Abstract
Background
The purpose of this study was to design and evaluate fluorescent in situ hybridization (FISH) probes for the single-cell detection and enumeration of lactic acid bacteria, in particular organisms belonging to the major phylogenetic groups and species of oral lactobacilli and to Abiotrophia/Granulicatella.
Results
As lactobacilli are known for notorious resistance to probe penetration, probe-specific assay protocols were experimentally developed to provide maximum cell wall permeability, probe accessibility, hybridization stringency, and fluorescence intensity. The new assays were then applied in a pilot study to three biofilm samples harvested from variably demineralized bovine enamel discs that had been carried in situ for 10 days by different volunteers. Best probe penetration and fluorescent labeling of reference strains were obtained after combined lysozyme and achromopeptidase treatment followed by exposure to lipase. Hybridization stringency had to be established strictly for each probe. Thereafter all probes showed the expected specificity with reference strains and labeled the anticipated morphotypes in dental plaques. Applied to in situ grown biofilms the set of probes detected only Lactobacillus fermentum and bacteria of the Lactobacillus casei group. The most cariogenic biofilm contained two orders of magnitude higher L. fermentum cell numbers than the other biofilms. Abiotrophia/Granulicatella and streptococci from the mitis group were found in all samples at high levels, whereas Streptococcus mutans was detected in only one sample in very low numbers.
Conclusions
Application of these new group- and species-specific FISH probes to oral biofilm-forming lactic acid bacteria will allow a clearer understanding of the supragingival biome, its spatial architecture and of structure-function relationships implicated during plaque homeostasis and caries development. The probes should prove of value far beyond the field of oral microbiology, as many of them detect non-oral species and phylogenetic groups of importance in a variety of medical conditions and the food industry.
Background
It is estimated that more than 1010 bacteria per gram of dental plaque colonize the human oral cavity. More than half of them still remain uncultivable. Their existence is only known because a fingerprint in form of a sequence from a gene fragment, most often from 16S rDNA, could be traced in a clinical sample [1]. All oral microorganisms form biofilms on surfaces such as the oral mucosa, the tongue, or the surface of the teeth. Many supragingivally predominant bacteria belong to the Firmicutes phylum (Gram-positive rods and cocci of low G+C content) with the lactic acid producing bacteria (LAB) as the largest and clinically important subgroup [2,3]. Comprising streptococci, lactobacilli, and Granulicatella/Abiotrophia species (formerly described as nutritionally variant streptococci), LAB are main constituents of the commensal microbiota of the human oral cavity, but form also part of the biofilms colonizing the upper respiratory, intestinal and urinary tracts. In the oral cavity, they are thought to play major roles in dental plaque formation and oral biofilm homeostasis. However, under conditions of prolonged shifts of biofilm composition, LAB may induce dental caries through excessive lactic acid formation [4], and upon penetration into the blood stream LAB may cause in susceptible individuals a variety of life-threatening conditions such as endocarditis, septicemia, or meningitis [5,6].
In situ techniques that allow monitoring individual cells and cell populations within biofilms are important tools to investigate natural biofilm ecologies [7,8]. However, few probes for the detection and quantification by fluorescent in situ hybridization (FISH) of oral LAB species have been described so far [9,10]. Here we report the design, characterization and pilot evaluation of probes recognizing major phylogenetic clusters or species of oral lactobacilli, the Abiotrophia/Granulicatella group, and a few taxa of oral streptococci. Applied for validation to in situ formed supragingival biofilms, the probes detected high levels of both mitis group streptococci and Abiotrophia/Granulicatella species, and identified strains of Lactobacillus fermentum and the Lactobacillus casei group.
(The study is part of the requirements for BQ's Doctor degree of Dental Medicine.)
Results and Discussion
Probe design
In this study we relied for probe design on the species and phylotype description provided by the human oral microbiome database (HOMD) [11], which comprises a collection of 16S rRNA sequences of both cultivable and so far non-cultivable taxa representing the currently known width of bacterial diversity found in the human oral cavity [12]. Oligonucleotide probes were designed with specificity for phylogenetic groups or species of Lactobacillus, Streptococcus, Lactococcus, Granulicatella and Abiotrophia. Table 1 lists all probes with their sequence and optimum formamide concentration. The latter was determined by systematic optimization in experiments with both reference strains and clinical plaque samples.
Table 1.
Description and specificity of oligonucleotide probes
| Probe | Target sequence (5' to 3')a | Label | Positionb | Target bacteriac | Formamide concentrationd | Source |
|---|---|---|---|---|---|---|
| LGC358a | CCATTGTGGAAGATTCCCT | Cy3 | 358-76 | Most lactobacilli | 25-30 | [13], modified |
| LGC358b-comp | CCATTGCGGAAGATTCCCT | - | 358-76 | Competitor probe for LGC358a: Staphylococci, Gemella, Granulicatella, Abiotrophia |
25-30 | [13], modified |
| LAB759 | CTACCCATRCTTTCGAGCC | Cy3, FAM | 759-77 | Most lactobacilli without L. salivarius group | 30-35 | [8] |
| LAB759-comp | CTACCCACGCTTTCGAGCM | - | 759-77 | Competitor probe for LAB759: Many streptococci, β-Proteobacteria, but no lactobacilli |
30-35 | this study |
| L-Lbre466-2 | ACCGTCAACCCTTGAACAG | Cy3 | 466-84 | L. brevis | 30-55 | this study |
| L-Lbuc438-2 | CACCYGTTCTTCTCCAACA | FAM | 439-57 | L. buchneri (L. hilgardii, L. kefiri, L. parabuchneri) | 50-55 | this study |
| Lcas467 | CCGTCACGCCGACAACAG | Cy3, FAM | 467-84 | L. casei, L. paracasei subsp. paracasei, L. rhamnosus, L. zeae | 25-40 | this study |
| L-Lcol732-2 | GTTGCAAGCTAGACAGCC | Cy3 | 732-49 | L. coleohominis, L. reuteri (some strains) | ≥30 | this study |
| Lfer466 | CCGTCAACGTATGAACAG | Cy3 | 466-83 | L. fermentum | 25 | this study |
| Lfer466-H448 | TTACTCTCATACGTGTTC | - | 448-65 | Helper probe for Lfer466 | 25 | this study |
| Lfer466-H484 | GCCGTGACTTTCTGGTTAAATA | - | 484-505 | Helper probe for Lfer466 | 25 | this study |
| Lgas183 | GACATGCGTCTAGTGTTG | FAM | 183-200 | L. gasserii, L. johnsonii | 25-30 | this study |
| Lgas458 | ATAAAGGCCAGTTACTACC | FAM | 458-76 | L. acidophilus L. crispatus, L. gasserii, L. jensenii, L. johnsonii (L. amylolyticus, L. amylovorus, L. fornicalis, L. hamsteri, L. helveticus, L. kefiranofaciens, L. kitasatonis) | 25 | this study |
| Lpla759 | CTACCCATACTTTCGAGCC | FAM | 759-77 | L. paraplantarum, L. plantarum, L. pentosus | 20-30 | this study |
| Lpla990 | ATCTCTTAGATTTGCATAGTATG | Cy3 | 990-1012 | L. paraplantarum, L. plantarum, L. pentosus | 20-35 | this study |
| Lpla990-H1018 | CCCGAAGGGAACGTCTA | - | 1018-34 | Helper probe for Lpla990 | 20-35 | this study |
| Lreu986 | GCGCAAGATGTCAAGACC | Cy3, FAM | 986-1004 | L. coleohominis, L. fermentum, L. oris, L. reuterii, L. vaginalis(L. frumenti, L. gastricus, L. ingluviei, L. mucosae, L. panis, L. pontis, L. suebicus) | 25-30 | this study |
| Lreu986-H967 | TGGTAAGGTTCTTCGCGTA | - | 967-85 | Helper probe for Lreu986 | 25-30 | this study |
| Lsal574 | AAAGACCGCCTGCGTTCCC | Cy3, FAM | 574-92 | L. salivarius (L. acipiscis, L. animalis, L. apodemi, L. murinus, L. ruminis, L. satsumensis, L. vini) | 35-50 | this study |
| L-Lsal1113-2 | CTGGCAACTGACAACAAG | FAM | 1113-30 | L. salivarius (L. agilis, L. equi, L. saerimneri) | 35-45 | this study |
| Lvag222 | ACCGCGGGCCCATCCTGA | Cy3 | 222-39 | L. vaginalis | 35-50 | this study |
| STR405 | TAGCCGTCCCTTTCTGGT | Cy3 | 405-22 | Streptococci | ≤ 50 | [10,38] |
| LGC358c | CCATTGCCGAAGATTCCCT | FAM | 358-76 | Streptococci | 25-30 | [13], modified |
| MIT447 | CACYCGTTCTTCTCTTACA | FAM | 447-65 | Mitis group of streptococci | 25 | [10,38] |
| MUT590 | ACTCCAGACTTTCCTGAC | Cy3 | 590-607 | Streptococcus mutans | 30 | [10,38] |
| L-Ssob440-2 | CACACGTTCTTCCCCTAC | FAM | 440-57 | Streptococcus sobrinus | 45 | this study |
| L-Sco/int172-2 | CAGTAAATGTTCTTATGCGGTA | Cy3, FAM | 172-93 | Streptococcus constellatus, S. intermedius | 40-55 | [39] |
| ABI161 | TGCGGTTTTAGCATCCGT | Cy3 | 161-78 | Granulicatella adjacens, G. elegans | ≤ 30 | this study |
| ABI1246 | AGTTCGCTGCTCGTTGTA | Cy3 | 1246-63 | Abiotrophia defectiva, Granulicatella adjacens, G. elegans, L. coleohominis (Facklamia hominis, F. languida, F. miroungae) | ≤ 35 | this study |
| LCC1030 | CCTGTATCCCGTGTCCCG | Cy3, FAM | 1030-47 | Lactococcus lactis, L. garvieae | 40-55 | this study |
| EUB338 | GCTGCCTCCCGTAGGAGT | Cy3, FAM | 338-55 | Most Eubacteria | ≤ 50 | [40] |
a Bold printed bases indicate the position of locked-nucleic-acids.
b 16S rRNA target position (Escherichia coli numbering).
c Taxa in parentheses are detected by the probe but have not been described to colonize the human oral cavity [11].
d Optimum formamide concentration in hybridization buffer.
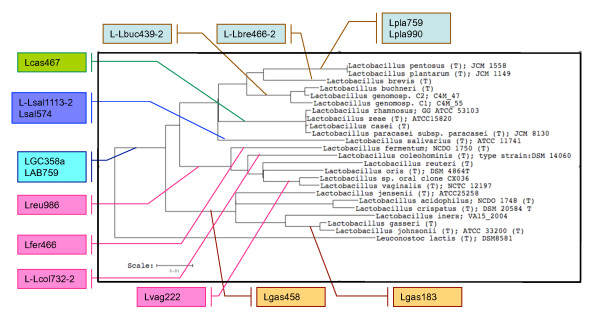
Figure 1 outlines the concept for the design of the probes targeting oral lactobacilli. Two broad Lactobacillus probes (LGC358a and LAB759) were generated with the idea that they should complement each other and thus limit the potential of misidentifications [7]. Elongated by one and shifted by four bases LGC358a is a derivative of probe LGC354a [13]. Probes LGC358b (staphylococci and related bacteria) and LGC358c (streptococci) are analogously related to LGC354b and LGC354c described by Meier et al. [13]. As observed often with probes to larger phylogenetic groups, initial experiments with both probes detected besides the targeted lactobacilli significant numbers of false positives (predominantly cocci) when applied to oral plaque samples (see below). In silico analyses suggested that these false hybridizations were due to single sequence mismatches and could possibly be avoided by the application of unlabeled competitor probes that are fully complimentary to the targeted 16S rRNA segment of the false positive organisms. Applied in excess together with the labeled FISH probe such competitor probes can increase the differentiation between true- and potential false positives [14]. Thus, LGC358a used in conjunction with LGC358b-comp should recognize selectively most Lactobacillaceae organisms and in addition detect parts of the non-oral families Leuconostocaceae and Carnobacteriaceae, whereas LAB759, when applied together with LAB759-comp (which should suppress recognition of Streptococcus mutans as well as Eikenella, Kingella, and Neisseria sp.) is supposed to identify all oral lactobacilli except Lactobacillus salivarius and the majority of L. fermentum strains. Application of these competitor probes to various types of plaques samples proved to be successful in providing specificity for lactobacilli (see below). The other probes for lactobacilli were designed to identify bacteria from all major deep branching clusters of the phylogenic tree (Figure 1). Three probes recognize deeply branched, individual species (L. fermentum, L. salivarius and Lactobacillus vaginalis), which, however, belong to the most frequently detected oral lactobacilli. Probes recognizing oral Granulicatella and Abtiotrophia species (ABI161 and ABI1246), Lactococcus lactis (LCC1030) and cocci belonging to Streptococcus constellatus/Streptococcus intermedius (L-Sco/int172-2) or Streptococcus sobrinus (L-Ssob440) complement the set of new probes.
Figure 1.
16S rRNA based phylogenetic tree of oral lactobacilli. The flags indicate the different oligonucleotide probes used in this study, their colored lines point to the respective phylogenic groups detected by the probes. All listed oral lactobacilli reference strains and phylotypes were retrieved from the Human Oral Microbiome Database [11]. The phylogenetic tree was constructed with Leuconostoc lactis as the outgroup using the Tree Builder algorithm of the Ribosomal Data Base Project (http://rdp.cme.msu.edu/index.jsp).
Permeabilization of lactobacilli for FISH
Uniform permeabilization for FISH of fixed lactobacilli (but not of streptococci or Abiotrophia/Granulicatella) is a known problem [9], in particular with certain 'notorious' strains. Like other authors before, we have evaluated several permeabilization protocols that precede hybridization and obtained the best results with a modification of a procedure proposed by Harmsen et al. [9] (data not shown). It was applied selectively to all Lactobacillus probes and consists of a 5 min exposure to lysozyme and achromopeptidase, followed by a 30 min incubation with lipase.
Fluorescence intensity and probe specificity
Lactobacillus probes were tested with 22 reference strains representing the different oral lactobacilli clusters as described by the HOMD (Table 2) and, with the exceptions of probe LAB759 and Lfer466, displayed the anticipated reactivity profile. As an example Figure 2A shows the staining of Lactobacillus rhamnosus AC 413 with Lcas467-Cy3. Pointing at one of the strengths of single cell analyses with FISH, strain Lactobacillus crispatus ATCC 33820 was found contaminated with L. fermentum and required recloning (Figure 2B). With several probes the fluorescence intensity was weak but could be significantly improved by adding non-fluorescent helper probes to the hybridization solution [15], or by employing probes containing locked-nucleic-acids (LNA) [16]. The former bind to regions of the 16S rRNA that are adjacent to the target sequence thereby contributing to the opening of the rRNA's 3-D structure and improving probe accessibility, whereas the latter contain one or two derivative nucleotide analogs with their ribose locked in a C3'-endo conformation which leads to a higher target selectivity of the probe. Unexpected from in silico data, LAB759 labeled the L. salivarius reference strain ATCC 11741 and Lfer466 bound to the Lactobacillus reuteri type strain CCUG 33624T. The reasons for these exceptional hybridizations remain to be determined. Generally, the LNA-probes yielded high fluorescence intensity but also required high formamide concentrations to display the predicted specificity. In particular, L-Lbre466 was cross-reactive with Lactobacillus colehominis and L-Lbuc438 was cross-reactive with some strains of both the L. casei and L. reuteri clusters if the formamide concentration was kept below 45%. Similarly, the L-Lcol732 probe was false positive with strains from the Lactobacillus buchneri cluster at < 40% formamide. Of the other probes listed in Table 1, ABI1246 was strongly positive with all four Abiotrophia/Granulicatella reference strains tested (Granulicatella adjacens CCUG 27809T and HE-G-R 613A, Granulicatella elegans CCUG 38949T and Abiotrophia defectiva CCUG 36937), whereas ABI161 labeled only the Granulicatella strains. Probe LCC1030 was positive with Lactococcus lactis subsp. lactis reference strain NCC2211 [17], and the S. mutans and S. sobrinus probes Smut590 and L-Lsob440 stained reference strains UA159T and OMZ 176, respectively, while none of the probes was positive with strains from other streptococcal species. Probe L-Ssob440-2 yielded better fluorescence intensity than the previously described probe SOB174 [10], but had to be used at high stringency. All these findings were as expected from in silico data.
Table 2.
Reactivity of FISH probes to lactobacilli with target and non-target strains
| 16S rRNA probes | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group, Strain | OMZ | LGC358a | LAB759 + LABB759-comp | Lpla759 | Lpla990 + H1018 | L-Lbre466-2 | L-Lbuc438-2 | Lcas467 | Lsal574 | L-Lsal1113-2 | Lreu986 + H1018 | Lfer466 + H448+ H484 | L-Lcol732-2 | Lvag222 | Lgas458 | Lgas183 |
| L. buchneri et rel. | ||||||||||||||||
| L. plantarum FAM 1638 | 945 | 2-4+*,a | 3-4+ | 3-4+ | 2-4+* | - | - | - | - | - | - | - | - | - | - | - |
| L. brevis ATCC 14869 | 625 | 3-4 + | 2-3 + | - | - | 4+ | - | - | - | - | - | ± | -b | - | - | - |
| L. brevis OMZ 1114 | 1114 | 2-4+ | 2-3+* | - | - | 3-4+ | - | - | - | - | - | - | -b | - | - | - |
| L. buchneri ATCC 4005 | 626 | 2-4 + | 1-2 + | - | - | - | 3-4 + | - | - | - | - | - | -b | - | - | - |
| L. buchneri | 1097 | 2-4 +* | 2-3 +* | - | - | - | 3+ | - | - | - | - | - | -b | - | - | - |
| L. casei et rel. | ||||||||||||||||
| L. casei ATCC 393 | 939 | 2-4+ | 3-4+ | - | - | - | -c | 3+ | - | - | - | - | - | - | - | - |
| L. casei Cl-16 | 638 | 3-4 + | 3-4 + | - | - | - | -c | 3-4 + | - | - | - | - | - | - | - | - |
| L. paracasei ATCC 25598 | 624 | 2-4 +* | 2-4 +* | - | - | - | -c | 3-4 +* | - | - | - | - | - | - | - | - |
| L. rhamnosus AC 413 | 629 | 2-4 + | 2-4 + | - | - | - | - | 3-4 + | - | - | - | - | - | - | - | - |
| L. rhamnosus ATCC 7469T | 602 | 2-4 + | 2-4 + | - | - | - | - | 3 + | - | - | - | - | - | - | - | - |
| L. salivarius | ||||||||||||||||
| L. salivarius ATCC 11741 | 525 | 3-4+ | 3-4+ | - | - | - | - | - | 2-4+ | 3-4+ | ± | - | - | - | - | - |
| L. salivarius OMZ 1115 | 1115 | 2-4+ | - | - | - | - | - | - | 3-4+ | 3-4+ | - | - | - | - | - | - |
| L. reuteri et rel. | ||||||||||||||||
| L. coleohominis DSM14060T | 1113 | 1-3 + | 2-4 + | - | - | -d | - | - | - | - | 3 + | - | 3-4 + | - | - | - |
| L. fermentum ATCC 14931 | 524 | 2-4 +* | 2 +*, e | - | - | - | - | - | - | - | 2-4 + | 3-4 + | - | - | - | - |
| L. fermentum OMZ 1116 | 1116 | 2-4 + | 2 +*, e | - | - | - | - | - | - | - | 2-4 + | 3-4 + | - | - | - | - |
| L. reuteri CCUG 33624T | 1100 | 2-4 + | 3-4 + | - | - | - | -c | ± | - | - | 2-4 + | 2-4 + | - | - | - | - |
| L. vaginalis UMCG 5837 | 1095 | 2-4 + | 3-4 + | - | - | - | -c | - | - | - | 1-3 +* | - | - | 3-4 + | - | - |
| L. gasseri et rel. | ||||||||||||||||
| L. acidophilus ATCC 4357 | 523 | 2-4+ | 3-4+ | - | - | - | - | - | - | - | ± | ± | - | - | 2-4+ | - |
| L. crispatus ATCC 33820 | 522 | 3-4 + | 3-4 + | - | - | - | - | - | - | - | - | - | - | 3-4 + | - | |
| L. gasseri ATCC 19992 | 520 | 2-4 + | 2-4 + | - | - | - | - | - | - | - | ± | 1 + | - | - | 1-3 + | 2-4 + |
| L. gasseri OMZ 549 | 549 | 2-4 + | 2-4 + | - | - | - | - | - | - | - | ± | - | - | - | 2-4 + | 2-4 + |
| L. jensenii UMCG 20557 | 1096 | 2-4 + | 3-4 + | - | - | - | - | - | - | - | - | - | - | - | - | - |
* The asterisk indicates that only a small percentage of the cells could be stained by the probe, in spite of enzymatic pretreatment to improve probe penetration.
a Fluorescence intensity was graded using an arbitrary five-step scale, where - (no fluorescence above background) and 1+ (very faint fluorescence) were considered negative signals, and 2+ (weak), 3+ (strong) and 4+ (brilliant fluorescence) were considered positive signals.
b Probe L-Lcol732-2 labeled L. brevis and L. buchneri strains at formamide concentrations below 40%.
c L-Lbuc438-2 cross-reacted with certain strains from the L. casei and L. reuteri groups at formamide concentrations of ≤ 45%.
d L-Lbre466-2 was positive with L. coleohominis at ≤ 45% formamide in the hybridization buffer.
e L. fermentum was stained with low intensity due to a weak mismatch at position 760.
Figure 2.
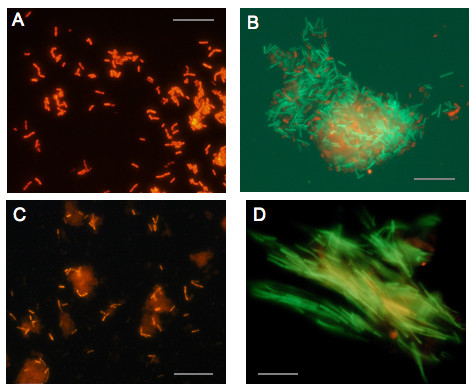
FISH staining of reference strains and biofilm samples with LAB probes. (A) L. rhamnosus strain AC 413 stained with Lcas467-Cy3 (40% formamide). (B) L. crispatus ATCC 33820 stained with both Lfer466-Cy3 (plus the corresponding helper probes) and Lgas458-FAM (25% formamide). The strain should be Lfer466-/Lgas458+, the FISH assay identified a previously unnoticed contamination with red-stained Lfer466+ cells, which had to be eliminated by recloning. (C) Identification of L. fermentum in biofilm 013 using probe Lfer466-Cy3 (plus helper probes; 25% formamide). Note the high proportion of L. fermentum in this in situ grown biofilm. (D) Sample from the dorsum of the tongue showing an aggregate of large unidentified filaments stained with the Lactococcus probe LCC1030-Cy3 and the streptococcal probe L-Sco/int172-2-FAM at 30% formamide. The bacteria are double false positive under these stringency conditions, whereby the detection of the Cy3 fluorescence is hampered by the much stronger FAM fluorescence. To prevent such false positive hybridization, the formamide concentration had to be increased to ≥ 40%. Bars: 10 μm.
Enumeration of lactic acid bacteria from in situ formed biofilms
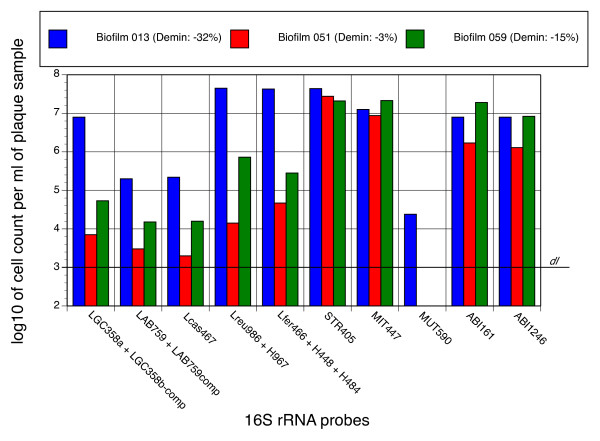
The applicability of the probes was tested with three in situ formed biofilm samples. The samples were harvested from bovine enamel discs carried in acrylic appliances on the buccal side of the mandibular premolar/molar regions [18] by three volunteers whose discs differed greatly in the extent of demineralization (-3%, -15%, and -32%) generated during the 10 days of intermittent extraoral exposure to a 5% glucose/5% sucrose solution. All samples were positive for lactobacilli as detected by the two broadly reactive Lactobacillus probes LGC358a and LAB759 (Figure 3). Total cell numbers and numbers of lactobacilli were very similar to findings from an earlier study investigating the microbiota associated with the in situ development of caries [19]. Both probes had to be used imperatively together with their respective competitor probes, since, without them, large numbers of cocci and other morphotypes were positive. In the biofilm from disc 013 (biofilm 013 in the following) LGC358a stained clearly two populations of rods that differed in length, whereas LAB759 identified only the shorter of the two morphotypes. The longer and predominant cell type had the probe reactivity profile LGC358a+/LAB759-/Lfer466+/Lreu986+/Lcas467- (Figure 2C), whereas the smaller one was LGC358a+/LAB759+/Lfer466-/Lreu986-/Lcas467+, indicating that the larger rods are L. fermentum and the smaller ones lactobacilli from the casei group. While the total number of L. casei, streptococci or Abiotrophia/Granulicatella seemed not to correlate with the extent of disc demineralization, the high concentration of L. fermentum in the biofilm of the extremely demineralized disc 013 was quite remarkable.
Figure 3.
Enumeration by FISH of lactic acid producing bacteria in three in situ grown biofilms. Biofilms were harvested from bovine enamel discs, carried in situ for 10 days and nights by three different volunteers. The discs differed greatly in the extent of demineralization indicated in the within legend of the plot. The detection limit (dl) of the FISH assay was approximately 103 bacteria per ml of sample.
All other lactobacillus probes gave negative results. Concerning Lsal574 and Lvag222 we found that both these probes had to be used at much higher stringency conditions (50% formamide) than expected from the in vitro experiments with reference strains to prevent cross-reactivity with other biofilm bacteria. In particular cells with the characteristic morphology of Selenomonas were often cross-reactive at conditions of insufficient stringency. Abiotrophia and Granulicatella could be detected in high numbers in all three samples. Both ABI161 and ABI1246 recognized cocci, which in double-labeling experiments stained always negative with the streptococcal probes LGC358c and MIT447 (data not shown). Finally, all samples contained high numbers of streptococci, mostly from the mitis group. S. mutans, however, was found with MUT590 in only one sample at low concentration, and the probes for S. sobrinus and S. constellatus/S. intermedius gave negative results. Identification by FISH of streptococci, in particular of the mitis group, is hindered severely by high conservation of the 16S rRNA gene sequence among these taxa [20,21] and therefore FISH detection of oral streptococci still relies mostly on phylogenic group-specific probes. A surprise finding, confirmed with supragingival plaque samples and scrapings from the dorsum of the tongue, was that both Lactococcus probe LCC1030 and S. constellatus/intermedius probe L-Sco/int172-2 triggered rather strong fluorescence of long filaments with blunted ends (Figure 2D), which could only be suppressed by applying formamide concentrations exceeding 40%. The results were confirmed when probes with exchanged fluorescence labels were used (Cy3 instead of 6-FAM and vice versa). Potential specificity problems associated with phylogenetic group-specific probes are a known phenomenon [7,22]. However, screening of the RDP10 database for oral bacteria with this type of morphology and ≤ 2 sequence mismatches within the gene fragments complementary to these probes, failed to reveal any hints about the possible identity of these filaments. Experiments aiming at their isolation by fluorescence activated cell sorting are ongoing.
Typing of Lactobacillus isolates from in situ grown oral biofilms
With the aim to verify the identification by FISH of the lactobacilli present in the three in situ grown biofilm samples (Figure 3), aliquots were cultured on LBS agar. Five strains (OMZ 1117-1121), representing the various colony types observed, were isolated and characterized by both FISH and partial sequencing of the 16S rDNA (Table 3). Sequence analysis identified two strains as L. fermentum (OMZ 1117 and 1121) [EMBL: FR667951] and two as L. casei/L. paracasei (OMZ 1118 and 1120) [EMBL: FR667952], based on 100% sequence similarity with respective reference strains. The fifth strain was typed as L. vaginalis (OMZ 1119) [EMBL: FR667953] with a sequence match score of 0.995 to reference strain Dox G3. L. vaginalis had not been detected by direct FISH analysis of the biofilms (Figure 3), presumably because the cell number was below the detection limit of approximately 103 bacteria per ml of sample suspension. Tested by FISH with the whole set of probes all five isolates showed the anticipated profile (Table 3). The two L. fermentum isolates were negative to weakly positive with LAB759 in repeated experiments. This is explained by L. fermentum strains having an adenine at position 760 of their 16S rRNA, as opposed to a cytosine at the corresponding position of probe LAB759. This peripheral mismatch may result sometimes in weak cross-reactivity (see also L. fermentum strains in Table 2). In summary, typing by gene sequencing corroborated the data obtained from the direct FISH analysis of the in situ grown biofilms.
Table 3.
Identification and FISH reactivity profiles of five isolates from in situ biofilms 013, 051 and 059
| Isolated strain (biofilm of origin) | |||||
|---|---|---|---|---|---|
| OMZ 1117 (013) | OMZ 1118 (013) | OMZ 1119 (051) | OMZ 1120 (051) | OMZ 1121 (059) | |
| 16S rRNA probes | |||||
| LGC358a | 2-4 + | 3-4 + | 3-4 + | 3-4 + | 3 + |
| LAB759 + LAB759-comp | - to 2 + | 3-4 + | 3-4 + | 3 + | - to 2 + |
| Lpla759 | - | - | - | - | - |
| Lpla990+ H1018 | - | - | - | - | - |
| L-Lbre466 | - | - | - | - | - |
| L-Lbuc438 | - | - a | - a | - a | - |
| Lcas467 | - | 4 + | - | 3-4 + | - |
| Lsal574 | - | - | - | - | - |
| L-Lsal1113 | - | - | - | - | - |
| Lreu986 + H967 | 2-4 + | - | 3-4 + | - | 3-4 + |
| Lfer466 + H448 + H484 | 2-4 + | - | - | - | 3-4 + |
| L-Lcol732 | - | - | - | - | - |
| Lvag222 | - | - | 3-4 + | - | - |
| Lgas458 | - | - | - | - | - |
| Lgas183 | - | - | - | - | - |
| Identificationc | L. fermentum | L. paracasei L.casei | L. vaginalis | L. paracasei L.casei | L. fermentum |
a Positive at ≤ 45% formamide.
b Scoring of fluorescence intensity is described in a footnote to Table 2.
c Species identification was based on ≥ 99.5% 16S rRNA gene sequence similarity with reference strains listed in RDP10 (http://rdp.cme.msu.edu/index.jsp). The respective partial 16S rRNA gene sequences of OMZ 1117 and 1121 [EMBL: FR667951], and of OMZ 1118 and 1120 [EMBL: FR667952] were identical. OMZ 1119 was identified as L. vaginalis [EMBL: FR667953].
Critical importance of several assay parameters
Lactobacilli are difficult targets for FISH because of their cell wall's resistance to probe penetration. The protocol used successfully in the present study to increase cell permeability evolved from the method of Harmsen et al. [9], which we supplemented with achromopeptidase, previously described to open cell walls of Actinomyces strains [23,24]. Systematic evaluation of this three-enzyme-pretreatment with 12 reference strains from seven Lactobacillus species showed its indispensability. However, a minority of strains proved to be particularly resistant, as up to 20% of the cells recognizable by phase contrast could not be stained. Of course this raised concerns that such false-negative results could also affect analyses of clinical samples. We cannot completely rule out this possibility, but after comprehensive analysis of many plaque samples we would like to hypothesize that there are differences in cell wall permeability between cultured and native lactobacilli and that false-negative cells are primarily seen after FISH with cultured lactobacilli.
With cell wall permeability remaining a potential reason for concern, maximum fluorescence intensity from penetrated probes is essential. Fluorescence intensity depends on cellular ribosome content, in situ probe accessibility to the probe target region, and rRNA stability [25]. Several procedures to maximize the performance of FISH probes have been described [15,16,26,27]. They alter the 3-dimensional structure of the target region by using helper probes, optimize probe length and hybridization conditions, improve binding affinity by modifying the probes' backbone with LNA substitutions, or inhibit enzymatic rRNA degradation. In this study we used all four procedures to improve fluorescence intensity of certain probes. For Lfer466, Lreu986, and Lpla990 one or two helper probes binding directly adjacent to the target site were added to the hybridization solution and in each case a clear-cut improvement of fluorescence intensity was observed. The same was the case when the LNA-substituted probe L-Ssob440-2 was compared to Ssob440. For five other probes the decision to opt for LNA insertions was taken solely based on own and published experience [25], suggesting limited accessibility of the probes' target site. All these LNA/DNA-probes displayed intensive fluorescence, but required strict adherence of very stringent hybridization conditions for sufficient specificity.
Conclusions
In this study we have described the application of 20 new phylogenetic group- or species-specific oligonucleotide probes for the single-cell detection of oral LAB in various clinical or experimental biofilms. The results show that the probes can be used for the rapid identification and quantification of a broad range of LAB for both research and diagnostic purposes, provided that restrictions concerning cell permeabilization (lactobacilli), hybridization stringency, fluorescence intensity optimization, and use of competitor probes are considered. Although designed to cover the diversity of oral lactobacilli, these probes should prove of value far beyond the field of oral microbiology, as many of them detect non-oral species and phylogenetic groups of importance to gastroenterology, gynecology, heart diseases, food industry, etc. Gene sequence typing of isolated strains confirmed the results obtained by analyzing biofilm samples directly by FISH. On a speculative note, the apparent correlation between the L. fermentum cell number and the extent of demineralization seen with the three samples from the in situ study could indicate that these bacteria have played a significant role in the carious process. The abundance of L. fermentum might be explained by high resistance to low pH giving these bacteria a selective ecological advantage during the formation of the biofilm.
Methods
Strains, plaque samples and in situ grown biofilms
Lactobacillus reference strains (listed in Table 2) were grown in 10% CO2 at 37 °C on LBS (Lactobacillus selection) agar and in LBS broth (Becton Dickinson). Lactococcus, Streptococcus, Abiotrophia and Granulicatella reference strains from the OMZ strain collection were propagated anaerobically on Columbia blood agar or in fluid universal medium [28]. They were harvested after 24-36 h during the late log-phase of growth. Supragingival plaque samples and scrapings from the dorsum of the tongue were collected from two of the authors, washed in 0.9% NaCl, fixed in 4% paraformaldehyde/PBS (20 min, 4 °C), and stored in 50% ethanol at -20 °C.
In situ grown biofilm samples were harvested from bovine enamel discs (6.8 mm Ø) carried for 10 days and nights by three volunteers in the course of a double-blind split-mouth de- and remineralization study carried out at the University of Bergen, Bergen, Norway [18]. The Regional Committee for Medical Research Ethics Western Norway approved the study protocol and the volunteers gave their informed written consent to participate in the study. Inclusion criteria for volunteers were normal salivary flow and a full dentition without non-restored caries lesions or evidence of moderate or severe gingivitis. Antibiotics, mouth rinses or tooth pastes containing antimicrobial agents (e.g. chlorhexidine, triclosan, SnF2, Zn2+, etc.) or drugs affecting the salivary flow rate should not have been used for the last three months. The appliances were kept in 0.9% NaCl during meals and tooth cleaning; in addition they were dipped seven times daily for 10 min in 5% glucose/5% sucrose solution to promote plaque formation. After 10 days in the oral cavity of the volunteers, the discs were removed from the appliances and the biofilms were harvested by scraping with plastic scalers (ZI 10; Deppeler, Rolle, Switzerland) into approximately 1 ml of sterile 0.9% NaCl as collecting fluid (exact volume determined for each sample). The samples were frozen at -80 °C and shipped to Zürich on dry ice for further analyses. There, freshly defrosted samples were vortexted for 1 min, sonicated for 5 s, aliquoted and assessed by FISH. Aliquots were also grown at 37 °C anaerobically and in 10% CO2 on LBS agar (Becton Dickinson) with the aim to isolate and type representative strains by partial 16S rDNA sequencing. Demineralization of discs was determined by quantitative light-induced fluorescence as described [29].
Preparation of multi-well slides for FISH
Overnight cultures of lactobacilli (LBS broth) were washed in 0.9% NaCl, diluted in coating buffer [30], spotted on 18- or 24-well slides (Cel-Line Associates), air-dried, and fixed in 4% paraformaldehyde/PBS (20 min, 4 °C). Analogously, in situ grown biofilm samples, supragingival plaque samples and tongue scrapings were vortexed at maximum speed for 60 s, diluted in coating buffer and coated to 18- or 24-well slides as described [30]. To improve cell wall permeability each well selected for FISH of lactobacilli was treated individually at room temperature first for 5 min with 9 μl of lysozyme (1 mg ml-1; Sigma-Aldrich L-7651) and achromopeptidase (1 mg ml-1; Sigma-Aldrich A-7550) in Tris-HCl (pH 7.5) with 5 mM EDTA, and then for 30 min with 9 μl of lipase (Sigma-Aldrich L-1754; at 25 mg ml-1 in water the lipase suspension was centrifuged for 5 min at 16'000 × g after which the supernatant was used). Thereafter, to limit unspecific FISH probe binding all wells were covered for 30 min at 37 °C with 9 μl of PBS containing Denhardt's solution (Fluka 30915; diluted 1:50) in the presence of protectRNA RNase inhibitor (Sigma-Aldrich R-7397; diluted 1:500) [15,16,26,27]. At the end of the respective incubation periods the solutions were carefully aspirated and the slides briefly washed in wash-buffer (0.9% NaCl, 0.05% Tween 20, 0.01% NaN3), dipped in water, and air-dried. All solutions were made with water of nano-pure quality.
Fluorescent in situ hybridization
The 16S rRNA targeted oligonucleotide probes used in this study are listed in Table 1. Custom-synthesized by Microsynth, they were labeled at 5'-end with Cy3 or 6-FAM, or in some cases at both ends with 6-FAM. Probes marked by "L-" in front of the probe name, contain one or two LNA to improve in situ hybridization efficiency [16]. Probes were designed as described previously [30] using the ARB software [31] with the SILVA rRNA database [32,33] and additional rRNA sequence information from 'The Ribosomal Data Base Project II' [34,35] and the 'National Center for Biotechnology Information' [36]. Probes LGC358a and LAB759 had to be used in conjunction with a competitor probe to suppress cross-reactivity with taxa other than lactobacilli and few probes were used together with helper probes [15] to improve fluorescence intensity (Table 1). Probes were used at final concentrations of 5 ng μl-1 (Cy3 conjugates) or 15 ng μl-1 (FAM conjugates, competitor and helper probes). EUB338 served as positive control. FISH was performed as described [30] using hybridization times of 2 or 4 h and probe-specific formamide concentrations as listed in Table 1. Optimum formamide concentrations were determined by varying the formamide concentrations systematically between 25% and 55% in FISH experiments with both reference strains and oral biofilm samples.
Scoring and enumeration of stained bacteria
Following FISH, air-dried multiwell slides were covered with mounting fluid (90% glycerol in PBS with 25 mg g-1 1,4-diazabicyclo[2,2,2]octan) and cover-slips. Bacteria stained by FISH were enumerated as described using an Olympus BX60 epifluorescence microscope (Olympus Optical [Schweiz]) [30]. Scoring of fluorescence intensity is described in a footnote to Table 2.
16S rDNA sequencing
Partial 16S rRNA gene sequences of five lactobacillus isolates (OMZ 1117-1121) from the three in situ grown biofilms were determined as described previously [35]. The sequences of 1393, 1360, 1366, 1371 and 1379 bp in length were compared to gene bank data of the The Ribosomal Data Base Project using the Seq Match algorithm [33]. Identification of isolates was based on ≥ 99.5% similarity. The sequences of OMZ 1117 - 1119 were deposited at EMBL with accession numbers FR667951 - FR667953.
Authors' contributions
BQ and HLS carried out and read FISH analyses. VZ and TT contributed to probe design and editing of the manuscript. EG and BG designed and carried out the in situ study and participated in editing the manuscript. RG designed the project and the probes, analyzed FISH experiments and wrote the manuscript. All authors read and approved the final manuscript. The authors declare no conflict of interest.
Contributor Information
Beatrice Quevedo, Email: b.quevedo@bluewin.ch.
Elin Giertsen, Email: Elin.Giertsen@odont.uio.no.
Vincent Zijnge, Email: vzijnge@yahoo.com.
Helga Lüthi-Schaller, Email: helga.luethi-schaller@zzm.uzh.ch.
Bernhard Guggenheim, Email: bernie@zzm.uzh.ch.
Thomas Thurnheer, Email: thomas.thurnheer@zzm.uzh.ch.
Rudolf Gmür, Email: rudolf.gmuer@zzm.uzh.ch.
Acknowledgements
The authors are grateful to Siren Hammer Østvold for excellent assistance with the in situ study carried out in Bergen, Norway. This work was supported in part by the University of Zürich and the Swedish Patent Revenue Fund for Research in Preventive Odontology.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian M. In: Topley and Wilson's Microbiology and Microbial Infections. Borriello P, Murray PR, Funke G, editor. London: Hodder Arnold; 2005. Streptococcus and Lactobacillus; pp. 833–881. [Google Scholar]
- Marsh PD, Martin MV. Oral Microbiology. 5. Edinburgh: Churchill Livingstone Elsevier; 2009. [Google Scholar]
- Marsh PD, Nyvad B. In: Dental Caries: The Disease and Its Clinical Management. 2. Fejerskov O, Kidd E. Chichester, editor. UK: Wiley-Blackwell; 2008. The oral microflora and biofilms on teeth; pp. 163–187. [Google Scholar]
- Baddour LM. Virulence factors among gram-positive bacteria in experimental endocarditis. Infect Immun. 1994;62:2143–2148. doi: 10.1128/iai.62.6.2143-2148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husni RN, Gordon SM, Washington JA, Longworth DL. Lactobacillus bacterimia and endocarditis: Review of 45 cases. Clin Infect Dis. 1997;25:1048–1055. doi: 10.1086/516109. [DOI] [PubMed] [Google Scholar]
- Amann R, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Micro. 2008;6:339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- Zijnge V, van Leeuwen MBM, Degener JE, Abbas F, Thurnheer T, Gmür R, Harmsen HJM. Oral biofilm architecture on natural teeth. PLoS ONE. 2010;5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen HJM, Elfferich P, Schut F, Welling GW. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in faecal samples by fluorescent in situ hybridization. Microb Ecol Health Dis. 1999;11:3–12. doi: 10.1080/089106099435862. [DOI] [Google Scholar]
- Thurnheer T, Gmür R, Giertsen E, Guggenheim B. Automated fluorescent in situ hybridization for the specific detection and quantification of oral streptococci in dental plaque. J Microbiol Methods. 2001;44:39–47. doi: 10.1016/S0167-7012(00)00226-8. [DOI] [PubMed] [Google Scholar]
- The human oral microbiome database. http://www.HOMD.org
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier H, Amann R, Ludwig W, Schleifer KH. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. System Appl Microbiol. 1999;22:186–196. doi: 10.1016/S0723-2020(99)80065-4. [DOI] [PubMed] [Google Scholar]
- Manz W, Amann R, Ludwig W, Wagner M, Schleifer KH. Phylogenetic oligonucleotide probes for the major subclasses of proteobacteria: problems and solutions. System Appl Microbiol. 1992;15:593–600. [Google Scholar]
- Fuchs BM, Glöckner FO, Wulf J, Amann R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 2000;66:3603–3607. doi: 10.1128/AEM.66.8.3603-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Ohashi A, Imachi H, Harada H. Improved in situ hybridization efficiency with locked-nucleic-acid-incorporated DNA probes. Appl Environ Microbiol. 2006;72:5311–5317. doi: 10.1128/AEM.03039-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comelli EM, Guggenheim B, Stingele F, Neeser JR. Selection of dairy bacterial strains as probiotics for oral health. Eur J Oral Sci. 2002;110:218–224. doi: 10.1034/j.1600-0447.2002.21216.x. [DOI] [PubMed] [Google Scholar]
- Giertsen E, Guggenheim B, Thurnheer T, Gmür R. Microbiological aspects of an in situ model to study effects of antimicrobial agents on dental plaque ecology. Eur J Oral Sci. 2000;108:403–411. doi: 10.1034/j.1600-0722.2000.108005403.x. [DOI] [PubMed] [Google Scholar]
- Thomas RZ, van der Mei HC, van der Veen MH, de Soet JJ, Huysmans MCDNJM. Bacterial composition and red fluorescence of plaque in relation to primary and secondary caries next to composite: an in situ study. Oral Microbiol Immunol. 2008;23:7–13. doi: 10.1111/j.1399-302X.2007.00381.x. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Whiley RA, Shu SE, Ezaki T, Hardie JM. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology. 1999;145:2605–2613. doi: 10.1099/00221287-145-9-2605. [DOI] [PubMed] [Google Scholar]
- Kilian M, Poulsen K, Blomqvist T, Håvarstein LS, Bek-Thomsen M, Tettelin H, Sørensen UBS. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS ONE. 2008;3:e2683. doi: 10.1371/journal.pone.0002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JJ, Blackall LL, Philip B. Further limitations of phylogenetic group-specific probes used for detection of bacteria in environmental samples. ISME J. 2010;4:1–3. doi: 10.1038/ismej.2010.37. [DOI] [PubMed] [Google Scholar]
- Bowden G, Johnson J, Schachtele C. Characterization of Actinomyces with genomic DNA fingerprints and rRNA gene probes. J Dent Res. 1993;72:1171–1179. doi: 10.1177/00220345930720080201. [DOI] [PubMed] [Google Scholar]
- Sekar R, Pernthaler A, Pernthaler J, Warnecke F, Posch T, Amann R. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl Environ Microbiol. 2003;69:2928–2935. doi: 10.1128/AEM.69.5.2928-2935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S, Rühland C, Inacio J, Huber H, Fonseca A, Spencer-Martins I, Fuchs BM, Amann R. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl Environ Microbiol. 2003;69:1748–1758. doi: 10.1128/AEM.69.3.1748-1758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz LS, Okten HE, Noguera DR. Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides. Appl Environ Microbiol. 2006;72:733–744. doi: 10.1128/AEM.72.1.733-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmür R, Lüthi-Schaller H. A combined immunofluorescence and fluorescent in situ hybridization assay for single cell analyses of dental plaque microorganisms. J Microbiol Methods. 2007;69:402–405. doi: 10.1016/j.mimet.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Gmür R, Guggenheim B. Antigenic heterogeneity of Bacteroides intermedius as recognized by monoclonal antibodies. Infect Immun. 1983;42:459–470. doi: 10.1128/iai.42.2.459-470.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmür R, Giertsen E, van der Veen MH, de Josselin de Jong E, ten Cate JM, Guggenheim B. In vitro quantitative light-induced fluorescence to measure changes in enamel mineralization. Clin Oral Invest. 2006;10:187–195. doi: 10.1007/s00784-006-0058-z. [DOI] [PubMed] [Google Scholar]
- Züger J, Lüthi-Schaller H, Gmür R. Uncultivated Tannerella BU045 and BU063 are slim segmented filamentous rods of high prevalence but low abundance in inflammatory disease-associated dental plaques. Microbiology. 2007;153:3817–3829. doi: 10.1099/mic.0.2007/010926-0. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Lüßmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis AP, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res. 2007. gkm864. [DOI] [PMC free article] [PubMed]
- Silva - Comprehensive Ribosomal RNA Database. http://www.arb-silva.de/
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribosomal Database Project. http://rdp.cme.msu.edu
- Basic Local Alignment Search Tool (BLAST) http://blast.ncbi.nlm.nih.gov/Blast.cgi [DOI] [PubMed]
- Gmür R, Munson MA, Wade WG. Genotypic and phenotypic characterization of fusobacteria from Chinese and European patients with inflammatory periodontal diseases. Syst Appl Microbiol. 2006;29:120–130. doi: 10.1016/j.syapm.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Bartoszyk IM, Dewhirst FE. Identification of oral streptococci using PCR-based, reverese-capture, checkerboard hybridization. Meth Cell Sci. 1998;20:223–231. doi: 10.1023/A:1009715710555. [DOI] [Google Scholar]
- Guggenheim B, Gmür R, Galicia JC, Stathopoulou P, Benakanakere MR, Meier A, Thurnheer T, Kinane D. In vitro modeling of host-parasite interactions: the 'subgingival' biofilm challenge of primary human epithelial cells. BMC Microbiol. 2009;9:280. doi: 10.1186/1471-2180-9-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]