Abstract
Early detection of oral squamous cell cancer (OSCC) is the key to improve the low 5-year survival rate. Using proteomic and genomic technologies we have previously discovered and validated salivary OSCC markers in American patients. The question arises whether these biomarkers are discriminatory in cohorts of different ethnic background. Six transcriptome (DUSP1, IL8, IL1B, OAZ1, SAT1, S100P) and three proteome (IL1B, IL8, M2BP) biomarkers were tested on 18 early and 17 late stage OSCC patients and 51 healthy controls with quantitative PCR and ELISA. Four transcriptome (IL8, IL1B, SAT1, S100P) and all proteome biomarkers were significantly elevated (p<0.05) in OSCC patients. The combination of markers yielded an AUC of 0.86, 0.85 and 0.88 for OSCC total, T1-T2, and T3-T4 respectively. The sensitivity/specificity for OSCC total was 0.89/0.78, for T1-T2 0.67/0.96, and for T3-T4 0.82/0.84. In conclusion, seven of the nine salivary biomarkers (3 proteins and 4 mRNAs) were validated and performed strongest in late stage cancer. Patient-based salivary diagnostics is a highly promising approach for OSCC detection. This study shows that previously discovered and validated salivary OSCC biomarkers are discriminatory and reproducible in a different ethnic cohort. These findings support the feasibility to implement multi-center, multi-ethnicity clinical trials towards the pivotal validation of salivary biomarkers for OSCC detection.
Keywords: Oral cancer, biomarkers, salivary diagnostics, proteome, transcriptome
Introduction
Globally there are 350 000 new cases of oral cancers each year, making it the thirteenth most common cancer in the US and the eighth most common cancer in Serbia 1, 2. Oral squamous cell cancer (OSCC) accounts for more than 90% of all oral cancers. While official data for Serbia is not available, in the US the 5-year survival rate remains low at 60%, which is mainly due to the fact that most OSCC are diagnosed at a late stage 1, 3–5. Although rather easily accessible compared to other cancers, the diagnosis of OSCC can be challenging since most lesions will be small and asymptomatic and are easily overlooked or misjudged. Early detection would have a great impact on survival, mortality and morbidity of OSCC.
One of the most easy to obtain and non-invasive source for disease biomarkers is saliva, being a mirror of the body and having shown high discriminatory power for pancreatic cancer 6, Sjögren’s syndrome 7, HIV 8, 9, Hepatitis (A, B, and C) 10–12, and OSCC 3, 13–16.
The gold standard for the diagnosis of OSCC is still a biopsy of the suspicious lesion. Obviously, taking a biopsy is not suited for screening purposes for early oral cancer detection due to its invasive nature, high cost, and need for specially trained medical personal and equipment. We have previously shown that salivary biomarkers are highly discriminatory for OSCC detection. The development from normal to OSCC cells can lead to altered expression of proteins 3, 13, 16 and mRNA 15 markers in saliva. One of the biggest challenges in the field of biomarker research is that initial studies find excellent biomarkers while subsequent studies fail to validate. Our aim is to evaluate if the previously reported transcriptomic (IL1B, IL8, SAT1, S100P, DUSP1, OAZ1) 15 and proteomic (IL1B, IL8, M2BP) 3, 13, 16 markers discovered and validated in American OSCC cohorts are valid in an independent cohort of OSCC patients from Serbia. Also, for the first time we will show the discriminatory power of the combination of transcriptomic and proteomic salivary biomarkers and performance in early (T1-T2) and late (T3-T4) stages OSCC.
Patients and Methods
Patient selection
OSCC patients were recruited from the Clinical Center of Serbia and Stomatology Faculty University of Belgrade, Belgrade, Serbia. 35 patients with recently diagnosed and untreated OSCC were included in this study (mean age 60.94±12.30 years, 60% smokers, 86% males), 18 of them with tumor stages T1-T2, 17 with stages T3-T4, and 51 healthy control subjects (mean age 38.24±12.50 years, 43% smokers, 55% males). No subject had a history of prior cancer, diabetes, autoimmune disorder, hepatitis, or HIV infection. The study was done according to harmonized FDA-EU Directive. All of the subjects signed the institutional review board-approved Informed Consent (IC). For detailed patient characteristics see Table 1 in Supplementary Data.
Saliva Collection and Processing
Unstimulated saliva was collected and processed as previously described separately for RNA 6 and protein portions 3. All PCR and ELISA assays were done at Dental Research Institute, University of California–Los Angeles, Los Angeles.
Primer Design
Nested PCR assays were designed using NCBI/Primer-BLAST software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Amplicons were intron-spanning whenever possible, with lengths of 77–132bp for the outer and 66–92bp for the inner products. For full gene names, gene accession numbers, and primer sequences see Table 4. The previously validated OSCC mRNA marker H3F3A was not used in this study due to the fact that the gene sequence has been updated several times since our first OSCC mRNA study 15. There is no specific primer for the recent third H3F3A sequence update (gene accession number NM 002107.3). Every possible primer would amplify a whole cluster of genes.
Table 4.
Gene accession number and primer sequences of all pre-amplification and qPCR primers
| Gene symbol | Full name | Accession number | Outer forward of pre-amplification | Outer reverse primer of pre-amplification | Inner forward primer of qPCR | Inner reverse primer of qPCR | Assay use |
|---|---|---|---|---|---|---|---|
| IL1B | Interleukin 1, beta | NM_000576.2 | GTACCTGTCCTGCGTGTTGAAAG | TTCTATCTTGTTGAAGACAAATCGCTT | TGTTGAAAGATGATAAGCCCACTCT | CAAATCGCTTTTCCATCTTCTTCT | OSCC marker |
| DUSP1 | Dual specificity phosphatase 1 | NM_004417.3 | CCTGTGGAGGACAACCACAAG | GCCTGGCAGTGGACAAACA | CAGACATCAGCTCCTGGTTCAA | CAAACACCCTTCCTCCAGCAT | OSCC marker |
| IL8 | Interleukin 8 | NM_000584.2 | TTTCTGATGGAAGAGAGCTCTGTCT | ATCTTCACTGATTCTTGGATACCACA | CCAAGGAAAACTGGGTGCAG | CTTGGATACCACAGAGAATGAATTTTT | OSCC marker |
| OAZ1 | Ornithine decarboxylase antizyme 1 | NM_004152.2 | TGCGAGCCGACCATGTC | CCCCGGTCTCACAATCTCAA | TCTTCATTTGCTTCCACAAGAACC | TCAAAGCCCAAAAAGCTGAAG | OSCC marker |
| S100P | S100 calcium binding protein P | NM_005980.2 | GCACGCAGACCCTGACCA | CGTCCAGGTCCTTGAGCAATT | GCTGATGGAGAAGGAGCTACCA | TTGAGCAATTTATCCACGGCAT | OSCC marker |
| SAT1 | Spermidine/spermine N1-acetyltransferase 1 | NM_002970.2 | GCACTGGACTCCGGAAGGACAC | ACTTGCCAATCCACGGGTCA | GGACTCCGGAAGGACACAGCATTG | GCCAATCCACGGGTCATAGGTAAA | OSCC marker |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | NM_002046.3 | CATTGCCCTCAACGACCACTT | ATGTGGGCCATGAGGTCCA | ACCACTTTGTCAAGCTCATTTCCT | CACCCTGTTGCTGTAGCCAAAT | Reference gene |
| ACTB | Actin, beta | NM_001101.3 | GATCATTGCTCCTCCTGAGC | CGGACTCGTCATACTCCTGC | CTCCTGAGCGCAAGTACTCC | ATACTCCTGCTTGCTGATCCAC | Reference gene |
| RPS9 | Ribosomal protein S9 | NM_001013.3 | GCGCCATATCAGGGTCCGCA | CCCCGTAGGGAGAGCGCAG | GCCATATCAGGGTCCGCAAGCA | TCTGGGAATCCAGGCGGACAA | Reference gene |
RT-PCR pre-amplification and quantitative PCR (qPCR)
The pre-amplification and qPCR were done as previously described 17. Only samples that showed specific qPCR products for the three saliva internal reference (SIR) genes GAPDH, ACTB, and RPS9 were included in this study. Ct values of all target genes (OAZ1, DUSP1, SAT1, IL1B, IL8, S100P) were normalized to arithmetic mean of the three SIR genes.
Immunoassays
ELISAs were done for Interleukin 1 Beta (IL1B), Interleukin 8 (IL8) (both from Thermo Fisher, Rockford, IL), and s90K/Mac-2 binding protein (M2BP) (IBL, Hamburg, Germany). All samples were measured in duplicate and calculated with the respective standard curves.
Statistical analysis
For each significant biomarker and also for biomarker combinations, receiver operating characteristic curve (ROC), area under the curve (AUC), sensitivity, and specificity were determined as previously described 6.
Results
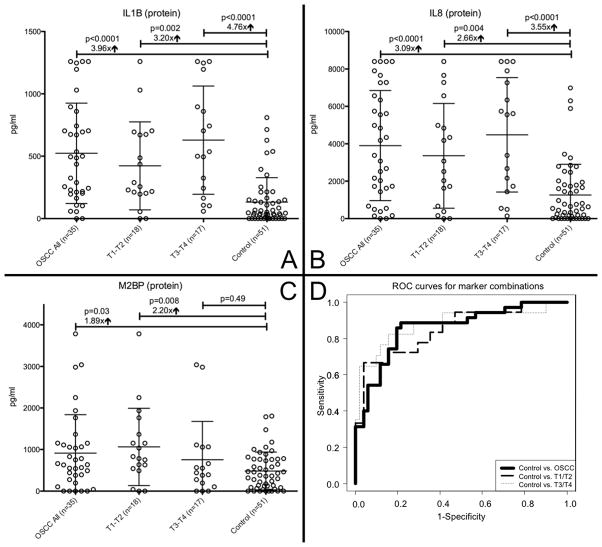
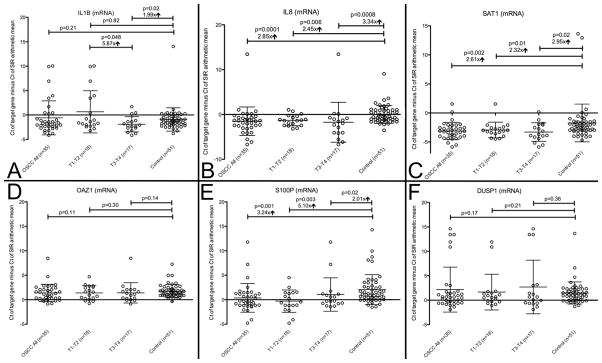
All 3 salivary protein markers (IL8, IL1B, M2BP) and 4 out of the 6 salivary mRNA markers (IL8, IL1B, SAT1, S100P) were elevated in OSCC patients and could discriminate (p<0.05) between cancer and control subjects as single markers (Table 2, Figure 1 and 2). All 35 OSCC patients and all 51 control subjects showed specific qPCR products for the 3 salivary internal reference (SIR) genes. M2BP was a highly significant marker for early stage oral cancer (T1/T2) (p=0.008) but was not discriminatory for late stage oral cancer (T3/T4) (p=0.49), while IL1B mRNA behaved in the opposite manner with p=0.82 for T1/T2 and p=0.02 for T3/T4 OSCC respectively. OAZ1 was the only marker that was not validated as a single marker and did not improve performance of combined markers in the Serbian cohort. Based on fold changes, IL1B protein and IL8 protein as well as S100P mRNA were increased the most between all OSCC patients and controls with a fold change of 3.96, 3.09, and 3.24 respectively. Combined markers proved to be the strongest discriminator of OSCC with an AUC of 0.86 for all cancer patients (IL1B protein + SAT1 mRNA + DUSP1 mRNA), 0.85 for T1-T2 (IL1B mRNA + SAT1 mRNA + DUSP1 mRNA), and 0.88 for T3-T4 (IL1B protein + DUSP1 mRNA) (Figure 1 and Table 3). The sensitivity/specificity for these groups were 0.89/0.78, 0.67/0.96, and 0.82/0.84 respectively. No biomarker could discriminate between male OSCC and female OSCC or smoking OSCC and non-smoking OSCC patients (all p-values > 0.05 with Mann-Whitney test). Although DUSP1 alone was not a significant single marker, it improved the performance for combined markers in the OSCC total group and in the T3-T4 group. In general, the stratification for early and late stages cancer showed that the salivary biomarkers were stronger discriminators for T3-T4 than for T1-T2 OSCC lesions.
Table 2.
Validation of saliva biomarkers in OSCC/T1-T2/T3-4 versus healthy control subjects
| Marker performance versus control group | P-value (OSCC total/T1-T2/T3-T4) | Mean fold increase (OSCC total/T1-T2/T3-T4) |
|---|---|---|
| Protein markers | ||
| IL1B | <0.0001/0.0002/<0.0001 | 3.96/3.20/4.76 |
| IL8 | <0.0001/0.004/<0.0001 | 3.09/2.66/3.55 |
| M2BP | 0.03/0.008/0.49 | 1.89/2.20/1.56 |
| RNA markers | ||
| IL8 | 0.0001/0.006/0.0008 | 2.85/2.45/3.34 |
| S100P | 0.001/0.003/0.02 | 3.24/5.10/2.01 |
| SAT1 | 0.002/0.01/0.02 | 2.61/2.32/2.95 |
| OAZ1 | 0.11/0.30/0.14 | 1.28/1.29/1.27 |
| IL1Ba | 0.21/0.82/0.02 | 0.80/0.34/1.99 |
| DUSP1 | 0.17/0.21/0.36 | 0.62/0.88/0.43 |
NOTE: P-values determined with Mann-Whitney test;
For IL1B mRNA, the difference between T1-T2 and T3-T4 was significant (p-value = 0.048) with T3-T4 mean 5.87 × higher than T1-T2
Figure 1. Protein markers and ROC curves.
Salivary protein levels (mean and standard deviation) for IL1B (panel A), IL8 (panel B), M2BP (panel C); p-values with Mann-Whitney U test; “↑” indicating mean fold change between cancer and control group; panel D: ROC curve analysis for the predictive power of combined salivary biomarkers. The final logistic model included for OSCC total 3 markers (IL1B protein + SAT1 mRNA + DUSP1 mRNA), for T1-T2 3 markers (IL1B mRNA+ IL8 mRNA+ SAT1 mRNA), and for T3-T4 2 markers (IL1B protein + DUSP1 mRNA) with an AUC of 0.86, 0.85, and 0.88 respectively
Figure 2. mRNA markers.
Salivary mRNA values (mean and standard deviation) for IL1B mRNA (panel A), IL8 (panel B), SAT1 (panel C), OAZ1 (panel D), S100P (panel E), and DUSP1 (panel F) normalized to SIR genes (GAPDH, ACTB, RPS9); p-values with Mann-Whitney U test; “↑” indicating mean fold change between cancer and control group; for IL1B mRNA, T3-T4 mean was 5.87 fold higher than T1-T2 mean
Table 3.
Receiver operator characteristic (ROC) curve analysis of OSCC-associated saliva biomarkers
| Marker performance versus control group | Area under ROC curve | Maximum Sensitivity | Maximum Specificity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein markers | OSCC total | T1-T2 | T3-T4 | OSCC total | T1-T2 | T3-T4 | OSCC total | T1-T2 | T3-T4 |
| IL1B | 0.83 | 0.79 | 0.88 | 0.83 | 0.83 | 0.76 | 0.76 | 0.78 | 0.84 |
| IL8 | 0.77 | 0.73 | 0.82 | 0.66 | 0.61 | 0.71 | 0.80 | 0.80 | 0.80 |
| M2BP | 0.64 | 0.71 | 0.56 | 0.37 | 0.83 | 0.29 | 0.90 | 0.59 | 0.92 |
| RNA markers | OSCC total | T1-T2 | T3-T4 | OSCC total | T1-T2 | T3-T4 | OSCC total | T1-T2 | T3-T4 |
| IL8 | 0.75 | 0.72 | 0.77 | 0.60 | 0.67 | 0.65 | 0.78 | 0.69 | 0.80 |
| S100P | 0.71 | 0.73 | 0.68 | 0.54 | 0.56 | 0.53 | 0.88 | 0.90 | 0.88 |
| SAT1 | 0.70 | 0.71 | 0.70 | 0.54 | 0.94 | 0.59 | 0.82 | 0.43 | 0.82 |
| OAZ1 | 0.60 | 0.58 | 0.62 | 0.40 | 0.39 | 0.41 | 0.92 | 0.92 | 0.41 |
| IL1B | 0.42 | 0.52 | 0.69 | 0.23 | 0.33 | 0.59 | 0.94 | 0.94 | 0.80 |
| DUSP1 | 0.41 | 0.40 | 0.43 | 0.14 | 0.11 | 0.29 | 0.98 | 0.98 | 0.92 |
| OSCC total | T1-T2 | T3-T4 | OSCC total | T1-T2 | T3-T4 | OSCC total | T1-T2 | T3-T4 | |
| Combination of Protein and RNA markers | 0.86a | 0.85b | 0.88c | 0.89a | 0.67b | 0.82c | 0.78a | 0.96b | 0.84c |
NOTE:
combination of IL1B protein + SAT1 + DUSP1;
combination of IL1B mRNA+ IL8 mRNA+ SAT1;
combination of IL1B protein + DUSP1
Discussion
The delayed diagnosis of OSCC at advanced stages is the main contributing factor to the poor 5-year survival rate. Currently used clinical strategies such as biopsy, vital tissue staining and exfoliative cytology will only be applicable to small patient groups and have clear limitations 18. The aim of using salivary biomarkers for OSCC detection is the facilitation of diagnostics at a point where OSCC is still small and treatment is very likely to be successful. In order to achieve this highly desirable goal, a saliva screening method must have sufficient sensitivity and specificity, be rather inexpensive, non-invasive, have high-throughput, and can be used by non-trained personnel. Using the latest advancements in technology could help put saliva in such a clinical context. Recent studies have shown that there is an abundance of accessible salivary biomarkers with highly discriminatory value for various diseases 3, 6–13, 15, 16. At the same time, the development of point of care devices such as the Oral Fluid NanoSensor Test (OFNASET) platform will provide easy to use saliva diagnostic technology 19. In this study, we used instant centrifugation at 4°C and adding of stabilizers followed by freezing samples at −80°C. Efforts are currently taken to develop a more convenient saliva RNA and protein preservation method, ideally even at room temperature. This would ease clinical logistics, sample preparation and transport of samples.
The results from this study show that our previously found salivary biomarkers are validatable and powerful discriminators between OSCC and controls in a Serbian cohort. This is of importance because it demonstrates that the OSCC salivary biomarkers are likely independent of ethnicity. The combination of markers from the proteome and transcriptome yielded the highest predictive power for OSCC total (IL1B protein + SAT1 mRNA + DUSP1 mRNA) with an AUC of 0.86, 0.89 sensitivity, and 0.78 specificity. When stratifying the OSCC patients for early and late stage cancer, the markers perform slightly stronger for late (T3-T4) than for early OSCC lesions (T1-T2) with an AUC of 0.88 and 0.85 respectively. Our findings underscore the correlation between the tested biomarkers and OSCC. The performance of the markers is slightly below but comparable to previously found results for the transcriptome with an AUC of 0.95 and for the proteome with an AUC of 0.93 in US cohorts 3, 15.
The question might be asked why there is not a single biomarker powerful enough to discriminate between OSCC and controls alone. Expanding the number of biomarkers takes into account the multi-factorial and heterogenic pathogenesis of OSCC, thus increasing the discriminatory power of single biomarkers to the high performance of combined biomarkers.
Some of the OSCC markers validated are inflammatory markers (IL1B, IL8) also found in the oral cavity in other conditions than OSCC. Studies are currently ongoing evaluating biomarkers for periodontitis, which is the strongest and most common inflammatory oral disease. A previous OSCC study already found a significant difference for salivary IL8 protein levels between OSCC and periodontitis patients 16. Our data shows that the panel of various salivary markers is truly discriminatory for OSCC.
Determining the source of OSCC related salivary biomarkers remains to be a challenging field. Exfoliate cancer cells, oral mucosa cells, alterations in the salivary gland secretion patterns (parotid, submandibular, sublingual as well as minor glands) or gingival crevice fluid could contribute to the saliva biomarker profile. Although the further stratification of biomarker sources remains an important scientific venue, only the focus on an efficacious screening method using easy to obtain whole saliva will allow wide spread clinical implementation.
While the results are promising and underline the power of the salivary transcriptome and proteome markers, further studies will be needed with larger patient numbers to allow a population-level clinical application. A nationwide OSCC saliva biomarker validation study is currently ongoing in a prospective-specimen collection, retrospective-blinded-evaluation (PRoBE) design manner 20 to meet the guidelines of the Early Detection Network (EDRN) of the National Cancer Institute 21.
Supplementary Material
Supplementary Data Table 1 – Patient Characteristics
Acknowledgments
Grant support: The work was supported by grant R01 DE017170 from the National Institute of Dental Research/National Institute of Health.
Footnotes
Conflict of interest statement
David T. Wong is co-founder of RNAmeTRIX Inc., a molecular diagnostic company.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ole Brinkmann, Email: ole.brinkmann@gmail.com.
Dragana A. Kastratovic, Email: drmaca.kastratovic@gmail.com.
Milovan V. Dimitrijevic, Email: drmilovan@yahoo.com.
Vitomir S. Konstantinovic, Email: vksvita@sbb.rs.
Drago B. Jelovac, Email: jbdrago@gmail.com.
Jadranka Antic, Email: jadranka.antic@yahoo.com.
Vladimir S. Nesic, Email: snesic@sbb.rs.
Srdjan Z. Markovic, Email: sergiusmarkione@gmail.com.
Zeljko R. Martinovic, Email: drmaca.kastratovic@gmail.com.
David Akin, Email: dakin@dentistry.ucla.edu.
Nadine Spielmann, Email: spielmann.nadine@gmail.com.
Hui Zhou, Email: hzhou@dentistry.ucla.edu.
David T. Wong, Email: dtww@ucla.edu.
References
- 1.Cancer facts and figures 2009. Atlanta: American Cancer Society; 2009. [Google Scholar]
- 2.Cancer Incidence and Mortality in Central Serbia 2006, Report No. 8. Belgrade, Serbia: Institute of Public Health of Serbia “ Dr Milan Jovanović - Batut”, Center for Prevention and Control of Noncommunicable Diseases, Department for Prevention and Control of Noncommunicable Diseases. 2009.
- 3.Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, et al. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008 Oct 1;14(19):6246–52. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkman BM, Wong DT. Disease mechanism and biomarkers of oral squamous cell carcinoma. Curr Opin Oncol. 2006 May;18(3):228–33. doi: 10.1097/01.cco.0000219250.15041.f8. [DOI] [PubMed] [Google Scholar]
- 5.Walker DM, Boey G, McDonald LA. The pathology of oral cancer. Pathology. 2003 Oct;35(5):376–83. doi: 10.1080/00310290310001602558. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, Park NH, et al. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology. 2010 Mar;138(3):949–57. e1–7. doi: 10.1053/j.gastro.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu S, Wang J, Meijer J, Ieong S, Xie Y, Yu T, et al. Salivary proteomic and genomic biomarkers for primary Sjogren's syndrome. Arthritis Rheum. 2007 Nov;56(11):3588–600. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emmons W. Accuracy of oral specimen testing for human immunodeficiency virus. Am J Med. 1997 Apr 1;102(4A):15–20. doi: 10.1016/s0002-9343(97)00033-8. [DOI] [PubMed] [Google Scholar]
- 9.Malamud D. Oral diagnostic testing for detecting human immunodeficiency virus-1 antibodies: a technology whose time has come. Am J Med. 1997 Apr 1;102(4A):9–14. doi: 10.1016/s0002-9343(97)00032-6. [DOI] [PubMed] [Google Scholar]
- 10.Ochnio JJ, Scheifele DW, Ho M, Mitchell LA. New, ultrasensitive enzyme immunoassay for detecting vaccine- and disease-induced hepatitis A virus-specific immunoglobulin G in saliva. J Clin Microbiol. 1997 Jan;35(1):98–101. doi: 10.1128/jcm.35.1.98-101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaita TM, Graham SM, Maxwell SM, Sirivasin W, Sabchareon A, Beeching NJ. Salivary sampling for hepatitis B surface antigen carriage: a sensitive technique suitable for epidemiological studies. Ann Trop Paediatr. 1995 Jun;15(2):135–9. doi: 10.1080/02724936.1995.11747761. [DOI] [PubMed] [Google Scholar]
- 12.El-Medany OM, El-Din Abdel Wahab KS, Abu Shady EA, Gad El-Hak N. Chronic liver disease and hepatitis C virus in Egyptian patients. Hepatogastroenterology. 1999 May–Jun;46(27):1895–903. [PubMed] [Google Scholar]
- 13.St John MA, Li Y, Zhou X, Denny P, Ho CM, Montemagno C, et al. Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2004 Aug;130(8):929–35. doi: 10.1001/archotol.130.8.929. [DOI] [PubMed] [Google Scholar]
- 14.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009 Sep 1;15(17):5473–7. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004 Dec 15;10(24):8442–50. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 16.Arellano-Garcia ME, Hu S, Wang J, Henson B, Zhou H, Chia D, et al. Multiplexed immunobead-based assay for detection of oral cancer protein biomarkers in saliva. Oral Dis. 2008 Nov;14(8):705–12. doi: 10.1111/j.1601-0825.2008.01488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Z, Zimmermann BG, Zhou H, Wang J, Henson BS, Yu W, et al. Exon-level expression profiling: a comprehensive transcriptome analysis of oral fluids. Clin Chem. 2008 May;54(5):824–32. doi: 10.1373/clinchem.2007.096164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epstein JB, Zhang L, Rosin M. Advances in the diagnosis of oral premalignant and malignant lesions. J Can Dent Assoc. 2002 Nov;68(10):617–21. [PubMed] [Google Scholar]
- 19.Gau V, Wong D. Oral fluid nanosensor test (OFNASET) with advanced electrochemical-based molecular analysis platform. Ann N Y Acad Sci. 2007 Mar;1098:401–10. doi: 10.1196/annals.1384.005. [DOI] [PubMed] [Google Scholar]
- 20.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008 Oct 15;100(20):1432–8. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001 Jul 18;93(14):1054–61. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data Table 1 – Patient Characteristics