Abstract
Defects in DNA replication are implicated as early and causal events in malignancy. However, the immediate effects of impaired DNA replication licensing on cell cycle progression of non-malignant human cells are unknown. Therefore, we have investigated the acute effects of Mcm7 ablation using synchronized cultures of untransformed Human Dermal Fibroblasts (HDF). Mcm7 ablation elicited a G1 delay associated with impaired activation of CDK4 and CDK2 and reduced Rb phosphorylation. The cell cycle delay of Mcm7-ablated cells was not associated with a DNA damage response. However, levels of cyclin D1 mRNA were specifically reduced and binding of RNA Polymerase II to the CYCD1 promoter was decreased in Mcm7-depleted cells. Similar to Mcm7-deficiency, Mcm2- or Cdc6-depletion led to impaired cyclin D expression. Ectopic overexpression of Cdc6 in quiescent cells promoted cyclin D1 expression, CDK4 activation and G1 progression. Therefore timely and efficient expression of cyclin D1 during G1 phase requires replication licensing. Reconstitution of cyclin D1 expression was insufficient to correct the G1 delay of Mcm7-depleted cells, indicating that additional cell cycle events during G1 are dependent on replication licensing. However, ectopic expression of the HPV-E7 oncoprotein, and the resulting bypass of the requirement for cyclin D1-Rb signaling enabled Mcm7-depleted cells to enter S-phase. HPV-E7-induced S-phase entry of Mcm7-depleted cells led to a DNA damage response, a hallmark of pre-malignancy. Taken together, our results suggest the existence of a ‘replication licensing restriction point’ that couples pre-RC assembly with G1 progression in normal cells to minimize replication stress, DNA damage and tumorigenesis.
Keywords: DNA replication, licensing, pre-RC, cyclin D1, G1 progression
Introduction
Accurate replication and repair of DNA is crucial for maintaining genomic stability. To ensure faithful genome duplication, eukaryotic DNA replication is a highly ordered process involving regulated assembly and disassembly of protein complexes at origins of DNA replication. The Origin Recognition Complex (ORC), comprising ORC subunits 1–6 binds to origins of replication throughout the cell cycle. During late M-phase and G1 the mini-chromosome maintenance (Mcm) complex (Mcm2-7) is recruited to origins by the molecular chaperones Cdt1 and Cdc6, to form pre-replicative complexes or pre-RC (a process termed ‘replication licensing’).1-3 At the onset of S-phase, Cyclin-Dependent Kinases (CDKs) and Cdc7 convert pre-RC into Initiation Complexes (ICs). IC formation involves recruitment of initiation factors such as Mcm10 and Cdc45, melting of origin DNA, association of ssDNA-binding proteins, and loading of the Pol α-primase complex to synthesize the RNA primer necessary for subsequent DNA replication.1
Appropriate control of DNA replication and its integration with other cell cycle events is critical for maintaining genome stability and preventing transformation. Precisely how replication licensing is coordinated with other cell cycle events during G1 is not fully understood. A key event required for normal G1 to S-phase transition is phosphorylation-mediated inactivation of the retinoblastoma (Rb) tumor suppressor protein. Rb is a transcriptional repressor that negatively regulates G1 progression by inhibiting the E2F transcription factors. E2F family members regulate expression of genes important for S-phase entry and progression (including CYCA2, DHFR and CDC45).4 Thus, active Rb negatively regulates cell cycle progression through G1 (at least in large part) by repressing E2F. Inhibition of Rb by mitogenic stimuli or oncogenes de-represses E2F activity thereby inducing E2F-responsive genes and promoting cell cycle progression.5,6
Rb inactivation is mediated by the concerted action of G1-phase CDKs.7,8 CDKs phosphorylate multiple sites on Rb, resulting in its displacement from chromatin and de-repression of E2F. The CDKs responsible for phosphorylation and inactivation of Rb are CDKs 4 and 6 (which are cyclin D-dependent) and CDK2 (which is cyclin E-dependent). During mid-G1, CDK4/6-dependent phosphorylation of Rb at multiple sites results in E2F-mediated transcriptional induction of the Cyclin E gene, CYCE.9,10 Expression of Cyclin E and the ensuing activation of CDK2 induces hyper-phosphorylation of Rb and further de-repression of E2F activity. The G1-S transition is triggered when E2F activity reaches a critical threshold level. Regulation of CDKs occurs via multiple mechanisms, including assembly of CDK complexes, inhibitory and activating phosphorylation events, and association with cyclin-dependent kinase inhibitors (CKIs). Moreover, CDK inhibitors of the Cip/Kip and Ink4 families play key roles in regulating CDK activity and controlling the G1-Sphase transition. In addition to promoting expression of E2F-induced genes, CDK activities play important roles in regulating the licensing and initiation stages of DNA synthesis. For instance, the stabilization and accumulation of Cdc6 in G1 is CDK2-dependent,11 and recruitment of initiation factors and the onset of DNA synthesis is triggered by cyclin E-CDK2 and cyclin A-CDK2,2 (and also Cdc7).
Thus it is clear that G1 progression and S-phase entry require CDK activities. Most likely, it is important for cells to enter S phase only after sufficient numbers of origins have been licensed. S-phase entry of cells containing too few licensed origins could necessitate more DNA synthesis per licensed origin, potentially leading to replication stress, incomplete DNA synthesis, or other causes of genomic instability. Indeed, transgenic mice harboring a hypomorphic Mcm4 allele are cancer-prone,12 suggesting that defective replication licensing promotes genomic instability and leads to cancer.
The effects of impaired Mcm2-7 helicase expression or activity on cell cycle regulation of untransformed human cells have not been characterized. However, Blow and colleagues have shown that inhibiting licensing using degradation-resistant geminin results in reduced Cyclin E-CDK2 activity and G1 arrest.13 Similarly, Dutta and colleagues have shown that a ‘replication licensing checkpoint’ due to ORC-deficiency elicits p21/p27 induction and inhibits G1/S progression.14 It is unknown whether p27 induction and the resulting inhibition of Cyclin E-CDK2 represent the only mechanisms for inhibiting G1 progression in response to impaired replication licensing. Moreover, the previous studies of replication licensing checkpoints were performed using asynchronous cells,13,14 potentially complicating analysis and interpretation of events in G1. Therefore, we have investigated the relationship between Mcm7, replication licensing and mitogenic signaling events during G1 using synchronized cultures of untransformed human cells. We demonstrate that untransformed cells respond to impaired replication licensing by inhibiting cyclin D1 expression very early during G1. The effect of impaired replication licensing on cyclin D1 is dissociable from the p27/cyclin E-CDK2-mediated mechanisms described previously.13,14 We conclude that regulation of cyclin D1 expression represents a novel mechanism for cordinating replication licensing with G1 progression.
Results
Downregulation of Mcm7 inhibits S-phase entry of synchronized HDFs
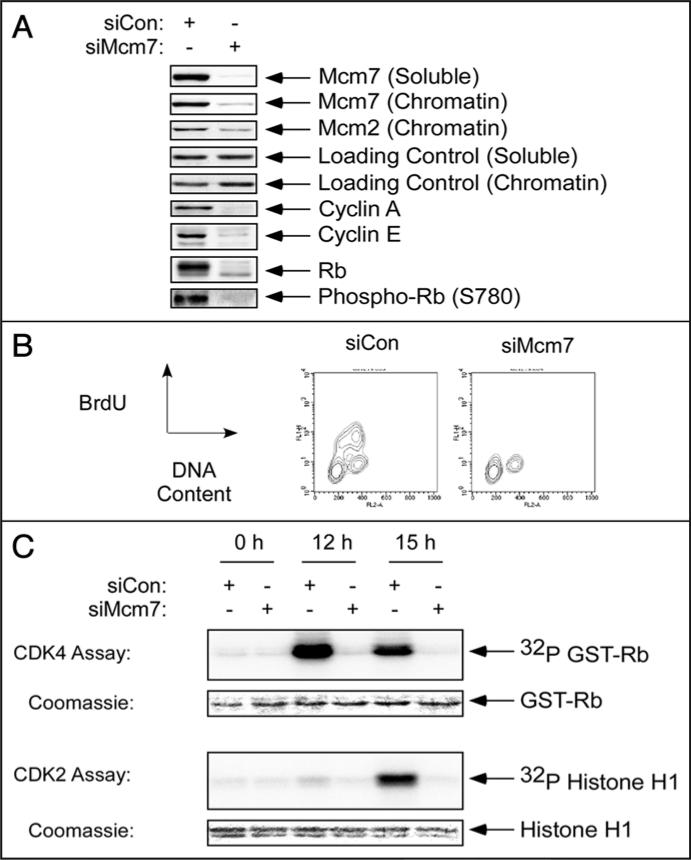
To determine the effects of reduced replication licensing on cell cycle progression, we used siRNA to deplete Mcm7 in quiescent Human Dermal Fibroblasts (HDF). Mcm7-depleted cells (or cells transfected with control siCon RNA duplexes) were stimulated to re-enter the cell cycle for 24 hr. As shown in Figure 1A, a significant reduction of Mcm7 expression was achieved in both soluble and chromatin fractions after siRNA treatment. Depletion of Mcm7 also resulted in reduced levels of Mcm2 on chromatin (Fig. 1A), indicating that Mcm7 knockdown interfered with assembly of the Mcm2-7 complex and prevented pre-RC formation. We next asked whether the levels of Mcm7 depletion attained under our experimental conditions affected cell cycle progression or DNA replication. Therefore, numbers of cells actively synthesizing DNA were determined using BrdU incorporation and flow cytometry. As shown in Figure 1B, Mcm7-depleted cells failed to enter S-phase as evidenced by the absence of BrdU-positive populations. Therefore, Mcm7-depletion prevented cell cycle progression of synchronized HDF.
Figure 1.
Acute depletion of Mcm7 inhibits S-phase entry in primary HDF. Quiescent hTERT-expressing HDF were transfected with siMcm7 or siCon oligonucleotides, then stimulated to enter the cell cycle. 24 hours after serum-stimulation, chromatin and soluble fractions were prepared and analyzed by SDS-PAGE and immunoblotting for expression of various proteins as indicated (A). Parallel cultures of siCon and siMcm7-transfected cells were incubated with 25 μM BrdU for 1 hr. The cell cycle distributions of the resulting HDF were analyzed by flow cytometry, as described under ‘Materials and Methods’ (B). In a similar experiment, quiescent HDF were transfected with siMcm7 or siCon oligonucleotides, then stimulated to enter the cell cycle. After 0, 12 and 15 hours of serum stimulation, cells were lysed and CDK4 and CDK2 complexes were immunoprecipitated for in vitro kinase assays. Rb-GST and histone H1 were used as substrates for CDK4 and CDK2 respectively (C).
We determined the effect of Mcm7-deficiency on the integrity of known mitogenic signaling events during G1-S progression. Our immunoblotting experiments showed that expression of cyclins E and A (which are encoded by E2F-inducible genes) were reduced in Mcm7-depleted cells relative to controls (Fig. 1A). Rb phosphorylation at S780 (a CDK4-specific site) was undetectable in Mcm7-depleted cells (Fig. 1A). Our failure to detect S780 phosphorylation also resulted in part from the reduced expression of total Rb in Mcm7-depleted cells. Similar to cyclins E and A, the RB gene is transcriptionally regulated by E2F.5 Therefore, the reduced levels of cyclin E, cyclin A and Rb in Mcm7-depleted HDF may be consistent with reduced de-repression of E2F as a result of Mcm7-deficiency.
The initial phosphorylation of Rb and de-repression of E2F during mid-G1 are due to cyclin D1-CDK4 activity. Therefore, we determined the effect of Mcm7 depletion on CDK4 activity in serum-stimulated HDF. CDK4 was immunoprecipitated from control and Mcm7-depleted HDF at different times after serum treatment. The CDK4 activity of the resulting immune complexes was measured in vitro using GST-Rb as a substrate. As expected, CDK4 activity was detectable at 12 hr following serum stimulation (a time point corresponding to mid-G1) in control HDF (Fig. 1C). Interestingly, CDK4 activity was reduced by >80% in Mcm7-depleted cells relative to controls (Fig. 1C).
Since CDK4-dependent Rb inactivation contributes to subsequent CDK2 activation in late G1 (due to induction of cyclins E and A), we also determined the effect of Mcm7-depletion on CDK2 activity. As shown in Figure 1C, CDK2 activity was first detectable in control HDF after 15 hours of serum stimulation, a time point subsequent to onset of CDK4 activity. However, similar to CDK4 activity, CDK2 activation was reduced in Mcm7-depleted cells (Fig. 1C). Therefore, appropriate levels of Mcm7 are necessary for activation of CDK4 and CDK2 during G1. While it is reported that replication licensing is required for CDK2 activation,13,14 a role for pre-RC assembly in CDK4 activation has not previously been described. Therefore we performed further experiments to investigate the novel link between replication licensing and CDK4.
Mcm7-depletion prevents cyclin D1 expression during early G1
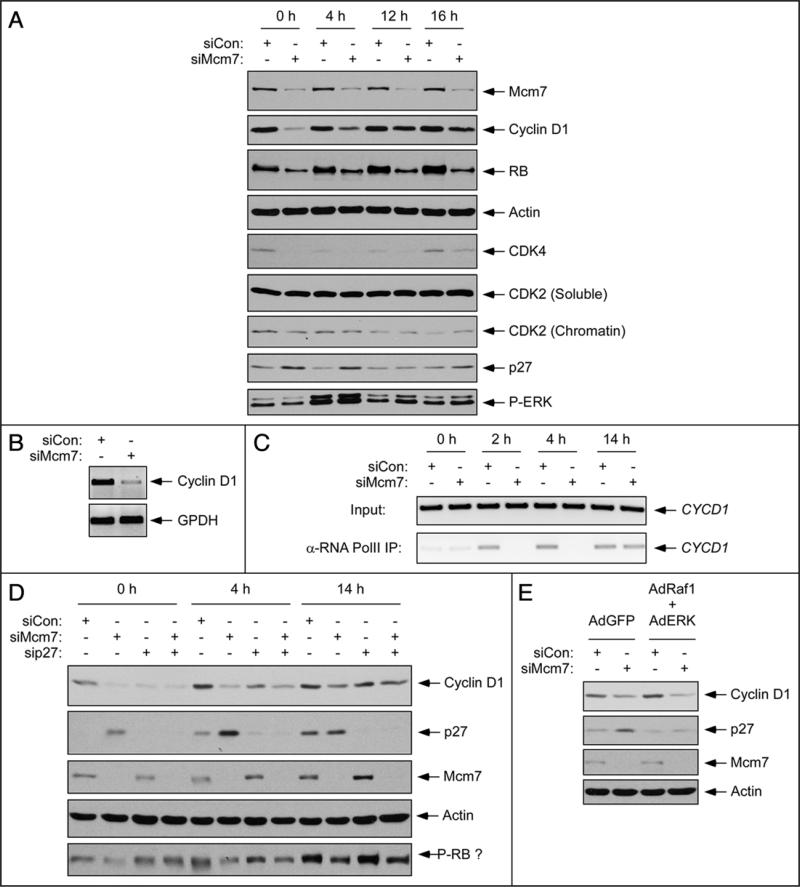
Full activation of CDK4 during G1 requires cyclin D1. To test whether altered cyclin D1 expression contributes to the reduced CDK4 activity of Mcm7-depleted cells we determined the effects of Mcm7-depletion on cyclin D1 levels. As shown in Figure 2A, levels of cyclin D1 protein in Mcm7-depleted cells were reduced relative to control (siCon) cultures. The differences in cyclin D1 expression between control and Mcm7-depleted cells were most evident at times corresponding to early G1 (0–4 h time point after serum treatment). Interestingly, levels of CDK4 and Rb protein were also reduced in response to Mcm7-depletion (Fig. 2A).
Figure 2.
Mcm7 depletion inhibits cyclin D1 expression independently of p27. (A) Quiescent hTERT-expressing HDF were transfected with siMcm7 or siCon oligonucleotides, then stimulated to enter the cell cycle. After 0 hr, 4 hr, 12 hr and 16 hr of serum stimulation chromatin and soluble fractions were prepared and analyzed for expression of various proteins (as indicated) using SDS-PAGE and immunoblotting. (B) Quiescent hTERT-expressing HDF were transfected with siMcm7 or siCon oligonucleotides, then stimulated to enter the cell cycle. After 4 hr, total RNA was extracted from the cells and analyzed for cyclin D1 and GPDH mRNAs using RT-PCR as described under ‘Materials and Methods’. (C) For ChIP analyses, control and Mcm7-depleted cells were synchronized as described in (A). At various times after stimulating cell cycle entry, chromatin fractions were harvested and immunoprecipitated using α-RNA Pol II antibody. Total chromatin (‘Input’) and α-RNA-Pol II immune complexes were analyzed by PCR using primers designed to amplify a fragment of the CYCD1 promoter. (D) Quiescent hTERT-expressing HDF were transfected with siCon, siMcm7 and sip27 individually or in combination. Transfected cells were stimulated with serum and after 0 hr, 4 hr and 14 hr soluble fractions were prepared and analyzed by SDS-PAGE and immunoblotting for expression of various proteins as indicated. (E) Quiescent hTERT-expressing HDF were transfected with siCon or siMcm7 for 24 hr. Transfected cells were then infected with AdGFP or AdRaf1 + AdERK as indicated. Cells were stimulated with serum and 4 hr after release from quiescence soluble fractions were prepared and analyzed by SDS-PAGE and immunoblotting as indicated.
We investigated the mechanism(s) by which Mcm7-depletion led to reduced expression of cyclin D1. Levels of cyclin D1 in Mcm7-depleted cells were not increased in response to proteasome inhibition (data not shown), suggesting that reduced D1 levels were not due to increased protein turnover. However, cyclin D1 mRNA levels were decreased as a result of Mcm7-depletion (Fig. 2B). Moreover, using ChIP assays we failed to detect serum-induced association of RNA PolII with the CYCD1 promoter in Mcm7-depleted cells (Fig. 2C). Reduced cyclin D1 transcription was not accompanied by a defect in growth factor signaling to ERK activation (Fig. 2A). Taken together, our results suggest that efficient transcriptional induction of cyclin D1 during G1 is dependent on Mcm7 at a step subsequent to ERK activation which involves recruitment of RNA PolII to the CYCD1 promoter.
To determine whether the effect of Mcm7 siRNA on the serum inducibility of Cyclin D might be due to a general perturbation of the serum response in Mcm7-depleted fibroblasts, we performed genome-wide gene-expression profiling of the serum response of Mcm7-depleted and control-siRNA-treated cells using Affymetrix Gene Arrays. We specifically focused our analysis on the expression of genes whose expression is induced by serum in primary human fibroblasts isolated from diverse anatomic sites (www.ncbi.nlm.nih.gov/sites/entrez?CrntRpt=DocSum&cmd=search&db=pubmed&term=14737219). We retrieved the gene symbols corresponding to 205 of these genes from the MSigDB database (www.pnas.org/content/102/43/15545.abstract), of which we were able to match 195 with probesets on the Affymetrix Gene Array. In cells treated with control siRNA, 75.4% of these 195 genes showed higher expression following addition of serum to serum-starved cells (binomial test p-value << 0.001). Similarly, in cells treated with Mcm7-targeting siRNA, 79.5% of these genes were induced following addition of serum (binomial test p-value << 0.001). Induction of the CYCD1 promoter is largely SRF-mediated, yet our microarray analyses indicated that Mcm7-depletion did not inhibit the serum-inducible expression of SRF-containing genes. Taken together, these results suggest that Mcm7-depleted fibroblasts have a generally intact gene-expression response to serum. We conclude that the effect of Mcm7-deficiency on cyclin D1 expression is relatively specific.
Mcm7-deficiency affects expression of cyclin D1 and p27 via independent mechanisms
Reportedly, impaired replication licensing during G1 inhibits CDK2 via a p27-dependent mechanism.13,14 Consistent with those studies, p27 expression was induced in Mcm7-depleted HDF concomitant with reduction of cyclin D1 (Fig. 2A). To determine whether the effect of Mcm7-deficiency on Cyclin D1 expression was related to the induction of p27, we performed co-silencing experiments in which we ablated expression of Mcm7 and p27 individually or in combination. As shown in Figure 2D, silencing of p27 did not restore expression of cyclin D1 in Mcm7-depleted cells. Therefore, the decreased expression of cyclin D1 in Mcm7-depleted HDF is not due p27 induction.
We asked whether the changes in expression of cyclin D1 and p27 in Mcm7-depleted HDF could be reversed by mitogenic stimuli that promote G1 progression. Therefore, we investigated the effect of constitutive Raf1/ERK signaling on levels of cyclin D1 and p27 in Mcm7-depleted cells. As shown in Figure 2E, ectopic overexpression of Raf1 and ERK prevented the induction of p27, but did not affect the repression of cyclin D1 in Mcm7-depleted cells. Therefore, Mcm7-depletion induces p27 expression and reduces cyclin D1 levels via dissociable mechanisms. Taken together, our results demonstrate the existence of at least two independent mechanisms (involving cyclin D1 repression and p27 induction) for coupling restriction licensing with G1 progression in untransformed cells.
Mcm7-deficiency does not elicit a DNA damage-induced G1 checkpoint response
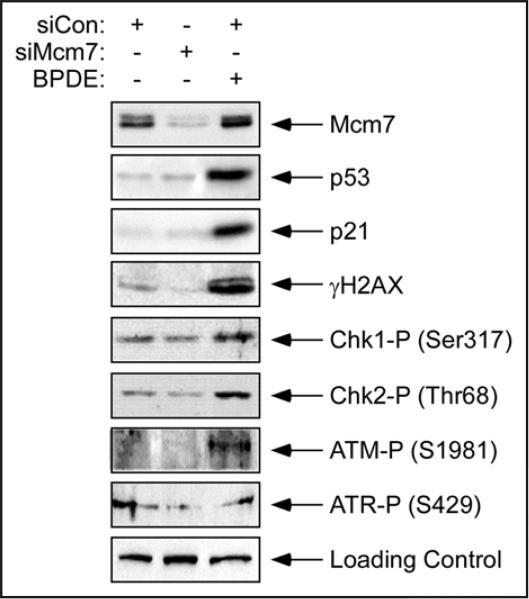
Mitogenic signaling events in G1 are often subject to negative regulation by checkpoint signaling.15-19 We considered the possibility that the reduced activation of CDK4 in Mcm7-ablated cells was the result of a checkpoint response to reduced replication licensing. To address this possibility, we determined the activation status of known checkpoint pathways following Mcm7 ablation. As shown in Figure 3, Mcm7-deficiency did not affect p53 or p21 levels, indicating that p53 was not activated in response to reduced replication licensing. We observed no increase in levels of γH2AX, an ATM and ATR substrate which is phosphorylated in response to bulky DNA lesions and stalled replication forks. Moreover, we did not detect changes in the phosphorylation status of Chk1 and Chk2 (DNA damage-activated checkpoint kinases and effectors of ATR and ATM respectively). As expected, treatment with BPDE (a bulky adduct-forming genotoxin) elicited checkpoint kinase activation and p53 accumulation, indicating that our failure to detect checkpoint signaling in Mcm7-depleted HDF was not due to defective checkpoint activation in these cells. Taken together, our analyses do not suggest that checkpoint signaling is induced (within the sensitivity limits of our assays) in response to Mcm7 ablation during G1.
Figure 3.
Acute Mcm7 depletion does not trigger a checkpoint response. Quiescent hTERT-expressing HDF were transfected with siMcm7 or siCon oligonucleotides and then stimulated to enter the cell cycle. Some cultures of siCon-transfected cells were treated with 600 nM BPDE to induce DNA damage. 12 hours after serum stimulation cells were lysed and analyzed by SDS-PAGE and immunoblot using the indicated antibodies.
Cyclin D1 expression is sensitive to Cdc6 status
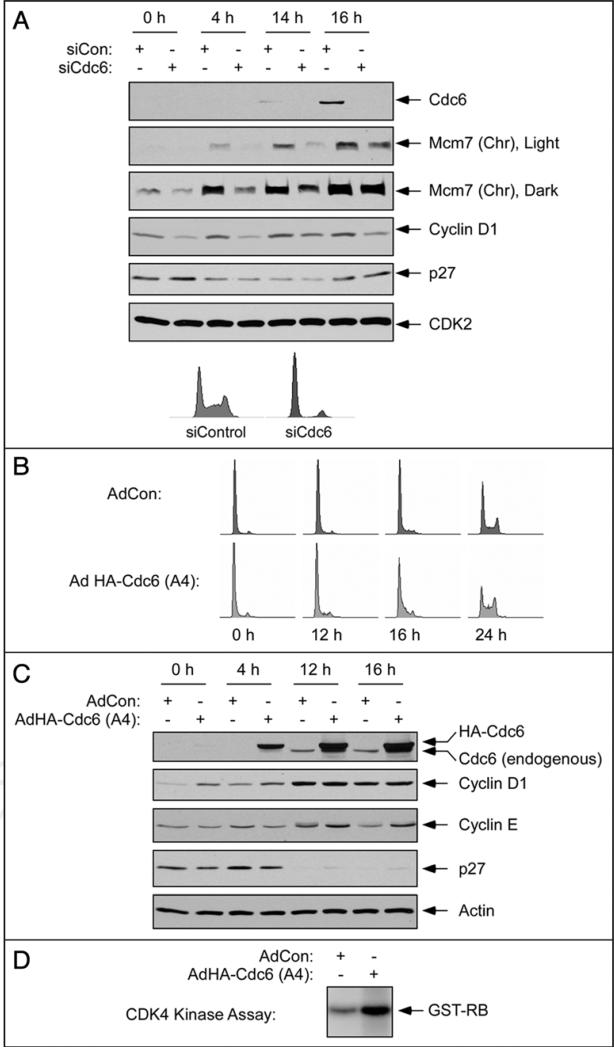
Similar to the effects of Mcm7-depletion, siRNA-mediated ablation of Mcm2 inhibited cyclin D1 expression during early G1 (data not shown), consistent with a role for replication licensing in promoting cyclin D1 induction. However, Mcm2-7 proteins are reported to participate in processes unrelated to DNA replication.20,21 Potentially, the effects of Mcm2- or Mcm7-deficiency on Cylin D1 (and other G1 signaling events) might result from a replication licensing-independent role of Mcm proteins. To further test a hypothetical link between replication licensing and G1 progression we investigated the effects of Cdc6 status on cyclin D1 expression. First we tested the effects of Cdc6-depletion on Cyclin D1 levels. Using siRNA we achieved good ablation of Cdc6 expression which also led to reduced levels of Mcm7 on chromatin as expected (Fig. 4A).
Figure 4.
Cdc6 is required for efficient cyclin D expression and G1-progression. (A) Quiescent hTERT-expressing HDF were transfected with siCdc6 or siCon oligonucleotides, then stimulated with serum. After 0 hr, 4 hr, 14 hr and 16 hr of serum stimulation soluble fractions were prepared and analyzed for expression of various proteins using SDS-PAGE and immunoblotting (upper). In a parallel experiment, siCdc6- or siCon-transfected cells were stimulated with serum for 20 hr, then analyzed by flow cytometry (lower). (B) Quiescent HDF were infected with adenovirus encoding HA-tagged Cdc6-A4 or with an empty-vector control adenovirus (AdCon). 24 hours after infection, cells were stimulated with serum. At different times after serum stimulation cells were harvested for PI staining and FACS analysis to determine cell cycle distributions. (C) Quiescent HDF were infected with Ad HA-Cdc6-A4 or AdCon. 24 hours after infection, cells were stimulated with serum. After 0 hr, 4 hr, 12 hr and 16 hr of serum stimulation soluble proteins were extracted and analyzed by SDS-PAGE and immunoblotting for expression of various proteins as indicated. (D) Quiescent HDF were infected with Ad HA-Cdc6-A4 or with an empty-vector control adenovirus (AdCon). 24 hours after infection, cells were stimulated with serum for 4 hr. Lysates from the resulting cells were immunoprecipitated with anti-cyclin D1 antibodies. Immune complexes were assayed for Rb-directed kinase activity in vitro.
Similar to the effects of Mcm7- and Mcm2-deficiency, Cdc6-depletion prevented S-phase entry and led to reduced expression of cyclin D1 protein during early G1 (Fig. 4A). Since depletion of either Mcm7, Mcm2 or of Cdc6 impaired cyclin D1 expression, it is most likely that cyclin D1 expression requires prior replication licensing, and that cyclin D1 expression is not specifically dependent on Mcm7.
Next we asked whether induction of replication licensing in quiescent cells would stimulate mitogenic events during G1. Ectopic expression of Cdc6 in quiescent fibroblasts is sufficient to induce replication licensing.22 Therefore, we used an adenovirus vector to express Cdc6 (or ‘empty’ vector for control) in quiescent HDF. A mutant Cdc6 harboring four Ser > Ala substitutions at CDK phosphorylation sites (‘Ad HA-Cdc6-A4’) was used to promote nuclear retention of the overexpressed licensing factor.23 As expected, ectopic expression of HA-Cdc6-A4 in quiescent HDF accelerated rates of G1/S progression (shown by the FACS profiles and DNA synthesis assays in Fig. 4B and C respectively). Interestingly, cyclin D1 expression was induced prematurely in quiescent cells as a result of Ad HA-Cdc6-A4 expression (Fig. 4C, compare lanes 1 and 2). Moreover, CDK4 activity (measured in vitro) was elevated in HA-Cdc6-A4-expressing cells relative to controls (Fig. 4D). Taken together, the results of our Cdc6 ablation and overexpression studies suggest that replication licensing promotes cyclin D expression.
Perturbation of the Rb pathway confers S-phase entry of Mcm7-depleted cells
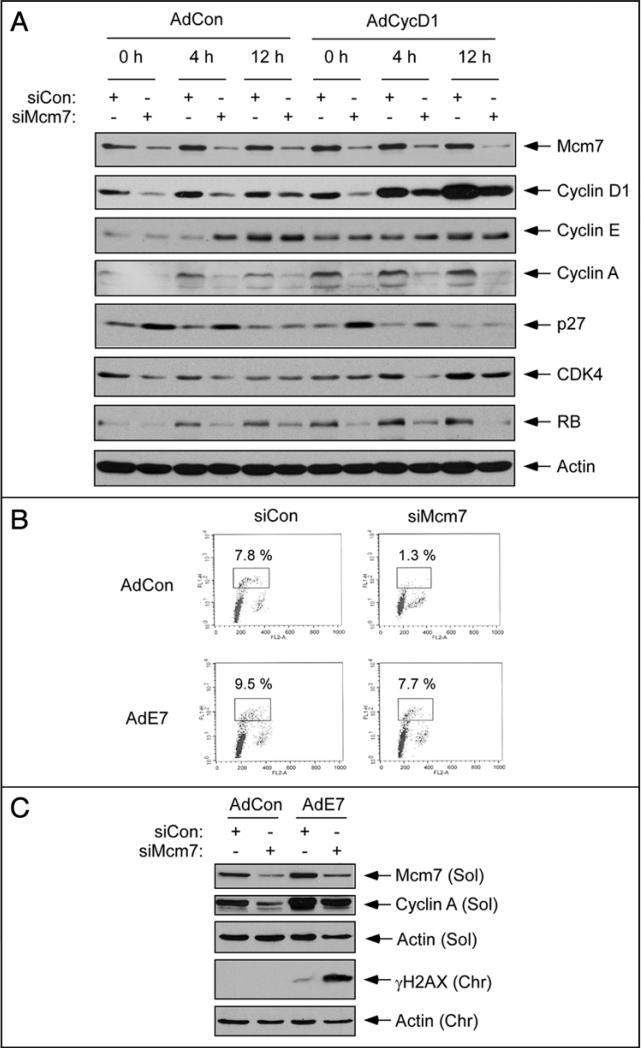
The results of Figures 2 and 4 indicated that efficient expression of cyclin D is promoted by replication licensing. Therefore, we asked whether reconstitution of cyclin D1 expression was sufficient to allow G1 progression and S-phase entry of Mcm7-depleted cells. We used an adeno-virus vector to express cyclin D1 in control and Mcm7-depleted HDF. As shown in Figure 5A, adenovirus-mediated gene delivery allowed us to achieve levels of cyclin D1 in Mcm7-depleted cells that were equivalent to those present in serum-stimulated control HDF during G1. However, using FACS analyses and [3H]-thymidine incorporation synthesis assays, we were unable to detect any effect of ectopically-expressed cyclin D1 on S-phase entry of Mcm7-depleted cells (data not shown). Consistent with this result, overexpressed cyclin D1 failed to correct the reduced expression of cyclin A (a necessary factor for DNA replication) in Mcm7-depleted cells (Fig. 5A). The induction of p27 in Mcm7-ablated cells was unaffected by Cyclin D1 overexpression, further suggesting that p27 induction and Cyclin D1 repression are independent responses to impaired replication licensing. Interestingly, the reduced expression of CDK4 and the reduced expression and phosphorylation of Rb in Mcm7-depleted cells (also evident in Fig. 2A) were not corrected by overexpressed cyclin D1 (Fig. 5A). We conclude that replication licensing is required not only for efficient expression of cyclin D1, but also for other key mitogenic events involving the CDK4-RB signaling axis during G1.
Figure 5.
HPV E7 expression allows S-phase entry of Mcm7-depleted cells. (A) Quiescent HDF were transfected with siCon or siMcm7 for 24 hr. Transfected cells were then infected with AdCon or AdCyclin D1 as indicated. Cells were stimulated with serum for 0 hr, 4 hr and 12 hr. Extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting with various antibodies as indicated. (B) Quiescent HDF were transfected with siCon or siMcm7, then infected with AdCon or AdE7 as described above. The resulting cells were incubated with 10 μM BrdU for 1 hr, then analyzed by flow cytometry to determine cell cycle distributions. (C) Quiescent HDF were transfected with siCon or siMcm7 for 24 hr. Transfected cells were then infected with AdCon or AdE7 as indicated. After 20 hr of serum stimulation cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting with various as indicated.
Activated oncogenes frequently enable cells to overcome restriction points that are otherwise imposed on normal cell cycle progression. Therefore, we asked whether ectopic expression of the HPV E7 oncoprotein (which inactivates Rb and other pocket proteins involved in regulating transcriptional events during G1) would permit S-phase entry of cells with impaired replication licensing. Quiescent cells were sequentially transfected with siMcm7 (or siCon), infected with AdE7 (or an ‘empty’ control adenovirus vector, AdCon), then stimulated with serum. We performed FACS analyses to quantify the early S-phase (BrdU-positive) populations of the cells after serum treatment. In AdCon-infected cells, Mcm7-depletion resulted in an 84% reduction of the early S-phase population (Fig. 5C, upper). Interestingly however, in E7-expressing cells, S-phase entry was only inhibited by 19% as a result of Mcm7-depletion (Fig. 5B, lower). In E7-expressing and Mcm7-depleted HDF, the BrdU-positive cells did not advance beyond early S-phase (data not shown), presumably due to insufficient replicative helicase activity in Mcm7-depleted cells. Nevertheless, our results clearly demonstrate that the E7 oncoprotein confers G1 progression and S-phase entry of Mcm7-depleted cells.
As expected, cyclin A expression (a marker of early S-phase) was attenuated as a result of Mcm7 depletion in AdCon-infected HDF (Fig. 5C). In contrast, in E7-expressing cells, cyclin A levels were only modestly reduced as a result of Mcm7 depletion (Fig. 5C).
We predicted that there would be detrimental consequences of S-phase entry in licensing factor-depleted cells containing insufficient pre-RC. Therefore, we asked whether S-phase entry in Mcm7-depleted cells was associated with markers of replication fork collapse and DSB. As shown in Figure 5C, Mcm7 ablation led to induction of γH2AX in AdE7-infected (but not AdCon-infected) cells concomitant with S-phase entry. Taken together, our results suggest that in untransformed cells cyclin D1 expression and other key mitogenic events in G1 require Mcm7 expression and replication licensing. Moreover, our HPV-E7 expression results show that oncogene-induced bypass of the replication licensing restriction point results in inappropriate S-phase entry leading to replication stress and DNA damage-signaling.
Discussion
We show here that in untransformed human cells, the appropriate and timely expression of cyclin D1 and the subsequent execution of subsequent signaling events during G1 is promoted (at least in part) by replication licensing (Fig. 6). Potentially, direct associations between pre-RC components and the transcriptional apparatus could contribute to induction of important cell cycle genes including CYCD1. It has previously been suggested that licensing factors cooperate with transcriptional activators to regulate gene expression. For example, Mcm5 interacts directly with the transcriptional activator domain of Stat1α.24 Moreover, Mcm5 is recruited to Stat1 target genes25 and is essential for IFNγ-induced gene expression.26 Since the cyclin D1 promoter is responsive to the Jak/Stat pathway, regulation of Stat family members by Mcm7 could explain the effects of Mcm7 and Cdc6 status on cyclin D1 expression which we have observed.
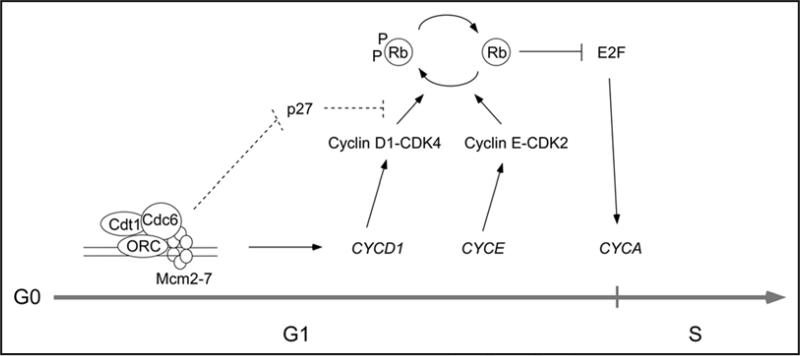
Figure 6.
Replication licensing promotes G1 progression of untransformed human cells. In untransformed human cells pre-RC assembly contributes to activation of G1 CDKs via independent mechanisms involving Cyclin D1 induction and downregulation of p27. See ‘Discussion’ for details.
Reduced cyclin D1 expression alone is insufficient to account fully for the G1 delay experienced by Mcm7-depleted cells. As shown in Figure 1C, at time points when cyclin D1 expression finally resumes in Mcm7-depleted cells, CDK4 activity and subsequent CDK2 activity remained attenuated. Moreover, Cyclin D1 reconstitution was insufficient to confer S-phase entry (Fig. 5A). These data indicate the existence of additional Mcm7-dependent mitogenic events during G1. Therefore, it is likely that licensed origins facilitate additional cell cycle events via transcriptional (or nontranscriptional) mechanisms. The CDK4 gene may also be negatively regulated in Mcm7-deficient cells, as shown by the inhibitory effect of Mcm7-depletion on CDK4 mRNA and protein levels. Potentially, direct interactions between pre-RC components and transcriptional machinery might be involved in regulating the expression of CYCD1, CDK4, and perhaps other cell cycle genes. Consistent with this possibility, higher order complexes of mammalian RNA Pol II have been found to contain Mcm proteins and specific interactions of the Mcm2-7 complex with RNA Pol II holoenzyme are mediated at least in part via Mcm2.27 Moreover, antibodies against Mcm2 inhibit transcription in Xenopus oocytes.28
The use of dual-function proteins which participate in DNA replication and transcription is not unique to vertebrate cells.29 For example, Mcm7 modulates expression of several cell cycle genes (MCM7, MCM5, CDC6) in S. cerevisiae by facilitating association of the transcriptional regulator Mcm1 with specific promoter elements.21 Interestingly, the histone chaperone FACT (facilitates active transcription), has been identified in Mcm2-7 helicase complexes in yeast30 and in mammalian cells.31 FACT allows transcriptional elongation by RNA Pol II and might also promote formation of transcriptional initiation complexes by stimulating recruitment of TBP to TATA boxes.32 Because FACT is present in Mcm2-7 complexes during G1,30,31 it is conceivable that licensed origins stimulate gene transcription via recruitment of FACT to promoters.
Communication between licensed origins and the transcriptional machinery may even occur in a bi-directional manner. For example, RNA Pol II is recruited to ARS and artificially tethering Pol II CTD to origins of replication stimulates minichromosome stability.33 Therefore, replication licensing is clearly coupled to transcriptional events in many instances. Many components of the G1 cell cycle machinery are reported to interact directly with the pre-RC, including cyclin-CDK complexes and Rb.11,34-39 Thus the pre-RC may serve as a central ‘hub’ for bi-directional communication between the DNA replication apparatus, the transcriptional machinery and cyclin-CDKs, thereby coordinating DNA synthesis with other cell cycle events.
Coordinating G1 cyclin expression and CDK activation with pre-RC assembly provides a potential mechanism for ensuring that replication competency is attained prior to initiation of DNA synthesis. G1 CDK activity is sufficient to induce DNA synthesis in quiescent human fibroblasts.40 Uncoupling of CDK activity from pre-RC formation (a likely event in tumor cells with defective Rb signaling) could result in S-phase entry with insufficient licensed origins. Completion of DNA replication in cells containing suboptimal levels of Mcm2-7 helicase complexes could impose a greater work load per replicon, thereby increasing replication-associated errors and replication stress. For example, sub-optimal recruitment of helicase complexes could slow DNA replication due to inefficient template unwinding. The resulting persistence of replication forks could increase the risk of breakage of fragile and unstable DNA replication intermediates or could result in more frequent collisions with other chromatin-associated complexes (such as the transcriptional apparatus), another potential cause of DNA damage. Indeed, our results show that Mcm7-ablation in HPV-E7 oncogene-expressing cells elicits checkpoint signaling via Chk2 and a γH2AX, both hallmarks of a DNA damage response.
The Mcm2-7 complex (together with GINS and Cdc45) constitutes the replicative helicase, which is responsible for establishing checkpoints during S phase. Therefore, S phase in cells with reduced levels of helicase activity would likely lead to impaired checkpoint activation in response to intrinsic/extrinsic sources of DNA damage. S phase checkpoints play key roles in stabilizing replication structures, and regulating DNA repair (e.g., via HR and PRR). Therefore, it is likely that defects resulting from reduced replication licensing might contribute to chromosome instability and cancer. Blow and colleagues have suggested that the cellular pool of Mcm2-7 exceeding that required for DNA synthesis during an unperturbed cell cycle is necessary to allow re-licensing and re-priming events downstream of stalled forks.41 Consistent with a role for excess Mcm in replication fork recovery, a Mcm4 hypomorph mouse that exhibits a severe reduction in licensing shows significant chromosomal abnormalities after treatment with aphidicolin.12
While other published studies have examined the consequence of depleting pre-RC components in human cells.14,42 Our work is the first to suggest that Cyclin D-CDK4 is linked to replication licensing. Dutta and colleagues showed that Orc2 depletion affects G1 progression in cancer cells via a mechanism involving p27 stabilization and cyclin E-CDK2 inhibition.14
Our results are fully consistent with those of the Dutta group, but additionally we demonstrate that cyclin D expression (an early-mid G1 event) is inhibited in response to Mcm7-depletion independently of changes in p27 expression, at least in untransformed cells (Fig. 6). It is possible that the G1 licensing restriction point targeting cyclin D1 in primary untransformed cells is absent in tumor cells. Indeed, defects in G1 checkpoints are frequently observed in cancer cells. Moreover, our Mcm7-ablation studies were conducted in synchronized HDF undergoing G0/G1 progression, whereas the effects of Orc2-deficiency were examined using asynchronous populations. Cancer cells typically fail to enter G0 and have a relatively short G1-phase. Therefore, it is likely that negative regulation of G1 events (such as cyclin D1 expression) is not as readily observed in cancer cell lines. Finally, it is possible that Orc2-depletion and Mcm7-deficiency trigger distinct restriction points that target CDK2 and CDK4 respectively. Taken together, it appears that multiple events involved in efficient CDK activation and cell cycle progression through G1 require appropriate replication licensing.
Most DNA replication factors were initially identified and characterized in yeast. The processes of cell cycle and DNA replication are very similar in all eukaryotes, though there much complexity has been acquired during metazoan evolution. Yeast possess functional equivalents of cyclins and CDKs, and Rb. Interestingly however, the response to replication licensing factor depletion observed in yeast differs from what we and others have observed in cultured mammalian cells. In S. cereviseae, loss of Cdc6 or Mcm resulted in bypass of S-phase and direct entry from G1 into M, followed by a form of reductional mitosis.43,44 Thus, budding yeast lack a G1 licensing restriction point. Reduced Orc2 protein levels in the ORC2-1 temperature-sensitive mutant result in reduced replication licensing. The incompletely-licensed ORC2-1 mutants enter S-phase at the same rate as wild-type cells but require more time to complete DNA synthesis.45-47 The incidence of irreversible cell cycle arrest and apoptosis is also higher in ORC2 mutants suggesting that irreparable DNA damage is acquired in cells with insufficient numbers of licensed origins.48-50 Therefore, the G1 licensing restriction point we have identified in mammalian cells may have evolved in multicellular organisms to coordinate growth and proliferation during various stages of development, and to guard against tumorigenesis.
It will be important to determine whether the replication licensing restriction point targeting cyclin D/CDK4 is commonly lost in cancer cells. Since the Rb signaling axis is defective in many tumors, we anticipate that oncogene-expressing cells will enter S-phase inappropriately when replication licensing is impaired (as we have shown for E7-expressing HDF). Moreover, it is likely that replication-associated stress and DNA damage incurred due to the inappropriate S-phase entry of pre-malignant cells with impaired replication licensing contributes to tumorigenesis. Indeed, it has been suggested that reduced Mcm2 expression results in stem/progenitor cell deficiency and cancer.51 Clearly further studies are necessary to elucidate the mechanisms that sense pre-RC formation and couple replication licensing with G1 progression. Additionally, it will be interesting to determine whether other partially redundant or failsafe restriction point mechanisms prevent CDK activation in response to incomplete replication licensing.
Materials and Methods
Cells and culture
Human Dermal Fibroblasts (HDF) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, streptomycin sulfate (100 μg/ml) and penicillin (100 units/ml). Cells were routinely passaged every third day at ratios of 1:3–1:5. Primary HDF were typically used prior to passage 10. In some experiments, HDF were retrovirally transduced with the human telomerase (hTERT) cDNA to prolong their replica-tive lifespan. Qualitatively similar results were obtained when using HDF-hTERT and primary untransduced HDF although consistent with previous reports, cyclin D1 levels were downregulated less efficiently following serum-starvation in TERT-expressing cells compared with untransduced HDF.52 Qualitatively similar results were also obtained when comparing multiple genetically unrelated hTERT-transduced HDF lines.
HDF synchronization protocols
To synchronize cells in quiescence, 10-cm dishes of HDFs were grown to confluence in DMEM containing 10% FBS. Confluent cultures of HDF were placed in medium containing 0.1% serum for 16–24 hrs. The following day, cells were trypsinized and replated at 40–50% confluence and maintained in 0.1% FBS for an additional 24 hours prior to adenoviral transduction or siRNA transfection. Cells were maintained in 0.1% FBS throughout the course of siRNA transfections or viral transductions. To stimulate cell cycle re-entry, cells were treated with fresh medium containing 10% FBS. At various times after cell cycle entry cells were harvested for analysis of protein markers (using immuno-blotting and kinase assays), RNA (using microarray), or cell cycle parameters (using flow cytometry or [3H]-thymidine incorporation).
siRNA transfections
For licensing factor depletion experiments quiescent HDF were transfected with siRNA duplexes (Dharmacon Research) twice on consecutive days according to standard protocols (Dharmacon Research). Target sequences of oligonucleotides used were as follows: Mcm7, 5'-AUC GGA UUG UGA AGA UGA A-3'; Control (Non-Targeting), 5'-UAG CGA CUA AAC ACA UCA A-3'; Cdc6, 5'-ACA AUU AAG UCU CCU AGC A-3'; p27, 5'-GCA ACC GAC GAU UCU UCU A-3'. The Mcm2 siRNA was a ‘SMARTPOOL’. In some experiments, quiescent siRNA-transfected cells were also infected with adenoviruses (1 × 1010/ml) for an additional 24 hrs, prior to serum stimulation.
mRNA extraction and analysis
Synchronized HDF were lysed with TRIzol reagent (Invitrogen) and total RNA was purified according to the manufacturer's protocol. The concentration and purity of the RNA were determined by measurements of absorbance at 260 and 280 nm. One microgram of RNA was used for each RT-PCR reaction. RT-PCR was performed using the ThermoScript RT-PCR system (Invitrogen) according to the manufacturer's instructions. 2 μl of the resulting reaction products were used as templates in PCR reactions to amplify fragments of the human Cyclin D1 and GAPDH cDNAs. Sequences of the oligonucleotide primer pair used to amplify Cyclin D1 (750 bp) are: upstream primer 5' TGT TTG CAA GCA GGA CTT TG 3'; downstream primer 5' ACG TCA GCC TCC ACA CTC TT 3'. The sequences of upstream and downstream primer used to amplify GAPDH are: 5' GAA GGT GAA GGT CGG AGT 3' and GAA GAT GGT GAT GGG ATT TC 3'. The amplified PCR products were separated on 1% agarose gels and visualized under an UV transilluminator.
Chromatin immunoprecipitation (ChIP) analysis of RNA polymerase II
Quiescent HDF were transfected with siRNA oligonucleotides as described above. After stimulation with serum for various times, formaldehyde was added directly into culture media to a final concentration of 1%, and the plates were rocked gently for 10 minutes to generate cross-linked protein-DNA complexes. The cross-linking reaction was quenched by adding glycine to a final concentration of 125 mM. After 10 minutes, the glycine-containing medium was aspirated, and the cells were washed twice with ice-cold PBS containing protease inhibitors and 1 mM phenylmethylsulfonyl fluoride. The cells were scraped into a conical tube and collected by brief centrifugation. The resulting cell pellets were resuspended in 1 ml of 0.1 M Tris-HCl (pH 9.4) and 0.1 M dithiothreitol, placed on ice for 5 min, and incubated at 30°C for 15 min. The cells were then collected by centrifugation and washed sequentially with 1 ml of ice-cold PBS; 1 ml of 10 mM HEPES (pH 6.5), 0.25% Triton X-100, 10 mM EDTA and 0.5 mM EGTA; and finally 1 ml of 10 mM HEPES (pH 6.5), 1 mM EDTA, 0.5 mM EGTA and 200 mM NaCl. The resulting cells were resuspended in 0.35 ml of buffer containing 50 mM Tris-HCl (pH 8.0), 1% SDS and 10 mM EDTA supplemented with 1x protease inhibitor mixture (Roche) and incubated on ice for 15 min. The resulting lysates were sonicated on ice to shear the DNA into 1–2-kb fragments. Sonicated lysates were centrifuged for 10 min at 10,000 xg. The clarified supernatants containing chromatin fragments were transferred to new tubes and normalized for protein content. 5% of each sheared chromatin sample was saved to provide an “input” control. The remainder of each chromatin solution was diluted to 1 ml with 20 mM Tris-Cl (pH 8.0), 1% Triton X-100, 2 mM EDTA and 150 mM NaCl freshly supplemented with protease inhibitor mixture. Each sample was supplemented with 400 μg/ml salmon sperm DNA and 1% bovine serum albumin. Samples were then pre-cleared with 40 μl of a 50% protein A-agarose slurry for 2 h at 4°C. Pre-cleared samples were immunoprecipitated with 2 μg of rabbit anti-RNAPII antibody (Santa Cruz). After overnight incubation at 4°C, 40 μl of pre-blocked protein G-agarose beads was added to each sample. 3 hr later, immune complexes were collected by centrifugation. Beads were washed sequentially with 1 ml of buffer containing 20 mM Tris-Cl (pH 8.0), 0.1% SDS, 1% Triton X-100, 2 mM EDTA and 150 mM NaCl; 1 ml of buffer containing 20 mM Tris-Cl (pH 8.0), 0.1% SDS, 1% Triton X-100, 2 mM EDTA and 500 mM NaCl; 1 ml of buffer containing 10 mM Tris-Cl (pH 8.0), 0.25 M LiCl, 1% Nonidet P-40, 1% deoxycholate and 1 mM EDTA; and finally 1 ml of Tris/EDTA buffer. Chromatin was eluted from the beads using 250 μl of 1% SDS in 0.1 M NaHCO3. Reverse cross-linking was performed by overnight incubation at 45°C. After proteinase K (10 μg/ml) and RNaseA (10 μg/ml) digestion, phenol/ chloroform extraction and ethanol precipitation, immunoprecipitated DNA was re-suspended in 50 μl of distilled water and analyzed by PCR. The PCR primers for the human Cyclin D1 promoter region are: upstream primer 5' CCT TGG GCA TTT GCA ACG AC 3'; downstream primer 5' CGC ATT TCC AAG AAC GCC AC 3'. The amplified DNA products were separated on 1% agarose gels and visualized under a UV transilluminator.
RNA extraction and microarray analysis
Quiescent HDF were transfected sequentially with siCon and siMcm7 oligonucleotides. 48 hrs after transfection, cells were stimulated with 10% FBS for 4 hrs (or were left unstimulated for controls). The resulting cells were lysed with TRIzol reagent (Invitrogen) and total RNA was harvested according to the manufacturer's instructions. The resulting RNA samples were submitted for microarray analysis by the Boston University Microarray Core Facility (http://www.bumc.bu.edu/busm-pathology/pathology-core-services/boston-university-microarray-resource/) using the Affymetrix 1.0 ST Human Transcriptome array.
Flow cytometry
For BrdU labeling assays, cells were pulsed for one hour with 10 μM BrdU to label the S-phase population. Cells were trypsinized, recovered by centrifugation for 30 s at 10,000 g and fixed in PBS containing 35% ethanol for at least 1 hr at 4°C. Fixed cells were incubated in 2N HCl for 20 minutes at room temperature. The HCl-treated cells were neutralized by washing in 0.1 M borax (pH 8.5) then washed in PBS. The resulting cell pellets were resuspended in 50 μl of antibody labeling solution comprising 30 μl PBS containing 0.5% Tween-20 and 0.5% BSA plus 20 μl FITC-conjugated BrdU antibody (BD Biosciences). The cells were incubated for 30 minutes in the dark with frequent mixing. Labeled cells were re-suspended and washed in 1 ml PBS to remove free antibody. Washed nuclei were re-suspended in 1 ml PBS containing 8 μg RNAse A and 50 μg Propidium Iodide. The nuclear suspensions were incubated in the dark at room temperature for 30 min prior to FACS analysis. Flow cytometry was performed on a Becton Dickinson instrument, exciting at 488 nm and measuring the BrdU-linked green fluorescence (FITC) through a 514 nm bandpass filter and the DNA-associated red fluorescence (PI) through a 600 nm wave-length filter. Analysis was performed on Cellquest software. In some experiments, the BrdU-labeling was omitted and the ethanol-fixed cells were resuspended directly in 1 ml PBS containing 8 μg RNAse A and 50 μg Propidium Iodide.
SDS PAGE and western blotting
Total cell lysates were prepared in lysis buffer containing 50 mM HEPES, pH 7.4, 0.1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 80 mM β-glycerophosphate and 1x protease inhibitor mixture (Roche Applied Science). In some experiments, cells were fractionated to generate soluble and chromatin-enriched nuclear fractions, exactly as described previously by Liu et al.53 In brief, monolayers of cells were washed three times in ice-cold phosphate-buffered saline (PBS) and scraped into 500 μl of ice-cold cytoskeleton buffer (CSK buffer: 10 mM PIPES, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1 mM ATP, 1 mM Na3VO4, 10 mM NaF and 0.1% Tritron X-100) freshly supplemented with protease inhibitor cocktail. CSK lysates were transferred to microcentrifuge tubes and incubated on ice for 5 minutes. The cell lysates were centrifuged at 1,000 g for 2 minutes. The supernatants were removed and further clarified by centrifugation at 10,000 g for 10 minutes to obtain Triton X-100-soluble fractions. The Triton-extracted insoluble nuclear fractions were washed once with 1 ml of CSK buffer and then re-suspended in a minimal volume of CSK. In some experiments as indicated, chromatin-bound proteins were released by digestion of nuclei in CSK with 1000 U/ml of RNAse-free DNAse I (Roche) at 25°C for 30 minutes. After digestion, soluble and insoluble materials were separated by centrifugation at 10,000 g for 10 minutes. Total cell extracts and soluble or nuclear protein samples were separated by SDS-PAGE, transferred to nitrocellulose using Amersham Pharmacia Multiphor II NovaBlot semi-dry transfer apparatus, and analyzed by immunoblotting with indicated antibodies. Anti-Cdc6 (sc-9964), anti-Cdk2 (sc-163), anti-Cdk4 (sc-260), anti-Chk1 (sc-7898), anti-Chk2 (sc-9064), anti-Cyclin D1 (sc-753), anti-Cyclin E (sc-247), anti-Cyclin A (sc-596), anti-Mcm2 (sc-10771), anti-Mcm7 (sc-9966), anti-phospho S780-Rb (sc-12901), anti-Rb (sc-050), anti-p53 (sc-6243) were from Santa Cruz Biotechnology. Anti-β-actin antibody (A-5441) was purchased from Sigma-Aldrich. Anti-p27 was from BD Biosciences.
In vitro kinase assays
To measure CDK4 and CDK2 kinase activity, cells were lysed for 30 min at 4°C in 0.5 ml of buffer containing 50 mM HEPES (pH 7.5), 10 mM MgCl2, 150 mM NaCl, 0.1% Tween-20, 1 mM dithiothreitol and 1x protease inhibitor cocktail. Cell lysates were then sonicated twice on ice, 5 seconds each pulse, at 30% power. Lysates were cleared by centrifugation at 10,000 xg for 10 minutes at 4°C. Supernatants were assayed for protein concentration. Protein extracts (approximately 500 μg per sample), were precleared for 1 hour with 20 μl of a protein A/G agarose bead suspension (Santa Cruz) and 0.5 μg normal rabbit IgG (Santa Cruz). Precleared lysates were then immunoprecipitated with 1 μg of either CDK2 or CDK4 antibody (Santa Cruz). Antibody-antigen complexes were allowed to form overnight at 4°C, with rotation. After extensive washing, immunoprecipitates were resuspended in kinase buffer (50 mM HEPES [pH 8.0], 10 mM MgCl2, 2.5 mM EGTA, 1 mM dithiothreitol) supplemented with 25 μM unlabeled ATP, 2.5 μg of either glutathione S-transferase-Rb protein (for CDK4, Santa Cruz) or histone H1 (for CDK2, Sigma) as substrate and 10 μCi [γ-32P]ATP (6000 Ci/mmol, Perkin Elmer) per sample. Reactions were incubated for 30 min at 30°C, and then terminated by the addition of 2x-Laemmli buffer. Labeled proteins were resolved on an SDS 12.5% polyacrylamide gel, Coomassie stained, dried onto Whatmann-filter paper, detected and quantified using a Bio-Rad Molecular Imager FX phosphoimager.
Reproducibility
All data shown are from experiments that were performed at least three times with similar results on each occasion.
Acknowledgements
This work was supported by National Institutes of Health grants ES099558 and ES12917 (C.V.) and K01-CA094907 (J.G.C.), an Environmental Pathology Training Grant T32-ES07017 (K.R.N.), and an American Cancer Society Research Scholars Grant GMC-111880 (J.G.C.).
Abbreviations
- ATM
ataxia telangiectasia-mutated
- ATR
ATM- and Rad3-related
- Cdc6
cell division cycle 6
- CDK2
cyclin dependent kinase 2
- CDK4
cyclin dependent kinase 4
- ChIP
chromatin immunoprecipitation
- Chk1
checkpoint kinase 1
- Chk2
checkpoint kinase 2
- HDF
human dermal fibroblasts
- HPV
human papilloma virus
- IC
initiation complex
- Mcm
mini-chromosome maintenance
- ORC
origin recognition complex
- pre-RC
pre-replicative complex
- Rb
retinoblastoma
- RNA Pol II
RNA polymerase II
- siRNA
small interfering RNA
References
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–74. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–43. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem Sci. 2005;30:437–44. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 4.DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–24. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, et al. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–30. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frolov MV, Dyson NJ. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci. 2004;117:2173–81. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- 7.Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–69. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 8.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–61. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitagawa M, Higashi H, Jung HK, Suzuki-Takahashi I, Ikeda M, Tamai K, et al. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. Embo J. 1996;15:7060–9. [PMC free article] [PubMed] [Google Scholar]
- 10.Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, et al. Dissociation among in vitro telomerase activity, telomere maintenance and cellular immortalization. Proc Natl Acad Sci USA. 1998;95:14723–8. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122:915–26. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–8. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- 13.Shreeram S, Sparks A, Lane DP, Blow JJ. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene. 2002;21:6624–32. doi: 10.1038/sj.onc.1205910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machida YJ, Teer JK, Dutta A. Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J Biol Chem. 2005;280:27624–30. doi: 10.1074/jbc.M502615200. [DOI] [PubMed] [Google Scholar]
- 15.Weinert T, Lydall D. Cell cycle checkpoints, genetic instability and cancer. Seminars in cancer biology. 1993;4:129–40. [PubMed] [Google Scholar]
- 16.Agami R, Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- 17.Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–88. [PMC free article] [PubMed] [Google Scholar]
- 18.Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–7. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 19.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 20.Dziak R, Leishman D, Radovic M, Tye BK, Yankulov K. Evidence for a role of MCM (mini-chromosome maintenance)5 in transcriptional repression of sub-telomeric and Ty-proximal genes in Saccharomyces cerevisiae. J Biol Chem. 2003;278:27372–81. doi: 10.1074/jbc.M301110200. [DOI] [PubMed] [Google Scholar]
- 21.Fitch MJ, Donato JJ, Tye BK. Mcm7, a subunit of the presumptive MCM helicase, modulates its own expression in conjunction with Mcm1. J Biol Chem. 2003;278:25408–16. doi: 10.1074/jbc.M300699200. [DOI] [PubMed] [Google Scholar]
- 22.Cook JG, Park CH, Burke TW, Leone G, DeGregori J, Engel A, et al. Analysis of Cdc6 function in the assembly of mammalian prereplication complexes. Proc Natl Acad Sci USA. 2002;99:1347–52. doi: 10.1073/pnas.032677499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delmolino LM, Saha P, Dutta A. Multiple mechanisms regulate subcellular localization of human CDC6. J Biol Chem. 2001;276:26947–54. doi: 10.1074/jbc.M101870200. [DOI] [PubMed] [Google Scholar]
- 24.DaFonseca CJ, Shu F, Zhang JJ. Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFNgamma. Proc Natl Acad Sci USA. 2001;98:3034–9. doi: 10.1073/pnas.061487598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang JJ, Zhao Y, Chait BT, Lathem WW, Ritzi M, Knippers R, et al. Ser727-dependent recruitment of MCM5 by Stat1alpha in IFNgamma-induced transcriptional activation. Embo J. 1998;17:6963–71. doi: 10.1093/emboj/17.23.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder M, He W, Zhang JJ. The DNA replication factor MCM5 is essential for Stat1-mediated transcriptional activation. Proc Natl Acad Sci USA. 2005;102:14539–44. doi: 10.1073/pnas.0507479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holland L, Gauthier L, Bell-Rogers P, Yankulov K. Distinct parts of minichromosome maintenance protein 2 associate with histone H3/H4 and RNA polymerase II holoenzyme. Eur J Biochem. 2002;269:5192–202. doi: 10.1046/j.1432-1033.2002.03224.x. [DOI] [PubMed] [Google Scholar]
- 28.Yankulov K, Todorov I, Romanowski P, Licatalosi D, Cilli K, McCracken S, et al. MCM proteins are associated with RNA polymerase II holoenzyme. Mol Cell Biol. 1999;19:6154–63. doi: 10.1128/mcb.19.9.6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawyer SL, Cheng IH, Chai W, Tye BK. Mcm10 and Cdc45 cooperate in origin activation in Saccharomyces cerevisiae. J Mol Biol. 2004;340:195–202. doi: 10.1016/j.jmb.2004.04.066. [DOI] [PubMed] [Google Scholar]
- 30.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–66. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 31.Auth T, Kunkel E, Grummt F. Interaction between HP1alpha and replication proteins in mammalian cells. Exp Cell Res. 2006;312:3349–59. doi: 10.1016/j.yexcr.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Laribee RN, Fuchs SM, Strahl BD. H2B ubiquitylation in transcriptional control: a FACT-finding mission. Genes Dev. 2007;21:737–43. doi: 10.1101/gad.1541507. [DOI] [PubMed] [Google Scholar]
- 33.Gauthier L, Dziak R, Kramer DJ, Leishman D, Song X, Ho J, et al. The role of the carboxyterminal domain of RNA polymerase II in regulating origins of DNA replication in Saccharomyces cerevisiae. Genetics. 2002;162:1117–29. doi: 10.1093/genetics/162.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinreich M, Liang C, Chen HH, Stillman B. Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proc Natl Acad Sci USA. 2001;98:11211–7. doi: 10.1073/pnas.201387198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furstenthal L, Kaiser BK, Swanson C, Jackson PK. Cyclin E uses Cdc6 as a chromatin-associated receptor required for DNA replication. J Cell Biol. 2001;152:1267–78. doi: 10.1083/jcb.152.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mimura S, Seki T, Tanaka S, Diffley JF. Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature. 2004;431:1118–23. doi: 10.1038/nature03024. [DOI] [PubMed] [Google Scholar]
- 37.Jallepalli PV, Brown GW, Muzi-Falconi M, Tien D, Kelly TJ. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 1997;11:2767–79. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honey S, Futcher B. Roles of the CDK phosphorylation sites of yeast Cdc6 in chromatin binding and rereplication. Mol Biol Cell. 2007;18:1324–36. doi: 10.1091/mbc.E06-06-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duursma AM, Agami R. CDK-dependent stabilization of Cdc6: linking growth and stress signals to activation of DNA replication. Cell Cycle. 2005;4:1725–8. doi: 10.4161/cc.4.12.2193. [DOI] [PubMed] [Google Scholar]
- 40.Connell-Crowley L, Elledge SJ, Harper JW. G1 cyclin-dependent kinases are sufficient to initiate DNA synthesis in quiescent human fibroblasts. Curr Biol. 1998;8:65–8. doi: 10.1016/s0960-9822(98)70021-1. [DOI] [PubMed] [Google Scholar]
- 41.Abbas T, Jha S, Sherman NE, Dutta A. Autocatalytic phosphorylation of CDK2 at the activating Thr160. Cell Cycle. 2007;6:843–52. doi: 10.4161/cc.6.7.4000. [DOI] [PubMed] [Google Scholar]
- 42.Teer JK, Machida YJ, Labit H, Novac O, Hyrien O, Marheineke K, et al. Proliferating human cells hypomorphic for origin recognition complex 2 and pre-replicative complex formation have a defect in p53 activation and Cdk2 kinase activation. J Biol Chem. 2006;281:6253–60. doi: 10.1074/jbc.M507150200. [DOI] [PubMed] [Google Scholar]
- 43.Labib K, Kearsey SE, Diffley JF. MCM2-7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol Biol Cell. 2001;12:3658–67. doi: 10.1091/mbc.12.11.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. Embo J. 1995;14:3788–99. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, et al. Regulation of DNA-replication origins during cell cycle progression. Nature. 1998;395:618–21. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 46.Weinberger M, Trabold PA, Lu M, Sharma K, Huberman JA, Burhans WC. Induction by adozelesin and hydroxyurea of origin recognition complex-dependent DNA damage and DNA replication checkpoints in Saccharomyces cerevisiae. J Biol Chem. 1999;274:35975–84. doi: 10.1074/jbc.274.50.35975. [DOI] [PubMed] [Google Scholar]
- 47.Shimada K, Pasero P, Gasser SM. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 2002;16:3236–52. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dillin A, Rine J. Roles for ORC in M phase and S phase. Science. 1998;279:1733–7. doi: 10.1126/science.279.5357.1733. [DOI] [PubMed] [Google Scholar]
- 49.Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito S, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. Embo J. 2002;21:5195–205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinberger M, Ramachandran L, Feng L, Sharma K, Sun X, Marchetti M, et al. Apoptosis in budding yeast caused by defects in initiation of DNA replication. J Cell Sci. 2005;118:3543–53. doi: 10.1242/jcs.02477. [DOI] [PubMed] [Google Scholar]
- 51.Pruitt SC, Bailey KJ, Freeland A. Reduced Mcm2 Expression Results in Severe Stem/Progenitor Cell Deficiency and Cancer. Stem Cells. 2007 doi: 10.1634/stemcells.2007-0483. [DOI] [PubMed] [Google Scholar]
- 52.Jagadeesh S, Banerjee PP. Telomerase reverse transcriptase regulates the expression of a key cell cycle regulator, cyclin D1. Biochem Biophys Res Commun. 2006;347:774–80. doi: 10.1016/j.bbrc.2006.06.172. [DOI] [PubMed] [Google Scholar]
- 53.Liu P, Barkley LR, Day T, Bi X, Slater DM, Alexandrow MG, et al. The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25a/Cdk2-independent mechanism. J Biol Chem. 2006 doi: 10.1074/jbc.M602982200. [DOI] [PubMed] [Google Scholar]