrPDis) fragments associated with Gerstmann-Sträussler-Scheinker disease (GSS)-A117V. All brain homogenates were treated with increasing concentrations of PK and with peptide N-glycosidase F (PNGase F). (A) With 1E4, all immunoblots from cases with the 129VV, 129MM, and 129MV genotypes essentially show variably protease-sensitive prionopathy (VPSPr)-20, VPSPr17, and VPSPr7. However, the PK resistance of these bands varies according to the 129 genotype and the individual bands within the same genotype in a way roughly similar to that shown in Figure 3. (B) The immunoblots probed with 3F4 reveal 2 major bands in the 129MM and 129MV genotypes, which, however, exhibit a quite different pattern of resistance to PK in the 2 genotypes. As expected, no PK-resistant PrP bands are detected in the 129VV genotype. (C) Additional analysis of the core fragments following treatment with 25µg/ml of PK and PNGase, using the antibody anti-C to the PrP C-terminal residues 220–231. Compared to 1E4, anti-C demonstrates 4 bands of 20kDa, 18kDa, 12–13kDa, and 8kDa, respectively, of which only the 20kDa band has the same gel mobility as VPSPr20 detected with 1E4. The other 3 bands, including 18kDa, 12–13kDa, and 8kDa, do not match the bands detected with 1E4. (See text for explanation). (D) Brain homogenates from VPSPr-129MV, GSS117, and VPSPr-129 were treated with PK and PNGase F prior to Western blotting and probed with 1E4 (left panel), 3F4 (middle panel), and anti-C (right panel) antibodies. With monoclonal antibody 1E4, bands matching VPSPr20, VPSPr17, and VPSPr7 are detected in the GSS-A117V preparations, but the GSS-V117V bands immunoreact much less with 1E4 than the bands of VPSPr-129MV and VPSPr-129VV. The 23kDa band is seen more prominently in GSS-A117V. With 3F4, the VPSPr17 and VPSPr7 bands are shared by GSS-A117V and VPSPr-129MV, but the 20kDa band is missing in GSS-A117V. The VPSPr7 band is much less reactive in 129MV. As previously, the 129VV is not reactive with 3F4. Anti-C reveals apparently the same bands in all 3 preparations, but with significantly different ratios. M = methionine; V = valine.
Further characterization of the core fragments with the antibody anti-C (C-terminal residues 220–231) demonstrated 4 PrP bands migrating at approximately 20kDa, 18kDa, 12–13kDa, and 8kDa, of which only the 20kDa band matched VPSPr20 detected with 1E4 and 3F4 (see Fig 4C). The 3 fragments undetected by 1E4 and 3F4 must comprise the C-terminal region (reactive with anti-C) and must lack the 97–112 sequence containing the 1E4 and 3F4 epitopes. Therefore, 6 core fragments of relative molecular weights between 20kDa and 7kDa were identified by the combined use of 1E4 and anti-C. Several PrP C-terminal fragments of similar relative molecular weight have been previously reported.18–20 Considerable similarities were observed in the electrophoretic mobilities of the PK-resistant core fragments from the 129MV, 129VV, and 129MM genotypes and GSS-A117V (see Fig 4D). Also in GSS-A117V, the mAb 1E4 demonstrated the presence of 3 bands of 20kDa, 17kDa, and 7kDa described in VPSPr, which, however, displayed different immunoreactivities. Comparable variations in antibody immunoreactivity and band representation were seen with 3F4 and anti-C (see Fig 4D). Therefore, most of the PK-resistant PrPDis fragments appeared to have similar sizes in VPSPr and GSS-A117V, but different ratios and antibody reactivities.
Discussion
At variance with the series of 11 cases of PSPr described in 2008 and 2 cases subsequently published by others—all 13 of which were 129VV homozygous at the PrP gene—the 15 cases reported here also include affected subjects who are 129MV and 129MM, in addition to new 129VV subjects.3–5 Comparative analyses indicated that all these cases are affected by the same disease process, and that most of the heterogeneity that we observed results from distinct 129 genotypes.
These cases are likely to be affected by the same disease because of the overall similarity in major phenotypical characteristics, including the clinical features, which prominently exhibit aphasia, ataxia, and parkinsonian signs; SD, displaying vacuoles comparable in size in the 3 genotypes but otherwise different from the vacuoles of other common prion diseases; and finally PrP immunostaining patterns, which also display comparable general features in all these cases. However, the 2 most striking similarities reside in the ladderlike electrophoretic pattern of the PK-resistant fragments and in the unique immunoreactivity of PrPDis with mAb 1E4. Cumulatively, these findings suggest that all these cases share a similar molecular mechanism of PrPDis formation.
Significant clinical differences among the 129VV and 129MV groups (only 2 129MM symptomatic subjects were available) occurred in the mean age at onset and in disease duration (see Table). PrP immunostaining patterns were also distinguishable in the 3 groups. An additional difference might lay in disease prevalence, which appeared to be highest in 129VV subjects (65% of the cases), followed by the 129MV (23%) and 129MM subjects (12%) (see Table). However, 2 distinctive features were evident among the 3 groups; these were: (1) the apparent resistance to PK digestion, which was generally much lower in the 129VV cases than in the 129MM and 129MV cases; and (2) the immunoreactivity of the PK-resistant PrPDis with mAb 3F4, which was strong in the 129MM cases, weak in the 129MV cases, and lacking in the 129VV cases. Cumulatively, these findings argue that, although PrPDis may be formed by a similar mechanism in the 3 genotypes, the conformation or aggregation is likely different, and this difference results in variable resistance to PK, variable accessibility by 3F4, or both.
These findings also indicate that, in the present series of cases, it is the 129 genotype that modifies the phenotypic characteristics, including PK resistance and antibody immunoreactivity of PrPDis. However, the possibility that phenotypic heterogeneity is caused by other variations in PrPDis among the 3 groups or by a combination of different 129 genotypes and PrPDis characteristics, as is the case with sCJD, cannot be excluded.
The variations in prevalence are likely to be associated with the 129 genotype as well. This also is a feature of sCJD, in which 129MM cases account for about 70% of the total, 129MV for 11%, and 129VV for 17%.1,12 It is remarkable that the effect of the 129 genotype on disease prevalence in our series of cases appears to be the opposite of that in sCJD. The high percentage of 129VV subjects described to date (20 of 28 known subjects, including the 2 cases reported elsewhere) and the apparent rarity of 129MM subjects (only 3 of 28 subjects) suggest that the prevalence of VPSPr is directly related to the presence of the 129V allele.3–5 Indeed, at least 1 129V allele is present in 25 of the 28 known cases of VPSPr. The prevalence of the 3 129 genotypes in VPSPr is quite different from that in normal Caucasian populations, in which the 129MM genotype accounts for 43% of subjects, the 129MV for 49%, and the 129VV for 8%.21
The present findings raise a number of questions concerning the nature of VPSPr and its place within the group of known prion diseases.
In our series of 26 VPSPr cases collected to date, 8 subjects apparently had familial dementia; they were all 129VV except for 1 129MM. One of the 2 VPSPr-129VV cases reported by others also had a definitive family history of neurodegenerative disease.4 This raises the possibility that VPSPr is a familial disease with a locus other than the ORF of the PrP gene (which is free of mutations), a condition analogous to that of familial Alzheimer disease. 3,22 Whether the VPSPr subjects reported to date also include inherited cases belonging solely to the 129VV and 129MM genotype remains to be determined.
Well-recognized prion diseases, which are associated with the classic PrPDis commonly identified as PrP27–30, such as all sCJD subtypes and several subtypes of familial CJD and sporadic and familial fatal insomnia, are transmissible with relative ease to receptive animals. Inoculated animals develop a full-blown disease with clinical signs, SD, and presence of a PrP27–30 that generally reproduces the characteristics of the PrP27–30 present in the inoculum.1,23,24 In contrast, other prion diseases, especially GSS, a rare phenotype that to date has been reported as exclusively associated with PrP gene mutations, have been more difficult to transmit or have been reported not to be transmissible at all.25–28 For example, inoculation of brain homogenate from a subtype of GSS linked to the P102L mutation and characterized by the immunoblot presence of only a PK-resistant fragment of 7kDa similar to that present in VPSPr did not cause a symptomatic disease in recipient transgenic mice, but elicited the formation of PrP amyloid deposits in the absence of abnormal PrP.26 Similarly, inoculation of PK-sensitive recombinant PrP polymerized into amyloid fibers generated a prion disease in PrP overexpressing transgenic mice that was apparently asymptomatic but caused SD and deposition of PK-sensitive abnormal PrP, 2 features shared by the 129VV genotype of VPSPr, only late in the life of the inoculated animals, consistent with very long incubation times.29 Furthermore, similar transmission patterns on inoculation of brain homogenates from affected animals or humans have been observed in other neurodegenerative diseases, such as Alzheimer disease and diseases of the tau protein or tauopathies.30,31 Experiments on the transmissibility of VPSPr are ongoing. Preliminary data indicate that VPSPr transmissibility, if it occurs at all, is not efficient, and it could be more like that of GSS-P102L associated with PrPDis 7kDa or of PK-sensitive PrP amyloid fibers, which require long incubation times and do not shorten the life span of the affected animals.26,29
It is intriguing that GSS also shows characteristics of the phenotype and of the PrPDis associated with some of the mutations that resemble those of VPSPr.25 They include long disease duration, multiple PK-resistant fragments, and variable PK resistance of PrPDis. Our comparative analysis of the electrophoretic profiles in VPSPr and GSS-A117V reveals provocative similarities. This finding raise the issue of whether VPSPr might be viewed as the sporadic form of GSS.
Regardless of its relationship with GSS, the finding that VPSPr affects all 3 129 genotypes, resulting in distinct disease phenotypes and PrPDis characteristics, establishes VPSPr as the second “sporadic” prion protein disease, after sCJD.
Acknowledgment
This study was supported by the NIH (grants NIA AG14359 and AG08702, P.G.; NINDS R01NS062787, W.-Q.Z.), Centers for Disease Control and Prevention (grant CCU 515004, P.G.), Britton Fund (P.G.), CJD Foundation (W.-Q.Z.), Alliance BioSecure(W.-Q.Z.), University Center on Aging and Health with the support of the McGregor Foundation and President’s Discretionary Fund (Case Western Reserve University) (W.-Q.Z.), and National Institute on Aging (grant AG05681, J.M.).
We thank the families of the patients for their generous cooperation; the CJD Foundation for its invaluable help; Drs N. Rance, P. Kirby, B. Zabel, M. DeVries, and J. Powers for help in obtaining the cases; D. Kofsky and P. Scalzo for skillful histologic and immunohistochemical preparations; Y. Cohen and W. Chen for insightful diagnostic Western blot preparations; Dr M. Zheng for contributing to sequencing of the PrP gene; and S. Berri, J. Blevins, and K. Glisic for ably assisting in managing the cases and preparing the manuscript.
Footnotes
Potential Conflicts of Interest
Nothing to report.
References
- 1.Gambetti P, Kong Q, Zou WQ, et al. Sporadic and familial CJD: classification and characterization. Br Med Bull. 2003;66:213–239. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gambetti P, Dong Z, Yuan J, et al. A novel human disease with abnormal prion protein sensitive to protease. Ann Neurol. 2008;63:677–708. doi: 10.1002/ana.21420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jansen C, Head MW, van Gool WA, et al. The first case of protease-sensitive prionopathy (PSPr) in The Netherlands: a patient with an unusual GSS-like clinical phenotype. J Neurol Neurosurg Psychiatry. 2010 Jun 14; doi: 10.1136/jnnp.2009.175646. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Head MW, Knight R, Zeidler M, et al. A case of protease sensitive prionopathy in a patient in the UK. Neuropathol Appl Neurobiol. 2009;35:628–632. doi: 10.1111/j.1365-2990.2009.01040.x. [DOI] [PubMed] [Google Scholar]
- 6.Gambetti P, Puoti GF, Kong Q, et al. Novel human prion disease affecting 3 prion codon 129 genotypes: the sporadic form of Gerstmann-Sträussler-Scheinker disease? J Neuropathol Exp Neurol. 2009;68:554. [Google Scholar]
- 7.Gambetti P, Kong Q, Zou WQ. A novel human prion disease affecting subjects with three prion protein codon 129 genotypes: could it be the sporadic form of Gerstmann-Sträussler-Scheinker disease? Prion. 2008;124 [Google Scholar]
- 8.Gambetti P. A novel human prion disease affecting subjects with three prion protein codon 129 genotypes: could it be the sporadic form of Gerstmann-Sträussler-Scheinker disease? Prion. 2009:45. [Google Scholar]
- 9.Capellari S, Powers JM, Petersen RB, et al. A novel molecular and clinico-pathologic phenotype of prion disease in an American. Neurology. 1998;50:A235. [Google Scholar]
- 10.Giaccone G, Di Fede G, Mangieri M, et al. A novel phenotype of Creutzfeldt-Jakob Disease. J Neurol Neurosurg Psychiatry. 2007;78:1379–1382. doi: 10.1136/jnnp.2007.115444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parchi P, Castellani R, Capellari S, et al. Molecular basis of phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 1996;39:767–778. doi: 10.1002/ana.410390613. [DOI] [PubMed] [Google Scholar]
- 12.Parchi P, Giese A, Capellari S, et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol. 1999;46:224–233. [PubMed] [Google Scholar]
- 13.Kong Q, Huang S, Zou WQ, et al. Chronic wasting disease of elk: transmissibility to humans examined by transgenic mouse models. J Neurosci. 2005;25:7944–7949. doi: 10.1523/JNEUROSCI.2467-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cali I, Castellani R, Alshekhlee A, et al. Co-existence of scrapie prion protein types 1 and 2 in sporadic Creutzfeldt-Jakob disease: its effects on the phenotype and prion type characteristics. Brain. 2009;132:2643–2658. doi: 10.1093/brain/awp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan J, Xiao X, McGeehan J, et al. Insoluble aggregates and protease-resistant conformers of prion protein in uninfected human brains. J Biol Chem. 2006;281:34848–34858. doi: 10.1074/jbc.M602238200. [DOI] [PubMed] [Google Scholar]
- 16.Yuan J, Dong Z, Guo JP, et al. Accessibility of a critical prion protein region involved in strain recognition and its implications for the early detection of prions. Cell Mol Life Sci. 2008;65:631–643. doi: 10.1007/s00018-007-7478-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou WQ, Langeveld J, Xiao X, et al. PrP conformational transitions alter species preference of a PrP-specific antibody. J Biol Chem. 2010;285:13874–13884. doi: 10.1074/jbc.M109.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou WQ, Capellari S, Parchi P, et al. Identification of novel proteinase K-resistant C-terminal fragments of PrP in Creutzfeldt-Jakob disease. J Bio Chem. 2003;278:40429–40436. doi: 10.1074/jbc.M308550200. [DOI] [PubMed] [Google Scholar]
- 19.Colucci M, Moleres F, Xie Z, et al. Gerstmann-Straussler-Scheinker: a new phenotype with “curly” PrP deposits. J Neuropathol Exp Neurol. 2006;65:642–651. doi: 10.1097/01.jnen.0000228198.81797.4d. [DOI] [PubMed] [Google Scholar]
- 20.Taguchi Y, Shi ZD, Ruddy B, et al. Specific biarsenical labeling of cell surface proteins allows fluorescent- and biotin-tagging of amyloid precursor protein and prion proteins. Mol Biol Cell. 2009;20:233–244. doi: 10.1091/mbc.E08-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmermann K, Turecek PL, Schwarz HP. Genotyping of the prion protein gene at codon 129. Acta Neuropathol. 1999;97:355–358. doi: 10.1007/s004010050998. [DOI] [PubMed] [Google Scholar]
- 22.Selkoe DJ, Podlinsky MB. Deciphering the genetic basis of Alzheimer’s disease. Ann Rev Genom Hum Genet. 2002;3:67–99. doi: 10.1146/annurev.genom.3.022502.103022. [DOI] [PubMed] [Google Scholar]
- 23.Korth C, Kaneko K, Groth D, et al. Abbreviated incubation times for human prions in mice expressing a chimeric mouse-human prion protein transgene. Proc Natl Acad Sci USA. 2003;100:4784–4789. doi: 10.1073/pnas.2627989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asante EA, Linehan J, Desbruslais M, et al. BSE prions propagate as either variant CJD-like or sporadic CJD-like prion strains in transgenic mice expressing human prion protein. EMBO J. 2002;21:6358–6366. doi: 10.1093/emboj/cdf653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong Q, Surewicz WK, Peterson RB, et al. Inherited prion diseases. In: Prusiner SB, editor. Prion biology and disease. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2004. pp. 673–775. [Google Scholar]
- 26.Piccardo P, Manson JC, King D, et al. Accumulation of prion protein in the brain that is not associated with transmissible disease. Proc Nat Acad Sci USA. 2007;104:4712–4717. doi: 10.1073/pnas.0609241104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tateishi J, Kitamoto T, Hoque MZ, et al. Experimental transmission of Creutzfeldt-Jakob disease and related diseases to rodents. Neurology. 1996;46:532–537. doi: 10.1212/wnl.46.2.532. [DOI] [PubMed] [Google Scholar]
- 28.Hegde RS, Tremblay P, Groth D, et al. Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature. 1999;402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]
- 29.Colby DW, Wain R, Baskakov IV, et al. Protease-sensitive synthetic prions. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000736. e1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer-Luehmann M, Coomaraswamy J, Bolmont T. Exogenous induction of cerebral β-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 31.Clavaguera F, Bolmont T, Crowther RA. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11:909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krasnianski A, Kallenberg K, Collie DA, et al. MRI in the classical MM1 and the atypical MV2 subtypes of sporadic CJD: an inter-observer agreement study. Eur J Neurol. 2008;15:762–771. doi: 10.1111/j.1468-1331.2008.02209.x. [DOI] [PubMed] [Google Scholar]
- 33.Heinemann U, Krasnianski A, Meissner B, et al. Creutzfeldt-Jakob disease in Germany: a prospective 12-year surveillance. Brain. 2007;130:1350–1359. doi: 10.1093/brain/awm063. [DOI] [PubMed] [Google Scholar]
- 34.Castellani RJ, Colucci M, Xie Z, et al. Sensitivity of 14-3-3 protein test varies in subtypes of sporadic Creutzfeldt-Jakob disease. Neurology. 2004;63:436–442. doi: 10.1212/01.wnl.0000135153.96325.3b. [DOI] [PubMed] [Google Scholar]