Summary
Proper centrosome separation is a prerequisite for establishing and positioning the bipolar spindle. While studies from a diversity of cell types demonstrate that microtubules (MTs) and their associated motors are essential for driving centrosome separation [1], the role of actin in driving centrosome separation remains less clear. Studies in tissue culture cells have led to a model in which actin/myosin based cortical flow is primarily responsible for driving late centrosome separation [2], while other in vivo studies suggest actin plays a more passive role by serving as an attachment site of astral MTs to pull centrosomes apart [3–6]. Here, we demonstrate that prior to nuclear envelope breakdown (NEB) in Drosophila embryos, proper centrosome separation does not require myosin II, but requires dynamic actin rearrangements at the growing edge of the interphase cap. Both Arp2/3- and Formin-mediated actin remodeling are required for separating the centrosome pairs before NEB. The Apc2-Armadillo complex appears to link cap expansion to centrosome separation. In contrast, the mechanisms driving centrosome separation after NEB are independent of the actin cytoskeleton and can compensate for earlier separation defects. Our studies show that the dynamics of actin polymerization drive centrosome separation and this has important implications for centrosome positioning during processes such as cell migration [7, 8], cell polarity maintenance [9, 10] and asymmetric cell division [11, 12].
Results and Discussion
Centrosome separation is concomitant with actin cap expansion
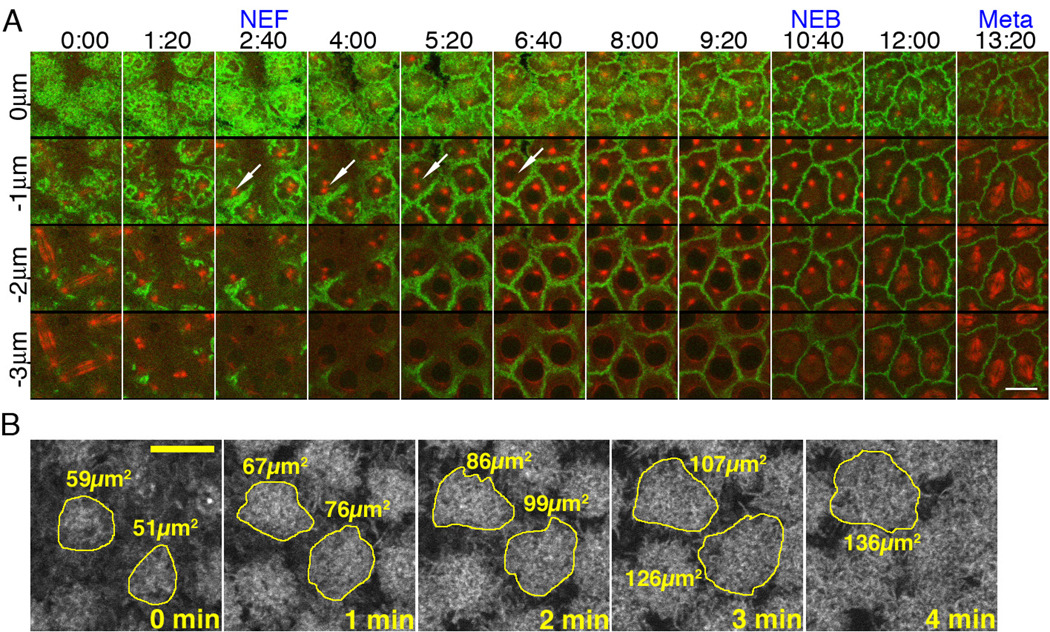
To define the role of the actin cytoskeleton in centrosome separation, we examined centrosome separation in early Drosophila embryos. During the rapid synchronous divisions in the syncytial Drosophila embryo, the nuclei divide on a plane just beneath the plasma membrane, providing a means to simultaneously follow centrosomes, MTs and actin dynamics (Fig.1A). During these divisions, centrosomes duplicate during telophase when actin caps form directly above each centrosome pair. Centrosome pairs migrate along the nuclear envelope at nuclear envelope formation (NEF) and move to the opposite poles (close to 180 degrees) before nuclear envelope breakdown (NEB) (Fig.1A arrows, Fig.S1A–E, and MovieS1). During this time, lateral expansion of the actin caps occurs (Fig.1A–B). Centrosome separation is concomitant with actin cap expansion (Fig.1A).
Figure 1. Centrosome separation is concomitant with actin cap expansion.
(A) Cycle-12 syncytial blastoderm. Green: GFP-Moesin. Red: Rhodamine-tubulin. X-axis: time Y-axis: depth. Z-series are shown, starting from the cortical surface (z=0) to 3µm below the surface (z=−3µm) at 1µm increments. At telophase, actin caps form at the cortical surface and initiate lateral expansion. This cap expansion is shown in the top panel, where gaps between caps are seen initially from time 0:00 to 6:40, after which GFP-Moesin marked actin has filled the entire frame. Concurrently, centrosome pairs separate from each other (arrows). See also Figure S1. (B) Panels depict the method of measuring actin cap area in wild-type embryos expressing GFP-Moesin. The freehand tool in ImageJ was used to outline individual caps at the beginning of cycle 12. Each cap is measured every minute until it can no longer be distinguished from its neighbors. Scale bars: 10µm.
Disruption of F-actin cytoskeleton prevents centrosome separation before, but not after, NEB
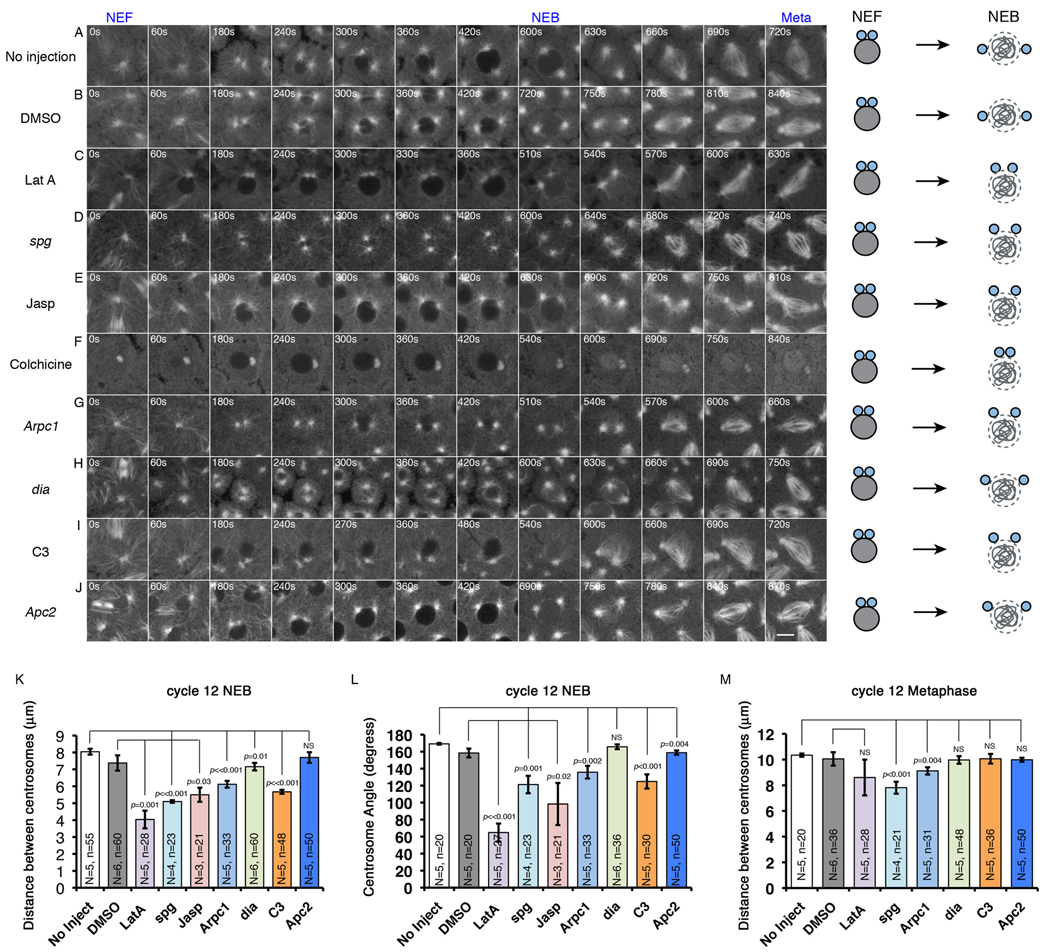
To investigate the roles of the cortical actin cytoskeleton in centrosome separation and spindle assembly, embryos expressing GFP-Tubulin were injected with Latrunculin A (LatA) just prior to NEF. Since F-actin is constantly turning over in the furrows [13] and caps (Fig.3C), LatA injection resulted in a rapid loss of F-actin from both these structures and prevented furrow invagination in the following cell cycle (Fig.S2). In wild-type uninjected cycle-12 embryos, the distance between centrosome pairs at cycle 12 NEB is about 8µm (Fig.2A and 2K). DMSO injection had very little effect on centrosome separation (Fig.2B and Fig.2K–L). In LatA injected embryos, approximately a quarter of the nuclei clustered during early interphase, which resulted in failed centrosome separation and multipolar spindles (Fig.S3 and MovieS2). To avoid secondary effects on centrosome separation due to LatA-induced clustering of nuclei, we only quantified centrosome separation in nuclei that did not cluster (the same criteria also applies to the other genetic or drug manipulations). For the unclustered nuclei, LatA did not appear to affect centrosome splitting, as the centrosome pairs were clearly distinguishable and detached from each other after NEF (Fig.2C and MovieS2). However, during the interval between centrosome splitting and NEB, centrosomes failed to separate normally (Fig.2C, 2K–L and MovieS2). The distance between centrosomes (4.0±0.5µm) was significantly shorter and the separation angle (65±11°) was also significantly smaller at NEB of cycle-12 than in control embryos injected with DMSO (7.4±0.5µm and 158±5°), indicating a role for actin in early separation of centrosomes (Fig.2C and MovieS2). Defects in early centrosome separation were also observed in embryos derived from females homozygous for the sponge (spg) maternal-effect mutation (Fig.2D and 2K–L), which lack both actin caps and furrows [14]. However, following NEB in LatA treated embryos and embryos laid by sponge mutant females, sister centrosomes separated fully to ultimately establish a bi-polar spindle during prometaphase-metaphase (Fig.2C–D, 2M, and MovieS2), indicating that a nuclear envelope and actin independent pathway compensates for the earlier actin-based separation defects.
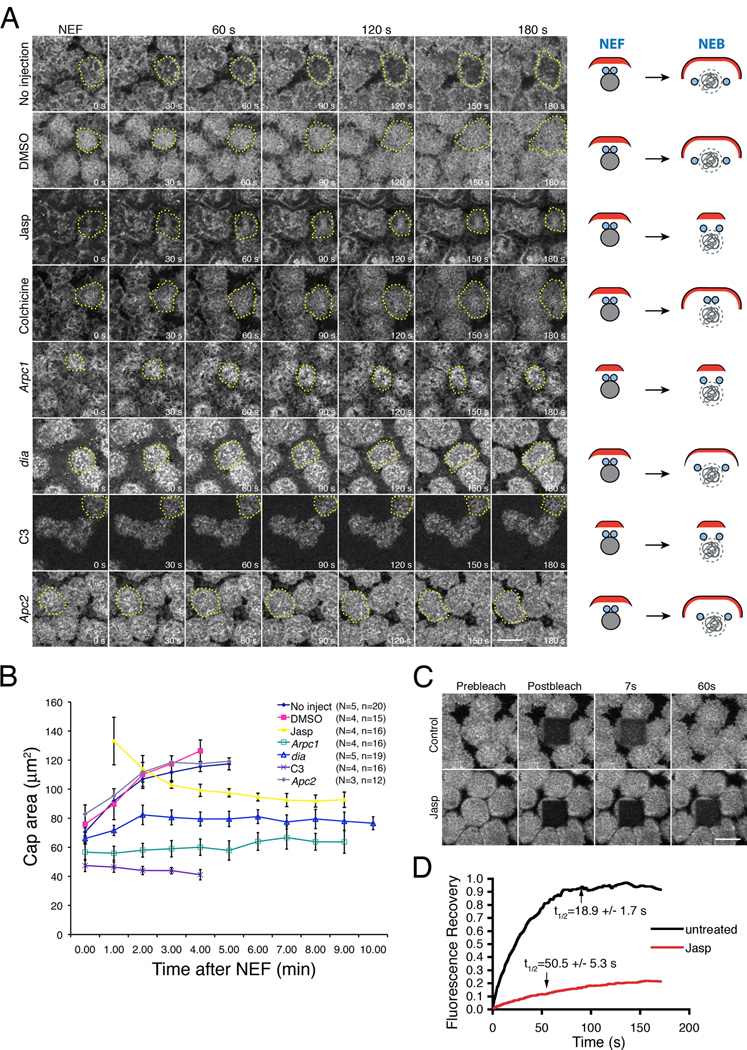
Figure 3. Actin cap expansion is driven by Arp2/3 and RhoA-Diaphanous mediated actin remodeling.
(A) Actin-based cap expansion was imaged after NEF in wild-type untreated, DMSO-treated, Jasp-treated, Colchicine-treated, C3-treated, Arpc1, dia, and Apc2 embryos. In each row, cap expansion is illustrated by dotted lines. Schematic drawings on the right side of each image series illustrate actin cap expansion (actin in red) relative to centrosome separation from NEF to NEB. See also Figure S2, Movie S4 and Movie S5. (B) The rate of actin cap area expansion after NEF in the drug-treated and mutant embryos imaged in (A). N=Total number of embryos; n=total number of caps counted. (C, D) FRAP analysis of actin turnover at the interphase cap in untreated and Jasp-treated embryos, showing relative fluorescence intensities of Rhodamine-actin at the caps after photobleaching. The prebleach intensities were arbitrarily set to 1. Scale bars: 10µm.
Figure 2. Cortical actin reorganization facilitates centrosome separation before NEB.
Time lapse images of: (A) Uninjected GFP-Tub embryo. See Movie S1. (B) DMSO injected GFP-Tub embryo. (C) 10mM LatA injected GFP-Tub embryo. See also Figure S3 and Movie S2. (D) Rhodamine-Tubulin-injected spg embryo. (E) 1mM Jasp injected GFP-Tub embryo. See Movie S3. (F) 0.5mM colchicine-injected GFP-Tub embryo. (G) Rhodamine-Tubulin-injected Arpc1 embryo. (H) Rhodamine-Tubulin-injected dia embryo. Centrosomes in dia and Arpc1 embryos were imaged more apically than controls, indicating a slowed migration towards the midline of the nucleus. (I) 1mg/mL C3 exotransferase injected GFP-Tub embryo. (J) Rhodamine-Tubulin-injected Apc2 embryo. Schematic drawings on the right side of each image series (A–J) illustrate the degree of centrosome separation from NEF to NEB. (K) Mean distance between centrosome pairs at NEB. (L) Mean angle between centrosome pairs at NEB with respect to the nuclear center. The centrosome pairs of dia embryos failed to migrate basally but were able to separate from each other normally at a focal plane above the equator of each nucleus. (M) Mean centrosome pair distances at metaphase. N=total number of embryos counted; n=total number of nuclei counted. NS: not statistically significant.
Actin turnover is required for centrosome separation before NEB
To determine whether actin dynamics are required for centrosome separation, the actin-stabilizing drug, Jasplakinolide (Jasp), was injected into embryos immediately prior to NEF. Similar to LatA-mediated inhibition of actin polymerization, Jasp-mediated actin stabilization strongly inhibited centrosome migration before NEB (Fig.2E, 2K–L and MovieS3). The respective pole-pole distance and separation angle were 5.5±0.4µm and 98±25° in Jasp-treated embryos, compared to 7.4±0.5µm and 158±5° in DMSO treated control embryos. Since both disruption and stabilization of F-actin inhibit early centrosome separation, these data suggest that actin turnover is important for proper centrosome separation before NEB.
Inhibition of actin turnover during interphase prevents actin cap expansion
Injecting Jasp before NEF resulted in strong actin accumulation at the cap (Fig.S2). In control embryos, actin caps expanded laterally from NEF through early interphase and eventually made contact with one another (Fig.3A–B and MovieS4). In contrast, actin caps failed to expand and actually shrunk over time after Jasp treatment (Fig.3A–B and MovieS5). FRAP analysis indicated the Jasp-induced defects in actin cap expansion could be due to failed actin turnover at the cap. Actin turnover rates (t1/2) at the interphase caps in untreated (N=10 embryos) or DMSO (N=10 embryos) injected embryos were 18.9±1.7s and 17.9±2.2s, with most of the actin (87.3±2.4% and 85.0±4.3%, respectively) recovered in the photobleached region after 80s (Fig.3C–D). However, in Jasp-treated embryos, only 25.5±3.0% of the total actin recovered after 80s (N=10 embryos), with a very slow turnover halftime of 50.5±5.3s (Fig.3C–D).
Thus inhibition of actin turnover results in failed actin cap expansion and failed centrosome separation. Previous studies have demonstrated that centrosome separation is not required for actin cap expansion: centrosome separation fails in colchicine-treated embryo but there is very little effect on actin cap expansion [6]. Our experiments confirm this finding (Fig.2F and Fig 3A).
Disruption of Arp2/3, an actin branching complex, strongly inhibits actin cap expansion and centrosome separation
To test the converse relationship, whether actin-cap expansion is required for centrosome separation, we analyzed mutants in Arpc1, a key component of the Arp2/3 complex. The Arp2/3 complex has been shown to localize to the margins of actin caps and promote cap expansion, presumably through its actin branching activity [15]. In control embryos, actin caps expanded to their maximum size (117.4±4.1µm2) 5 min after NEF. However, the cap size in Arpc1 embryos (the progeny of Arpc1 mutant maternal germline clones) had only increased slightly 5 min after NEF and from then on maintained an almost constant size until NEB (63.7±7.8µm2, Fig.3A–B). This is about half of the maximum cap size observed in wild-type controls. These data are consistent with the published Arpc1 phenotype [15]. Concomitantly, Arpc1 mutant embryos displayed a significant reduction in the distance and angle of separation between centrosome pairs at NEB (6.1±0.2µm and 136±7°, compared to 8.0±0.2µm and 169±1° in wild-type controls, Fig.2G and 2K–L).
Blocking Formin/Diaphanous and RhoA activities also interferes with cap expansion and centrosome separation before NEB
Cap expansion driven by Formin-mediated actin bundling is also required for centrosome separation before NEB. Diaphanous (Dia), the Drosophila Formin homolog, is required for actin bundling and metaphase furrow formation [16]. Initial cap size was normal in dia embryos (the progeny of dia mutant maternal germlines) but cap expansion was strongly inhibited, with the cap size at NEB (76.6±4.4µm2) reaching only 65% of the normal cap size (Fig.3A–B). Inhibition of RhoA, the upstream regulator of Diaphanous [17], through C3 exotransferase (C3), resulted in stronger defects in cap expansion (Fig.3A–B). Corresponding decreases in centrosome separation before NEB were observed in dia and C3 treated embryos (Fig.2H–I and 2K–L). Together these studies demonstrate that cap expansion is required for centrosome separation.
Apc2 mutant embryos exhibit relatively normal cap expansion
The Apc2-Armadillo complex in the fly early embryo has been shown to regulate centrosome separation [3]. Based on its localization to cortical sites where actin and MTs interact [18], it was proposed that the Apc2-Armadillo complex facilitates centrosome separation through stabilizing the interaction between the actin cortex and astral MTs [3]. Alternatively, since we have shown actin cap expansion is required for centrosome separation before NEB, it is possible that the Apc2-Armadillo complex facilitates centrosome separation by directly promoting actin cap expansion. To differentiate these two possibilities, cap expansion was examined in Apc2 mutant embryos. The initial cap size and the cap expansion rate in Apc2 embryos are very similar to that observed in uninjected control embryos (Fig.3A–B). To confirm that these Apc2 embryos were defective in centrosome separation, Rhodamine-labeled tubulin was injected into these embryos to follow centrosome movements. Our data showed very similar centrosome separation defects before NEB (159±2° compared to 169±1° in wild-type controls, Fig.2J and 2L) as previously published results [3]. Because Apc2 embryos do not disrupt actin cap organization [18, 19], the effects on centrosome separation were not as dramatic as those observed for the actin inhibitors and thus we did not observe significant differences in distance in which the centrosomes separated (Fig.2K). In addition, after NEB, the incomplete separated centrosome pairs were able to correct the earlier defects and achieve full separation by metaphase (Fig.2M). Taken together, these suggest that Apc2-Armadillo complex functions downstream of actin cap expansion to regulate centrosome separation before NEB.
Myosin II-driven cortical flow is not required for centrosome separation before NEB
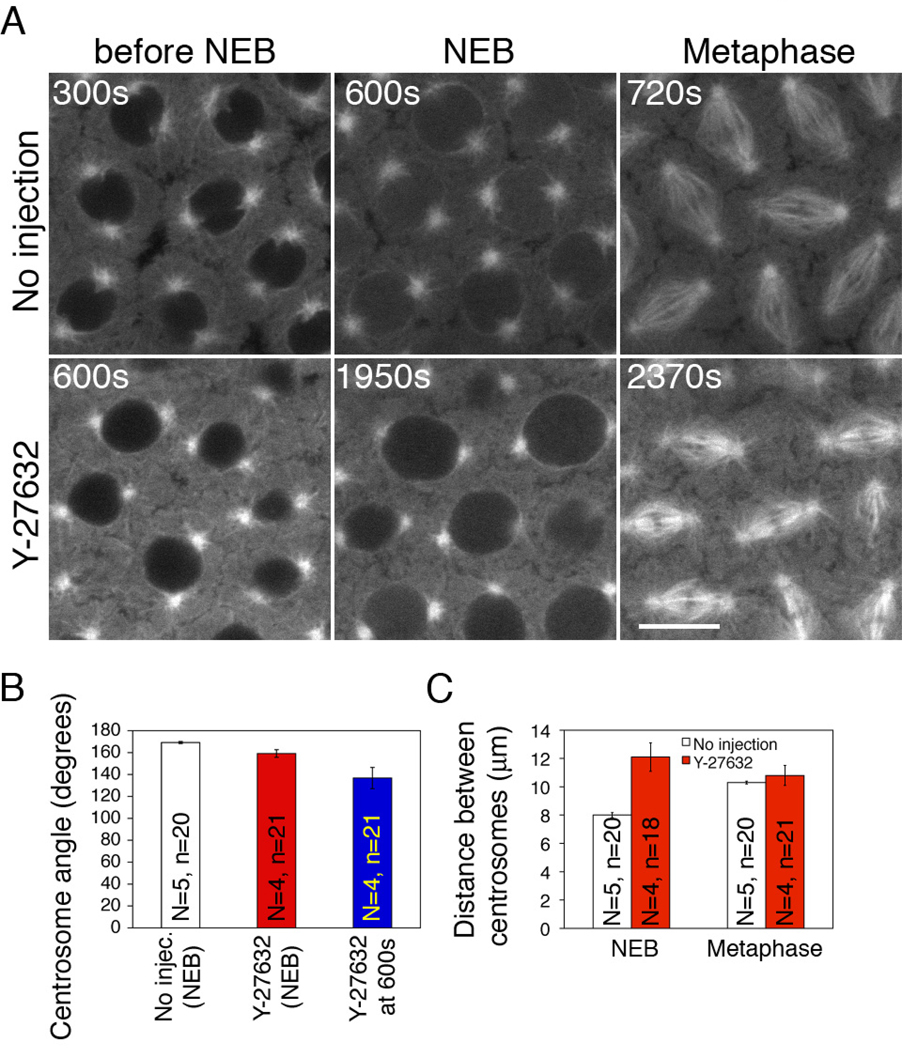
Actin-myosin II driven cortical flow has been proposed to separate centrosomes after NEB in mammalian cultured cells [2]. To determine whether myosin II, similar to actin, facilitates centrosome separation during the cortical divisions in Drosophila embryos, we relied on the small molecule inhibitor Y-27632, a drug that inhibits Rho kinase, which in turn blocks myosin II light chain kinase and thus myosin II activity. We used a drug concentration (50mM) that has been proven to effectively block myosin II activity in our system [20]. Significant delay into mitosis was often observed after Y-27632 injection (from cycle 12 NEF to NEB, 1455 ± 296 s versus 588 ± 15 s in control embryos). Unlike LatA injection, myosin II inhibition by Y-27632 had only a very mild effect on centrosome separation before NEB and the metaphase spindle length was about the same as that in control embryos (Fig.4, A–C). The relatively normal centrosome separation and spindle formation after inhibiting myosin II is consistent with previously published results [20]. Thus, it appears that centrosome separation during the cortical divisions in the Drosophila embryo does not rely on myosin II activity.
Figure 4. Disruption of myosin II by Y-27632 has very mild effect on centrosome separation before NEB.
(A) 50mM Y-27632 was injected into GFP-Tubulin embryos. Prolonged interphase, nuclear swelling and centrosome detachment from the nuclear envelope were observed. A lower drug concentration (2mM) failed to produce any of these phenotypes (data not shown). Scale bar: 10µm. (B) Mean angle of centrosome pairs at NEB of uninjected embryos and Y-27632-treated embryos. Since 50mM Y-27632 injection induced prolonged interphase, the centrosome angle of Y-27632-treated embryos was also measured at 600s after NEF, the equivalent timing of NEB in control embryos. (C) Centrosome distance at NEB and metaphase of uninjected embryos and Y- 27632-treated embryos. The increase in centrosome distance in Y-27632 treated embryos at NEB was due to nuclear swelling phenotype induced by this drug injected at 50mM. N=total number of embryos; n=total number of nuclei counted.
Conclusions
We have described an essential role for actin turnover in centrosome separation prior to NEB. During this stage, centrosome separation occurs along the nuclear envelope and requires Dynein, a minus-end directed motor protein [4, 21–23]. However additional mechanisms must be required as some separation of centrosomes still occurs in dynein mutants [4]. Work by Rosenblatt and colleagues demonstrated that actin-myosin II based cortical flow drives the late stages of centrosome separation after NEB [2], raising the possibility that similar cortical based mechanisms are driving the initial stages of centrosome separation. In fact, previous studies in both mammalian and Drosophila systems indicated that actin is necessary for centrosome separation before NEB [6, 24], but it remained unclear where and how actin functions to separate centrosomes and whether this is myosin-based. Our results demonstrate that in the Drosophila embryo, the initial stages of centrosome separation prior to NEB rely on dynamic actin turnover but are independent of myosin.
Specifically, our results demonstrate a role for both Arp2/3-mediated actin branching and Formin/Diaphanous-mediated actin bundling in cortical actin cap expansion, which in turn drives centrosome separation prior to NEB. There was also a strong positive correlation between the severity of the centrosome separation defects and the cap expansion defects. Among the drugs and mutations that didn’t remove actin completely, Jasp and C3 produce the greatest disruption in cap expansion and also the greatest disruption in centrosome separation. Arpc1 and dia produced progressively less severe defects in cap expansion and centrosome separation. Recent models propose that centrosome separation is driven by the dynein-dynactin complex associated with cortical actin [3–5]. Cortical dynein localizes at dynamic actin caps to pull astral MTs outward, and being opposed by an inward force by Ncd on interpolar MT bundles [22]. In addition, the MT depolymerase Klp10A also plays a role in the initial phase of centrosome separation [25]. Our results suggest that either the astral MT plus ends are constantly seeking some unknown cues at the growing edge of the cortical expanding caps or the actin-MT plus end interaction is stabilized preferentially at the growing edge of the expanding caps. Based on our results and previous studies [3], the Apc2-Armadillo complex may provide a link between the expanding actin cap and astral microtubules. It will be of interest to determine whether actin turnover at the cortex also plays a role in centrosome and spindle positioning in other cell types [12, 26, 27]. Our finding that the actin-based centrosome separation defects are fully corrected after NEB is particularly intriguing and may provide insight into mechanisms regulating spindle size.
Experimental Procedures
Fly Strains and Genetics
Germline clones of diaphanous5 or Arpc1R337st were generated using the FLP-DFS technique [15, 16, 28]. These two mutants and Apc2ΔSwere acquired from Bloomington Drosophila stock center. sponge335 was a gift from Eric Wieschaus. For live embryo imaging, we used the following stocks: GFP-alphaTubulin (a gift from Thomas Kaufman, [29]), GFP-Moesin (a gift from Daniel Kiehart, [30]) and GFP-Dlg (Discs Large) (FlyTrap Project, [31]). All stocks were raised at 25°C on standard corn meal/molasses media.
Live Embryo Analysis
Embryos were prepared for microinjection and time-lapse scanning confocal microscopy as previously described [32]. All the reagents were injected at 50% egg length and were diluted approximately 100 fold in the embryos [33]. For sponge, diaphanous and Arpc1 mutant embryos, Rhodamine-conjugated tubulin (10mg/ml, Cytoskeleton) or Rhodamine-conjugated actin (10mg/ml, Cytoskeleton) was injected into the embryos at late cycle 10 or early cycle 11. The following drugs were injected at cycle 11 anaphase: DMSO alone (Sigma-Aldrich), LatA (10mM in DMSO, Sigma-Aldrich), Y-27632 (50mM and 2mM, Tocris), Jasp (1mM in DMSO, Calbiochem), C3 exotransferase (1mg/ml, Cytoskeleton) and colchicine (0.5mM, Sigma-Aldrich). GFP-Dlg was used to mark the furrow membrane [13].
Confocal Microscopy and FRAP analysis
Confocal microscope images were captured on an inverted photoscope (DMIRB; Leitz) equipped with a laser confocal imaging system (TCS SP2; Leica) using an HCX PL APO 1.4 NA 63X oil objective (Leica). ImageJ software (National Institutes of Health, Bethesda, MD) was used to quantify the confocal images.
FRAP analysis were performed as previously described [13]. Imaging was controlled by the Leica Confocal Software Microlab. After five prebleach scans of an entire image, 10 bleaching scans (0.7s each) with 100% intensity of 488 nm and 543 nm over the region of interest in the actin caps (10µm × 10µm) were performed. After photobleaching, fluorescence recovery was monitored 10 times every 0.7s and 60 times every 2s, and 10 times every 5s. The recovery of fluorescence intensities was measured with Microlab. The intensity of the bleached cap area was normalized to the background nonbleached area. Recovery percentage was calculated as the final plateau intensity (IF) minus the first intensity after photobleaching (I0) all divided by the difference between prebleach (II) and postbleach (I0) intensities ([IF - I0]/[II - I0]). The fluorescence intensity of each time-point (It) was transformed into a 0–1 scale calculated by [It - I0]/[II - I0]). The values of relative intensities versus time were plotted using Excel (2004; Microsoft), and the recovery t1/2 was measured from the plots.
Image Quantifications and Statistics
Centrosome pair distances were quantified in the optical section with the strongest centrosome signal (GFP-Tubulin or injected Rhodamine-Tubulin). Centrosome pair angles were measured using the angle tool in ImageJ by placing the vertex at the approximate center of each nucleus. In Jasp-injected embryos, although centrosomes can separate further during subsequent prometaphase and metaphase, spindles tend to fuse with each other. To avoid secondary defects due to spindle fusion, we analyzed centrosomes only prior to NEB in Jasp-treated embryos.
For cap expansion analysis, time-lapse confocal images were taken of either GFP-Moesin or Rhodamine-actin injected embryos from NEF to NEB of cycle 12. A z-series was taken every 30 seconds during this time period with z-steps of 0.75µm starting at the very surface of the embryo. Cap expansion was measured using ImageJ. Confocal sections representing just the actin cap were used in all experiments. The freehand tool in ImageJ was used to encircle each cap, which allowed an area measurement. Four individual caps were measured in each embryo from the beginning of cycle 12. These four were followed every minute until the boundaries of each cap could no longer be differentiated from that of neighboring caps (Fig.1B). In C3 treated embryos, caps were tracked only until 4 minutes after NEF when the cap boundaries became indiscernible. At least three embryos were analyzed per genotype/drug treatment.
Student’s t-tests (two-tailed, equal variance) were performed to analyze the data. For each embryo, multiple mitotic apparati were quantified and averaged. These averaged values were then used for statistics to estimate the variance between embryos under the same treatment. Error bars represent the standard error of the mean (SEM) from at least three independent experiments. For videos, image series collected over time were cropped in ImageReady (v9.0; Adobe) and converted to QuickTime (Apple) videos using PNG lossless compression.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grant to W.S. (GM046409). We would like to thank Jon Scholey, Minx Fuller, and Laura Serbus for providing helpful feedback to the manuscript. We are grateful for the fly stocks provided by Thom Kaufman, Eric Wieschaus, Dan Kiehart, and FlyTrap.
References
- 1.Lim HH, Zhang T, Surana U. Regulation of centrosome separation in yeast and vertebrates: Common threads. Trends Cell Biol. 2009 doi: 10.1016/j.tcb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Rosenblatt J, Cramer LP, Baum B, McGee KM. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell. 2004;117:361–372. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- 3.Buttrick GJ, Beaumont LM, Leitch J, Yau C, Hughes JR, Wakefield JG. Akt regulates centrosome migration and spindle orientation in the early Drosophila melanogaster embryo. J Cell Biol. 2008;180:537–548. doi: 10.1083/jcb.200705085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson JT, Wojcik EJ, Sanders MA, McGrail M, Hays TS. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J Cell Biol. 1999;146:597–608. doi: 10.1083/jcb.146.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cytrynbaum EN, Sommi P, Brust-Mascher I, Scholey JM, Mogilner A. Early spindle assembly in Drosophila embryos: role of a force balance involving cytoskeletal dynamics and nuclear mechanics. Mol Biol Cell. 2005;16:4967–4981. doi: 10.1091/mbc.E05-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevenson VA, Kramer J, Kuhn J, Theurkauf WE. Centrosomes and the Scrambled protein coordinate microtubule-independent actin reorganization. Nat Cell Biol. 2001;3:68–75. doi: 10.1038/35050579. [DOI] [PubMed] [Google Scholar]
- 7.Schmoranzer J, Fawcett JP, Segura M, Tan S, Vallee RB, Pawson T, Gundersen GG. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol. 2009;19:1065–1074. doi: 10.1016/j.cub.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solecki DJ, Trivedi N, Govek EE, Kerekes RA, Gleason SS, Hatten ME. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai RA, Gao L, Raghavan S, Liu WF, Chen CS. Cell polarity triggered by cell-cell adhesion via E-cadherin. J Cell Sci. 2009;122:905–911. doi: 10.1242/jcs.028183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupin I, Camand E, Etienne-Manneville S. Classical cadherins control nucleus and centrosome position and cell polarity. J Cell Biol. 2009;185:779–786. doi: 10.1083/jcb.200812034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowan CR, Hyman AA. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu Rev Cell Dev Biol. 2004;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao J, Albertson R, Riggs B, Field CM, Sullivan W. Nuf, a Rab11 effector, maintains cytokinetic furrow integrity by promoting local actin polymerization. J Cell Biol. 2008;182:301–313. doi: 10.1083/jcb.200712036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Postner MA, Miller KG, Wieschaus EF. Maternal effect mutations of the sponge locus affect actin cytoskeletal rearrangements in Drosophila melanogaster embryos. J Cell Biol. 1992;119:1205–1218. doi: 10.1083/jcb.119.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevenson V, Hudson A, Cooley L, Theurkauf WE. Arp2/3- dependent pseudocleavage [correction of psuedocleavage] furrow assembly in syncytial Drosophila embryos. Curr Biol. 2002;12:705–711. doi: 10.1016/s0960-9822(02)00807-2. [DOI] [PubMed] [Google Scholar]
- 16.Afshar K, Stuart B, Wasserman SA. Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development. 2000;127:1887–1897. doi: 10.1242/dev.127.9.1887. [DOI] [PubMed] [Google Scholar]
- 17.Padash Barmchi M, Rogers S, Hacker U. DRhoGEF2 regulates actin organization and contractility in the Drosophila blastoderm embryo. J Cell Biol. 2005;168:575–585. doi: 10.1083/jcb.200407124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCartney BM, McEwen DG, Grevengoed E, Maddox P, Bejsovec A, Peifer M. Drosophila APC2 and Armadillo participate in tethering mitotic spindles to cortical actin. Nat Cell Biol. 2001;3:933–938. doi: 10.1038/ncb1001-933. [DOI] [PubMed] [Google Scholar]
- 19.Webb RL, Zhou MN, McCartney BM. A novel role for an APC2-Diaphanous complex in regulating actin organization in Drosophila. Development. 2009;136:1283–1293. doi: 10.1242/dev.026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Royou A, Field C, Sisson JC, Sullivan W, Karess R. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol Biol Cell. 2004;15:838–850. doi: 10.1091/mbc.E03-06-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitehead CM, Winkfein RJ, Rattner JB. The relationship of HsEg5 and the actin cytoskeleton to centrosome separation. Cell Motil Cytoskeleton. 1996;35:298–308. doi: 10.1002/(SICI)1097-0169(1996)35:4<298::AID-CM3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- 26.Burakov A, Nadezhdina E, Slepchenko B, Rodionov V. Centrosome positioning in interphase cells. J Cell Biol. 2003;162:963–969. doi: 10.1083/jcb.200305082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunda P, Baum B. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol. 2009;19:174–179. doi: 10.1016/j.tcb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grieder NC, de Cuevas M, Spradling AC. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development. 2000;127:4253–4264. doi: 10.1242/dev.127.19.4253. [DOI] [PubMed] [Google Scholar]
- 30.Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev Biol. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- 31.Quinones-Coello AT, Petrella LN, Ayers K, Melillo A, Mazzalupo S, Hudson AM, Wang S, Castiblanco C, Buszczak M, Hoskins RA, Cooley L. Exploring strategies for protein trapping in Drosophila. Genetics. 2007;175:1089–1104. doi: 10.1534/genetics.106.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tram U, Riggs B, Koyama C, Debec A, Sullivan W. Methods for the study of centrosomes in Drosophila during embryogenesis. Methods Cell Biol. 2001;67:113–123. doi: 10.1016/s0091-679x(01)67008-0. [DOI] [PubMed] [Google Scholar]
- 33.Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.