Abstract
Plant roots strongly influence C and N availability in the rhizosphere via rhizodeposition and uptake of nutrients. This study aimed at investigating the effect of resource availability on microbial processes and community structure in the rhizosphere. We analyzed C and N availability, as well as microbial processes and microbial community composition in rhizosphere soil of European beech and compared it to the bulk soil. Additionally, we performed a girdling experiment in order to disrupt root exudation into the soil. By this novel approach we were able to demonstrate that enhanced resource availability positively affected N mineralization and hydrolytic enzyme activities in the rhizosphere, but negatively affected nitrification rates and oxidative enzyme activities, which are involved in the degradation of soil organic matter. Both rhizosphere effects on N mineralization and oxidative enzyme activities disappeared in the girdling treatment. Microbial community structure in the rhizosphere, assessed by phospholipid fatty acid analysis, differed only slightly from bulk soil but was markedly altered by the girdling treatment, indicating additional effects of the girdling treatment beyond the reduction of root exudation. Differences in oxidative enzyme activities and nitrification rates between rhizosphere soil and bulk soil, however, suggest considerable differences in the (functional) microbial community composition.
Keywords: Rhizosphere, Root exudates, Tree girdling, Microbial community composition, Phospholipid fatty acids, Microbial processes, Extracellular enzymes
Research highlights
►Positive rhizosphere effect on N mineralization and hydrolytic enzyme activities. ► Negative rhizosphere effect on nitrification and oxidative enzyme activities. ► Girdling significantly alters microbial community structure in the rhizosphere
1. Introduction
It has long been recognized that plant roots exert a large influence on the soil environment and its microflora and that these interactions in turn have considerable impact on plant growth (Lambers et al., 2009; Pinton et al., 2007). The term ’rhizosphere’ has been coined to describe the soil adjacent to and influenced by plant roots, a zone of usually high microbial activity, clearly distinct from bulk soil with regard to availability of nutrients, water and oxygen, in pH and redox potential (Hinsinger et al., 2009). The main reason for the high abundance of microorganisms in the vicinity of plant roots is rhizodeposition, which may account for as much as 25% of belowground allocated C (or 10% of net fixed C) (Jones et al., 2009; Uren, 2007; Whipps, 1990) and comprises water soluble root exudates such as sugars, amino acids, organic acids and hormones, as well as mucilage, enzymes, sloughed root cells and C allocated to root-associated symbionts (e.g. mycorrhiza) (Dennis et al., 2010; Grayston et al., 1997; Uren, 2007). Due to the high C/N ratio of rhizodeposits and due to plant nutrient uptake, the rhizosphere environment is characterized by a surplus of easily available C and as a result a strong nutrient limitation (Kuzyakov, 2002).
Positive as well as negative effects of plant roots on microbial growth and microbial decomposition processes have been observed, which is probably due to differing soil N and C availabilities, as well as differences in the investigated soil types or soil horizons (Kuzyakov, 2002). Although competition for water and N may even result in reduced microbial activity in the vicinity of roots (Wang and Bakken, 1997), plant-derived labile C sources often stimulate microbial growth and are rapidly metabolized (Butler et al., 2004). Several studies have shown that microbial activation by plant C exudates and N limitation in the rhizosphere may cause enhanced microbial decomposition of SOM (Bird et al., in press; Bottner et al., 1999; Cheng and Coleman, 1990), presumably because microbes use energy and C supplied by easily assimilable compounds to synthesize enzymes to hydrolyze, in turn, more complex SOM to acquire additional N (Blagodatskaya and Kuzyakov, 2008; Kuzyakov, 2002). Such enhanced decomposition of recalcitrant compounds after the addition of easily assimilable compounds or regular polymers (e.g. cellulose) has been termed “rhizosphere priming” (Blagodatskaya and Kuzyakov, 2008; Fontaine et al., 2003; Kuzyakov, 2010). In some studies, however, the observed increase in C mineralization following the addition of labile C compounds was rather attributed to higher microbial turnover rates and breakdown of microbial C pools than to enhanced depolymerisation of SOM (De Nobili et al., 2001; Weintraub et al., 2007).
It is widely known that the rhizosphere is a zone of high microbial turnover and activity, and several studies investigated the influence of plant roots on specific microbial processes, such as extracellular enzyme activities (Kaiser et al., 2010b; Weintraub et al., 2007), N mineralization (Norton and Firestone, 1996), nitrification and denitrification (Priha et al., 1999a, 1999b). Differences in microbial community structure between rhizosphere and bulk soil have already been analyzed by means of phospholipid fatty acid profiles (Butler et al., 2003; Paterson et al., 2007; Steer and Harris, 2000), or by molecular techniques (Marilley and Aragno, 1999), but few studies have been reported so far that link microbial community structure to microbial processes.
The goal of the present study was to explore how rhizosphere processes and microbial community composition are affected by resource availability. Our hypothesis was that root exudates facilitate the establishment of a distinct microbial community in the rhizosphere, and that an interruption of plant C exudation would cause the rhizosphere community to become more similar to bulk soil community. We further hypothesized that differences in resource availability between rhizosphere and bulk soil would result in different microbial processes, either as an effect of distinct microbial communities or as a direct priming effect on microbial decomposition processes by root exudates. In order to experimentally alter the resource availability we reduced plant C exudation by tree girdling, a method which has successfully been applied to disrupt belowground carbon allocation in trees (Högberg et al., 2001; Kaiser et al., 2010b). The combination of a comprehensive analysis of microbial processes, community composition, and C and N availability in the rhizosphere on the one hand and an experimental approach to reduce the plant influence on the soil microbial community on the other hand, is novel and allows to dissect the effect of resource availability and community composition on microbial processes.
2. Material and methods
2.1. Study site
Our study site was located in a 70 year old beech forest (Fagus sylvatica) about 40 km southwest from Vienna (48°07’ N 16°03′ E, 510 m a.s.l.). Tree density at the study site is 710 stems ha−1, with a canopy height of 26 m and mean diameter at breast height of trees of 21.5 cm. The mean air temperature at the study site is approximately 8 °C and the mean annual precipitation is 730 mm. Soil type is a Dystric Cambisol over flysh (pH in CaCl2 4.5–5.1) with an organic carbon content of 7.45% and total nitrogen content of 0.48% in the A horizon.
2.2. Experimental setup
Three circular girdling plots (15 m in diameter) and 6 control plots (5 m × 5 m) were established at the study site. Girdling of trees was performed in May 2008 by removal of the bark over 10 cm sections around the circumference of the stems at about 1.5 m above ground. Understory vegetation was removed from girdling and control plots.
2.3. Soil sampling
The first sampling was performed prior to girdling in May 2008 (bulk soil only), the main sampling was performed in September 2008. Six replicate samples of mineral soil were taken from control and girdling plots, respectively (2 replicates per plot in case of girdling plots, taken only from the inner 5 m diameter zone of the girdling plots). Each replicate consisted of 5 subsamples (soil cores of 7 cm diameter and 14.5 cm depth). Rhizosphere soil and roots were separated from bulk soil by hand: the soil closely adhering to the roots (up to 2.5 mm around the root) was considered as rhizosphere soil. Bulk samples were sieved (<2 mm). Bulk and rhizosphere samples were stored at 4 °C.
2.4. Soil extracts, C and N content
Two aliquots of fresh soil (2 g) were extracted with 20 ml of water and 1 M KCl, respectively. After adding HgCl2-solution (to a final concentration of 10 μM) to water extracts, extracts were stored at −20 °C. Subsamples of soil were oven-dried, weighed and ground for determination of C and N content by elemental analysis (EA 1110, Carlo Erba Instruments, Milan, Italy).
2.5. Inorganic N, DON, DOC and microbial biomass
NO3− and NH4+ were determined from water extracts by chemically suppressed ion-chromatography (Dionex HPAEC on a AS11 column for anions and Dionex HPCEC on a CS16 column for cations) (Kaiser et al., 2005). Organic C and total dissolved N were analyzed in water extracts by a TOC/TN analyzer (TOC-V CPH E200V/TNM-1 220V, Shimadzu). Dissolved organic N was calculated by subtracting inorganic N from total dissolved N.
2.6. Gross N mineralization and gross nitrification
Gross N mineralization and gross nitrification were assessed using the pool dilution technique (Kaiser et al., 2005; Myrold and Tiedje, 1986): 500 μl of 15NH4Cl or K15NO3 (0.25 mM, 10 atom% 15N), respectively, were applied to 2 subsamples (2 g) of fresh soil, then incubated for 4 h and 24 h and finally extracted with 15 ml of 2 M KCl. For determination of mineralization 15N values of NH3 diffused into acid traps were analyzed by continuous-flow isotope ratio mass spectrometry (IRMS) using an elemental analyzer coupled to a gas IRMS system (DeltaPLUS, Finnigan MAT, Bremen, Germany). 15N/14N ratios of NO3− were determined according to the protocol from Stange et al. (2007). In the SpinMAS system NO3− in the soil extract was reacted to NO by the addition of acidic V(III)Cl3 solution. The produced NO was purged from H2O and CO2 with a cryotrap and 15N abundance of the NO was analyzed in a quadrupole mass spectrometer (GAM 400, InProcess Instruments GmbH, Bremen, Germany). Rates of gross N mineralization and gross nitrification were calculated according to the following equations:
where m is gross mineralization, At is the NH4+–N pool after time t, A0 is the initial NH4+–N pool, APE (atom percent excess) is at%15Nsample–at%15Nnatural abundance
where n is gross nitrification, Nt is the NO3−–N pool after time t, N0 is the initial NO3−–N pool.
2.7. Extracellular enzyme activities
Enzyme activities were assayed by microplate fluorimetric and photometric assays: 1 g of fresh, sieved soil was suspended in 100 ml sodium acetate buffer (100 mM, pH 5.5) and ultrasonicated at low-energy for 2 min. Peptidase and phosphatase were measured fluorimetrically (Marx et al., 2001; Saiya-Cork et al., 2002): 200 μl of soil suspension and 50 μl of substrate (0.5–1 mM l-leucine 7-amido-4-methyl coumarin and 2 mM 4-methylumbelliferyl-phosphate, respectively) were pipetted into black microtiter plates (3 analytical replicates). After incubation in the dark for 140 min fluorescence was measured at 450 nm emission and 365 nm excitation (Tecan Infinite M200 fluorimeter). Aminomethylcoumarin (AMC) was used as a calibration standard for peptidase whereas Methylumbelliferyl was used for calibration of phosphatase.
Phenoloxidase and peroxidase were measured according to standard methods (Sinsabaugh et al., 1999), with small modifications: Subsamples of the soil suspension were mixed with an L-3,4-dihydroxyphenylalanin (DOPA) solution (20 mM DOPA, 1:1), shaken for 10 min and centrifuged. Aliquotes were pipetted into transparent microtiter plates with 6 analytical replicates per sample. Half of the wells additionally received 10 μl of a 0.3% H2O2 solution for measurement of peroxidase activity. Absorption at 450 nm was measured at the starting time point and after an incubation time of 20 h. Enzyme activity was calculated from the difference in absorption divided by an extinction-factor of 0.62, which was determined in a pre-experiment.
2.8. PLFAs
Phospholipid fatty acids were analyzed using a modified procedure described by (Frostegard et al., 1991). Fresh soil samples (2 g) were extracted overnight with a mixture of citrate buffer (0.15 M, pH 4.0 with NaOH), chloroform and methanol at a ratio of 0.8: 1: 2 (v/v/v). Samples were centrifuged and the supernatant was transferred to new vials. After addition of chloroform and citrate buffer for separation of the phases, the organic phase was removed and dried under a stream of dry N2. Lipids were redissolved in chloroform and neutral lipids were separated from phospholipids on silica columns (Supelco) by subsequent elution with chloroform, acetone and methanol. The methanol–fraction was dried under N2. Phospholipids were subsequently converted to fatty acid methyl esters by alkaline methanolysis. After adding 100 μL of a solution of methyl-nonadecanoate (200 μg mL−1) as an internal standard, phospholipids were dissolved in 1 mL of methanolic 0.2 M KOH and 1 mL of methanol-toluene (1:1) and incubated for 15 min at 37°, then mixed with 2 mL of water, 300 μL of acetic acid and 2 mL of hexane-chloroform (4:1). The hexane–phase was removed and dried under N2. Dried FAMEs were redissolved in isooctane and analyzed by gas chromatography (HP G1530A) on a DB23 column (Agilent, Vienna, Austria). A mixture of bacterial FAMEs (Supelco, Vienna, Austria) was used as a qualitative standard. Concentrations of single FAMEs were calculated using the internal standard (19:0) peak as a reference. PLFAs were designated following standard nomenclature, i.e. the number before the colon refers to the number of C atoms, the number after the colon indicates the number of double bonds followed by the position of the double bonds from the methyl end (ω). The prefixes i, a, and cy indicate iso- and anteiso-branching, and cyclopropyl-groups, respectively. We used the sum of the fatty acids i15:0, a15:0, i16:0, i17:0, a17:0 as indicator of Gram-positive bacteria, the sum of 16:1ω9, 16:1ω7, 18:1ω7, 18:1ω5, cy17:0, cy19:0, cy18:0 as indicator of Gram-negative bacteria, and all these together with 17:0, 17:1ω6, 17:1ω7 as a measure for total bacteria (Leckie, 2005; Zelles, 1997, 1998). The quantity of 18:2ω6,9 was used as an indicator of fungal biomass. 18:2ω6,9 is also common in plant tissue. However, it has recently been shown that the contribution of plants to this biomarker was negligible (Kaiser et al., 2010a).
2.9. Statistical analysis
Data were transformed to their natural logarithms in order to achieve normality and homogeneity of variances before analysis. Differences between rhizosphere and bulk soil, as well as differences between treatments were assessed using student’s t-test. Microbial community composition was analyzed by a principal component analysis (PCA) of the relative abundances of PLFAs. Differences between microbial communities were estimated by an analysis of similarity (ANOSIM) using a distance matrix of Euclidian distances. ANOSIM is a procedure which generates an R test statistic from distances between and within groups of samplings. The higher the R-value, the greater is the difference between groups. Pearson correlations were used for relating microbial processes to abundances of PLFAs, and for relating PCA-axes to C and N pools and microbial processes, respectively. Univariate statistical analyses were performed using Statistica 6, multivariate analyses were performed by Primer 6.
3. Results
3.1. C and N availability
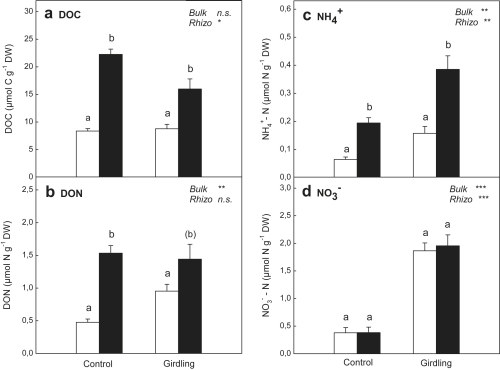
Concentrations of dissolved organic C and N were significantly higher in the rhizosphere than in bulk soil (Fig. 1a and b). Girdling significantly reduced DOC concentrations in the rhizosphere, whereas no effect of girdling on rhizospheric DON concentrations and a significant increase in DON concentrations in bulk soil was observed. Concentrations of NH4+ were significantly enhanced in rhizosphere soil compared to bulk soil (Fig. 1c), while no differences between rhizosphere and bulk soil were found for NO3− (Fig. 1d). Concentrations of inorganic N were markedly increased by girdling, especially concentrations of NO3−, suggesting a stronger effect of the girdling treatment on N than on C availability.
Fig. 1.
Concentrations of (a) DOC, (b) DON, (c) NH4+ and (d) NO3− in bulk soil (open bars) and rhizosphere soil (black bars) of control plots and girdling plots. Significant differences between rhizosphere and bulk soil are indicated by different letters (p < 0.05; letters in brackets indicate significance level of p < 0.1). Significance of differences between control plots and girdling plots for bulk soil and rhizosphere soil, respectively, is indicated by ∗∗∗ (p < 0.001), ∗∗ (p < 0.01), ∗ (p < 0.05) or n.s. (not significant). Error bars represent standard errors (n = 6).
3.2. Microbial processes
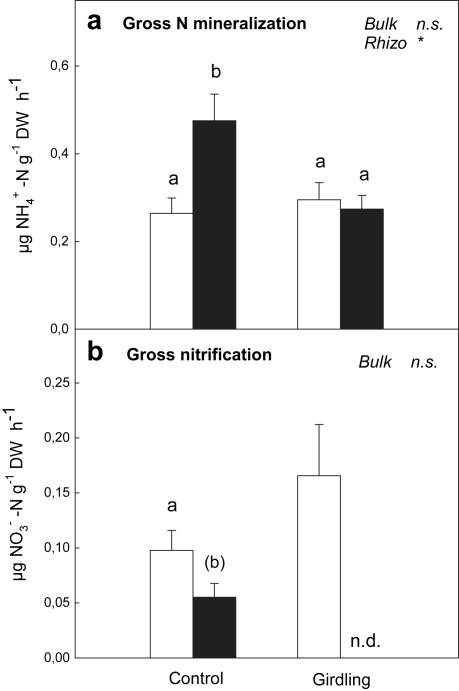
Gross N mineralization rates were significantly higher in the rhizosphere of control plots compared to bulk soil (Fig. 2a), while this rhizosphere effect was not observed in girdling plots. Gross nitrification rates, on the contrary, were lower in the rhizosphere compared to bulk soil (Fig. 2b). Nitrification accounted for only 10% of gross N mineralization in the rhizosphere, compared to 33% in bulk soil and 60% in bulk soil of girdling plots. Unfortunately, nitrification rates in the rhizosphere of girdling plots could not be measured due to problems during mass spectrometry.
Fig. 2.
(a) Gross N mineralization rates and (b) gross nitrification rates measured in bulk soil (open bars) and rhizosphere soil (black bars) of control plots and girdling plots. Significant differences between rhizosphere and bulk soil are indicated by different letters (p < 0.05; letters in brackets indicate significance level of p < 0.1). Significance of differences between control plots and girdling plots for bulk soil and rhizosphere soil, respectively, is indicated by ∗ (p < 0.05) or n.s. (not significant). Error bars represent standard errors (n = 6). n.d. means ‘not determined’.
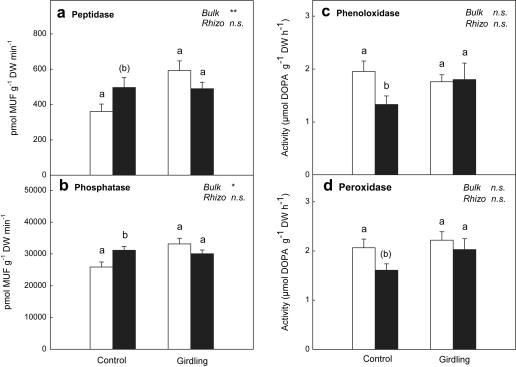
Extracellular enzyme activities separated into two groups: Peptidase and phosphatase activities (Fig. 3a and b) were increased in the rhizosphere of control plots and these enzyme activities were enhanced by the girdling treatment in bulk soil only. Oxidative enzyme activities (Fig. 3c and d), in contrast, were reduced in the rhizosphere compared to bulk soil in control but not in girdling plots.
Fig. 3.
Extracellular enzyme activities ((a) peptidase, (b) phosphatase, (c) phenoloxidase and (d) peroxidase) measured in bulk soil (open bars) and rhizosphere soil (black bars) of control plots and girdling plots. Significant differences between rhizosphere and bulk soil are indicated by different letters (p < 0.05; letters in brackets indicate significance level of p < 0.1). Significance of differences between control plots and girdling plots for bulk soil and rhizosphere soil, respectively, is indicated by ∗∗ (p < 0.01), ∗ (p < 0.05) or n.s. (not significant). Error bars represent standard errors (n = 6).
3.3. Microbial community composition
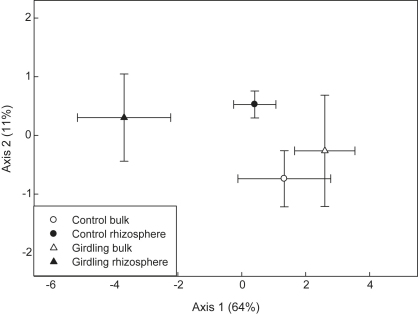
Surprisingly, a principal component analysis (PCA) and analysis of similarity (ANOSIM) of the relative abundances of phospholipid fatty acids (PLFAs) revealed that microbial community composition of the rhizosphere was not clearly distinct from bulk soil community composition in control plots (Fig. 4, Table 1). Girdling, however, lead to a marked shift in the rhizosphere community composition. A correlation of the results of the PCA with C and N pools and microbial processes exhibited that the first PCA-axis, separating the rhizosphere of girdling plots from the rhizosphere of control plots and from bulk soils was negatively correlated to NH4+ concentrations (p < 0.05). The second PCA-axis, mainly separating bulk soil from rhizosphere soil of control plots, showed a positive relationship to DOC and DON concentrations (p < 0.05), as well as correlations to oxidative enzyme activities and nitrification (p < 0.1).
Fig. 4.
Microbial community composition described by a principal component analysis of the relative abundances of PLFAs. Error bars represent standard errors (n = 6).
Table 1.
Results of ANOSIM describing differences between microbial communities by analysis of PLFA-abundances.
| R-value∗ | Significance Level % | |
|---|---|---|
| Control bulk–rhizo | 0.083 | 19.7 |
| Girdling bulk–rhizo | 0.445 | 1.9 |
| Bulk control–girdling | −0.084 | 81.7 |
| Rhizo control–girdling | 0.193 | 9.5 |
R-value indicates the degree of separation of two communities.
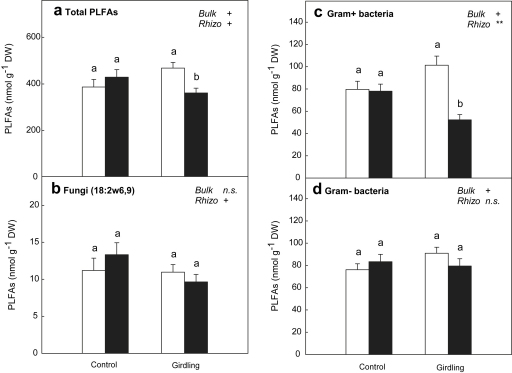
The results of the PCA are further supported by the absolute abundances of PLFAs specific for different microbial groups (Fig. 5b–d). No significant differences between rhizosphere and bulk soil of control plots were observed for any of the microbial groups. Girdling reduced fungal PLFAs in the rhizosphere (Fig. 5b). Marker fatty acids of both Gram-positive and Gram-negative bacteria were enhanced by girdling in bulk soil (Fig. 5c and d), while a significant decrease in rhizosphere abundance of Gram-positive bacteria was found. These patterns were reflected in the total abundance of PLFAs, used to estimate total microbial biomass (Fig. 5a), as we observed no significant differences between rhizosphere and bulk soil in control plots, but an increase in total PLFAs in bulk soil and a decrease in the rhizosphere by the girdling treatment.
Fig. 5.
Concentrations of (a) total PLFAs and marker PLFAs for (b) fungi, (c) Gram-positive bacteria and (d) Gram-negative bacteria in bulk soil (open bars) and rhizosphere soil (black bars) of control plots and girdling plots. Significant differences (p < 0.05) between rhizosphere and bulk soil are indicated by different letters. Significance of differences between control plots and girdling plots for bulk soil and rhizosphere soil, respectively, is indicated by ∗∗ (p < 0.01), +(p < 0.1) or n.s. (not significant). Error bars represent standard errors (n = 6).
Correlations between the abundances of selected group-specific PLFA–markers and microbial processes (Table 2) revealed that gross mineralization rates, as well as phosphatase and peptidase activities, which are all phylogenetically broad processes, i.e. processes related to a wide range of microbial taxa, were positively correlated to the abundance of marker fatty acids of various microbial groups and also to ubiquitous fatty acids. Gross nitrification rates and oxidative enzyme activities, on the contrary, showed mainly negative correlations to both bacterial and fungal marker fatty acids.
Table 2.
Correlations between microbial processes and abundances of selected marker PLFAs.
| PLFA-Markers | N-Mineral. | Nitrific. | Phosph. | Peptidase | Phenox. | Perox. | |
|---|---|---|---|---|---|---|---|
| Gram + bacteria | a15:0 | 0.05 | 0 | −0.11 | 0.09 | 0.26 | −0.14 |
| i16:0 | 0.47∗ | 0.1 | −0.02 | 0.08 | 0.27 | 0 | |
| i17:0 | 0.32 | 0.24 | 0.27 | 0.31 | 0.11 | 0.16 | |
| a17:0 | 0.11 | −0.46+ | 0.38+ | 0.43∗ | −0.57∗∗ | −0.5∗ | |
| Gram − bacteria | cy17:0 | 0.25 | −0.24 | 0.33 | 0.32 | −0.33 | −0.28 |
| cy18:0 | −0.19 | 0.31 | 0.34 | 0.58∗∗ | −0.34 | −0.2 | |
| 18:1ω7 | 0.17 | −0.5+ | 0.33 | 0.3 | −0.44∗ | −0.39+ | |
| 18:1ω5 | 0.39+ | −0.75∗∗∗ | 0.26 | 0.23 | −0.68∗∗∗ | −0.7∗∗∗ | |
| General bacteria | 15:0 | 0.38+ | −0.08 | −0.19 | 0.07 | 0.34 | −0.11 |
| 17:0 | 0.39+ | −0.26 | 0.37+ | 0.41+ | −0.45∗ | −0.35 | |
| Not specified | 16:1ω6 | 0.15 | −0.26 | 0.47∗ | 0.43∗ | −0.32 | −0.31 |
| i17:1ω8 | 0.2 | 0.14 | 0.67∗∗∗ | 0.26 | 0.12 | 0.24 | |
| Fungi | 18:2ω6,9 | 0.58∗∗ | −0.58∗ | 0.17 | 0.09 | −0.5∗ | −0.54∗∗ |
| 18:1ω9c | 0.41+ | −0.6∗ | 0.35 | 0.31 | −0.48∗ | −0.46∗ | |
| Ubiquitous | 16:0 | 0.52∗ | −0.29 | 0.34 | 0.41+ | −0.28 | −0.27 |
| 18:0 | 0.47∗ | −0.3 | 0.3 | 0.42+ | −0.22 | −0.3 | |
Presented are correlation coefficients. Significance levels are indicated by ∗∗∗ (p < 0.001), ∗∗ (p < 0.01), ∗ (p < 0.05), +(p < 0.1); n = 6.
4. Discussion
Despite the relatively small percentage that the rhizosphere occupies of the total soil volume, rhizosphere processes may be more important than bulk soil processes for soil functioning and may play a vital role in ecosystem nutrient cycling (Badalucco and Nannipieri, 2007). In this study we investigated microbial processes in the rhizosphere of European beech. We expected that differences in resource availability between rhizosphere and bulk soil would result in distinct microbial processes and community structure, and that these differences would be reduced by an interruption of belowground carbon allocation.
The rhizosphere soil was characterized by markedly higher C and N availability compared to bulk soil. Girdling significantly decreased concentrations of DOC in the rhizosphere (Fig. 1a). Decreased concentrations of DOC were also found in previous girdling experiments in bulk soils (Kaiser et al., 2010b; Weintraub et al., 2007). Although DOC concentrations in the rhizosphere were reduced in girdled plots, they were still higher than concentrations in the bulk soil, maybe due to the fact that DOC may also originate from degradation of SOM (Ekberg et al., 2007; Kalbitz et al., 2000). It should be kept in mind, however, that our data do not allow us to distinguish between enhanced production and decreased uptake of DOC or DON, for example by plant roots or microbes. The significant increase in concentration of DON in bulk soil of girdling plots corresponds to increased peptidase activities, indicating that the measured potential enzyme activities can be linked to the concentration of enzymatic products.
Concentrations of NH4+ were significantly enhanced in the rhizosphere, while no differences were found for NO3−, which is most likely due to the high mobility of NO3− in soil (Fig. 1c and d). The marked increase in concentrations of inorganic N by the girdling treatment, which has already been observed in a previous experiment (Kaiser et al., 2010b) is probably an effect of reduced plant and mycorrhizal N uptake, since uptake of nutrients is an energy requiring process and root C reserves were exhausted by the girdling treatment (data not shown).
High resource availability in the rhizosphere was reflected in the pattern of microbial processes: N mineralization rates were strongly increased in the rhizosphere (Fig. 2a), as already reported for rhizosphere soils of trees (Norton and Firestone, 1996). The fact that the marked difference in N mineralization rates between rhizosphere soil and bulk soil of control plots disappeared in the girdling treatment, clearly points to microbial activation by supply of labile C (“rhizosphere priming”) as suggested previously (Blagodatskaya and Kuzyakov, 2008; De Nobili et al., 2001; Weintraub et al., 2007).
Nitrification rates, on the contrary, tended to be lower in the rhizosphere (Fig. 2b). This could be interpreted as an effect of competition between nitrifiers and plant roots for NH4+. However, as nitrification rates were measured in incubated soils after separation from plant roots, it is also possible that low nitrification rates are the result of the dominance of fast growing heterotrophic bacteria in the rhizosphere.
Regarding extracellular enzyme activities, we found that phosphatase and peptidase activities, both phylogenetically broad processes, were generally enhanced in the rhizosphere (Fig. 3a and b). Enhanced phosphatase activities but no change in peptidase activities after the addition of low-molecular-weight organic compounds by a type of artificial root have been reported previously (Renella et al., 2007). Phosphatases and peptidases produced by mycorrhizal fungi probably contribute to enhanced enzyme activities in the rhizosphere, but the pattern of these enzyme activities also reflects the general abundance of bacteria (e.g. the increase in the bulk soil of the girdling treatment).
In contrast, oxidative enzyme activities involved in the degradation of lignin and soil organic matter (phenoloxidase and peroxidase) were lower in the rhizosphere than in bulk soil (Fig. 3c and d). This may be explained by the fact that oxidative enzymes are thought to be produced by slow growing K-strategists, mainly saprotrophic fungi and some specialized groups of bacteria (Buee et al., 2009), which have a competitive disadvantage against mycorrhizal fungi and fast growing copiotrophic bacteria using easily assimilable plant-derived C sources. The reduction in potential oxidative enzyme activities in the rhizosphere compared to bulk soil was not observed in the girdling plots. Our findings hence do not suggest an increase in decomposition of humified SOM by plant exudates (“priming”), as has been reported by others (Bottner et al., 1999; Ekberg et al., 2007; Kuzyakov, 2002). However, although plant exudates have led to reduced activities of oxidative enzymes, they have also accelerated the activities of proteolytic enzymes in our study, and thus may have enhanced the degradation of fresh SOM. Similar results have also been reported recently for cellulytic and other hydrolytic enzymes (Kaiser et al., 2010b). High levels of inorganic N in the girdling plots apparently did not affect the production of oxidative enzymes, contrary to results from a study on the effects of high NO3−-availability on oxidative enzyme activities in forest soils (Waldrop and Zak, 2006), which reported decreased production of phenoloxidases by white-rot fungi but increased production by bacteria or ascomycetes. The strong negative correlation of oxidative enzyme activities to the abundance of fungal PLFAs seems surprising at first glance (Table 2), but is probably caused by the fact that these biomarkers occur in both mycorrhizal and saprotrophic fungi.
While our results revealed clear rhizosphere effects on microbial processes, microbial community composition in the rhizosphere differed only slightly from bulk soil in controls (ANOSIM R = 0.083, p = 0.19). The girdling treatment, however, caused a markedly altered community structure at the level of resolution of PLFAs (Fig. 4). Only small rhizosphere effects on microbial community composition in mineral soils have already been reported for grass and tree rhizospheres (Butler et al., 2003; Priha et al., 1999b). We observed no significant differences in the abundance of Gram-positive and Gram-negative bacteria, although the relative abundance of Gram-negative bacteria in the rhizosphere tended to be slightly higher than the relative abundance of Gram-positive bacteria, while in bulk soil the opposite was the case (data not shown). This is consistent with the suggestion that Gram-negative bacteria are more frequent in the rhizosphere, preferably growing on plant labile C, while Gram-positive bacteria may be dominant in bulk soil (Bird et al., in press; Paterson et al., 2007; Treonis et al., 2004). The marked decrease in the abundance of Gram-positive bacteria in rhizosphere soils of girdling plots, which is mainly responsible for the strong shift in community composition, is therefore difficult to explain (Fig. 5c). Since the reduction in labile C supply is unlikely to be the cause, other effects of the girdling treatment on rhizospheric conditions, such as alterations in oxygen supply, pH and redox potential in consequence of reduced root respiration or uptake of nutrients by plants may be possible explanations. Data on fine root biomass (Kaiser et al., 2010b) do not point to increased root mortality or membrane damage of root cells four months after girdling, so these factors can be disregarded. The increase in the abundance of total PLFAs in the bulk soil of the girdling treatment (Fig. 5a), which is also supported by measurements of microbial biomass by the chloroform-fumigation-extraction method (data not shown), may be an effect of the enhanced supply of inorganic N. However, this is likely just a transient effect, since previous results (Kaiser et al., 2010b) showed a reduction of microbial biomass by the girdling treatment throughout most of the seasons.
We found a 28% reduction of the fungal biomarker 18:2ω6,9 in the rhizosphere by girdling as expected (Fig. 5b), but we did not find a reduction of this biomarker in the bulk soil as described in other girdling studies (Högberg et al., 2007), maybe due to an increase in saprotrophic fungi or due to seasonal aspects. The abundance of this fungal biomarker was also not significantly enhanced in the rhizosphere compared to the bulk soil.
Total microbial biomass estimated by total PLFAs was not significantly higher in the rhizosphere (Fig. 5a), although rhizosphere soils are generally considered to be enriched in microbial biomass compared to bulk soil (Cheng and Coleman, 1990; Norton and Firestone, 1991). Results from another study, however, also showed enhanced microbial biomass C only in rhizospheres of spruce, but not of pine and birch (Priha et al., 1999a). Our data may point to a higher microbial activity (activation state) in the rhizosphere at a similar biomass, maybe because of a high availability of easily assimilable carbon and nutrients.
By using a novel approach, i.e by combining the analysis of resource availability, microbial processes and community structure in rhizosphere soil with a tree girdling experiment, we were able to demonstrate that N mineralization rates and hydrolytic enzyme activities were enhanced in the rhizosphere by root exudates, probably an effect of microbial activation by increased resource availability, while activities of oxidative enzymes (involved in the degradation of humified soil organic matter and lignin) were reduced. Thus, we did not find indications of a positive priming effect of root exudates on the degradation of recalcitrant SOM in this study, but rather the opposite effect. Differences in oxidative enzyme activities and nitrification rates between rhizosphere soil and bulk soil suggest considerable differences in the functional microbial community composition, although rhizosphere and bulk soil communities differed only slightly at the level of resolution that we used here (PLFAs). The reduction of root exudations by girdling, on the other hand, led to strong changes of the microbial community in the rhizosphere.
Acknowledgements
We are grateful to Elisabeth Kogler, Sonja Leitner and Ralph Steingruber for valuable help in the field and to the Austrian Science Fund (FWF, P18495-B03) for financial support.
References
- Badalucco L., Nannipieri P. Nutrient transformations in the rhizosphere. In: Pinton R., Varanini Z., Nannipieri P., editors. The rhizosphere; Boca Raton: 2007. pp. 111–133. [Google Scholar]
- Bird, J.A., Herman, D.J., Firestone, M.K. Rhizosphere priming of soil organic matter by bacterial groups in a grassland soil. Soil Biology & Biochemistry, in press. doi:10.1016/j.soilbio.2010.08.010.
- Blagodatskaya E., Kuzyakov Y. Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: critical review. Biology and Fertility of Soils. 2008;45:115–131. [Google Scholar]
- Bottner P., Pansu M., Sallih Z. Modelling the effect of active roots on soil organic matter turnover. Plant and Soil. 1999;216:15–25. [Google Scholar]
- Buee M., De Boer W., Martin F., van Overbeek L., Jurkevitch E. The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant and Soil. 2009;321:189–212. [Google Scholar]
- Butler J.L., Bottomley P.J., Griffith S.M., Myrold D.D. Distribution and turnover of recently fixed photosynthate in ryegrass rhizospheres. Soil Biology & Biochemistry. 2004;36:371–382. [Google Scholar]
- Butler J.L., Williams M.A., Bottomley P.J., Myrold D.D. Microbial community dynamics associated with rhizosphere carbon flow. Applied and Environmental Microbiology. 2003;69:6793–6800. doi: 10.1128/AEM.69.11.6793-6800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W.X., Coleman D.C. Effect of living roots on soil organic-matter decomposition. Soil Biology & Biochemistry. 1990;22:781–787. [Google Scholar]
- De Nobili M., Contin M., Mondini C., Brookes P.C. Soil microbial biomass is triggered into activity by trace amounts of substrate. Soil Biology & Biochemistry. 2001;33:1163–1170. [Google Scholar]
- Dennis P.G., Miller A.J., Hirsch P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiology Ecology. 2010;72:313–327. doi: 10.1111/j.1574-6941.2010.00860.x. [DOI] [PubMed] [Google Scholar]
- Ekberg A., Buchmann N., Gleixner G. Rhizospheric influence on soil respiration and decomposition in a temperate Norway spruce stand. Soil Biology & Biochemistry. 2007;39:2103–2110. [Google Scholar]
- Fontaine S., Mariotti A., Abbadie L. The priming effect of organic matter: a question of microbial competition? Soil Biology & Biochemistry. 2003;35:837–843. [Google Scholar]
- Frostegard A., Tunlid A., Baath E. Microbial biomass measured as total lipid phosphate in soils of different organic content. Journal of Microbiological Methods. 1991;14:151–163. [Google Scholar]
- Grayston S.J., Vaughan D., Jones D. Rhizosphere carbon flow in trees, in comparison with annual plants: the importance of root exudation and its impact on microbial activity and nutrient availability. Applied Soil Ecology. 1997;5:29–56. [Google Scholar]
- Hinsinger P., Bengough A.G., Vetterlein D., Young I.M. Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant and Soil. 2009;321:117–152. [Google Scholar]
- Högberg M.N., Högberg P., Myrold D.D. Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia. 2007;150:590–601. doi: 10.1007/s00442-006-0562-5. [DOI] [PubMed] [Google Scholar]
- Högberg P., Nordgren A., Buchmann N., Taylor A.F.S., Ekblad A., Högberg M.N., Nyberg G., Ottosson-Lofvenius M., Read D.J. Large-scale forest girdling shows that current photosynthesis drives soil respiration. Nature. 2001;411:789–792. doi: 10.1038/35081058. [DOI] [PubMed] [Google Scholar]
- Jones D.L., Nguyen C., Finlay R.D. Carbon flow in the rhizosphere: carbon trading at the soil-root interface. Plant and Soil. 2009;321:5–33. [Google Scholar]
- Kaiser C., Frank A., Wild B., Koranda M., Richter A. Negligible contribution from roots to soil-borne phospholipid fatty acid biomarkers 18:2w6,9 and 18:1w9. Soil Biology & Biochemistry. 2010;42:1650–1652. doi: 10.1016/j.soilbio.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Koranda M., Kitzler B., Fuchslueger L., Schnecker J., Schweiger P., Rasche F., Zechmeister-Boltenstern S., Sessitsch A., Richter A. Belowground carbon allocation by trees drive seasonal pattern of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytologist. 2010;187:843–858. doi: 10.1111/j.1469-8137.2010.03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Meyer H., Biasi C., Rusalimova O., Barsukov P., Richter A. Storage and mineralization of carbon and nitrogen in soils of a frost-boil tundra ecosystem in Siberia. Applied Soil Ecology. 2005;29:173–183. [Google Scholar]
- Kalbitz K., Solinger S., Park J.H., Michalzik B., Matzner E. Controls on the dynamics of dissolved organic matter in soils: a review. Soil Science. 2000;165:277–304. [Google Scholar]
- Kuzyakov Y. Review: factors affecting rhizosphere priming effects. Journal of Plant Nutrition and Soil Science-Zeitschrift für Pflanzenernährung und Bodenkunde. 2002;165:382–396. [Google Scholar]
- Kuzyakov Y. Priming effects: interactions between living and dead organic matter. Soil Biology & Biochemistry. 2010;42:1363–1371. [Google Scholar]
- Lambers H., Mougel C., Jaillard B., Hinsinger P. Plant-microbe–soil interactions in the rhizosphere: an evolutionary perspective. Plant and Soil. 2009;321:83–115. [Google Scholar]
- Leckie S.E. Methods of microbial community profiling and their application to forest soils. Forest Ecology and Management. 2005;220:88–106. [Google Scholar]
- Marilley L., Aragno M. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Applied Soil Ecology. 1999;13:127–136. [Google Scholar]
- Marx M.C., Wood M., Jarvis S.C. A microplate fluorimetric assay for the study of enzyme diversity in soils. Soil Biology & Biochemistry. 2001;33:1633–1640. [Google Scholar]
- Myrold D.D., Tiedje J.M. Simultaneous estimation of several nitrogen-cycle rates using N-15-Theory and application. Soil Biology & Biochemistry. 1986;18:559–568. [Google Scholar]
- Norton J.M., Firestone M.K. Metabolic status of bacteria and fungi in the rhizosphere of Ponderosa pine-seedlings. Applied and Environmental Microbiology. 1991;57:1161–1167. doi: 10.1128/aem.57.4.1161-1167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton J.M., Firestone M.K. N dynamics in the rhizosphere of Pinus ponderosa seedlings. Soil Biology & Biochemistry. 1996;28:351–362. [Google Scholar]
- Paterson E., Gebbing T., Abel C., Sim A., Telfer G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytologist. 2007;173:600–610. doi: 10.1111/j.1469-8137.2006.01931.x. [DOI] [PubMed] [Google Scholar]
- Pinton R., Varanini Z., Nannipieri P. The rhizosphere; Boca Raton: 2007. [Google Scholar]
- Priha O., Grayston S.J., Pennanen T., Smolander A. Microbial activities related to C and N cycling and microbial community structure in the rhizospheres of Pinus sylvestris, Picea abies and Betula pendula seedlings in an organic and mineral soil. FEMS Microbiology Ecology. 1999;30:187–199. doi: 10.1111/j.1574-6941.1999.tb00647.x. [DOI] [PubMed] [Google Scholar]
- Priha O., Hallantie T., Smolander A. Comparing microbial biomass, denitrification enzyme activity, and numbers of nitrifiers in the rhizospheres of Pinus sylvestris, Picea abies and Betula pendula seedlings by microscale methods. Biology and Fertility of Soils. 1999;30:14–19. [Google Scholar]
- Renella G., Landi L., Valori F., Nannipieri P. Microbial and hydrolase activity after release of low molecular weight organic compounds by a model root surface in a clayey and a sandy soil. Applied Soil Ecology. 2007;36:124–129. [Google Scholar]
- Saiya-Cork K.R., Sinsabaugh R.L., Zak D.R. The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biology & Biochemistry. 2002;34:1309–1315. [Google Scholar]
- Sinsabaugh R., Klug M., Collins H., Yeager P., Petersen S. Characterizing soil microbial communities. In: Robertson G., Coleman D., Bledsoe C., Sollins P., editors. Standard Soil Methods for Long-Term Ecological Research. 1999. New York. [Google Scholar]
- Stange C.F., Spott O., Apelt B., Russow R.W.B. Automated and rapid online determination of N-15 abundance and concentration of ammonium, nitrite, or nitrate in aqueous samples by the SPINMAS technique. Isotopes. Environmental and Health Studies. 2007;43:227–236. doi: 10.1080/10256010701550658. [DOI] [PubMed] [Google Scholar]
- Steer J., Harris J.A. Shifts in the microbial community in rhizosphere and non-rhizosphere soils during the growth of Agrostis stolonifera. Soil Biology & Biochemistry. 2000;32:869–878. [Google Scholar]
- Treonis A.M., Ostle N.J., Stott A.W., Primrose R., Grayston S.J., Ineson P. Identification of groups of metabolically-active rhizosphere microorganisms by stable isotope probing of PLFAs. Soil Biology & Biochemistry. 2004;36:533–537. [Google Scholar]
- Uren N.C. In: Types, amounts and possible functions of compounds released into the rhizosphere by soil-grown plants. Pinton R., Varanini Z., Nannipieri P., editors. The rhizosphere; Boca Raton: 2007. pp. 1–21. [Google Scholar]
- Waldrop M.P., Zak D.R. Response of oxidative enzyme activities to nitrogen deposition affects soil concentrations of dissolved organic carbon. Ecosystems. 2006;9:921–933. [Google Scholar]
- Wang J.G., Bakken L.R. Competition for nitrogen during decomposition of plant residues in soil: effect of spatial placement of N-rich and N-poor plant residues. Soil Biology & Biochemistry. 1997;29:153–162. [Google Scholar]
- Weintraub M.N., Scott-Denton L.E., Schmidt S.K., Monson R.K. The effects of tree rhizodeposition on soil exoenzyme activity, dissolved organic carbon, and nutrient availability in a subalpine forest ecosystem. Oecologia. 2007;154:327–338. doi: 10.1007/s00442-007-0804-1. [DOI] [PubMed] [Google Scholar]
- Whipps J.M. In: Carbon Economy. Lynch J.M., editor. The rhizosphere; Chichester: 1990. pp. 59–97. [Google Scholar]
- Zelles L. Phospholipid fatty acid profiles in selected members of soil microbial communities. Chemosphere. 1997;35:275–294. doi: 10.1016/s0045-6535(97)00155-0. [DOI] [PubMed] [Google Scholar]
- Zelles L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biology and Fertility of Soils. 1998;29:111–129. [Google Scholar]