Abstract
Background
Several studies have mentioned network modularity—that a network can easily be decomposed into subgraphs that are densely connected within and weakly connected between each other—as a factor affecting metabolic robustness. In this paper we measure the relation between network modularity and several aspects of robustness directly in a model system of metabolism.
Methodology/Principal Findings
By using a model for generating chemical reaction systems where one can tune the network modularity, we find that robustness increases with modularity for changes in the concentrations of metabolites, whereas it decreases with changes in the expression of enzymes. The same modularity scaling is true for the speed of relaxation after the perturbations.
Conclusions/Significance
Modularity is not a general principle for making metabolism either more or less robust; this question needs to be addressed specifically for different types of perturbations of the system.
Introduction
Graph theoretical methods are useful to study the large-scale organization of biological systems [1]. One such system is the metabolism—the set of chemical reactions needed to sustain the normal, healthy state of an organism. We call a graph derived from a metabolic reaction system a metabolic network. One of the main findings from statistical studies of metabolic networks is that the metabolism has larger network modularity [2], [3] —the tendency for a network to be divisible into subgraphs that are densely connected within, and sparsely connected between each other—than expected [4]. However, metabolic networks are far from perfectly modular—no matter how the network modules are defined, there will be plenty of connections between them [4]–[8]. The network modules are often interpreted as biological modules—functionally independent subunits [9]. This interpretation is a natural consequence of interpreting edges as functional couplings of relatively equal strength. Despite the lack of comprehensive experimental evidence, metabolism is assumed to be robust to e.g. changes in concentration of metabolites [10]. Modularity is often thought to contribute to the robustness of various biological systems [11]–[13]. But if this is true for metabolism too, that modularity contributes to both functionality and robustness, then how come there are so many cross-modular couplings? One explanation could be that these couplings are inevitable—the laws of physics give no way of avoiding intermodular connections. Another explanation could be that the intermodular edges actually stabilize the system so that the organization we observe is a compromise where adding functionality increases modularity and adding robustness decreases modularity. Such a role of modularity relates to the concept of distributed robustness [14]—if a module fails, many other modules can collectively compensate for this loss, there need not be any replacement module. In terms of metabolic networks, this means that there will be many connections between the modules and thus that the network modularity will be comparatively low. In this paper we investigate the role of network modularity in large chemical reaction systems as directly as possible—by measuring the system's response to different types of perturbations in a model with tunable network modularity.
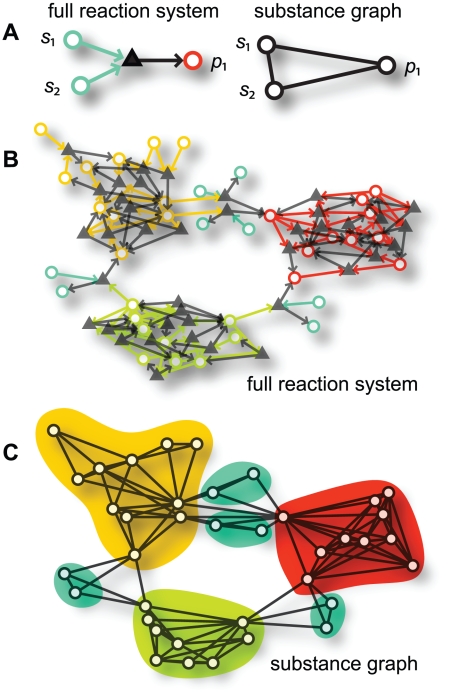
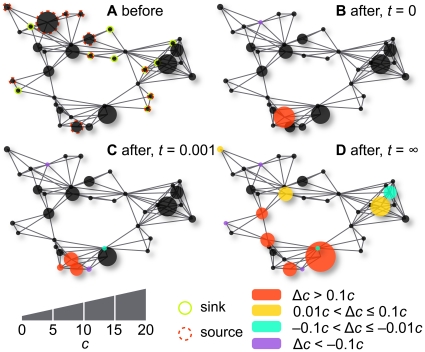
Our simulations start by generating a chemical reaction system. This generative algorithm is stochastic and by tuning the input parameters, we can control the expected network modularity (Fig. 1) [15]. Then we generate a random distribution of metabolites and relax the system to equilibrium (using mass-action kinetics with an implicit enzymatic control). From this state, we apply a certain type of perturbation to the system and let it relax to a new equilibrium. To quantify robustness, we measure how close the two equilibria are to each other. We also measure the relaxation time, i.e. how fast the system can respond to the perturbation (and for that reason, we do not employ faster calculations of the equilibrium state [16], [17]). In Fig. 2 we show an example of these steps. As the reaction system is generated by a stochastic method we repeat the procedure above to obtain averages. For each value of the input parameters, we measure average values over 500 realizations of all steps above of both the network modularity and the quantities characterizing robustness. From these data points we derive trends in the modularity-dependence of different aspects of robustness.
Figure 1. Example of the reduction from reaction systems to substance graphs and the generation of modular reaction systems.
In A we see how the two substrates and one product (circles) of a reaction
(triangle) gets reduced to a substance graph. An arrow going into a reaction
marks the substrate, an arrow going out marks the product. Panel B
illustrates a reaction system obtained with the method of the manuscript.
The parameter values for this reaction-system example are
 ,
, 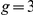 ,
,
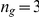 ,
, 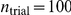 and
and
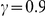 . The algorithm proceeds by assembling reactions and
metabolites in disjoint clusters (the three larger clusters of distinct
colors). Then we add a fraction of metabolites and reactions that can
connect to any parts of the system. The larger this fraction of global
reactions is, the lower is the network modularity of the projected
network.
. The algorithm proceeds by assembling reactions and
metabolites in disjoint clusters (the three larger clusters of distinct
colors). Then we add a fraction of metabolites and reactions that can
connect to any parts of the system. The larger this fraction of global
reactions is, the lower is the network modularity of the projected
network.
Figure 2. The procedure to measure robustness.
The figure illustrates a reaction system at equilibrium visualized by its reaction graph A, getting perturbed by redistributing the mass of (in this case two) metabolites B and how the system relaxes to another equilibrium (c,d). The concentration is illustrated by the size of the circles (the total mass, not the concentration is conserved, so the total areas of the circles are not the same in the different panels). The change in concentration is indicated by color. A metabolite unaffected by the perturbation is colored black.
Results
Robustness as a function of network modularity
Robustness is a broad concept that hardly can be condensed into one measure, even for a system as specific as metabolism. In general, robustness can be defined as a system's ability to remain unchanged when perturbed. One can imagine several types of perturbations. We investigate two rather different classes—changes in concentrations of metabolites and changes in the reaction system (new reactions replacing old) by genetic control. We refer to the first case as metabolic perturbations and the second as genetic perturbations. We will also distinguish between: if the perturbations are localized to one module, or if they can appear anywhere in the network. In total we consider four classes of perturbations—they can be either localized or global, and metabolic or genetic.
The main robustness measure, defined in the Methods section, is basically the relative change in the
concentration of a metabolite averaged over a set of metabolites. We consider
two such sets, either the whole set of metabolites, which gives the
system-wide robustness
 , or the metabolites that are perturbed giving the
focal robustness
, or the metabolites that are perturbed giving the
focal robustness
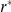 .
.
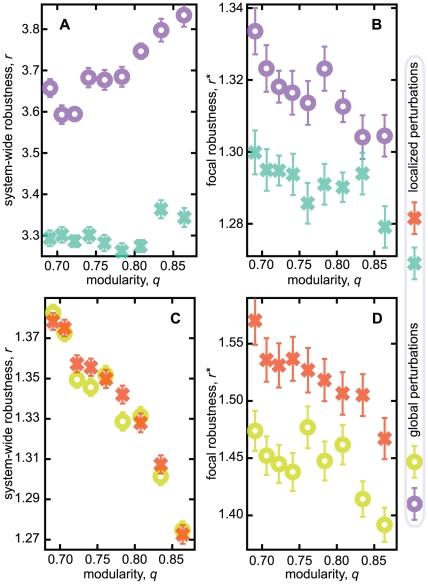
In Fig. 3, we plot the
average values of our robustness measures as functions of the average network
modularity 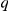 . The robustness to global metabolic perturbations
increases while the robustness to perturbations within a module remains fairly
constant (Fig. 3A). If one
looks only at the metabolites that were originally perturbed (Fig. 3A), the situation is
different—these metabolites are more affected by sudden shifts in the
concentrations the more modular the system is. This seems logical—if the
modularity is lower, the coupling to the rest of the network is stronger, so
there are more metabolites to influence the relaxation and to absorb the
perturbation. The fact that the system is more robust to global, compared to
localized, perturbations can be explained in a similar way—a localized
perturbation gives a larger impact on a restricted subsystem and this subsystem
cannot absorb that large impact as much as the whole system would. But why does
the system-wide robustness increase with modularity? One scenario is that
metabolic perturbations are better absorbed in a distributed fashion. With
global perturbations and high modularity each module handles its internal
perturbations and, if this fails, flows between the modules are too weak for the
perturbation to spread.
. The robustness to global metabolic perturbations
increases while the robustness to perturbations within a module remains fairly
constant (Fig. 3A). If one
looks only at the metabolites that were originally perturbed (Fig. 3A), the situation is
different—these metabolites are more affected by sudden shifts in the
concentrations the more modular the system is. This seems logical—if the
modularity is lower, the coupling to the rest of the network is stronger, so
there are more metabolites to influence the relaxation and to absorb the
perturbation. The fact that the system is more robust to global, compared to
localized, perturbations can be explained in a similar way—a localized
perturbation gives a larger impact on a restricted subsystem and this subsystem
cannot absorb that large impact as much as the whole system would. But why does
the system-wide robustness increase with modularity? One scenario is that
metabolic perturbations are better absorbed in a distributed fashion. With
global perturbations and high modularity each module handles its internal
perturbations and, if this fails, flows between the modules are too weak for the
perturbation to spread.
Figure 3. Robustness vs. modularity.
Panels A and B show data for the robustness against metabolic
perturbations. A displays robustness of the system as a whole; B shows
the robustness measured over the perturbed metabolites only. Panels C
and D show the corresponding plots for robustness against genetic
perturbations. Circles represent perturbations made in one module;
crosses indicate data for perturbations made in different modules. The
data is averaged over more than 500 runs (network realizations). The
errorbars in the average  are
smaller than the symbol size.
are
smaller than the symbol size.
For the genetic perturbations all curves are decreasing, meaning that modularity
makes the system less robust. These perturbations virtually add new reactions
and delete old. Even if the perturbations are designed not to affect the average
structure of the system (keeping e.g. the system size
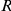 and the modularity
and the modularity 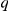 constant), they
obviously affect
constant), they
obviously affect  more than the
metabolic perturbations (cf. Fig.
3A and Fig. 3C).
A network module can presumably not handle a genetic perturbation as efficient
as a metabolic perturbation. Another factor for the decreasing
more than the
metabolic perturbations (cf. Fig.
3A and Fig. 3C).
A network module can presumably not handle a genetic perturbation as efficient
as a metabolic perturbation. Another factor for the decreasing
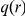 -curve could be that the interface between the modules
can change from the perturbations and that the interfaces get more influential
with increasing modularity. As seen in Fig. 3D, the localized perturbations
influence the directly affected metabolites (the ones that are involved in
reactions changed by the genetic perturbations) less strongly than the global
perturbations. From the changes at the interfaces, we can understand that
localized perturbations affect the rest of the system to a greater deal here
than compared with metabolic perturbations.
-curve could be that the interface between the modules
can change from the perturbations and that the interfaces get more influential
with increasing modularity. As seen in Fig. 3D, the localized perturbations
influence the directly affected metabolites (the ones that are involved in
reactions changed by the genetic perturbations) less strongly than the global
perturbations. From the changes at the interfaces, we can understand that
localized perturbations affect the rest of the system to a greater deal here
than compared with metabolic perturbations. 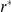 is larger for the
local compared with global genetic perturbations meaning that for metabolites
within a single module rewired by genetic perturbations the changes will be
larger than if the perturbations are more distributed.
is larger for the
local compared with global genetic perturbations meaning that for metabolites
within a single module rewired by genetic perturbations the changes will be
larger than if the perturbations are more distributed.
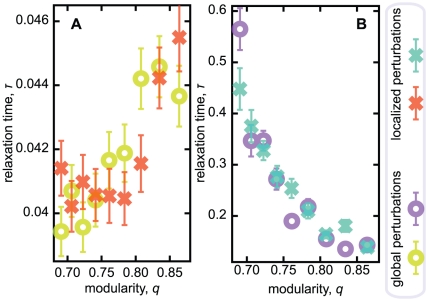
Relaxation time as a function of network modularity
In Fig. 4, we show the
relaxation time  as a function of
modularity. A small
as a function of
modularity. A small  value means that
the system reaches its new equilibrium fast. This dynamic response is different
for the two types of perturbations—the system reaches its new state faster
with higher modularity for the metabolic perturbations, but slower with the
genetic ones. The decreasing
value means that
the system reaches its new equilibrium fast. This dynamic response is different
for the two types of perturbations—the system reaches its new state faster
with higher modularity for the metabolic perturbations, but slower with the
genetic ones. The decreasing 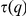 curves for
metabolic perturbations is in line with the above mentioned scenario that if
modules handle the perturbations independently, then the more modular the system
is the better (in this case faster) is the recovery. That, for genetic
perturbations, robustness increases with modularity is something we interpret as
an effect of the changed couplings across at the boundary. The stronger the
modularity is, the slower is the flow between the modules and the longer does
the system need to find a new equilibrium.
curves for
metabolic perturbations is in line with the above mentioned scenario that if
modules handle the perturbations independently, then the more modular the system
is the better (in this case faster) is the recovery. That, for genetic
perturbations, robustness increases with modularity is something we interpret as
an effect of the changed couplings across at the boundary. The stronger the
modularity is, the slower is the flow between the modules and the longer does
the system need to find a new equilibrium.
Figure 4. Relaxation time vs. modularity.
Panel A displays the corresponding data for perturbations in the concentrations of metabolites. Panel B shows the relaxation time for genetic perturbations within one module (circles) or the whole system (crosses). The data represents averages over more than 500 runs (the same runs as in Fig. 3).
Discussion
We have, in a model framework, directly measured the effects of network modularity on the robustness of chemical reaction systems. The main conclusion is that modularity does affect robustness but not in a unique way. Modularity is thus, it seems, not a general principle for either strengthening or weakening robustness, not even in such a specific system as metabolism. When relating robustness and modularity, one needs to specify what kind of perturbation we measure robustness against. For sudden changes icn concentration levels, in our model, more modular reaction systems are more robust and converge to an equilibrium state faster than less modular systems. If, on the other hand, the genetic control is altered—so that other enzymes are expressed—then modularity decreases robustness. In an evolutionary perspective, this essentially means that we need more detailed studies. Real metabolic networks are more modular (in the network-modularity sense) than random networks, but still far from, say, a system engineered by humans [18]. One scenario is that robustness is key driving force in evolution of metabolic-network structure and that this weakly modular structure above comes from trade-offs between robustness-increasing and robustness-decreasing changes in modularity. However, functionality and chemical constraint probably also play a major role in this evolution. Note that if one considers smaller feedback loops as modules, rather than network clusters, evolution is by necessity modular in the sense that adding the production of a new substance often needs the addition of its degradation (this is because many substances cannot penetrate the cell membrane and would be toxic if accumulated). The conclusion that modularity does not affect robustness in a single direction has further implications for synthetic biology that often, at least theoretically, strives to design functionality from combination of modules [19], [20]—our study hints the such an approach would not give robustness for free.
For the future, we anticipate more studies cataloguing the principles of robustness, and the effects of modularity. We believe model studies like the present are the best theoretical way to proceed. An alternative is to compare the modularity of different organisms [21] to find changes in the modularity over the course of evolution, but in such an approach it would be hard to tease apart fundamental physical constraints from evolutionary pressure. It would of course also be interesting to experimentally compare the response of different organisms, or cell types, with metabolism of different network modularity to perturbations. Further into the future, we hope for experimental methods to measure the dynamics of the entire chemical composition of cells.
Methods
Notations and mathematical framework
We consider a reaction system of 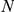 metabolites
metabolites
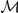 and
and 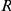 reactions
reactions
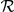 . A reaction
. A reaction 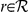 is characterized
by its substrates
is characterized
by its substrates 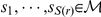 , their
multiplicities
, their
multiplicities  , its products
, its products
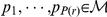 and their multiplicities
and their multiplicities  , and a reaction
coefficient
, and a reaction
coefficient 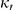 . Consider, for example, the reaction
2H
. Consider, for example, the reaction
2H + O
+ O
 2 H
2 H O. Then we have
O. Then we have
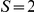 ,
, 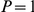
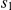 is H
is H ,
,
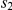 is O
is O ,
,
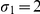 ,
, 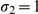 ,
,
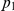 is H
is H O and
O and
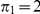 . From a reaction system one can derive a graph
. From a reaction system one can derive a graph
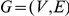 , where
, where 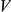 (
(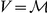 in this case) is the set of vertices of the graph and
in this case) is the set of vertices of the graph and
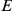 is the set of edges. One can define several types of
metabolic graphs. In this work we focus on substance graphs (claimed to encode
more functional information about the graphs than other simple-graph
representations [5], [15]), where the vertices are substances and there is an
(undirected) edge between two vertices if they are either substrates or products
of the same reaction (edges between a vertex to itself is not allowed). In the
example above, the reaction will contribute with three
edges—
is the set of edges. One can define several types of
metabolic graphs. In this work we focus on substance graphs (claimed to encode
more functional information about the graphs than other simple-graph
representations [5], [15]), where the vertices are substances and there is an
(undirected) edge between two vertices if they are either substrates or products
of the same reaction (edges between a vertex to itself is not allowed). In the
example above, the reaction will contribute with three
edges—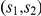 ,
,
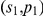 and
and 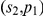 —to the
substance graph (see Fig.
1A).
—to the
substance graph (see Fig.
1A).
Network modularity
We will shortly discuss how network modularity is calculated. For a more
comprehensive review, see Refs. [2], [3]. Let the vertex set be
partitioned into groups and let 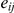 denote the
fraction of edges between group
denote the
fraction of edges between group  and
and
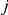 . The modularity of this partition is defined as
. The modularity of this partition is defined as
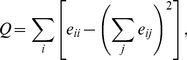 |
(1) |
where the sum is over all the vertex
groups. The term 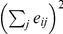 is the expectation
value of
is the expectation
value of  in a random graph. The measure for graph modularity that
we use is
in a random graph. The measure for graph modularity that
we use is 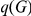 —
—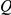 maximized over all
partitions (by a heuristics proposed in Ref. [3]). Comparing
maximized over all
partitions (by a heuristics proposed in Ref. [3]). Comparing
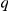 of graphs with different sizes and degree distributions
is not completely straightforward. Even for networks generated by one particular
model (that one would from construction expect to have the same modularity)
of graphs with different sizes and degree distributions
is not completely straightforward. Even for networks generated by one particular
model (that one would from construction expect to have the same modularity)
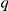 can vary with the network size [22]. Fortunately, for this
work, such changes are monotonous. This means that we can use
can vary with the network size [22]. Fortunately, for this
work, such changes are monotonous. This means that we can use
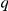 to detect changes in robustness in response to changes
in modularity even though the particular functional forms of the curves of
robustness vs.
to detect changes in robustness in response to changes
in modularity even though the particular functional forms of the curves of
robustness vs. 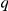 are hard to
interpret.
are hard to
interpret.
Model reaction systems with tunable network modularity
In this section, we will sketch the model of reaction systems with tunable
network modularity. The model we use treats atoms of the molecular species
explicitly. The set of all atoms is divided into 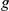 groups (or
proto-modules) of equal size
groups (or
proto-modules) of equal size 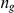 .
.
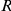 reactions are added to the system such that they obey
mass conservation (for all atom species, the number of individuals is the same
for substrates and products).
reactions are added to the system such that they obey
mass conservation (for all atom species, the number of individuals is the same
for substrates and products). 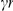 reactions are
added between molecules consisting of atoms from the same group. The remaining
reactions are
added between molecules consisting of atoms from the same group. The remaining
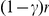 reactions are added between molecules of any atomic
composition. For low
reactions are added between molecules of any atomic
composition. For low  -values, relatively
few reactions will connect different groups and therefore the derived network
modularity will be low. If
-values, relatively
few reactions will connect different groups and therefore the derived network
modularity will be low. If  is close to one,
the derived graphs will be more modular. The molecules are constructed by
randomly combining atoms of a group. Reactions are generated by randomly
splitting and recombining molecules. If the mass conservation is broken, or the
reaction already exists in the data set, then the molecule construction is
repeated. If no reaction fulfilling mass conservation has been found after
is close to one,
the derived graphs will be more modular. The molecules are constructed by
randomly combining atoms of a group. Reactions are generated by randomly
splitting and recombining molecules. If the mass conservation is broken, or the
reaction already exists in the data set, then the molecule construction is
repeated. If no reaction fulfilling mass conservation has been found after
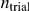 iterations, then this is done by defining new molecules.
With a larger value of
iterations, then this is done by defining new molecules.
With a larger value of 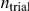 , the substance
graphs will thus be both denser and have fewer metabolites
(
, the substance
graphs will thus be both denser and have fewer metabolites
(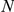 is, perhaps a little unusually, an output of the model,
whereas
is, perhaps a little unusually, an output of the model,
whereas 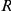 is a control parameter).
is a control parameter).
There are a number of other technicalities, like how the molecules are constructed from the atoms etc., that are explained in detail in Ref. [15]. We also modify the algorithm of Ref. [15] when it comes to inter-group reactions. In Ref. [15] these always act as sources and sinks (so that there is never a flow between modules); here all inter-group reactions are bridges between the modules (so that these reactions have at least one substrate in one group and one product in the other).
In this work we use the parameter values 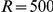 ,
,
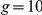 ,
, 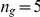 and
and
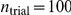 (the values of the other parameters, related to the
details in Ref. [15] are the same as in that paper).
(the values of the other parameters, related to the
details in Ref. [15] are the same as in that paper).
Reaction kinetics
To simulate the biochemical dynamics, we use simple mass-action kinetics. This
approach is, technically speaking, assuming all enzymatic effects can be encoded
into the reaction coefficients and the reaction system itself. The main reason
for this simplification is that, when speaking about network modularity, enzymes
are usually only included implicitly (via the active reactions), so to relate
the robustness to network modularity we need a kinetic description of the same
level of description. Given a reaction system generated by the scheme above we
assign a rate constant 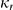 to each reaction
to each reaction
 drawn from a normal distribution
drawn from a normal distribution
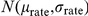 (the sign of
(the sign of 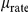 defines the
direction of the reaction) and initial concentration
defines the
direction of the reaction) and initial concentration
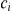 of a substance
of a substance  in
in
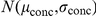 (setting negative concentrations to zero). From this
starting point, we use the kinetic equation
(setting negative concentrations to zero). From this
starting point, we use the kinetic equation
| (2) |
where the sum is over all reactions
 with
with  as a product,
where
as a product,
where 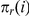 is
is  's
multiplicity in the reaction
's
multiplicity in the reaction  . To simulate the
metabolic flux we also add source and sink terms to Eq. 2 for some metabolites.
We let all the metabolites that are not substrates of any reaction be sinks
(otherwise their mass would just accumulate) and all metabolites that are not a
product of any reaction to be sources. In practice there will always be both
sources and sinks in the generated reaction systems. (If the reaction systems
would be generated in some other way one would need to put in sources and sinks
explicitly.) We model the outflux by letting the sink-metabolites flow out of
the system with a rate proportional to
. To simulate the
metabolic flux we also add source and sink terms to Eq. 2 for some metabolites.
We let all the metabolites that are not substrates of any reaction be sinks
(otherwise their mass would just accumulate) and all metabolites that are not a
product of any reaction to be sources. In practice there will always be both
sources and sinks in the generated reaction systems. (If the reaction systems
would be generated in some other way one would need to put in sources and sinks
explicitly.) We model the outflux by letting the sink-metabolites flow out of
the system with a rate proportional to  times the
concentration of the metabolite. In our simulations we use
times the
concentration of the metabolite. In our simulations we use
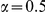 . We keep the inflow rate the same as the outflow rate so
that the total mass is conserved. The inflow is distributed to the inflow
metabolites in proportion to
. We keep the inflow rate the same as the outflow rate so
that the total mass is conserved. The inflow is distributed to the inflow
metabolites in proportion to 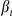 , a random variable
for each inflow metabolite drawn from a
, a random variable
for each inflow metabolite drawn from a 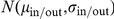 distribution when
the reaction system is generated.
distribution when
the reaction system is generated.
From the above setup, we run the system is until it converges (which it always
does for the dynamic systems in question). We integrate the system with the
Euler method (with time step 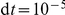 until the time
until the time
 when
when
| (3) |
We use 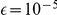 in this simulations. Higher precision in
in this simulations. Higher precision in
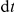 or
or  does not change
the outcome significantly. In this paper we use the parameter values
does not change
the outcome significantly. In this paper we use the parameter values
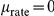 ,
, 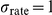 ,
,
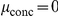 ,
, 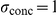 ,
,
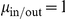 ,
, 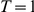 and
and
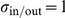 .
.
Genetic perturbations
Since we exclude genetic control and explicit enzymes in our reaction-system
kinetics, we have to model the genetic perturbations indirectly. This is on the
other hand quite straightforward. We replace 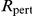 randomly chosen
reactions following the same rules as when the reaction system was first
constructed. For local perturbations, the reactions are chosen from one randomly
selected cluster (identified by the cluster-detection algorithm above). A
reaction is associated to the module to which a majority of its metabolites are
categorized (if there is a tie, we select a cluster randomly). In this process,
new metabolites will inevitably be generated and others possibly deleted. To
conserve mass in case the number of metabolites changes, we split the mass of
the deleted metabolites equally among the new. We also go over the system and
update the sources and sinks in the same way as when the reaction system was
constructed.
randomly chosen
reactions following the same rules as when the reaction system was first
constructed. For local perturbations, the reactions are chosen from one randomly
selected cluster (identified by the cluster-detection algorithm above). A
reaction is associated to the module to which a majority of its metabolites are
categorized (if there is a tie, we select a cluster randomly). In this process,
new metabolites will inevitably be generated and others possibly deleted. To
conserve mass in case the number of metabolites changes, we split the mass of
the deleted metabolites equally among the new. We also go over the system and
update the sources and sinks in the same way as when the reaction system was
constructed.
Metabolic perturbations
Analogously to the genetic perturbations, we also require the metabolic
perturbations to conserve the total mass. We control the magnitude of the
perturbation by a parameter 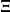 by requiring that
by requiring that
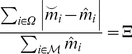 |
(4) |
where 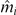 is the total mass
of metabolite
is the total mass
of metabolite  before the perturbation and
before the perturbation and
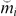 is the total mass after, and
is the total mass after, and
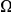 is a set of metabolites. In practice the masses have a
right-skewed, heavy tailed distribution (as observed in real systems [23]). This
means that if we just continue adding metabolites randomly until the condition
Eq. (4) is fulfilled, and
is a set of metabolites. In practice the masses have a
right-skewed, heavy tailed distribution (as observed in real systems [23]). This
means that if we just continue adding metabolites randomly until the condition
Eq. (4) is fulfilled, and 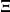 is not very small
(we use
is not very small
(we use 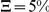 ), we will have to perturb a rather large fraction of the
metabolites. To get around this problem, consider a set
), we will have to perturb a rather large fraction of the
metabolites. To get around this problem, consider a set
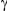 of metabolite pairs. For the local perturbations, we
choose a cluster (as detected by the algorithm above) at random as
of metabolite pairs. For the local perturbations, we
choose a cluster (as detected by the algorithm above) at random as
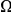 and add pairs of metabolites picked at random to
and add pairs of metabolites picked at random to
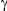 until the condition is met or all there are no
metabolites left in the cluster. For the global perturbations we let
until the condition is met or all there are no
metabolites left in the cluster. For the global perturbations we let
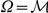 and split the metabolites into two sets
and split the metabolites into two sets
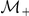 and
and 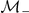 where the total
mass of any metabolite in
where the total
mass of any metabolite in 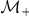 is larger than any
metabolite in
is larger than any
metabolite in 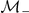 and
and 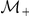 is as small as
possible such that the total mass of
is as small as
possible such that the total mass of 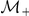 is larger than
is larger than
 . In our simulations
. In our simulations 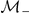 always has more
elements than
always has more
elements than 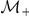 . Then we add pairs where one metabolite is randomly
selected from
. Then we add pairs where one metabolite is randomly
selected from 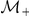 and one is randomly selected from
and one is randomly selected from
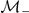 until Eq. (4) is true.
until Eq. (4) is true.
Robustness measures
Any measure of robustness should increase the more similar the system is before
and after a perturbation. For biological functionality, it could be just as
important to keep the concentrations of rare metabolites steady as those of the
most abundant ones. Let 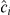 be the
concentration of metabolite
be the
concentration of metabolite  before the
perturbation and
before the
perturbation and 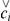 be the
concentration after. A natural choice would be to take the average over the
metabolites of the change
be the
concentration after. A natural choice would be to take the average over the
metabolites of the change 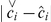 rescaled by the
typical concentration of
rescaled by the
typical concentration of  as a measure of
unrobustness (and thus its reciprocal value as a measure of robustness). As
“typical concentration” one choice is the average. In practice, the
metabolites that are very close to zero in concentration can give a rather large
signal due just to numerical errors. To suppress such numerical noise, we rather
use the quadratic mean, which decreases the expression's sensitivity to
fluctuations in the denominator in the frequent situation that the
concentrations are close to zero, thus putting a lower weight on the more
uncertain terms. Our robustness measure thus becomes
as a measure of
unrobustness (and thus its reciprocal value as a measure of robustness). As
“typical concentration” one choice is the average. In practice, the
metabolites that are very close to zero in concentration can give a rather large
signal due just to numerical errors. To suppress such numerical noise, we rather
use the quadratic mean, which decreases the expression's sensitivity to
fluctuations in the denominator in the frequent situation that the
concentrations are close to zero, thus putting a lower weight on the more
uncertain terms. Our robustness measure thus becomes
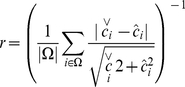 |
(5) |
where 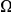 is a set of
metabolites and
is a set of
metabolites and  denotes the
absolute value of a number or the number of elements of a set. We consider two
versions of this measure, one averaged over the whole set of metabolites, which
we call system-wide perturbations
denotes the
absolute value of a number or the number of elements of a set. We consider two
versions of this measure, one averaged over the whole set of metabolites, which
we call system-wide perturbations  , and one averaged
over the metabolites directly affected by the perturbations (the metabolites
participating in a reaction catalyzed by a perturbed enzyme in the case of
genetic perturbations or, trivially, the perturbed metabolites of a metabolic
perturbation), which we refer to as focal robustness
, and one averaged
over the metabolites directly affected by the perturbations (the metabolites
participating in a reaction catalyzed by a perturbed enzyme in the case of
genetic perturbations or, trivially, the perturbed metabolites of a metabolic
perturbation), which we refer to as focal robustness
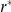 .
.
Footnotes
Competing Interests: The author has declared that no competing interests exist.
Funding: This work was supported by the Swedish Foundation for Strategic Research, the Swedish Research Council, and the WCU program through the National Research Foundation (NRF) of the Republic of Korea funded by the Ministry of Education, Science and Technology (MEST) (R31-2008-000-10029-0). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Barabási AL. Network biology: Understanding the cell's functional organization. Nature Reviews Genetics. 2004;4:101–114. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 2.Fortunato S. Community detection in graphs. Physics Reports. 2010;486:75–174. [Google Scholar]
- 3.Newman MEJ. Modularity and community structure in networks. Proc Natl Acad Sci USA. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huss M, Holme P. Currency and commodity metabolites: Their identification and relation to the modularity of metabolic networks. IET Systems Biology. 2007;1:280–285. doi: 10.1049/iet-syb:20060077. [DOI] [PubMed] [Google Scholar]
- 5.Holme P, Huss M. Substance graphs are optimal simple-graph representations of metabolism. Chinese Science Bulletin. 2010;55:3161–3168. [Google Scholar]
- 6.Zhao J, Yu H, Luo JH, Cao ZW, Li YX. Hierarchical modularity of nested bow-ties in metabolic networks. BMC Bioinformatics. 2006;7:386. doi: 10.1186/1471-2105-7-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1553–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J, Ding GH, Tao L, Yu H, Yu ZH, et al. Modular co-evolution of metabolic networks. BMC Bioinformatics. 2007;8:311. doi: 10.1186/1471-2105-8-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alon U. Biological networks: The tinkerer as an engineer. Science. 2003;301:1866–1867. doi: 10.1126/science.1089072. [DOI] [PubMed] [Google Scholar]
- 10.Smart AG, Amaral LAN, Ottino JM. Cascading failure and robustness in metabolic networks. Proc Natl Acad Sci USA. 2008;105:13223–13228. doi: 10.1073/pnas.0803571105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eum S, Arakawa S, Murata M. New York: Association of Computing Machinery; 2007. Toward bio-inspired network robustness - step 1. modularity. In: Bio-Inspired Models of Network, Information and Computing Systems 2. pp. 84–87. [Google Scholar]
- 12.Viana MP, Tanck E, Beletti ME, Costa LF. Modularity and robustness of bone networks. Mol Biosyst. 2009;5:255–261. doi: 10.1039/b814188f. [DOI] [PubMed] [Google Scholar]
- 13.Hinze A, Adami C. Evolution of complex modular biological networks. PLoS Comput Biol. 2008;4:e23. doi: 10.1371/journal.pcbi.0040023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner A. Princeton, NJ: Princeton University Press; 2005. Robustness and Evolvability in Living Systems. [Google Scholar]
- 15.Holme P. Model validation of simple-graph representations of metabolism. J Roy Soc Interface. 2009;40:1027–1034. doi: 10.1098/rsif.2008.0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fell D. Metabolic control analysis: a survey of its theoretical and experimental development. Biochem J. 1992;1:313–330. doi: 10.1042/bj2860313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuster S, Dandekar T, Fell D. Detection of elementary ux modes in biochemical networks: A promising tool for pathway analysis and metabolic engineering. Trends Biotech. 1999;17:53–60. doi: 10.1016/s0167-7799(98)01290-6. [DOI] [PubMed] [Google Scholar]
- 18.Snel B, Huynen MA. Quantifying modularity in the evolution of biomolecular systems. Genome Research. 2004;14:391–397. doi: 10.1101/gr.1969504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrianantorando E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Molecular systems biology. 2006;2:2006.0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keasling JD. Manufacturing molecules through metabolic engineering. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Erten S, Bebek G, Koyuturk M, Li J. 2009 Ohio Collaborative Conference on Bioinformatics; 2009. Comparative analysis of modularity in biological systems. pp. 104–109. [Google Scholar]
- 22.Guimerà R, Sales-Pardo M, Nunes Amaral LA. Modularity from uctuations in random graphs and complex networks. Phys Rev E. 2004;70:025101. doi: 10.1103/PhysRevE.70.025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko K. Berlin: Springer; 2006. Life: An introduction to complex systems biology. [Google Scholar]