Abstract
The comparison of eight tools applicable to ligand-binding site prediction is presented. The methods examined cover three types of approaches: the geometrical (CASTp, PASS, Pocket-Finder), the physicochemical (Q-SiteFinder, FOD) and the knowledge-based (ConSurf, SuMo, WebFEATURE). The accuracy of predictions was measured in reference to the catalytic residues documented in the Catalytic Site Atlas. The test was performed on a set comprising selected chains of hydrolases. The results were analysed with regard to size, polarity, secondary structure, accessible solvent area of predicted sites as well as parameters commonly used in machine learning (F-measure, MCC). The relative accuracies of predictions are presented in the ROC space, allowing determination of the optimal methods by means of the ROC convex hull. Additionally the minimum expected cost analysis was performed. Both advantages and disadvantages of the eight methods are presented. Characterization of protein chains in respect to the level of difficulty in the active site prediction is introduced. The main reasons for failures are discussed. Overall, the best performance offers SuMo followed by FOD, while Pocket-Finder is the best method among the geometrical approaches.
Electronic supplementary material
The online version of this article (doi:10.1007/s10822-010-9402-0) contains supplementary material, which is available to authorized users.
Keywords: Active site, Hydrolase, Ligand-binding site prediction, Receiver operating characteristic
Introduction
Understanding of how biological systems function is the salient motivation for the research in the field of biochemistry and molecular biology. The most comprehensive approach that aims at gaining insight into molecular function and mechanism of thousands of proteins relies on structural genomics initiatives [1–3]. One of the major challenges in structural genomics is identifying the function and evaluating the functional integrity of proteins [4]. Another goal, justifying the huge investments already made in structural genomics initiatives, is the ability to predict druggability of a particular protein based solely on its 3D structure [5, 6]. Accordingly, experimental as well as computational methods for identifying and characterizing ligand-binding sites on protein targets are being intensively developed nowadays [7, 8]. Herein we focus on in silico methods, as promising tools for finding and annotating functional sites in novel structures from structural genomics.
The strategies for prediction of ligand-binding sites that have already been developed, can be roughly divided into three groups. Methods that are tailored to detect pockets and clefts on the basis of pure geometric criteria such as POCKET [9], SurfNet [10], APROPOS [11], CAST [12, 13], LIGSITE [14] or PASS [15] constitute the first group. The methods in the second group, in addition to structural data use biophysical and/or chemical properties, such as pKa [16], electrostatic energy [17], solvent mapping [7, 18, 19], physical potential [20, 21], favourable regions for van der Waals probes on the protein surface [22] or hydrophobicity deficiency [23]. The third group of methods relies on knowledge derived from biochemical data and different types of databases. Many of them search for clusters or patterns of conserved residues [24–33], and therefore may be applicable to proteins that have homologues. There is also a series of tools that exploit various pattern matching approaches. They generally search for local structural similarity of a protein structure to known functional sites [34–48]. The main limitation of these methods is a finite set of functional sites they can identify, and therefore they are not suited to annotate new functional motifs that may be present in novel folds. Moreover, there are many methods that rely on statistical approaches [49, 50] and machine learning techniques based on neural networks [51–53], support vector machines [54–56] or Naive Bayes classifications [57]. They exploit a wealth of knowledge included in a training set, and aim at predicting specific functional roles of residues rather than broadly defined ligand-binding sites. Apart from the methods devoted to prediction of functional sites, alternative approaches such as molecular dynamics simulations [58] or docking [59–61] were successfully employed to identify ligand-binding sites. A thorough review of strategies for ligand-binding site detection is presented elsewhere [62].
Here we present the extensive comparison of eight methods designed for ligand-binding site prediction in order to reveal limitations that should be overcome in the future. The tools examined cover all three groups of approaches briefly described above. As representatives of the geometry-based approaches CASTp [13, 63], Pocket-Finder (an implementation of LIGSITE [14]) and PASS [15] were chosen. From the second group Q-SiteFinder [22] and FOD [23] were selected. The knowledge-based methods are represented by ConSurf [64], SuMo [45, 46] and WebFEATURE [49, 65, 66]. Short description of each of the eight methods is presented in Table 1. All the methods require protein’s 3D structure to make predictions, but only ConSurf exploits its sequence by means of calculation of an evolutionary conservation. Moreover all of them are freely available as web services or standalone executables and exhibit relatively short time of calculations. The results returned are straightforward, mainly in the form of a list of atoms/residues predicted as binding site(s). The exception is FOD and PASS, but their primary results can be easily transformed into the aforementioned list. Predictability of the methods was tested on the single chains of hydrolases, one of the best studied class of enzymes. The selection of this set has two reasons. Firstly, active sites of enzymes are the best characterized group of binding sites, even though many ligand-binding site databases exist [67–72]. Secondly, this class of enzymes is very extensively studied and therefore much is known about their mechanisms of catalysis [73–77] and dynamics of its structures [78–80]. Moreover this class is structurally and functionally miscellaneous [81, 82]. The accuracy of predictions was measured in reference to the catalytic residues documented in the Catalytic Site Atlas (CSA) [83]. The choice of a reference set is dictated by its reliability, clarity and an easy access (CSA). Even though the tested methods are more suitable to detect ligand-binding sites, the evaluation based on catalytic sites is justified as active sites are located within binding sites and provided that results are taken with the awareness. The measure of accuracy was another aspect we have focused on. Therefore, to designate the best method, different measures and criteria were employed.
Table 1.
Short description of each of the eight methods applicable to ligand-binding site prediction
| Method | Description | Availability |
|---|---|---|
| ConSurf | Calculates an evolutionary coneservation scores and maps them on protein structures | http://consurf.tau.ac.il/ |
| CASTp | Locates and measures pockets and voids on 3D protein structures based on the alpha shape and the pocket algorithm | http://castp.engr.uic.edu/cast |
| FOD | Calculates hydrophobicity differences of idealized hydrophobicity modeled by 3D Gauss function and observed hydrophobicity modeled by function introduced by Levitt [108] | http://www.bioinformatics.cm-uj.krakow.pl/activesite a |
| PASS | Identifies buried volumes in protein structures based on the algorithm that coats the protein with probe spheres and iteratively selects probes with many atom contacts | Standalone executableb |
| Pocket-finder | Detects pockets on the surface of a protein based on a series of simple operations on a cubic grid in the search for protein-solvent-protein events | www.modelling.reeds.ac.uk/pocketfinder |
| Q-SiteFinder | Locates energetically favourable binding sites using interaction energy between a protein and a simple van der Waals probe | www.modelling.reeds.ac.uk/qsitefinder |
| SuMo | Detects 3D sites in proteins using representation of a protein structure by a set of stereochemical groups and heuristic for finding similarities that uses groups of triangles of these chemical groups | http://sumo-pbil.ibcp.fr |
| WebFEATURE | Scans query structures for functional sites using a supervised learning algorithm that creates and identifies 3D physicochemical motifs, and predefined statistical models of functional sites | http://feature.stanford.edu/webfeature |
Means of access are given as well
aAn upgraded version as standalone executable is available from authors
Materials and methods
Preparation of the test set
The research was limited to one particular enzyme class: hydrolases. The polypeptide chains were selected form the literature entries of CSA (version 2.2.9) [83]. Firstly, the Enzyme Commission (EC) numbers were found for the entries, using LinkDB method of KEGG API [84]. For the entries that failed to obtain an EC number, id mapping from the Protein Data Bank (PDB) to the UniProtKB (UP), available on the UniProt web page [85] was performed and LinkDB method was used again (UP to EC mapping). Concurrently information about EC numbers for all the entries was extracted from CSA and PDB. As a result all the entries with EC numbers denoting hydrolases were selected (325 entries). In order to avoid the redundancy of a data set, the selected sequences were clustered using BlastClust program [86], with a minimum length coverage equal to 1.0 and a similarity threshold equal to 80 for both sequences. The value of the latter is the lowest one ensuring that the sequences in one cluster have identical active sites according to CSA. The final set comprises 189 structures of single chains which have complete structural data (Supplementary material).
Ligand-binding site prediction
The computations were performed using standalone executables (FOD, PASS version 2.0.36), web services made in Ruby able to communicate with CASTp, ConSurf, Pocket-Finder, Q-SiteFinder, WebFEATURE and text queries offered by SuMo server. Additionally, the precalculated results were downloaded from ConSurfDB [87]. Only the highest ranked pockets/predictions were considered. The outputs of all the methods were transformed into a common format in the form of a list comprising the pointed residues. The results of PASS, which are in the form of coordinates of points filling pocket(s) (probe spheres) were transformed into residue numbers in two steps. Firstly, the probe spheres were clustered using hierarchical clustering with the single-linkage, the Euclidean distance and the distance cut-off equal to 2 Å. Next, the biggest cluster was selected and for each of its points the closest residue was determined (with the closest atom to the centre of the probe sphere). FOD produces the hydrophobicity differences (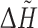 ) between the theoretical and the empirical hydrophobicities calculated for each residue in a chain. Simple preliminary tests revealed that residues with
) between the theoretical and the empirical hydrophobicities calculated for each residue in a chain. Simple preliminary tests revealed that residues with 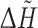 higher or equal to maximum
higher or equal to maximum 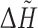 (for a chain), minus quarter of the
(for a chain), minus quarter of the 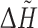 range (for a chain), are optimal predictions of the method. WebFEATURE results were interpreted using three z-score specificity cut-offs: 100, 99 and 95%. Similarly, ConSurf and ConSurfDB results were interpreted using three conservation grade cut-offs: 9, 8 and 7.
range (for a chain), are optimal predictions of the method. WebFEATURE results were interpreted using three z-score specificity cut-offs: 100, 99 and 95%. Similarly, ConSurf and ConSurfDB results were interpreted using three conservation grade cut-offs: 9, 8 and 7.
Measuring performance
We have considered a two-class prediction problem in which a method produces catalytic and non-catalytic residues. The results were compared with CSA as a golden standard. To assess the accuracy of prediction the following parameters were used: F-measure, MCC and points in the ROC space. Therefore, we consider the four possible outcomes: true positives (TPs, correctly classified catalytic residues), true negatives (TNs, correctly classified non-catalytic residues), false positives (FPs, non-catalytic residues incorrectly predicted as catalytic) and false negatives (FNs, catalytic residues incorrectly predicted as non-catalytic). The F-measure parameter is given by the equation:
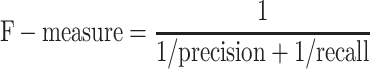 |
1 |
where
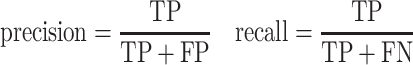 |
2 |
The formula for calculating MCC is shown in the equation:
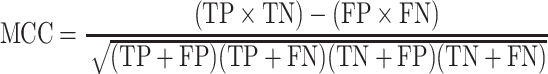 |
3 |
For each method a true positive rate (TPR) and a false positive rate (FPR) was calculated (Eq. 4) and plotted as a point in the ROC space.
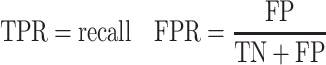 |
4 |
For the points in the ROC space, convex hull (ROCCH) was found. The ROCCH is the ‘north-west boundary’ of the points in the ROC space. This procedure allowed to determine the set of optimal methods [88]. The minimum expected cost analysis was performed. Therefore the slopes of the ROCCH segments were calculated, and the boundaries of minimum expected cost were assigned to the methods from ROCCH.
The accuracy of predictions was measured assuming two perspectives. First, we checked whether the predicted residues are in accordance with the catalytic residues from CSA, and we have called it the amino acid (AA) perspective. Since the analysed methods do not necessarily predict active sites but rather binding sites we decided to extend, in a specific way, the reference set to residues within the catalytic spheres [52]. Each active site has been assigned a sphere with its centre at the centroid of all the Cβ atoms of catalytic residues (Cα for glycine) and radius such that it contains all the Cβ atoms. Chains with one catalytic residue were set a radius of 3 Å. All the predicted residues which lie within an active site sphere were considered TPs, but residues not predicted and not catalytic lying within a sphere were still TNs. The second approach we have called the sphere overlapping (SO) perspective.
Residue analysis
The sets of residues were analysed according to polarity, secondary structure and relative solvent accessibility (RSA). In respect to polarity, following [89] we distinguished three groups of residues: charged (H, R, K, E, D), polar (Q, T, S, N, C, Y, W) and hydrophobic (G, A, V, L, I, M, P, F). Secondary structure was assigned using DSSP program [90], and we discriminated helices (H, G, I), β-strands (E) and coil regions (not helices and β-strands). Solvent accessibility was calculated using NACCESS program (an implementation of Lee and Richards method [91]) with a probe radius equal to 1.4 Å.
Results
Description of the test set
The test set contains 189 chains of hydrolases related to 184 entries of PDB [92]. As maintained by NC-IUBMB [93], it comprises 9 out of 13 subclasses of hydrolases. The highest number of representatives have hydrolases acting on ester bonds (EC 3.1), glycosylases (EC 3.2) and peptidases (EC 3.4). These three subclasses cover 78% of all selected chains.
The shortest chain has 79 amino acids, while the longest has 1023. The length of the majority of chains range from 100 to 400 amino acids. According to CATH structural classification [94], 184 chains have 284 domains assigned, however only 212 domains contain a catalytic residue. Majority of the latter belong to the α/β class (151 domains, 71%). The mainly alpha and the mainly beta classes constitute 12 and 16% of the catalytic domains, respectively. There is only one catalytic domain which belongs to the ‘few secondary structures’ class. The dominance of the α/β structures is in accordance with the distribution observed across all enzyme classes [89]. Nevertheless, the mainly alpha class is slightly under-represented in favour of the α/β class compared with all classes [89].
Almost all hydrolases in the test set have catalytic residues contained within just one subunit, with only 7 out of 184 enzymes having catalytic residues in at least two different subunits. Furthermore, as stated by PQS [95], 80 hydrolases are monomers, while the other are multimers. The research is focused on the single chains in order to ascertain an association between the performance and the quaternary structure.
Comparison of the predictions with the catalytic sites
Five out of the eight methods produced results for all chains. CASTp, FOD, PASS, Q-SiteFinder and SuMo constitute this group. Other methods gave fewer yields. Pocket-Finder failed in the case of 3 chains due to the large number of atoms forming these structures. ConSurf had problems with 5 and 23 chains depending on whether the precalculated database (ConsurfDB) or default parameters were used. In turn WebFEATURE gave no results for 1 chain using 95 and 99% specificity z-score cut-offs, and failed for 150 with this parameter set to 100% (WebFEATURE100). Therefore all statistics calculated for the predicted sites refer to the chains for which a method produced results by any means.
Size
Approximate evaluation of the predictions was made by means of comparison between the number of predicted and catalytic residues. The latter constitute only 1% of all residues in the test set, while the former variates depending on the method, and except WebFEATURE100 it is always higher than 1% (Table 2). Consequently, considering only the number of predicted residues it may be assumed that CASTp, FOD, PASS, Pocket-Finder and Q-SiteFinder tend to reveal binding sites rather than catalytic residues (Table 2). Conversely, SuMo and WebFEATURE (especially with z-score cut-off equal to 100) search for catalytic residues. ConSurf produces the largest set of residues regardless of the assumed conservation grade cut-off. Moreover, the smallest fraction of the predicted residues is for the highest possible cut-off (9) and equals 13.9 or 12.8% depending on the source (Table 2). The lower cut-offs (8, 7) produce the fraction of predicted residues above 20% (Data not shown). Therefore the latter predictions due to the small amount of information about catalytic residues were excluded from further analysis that takes into account polarity, secondary structure and solvent accessibility.
Table 2.
Statistics for catalytic and non-catalytic residues documented in CSA as well as residues predicted by the eight methods (WebFEATURE has three variants)
| Residue type | Secondary structure | Total | Total (%) | |||||
|---|---|---|---|---|---|---|---|---|
| Polar | Charged | Hydrophobic | Helix | β-strand | Coil | |||
| Non-catalytic | 28.7 | 23.3 | 48.0 | 33.4 | 21.4 | 45.3 | 60918 | 99 |
| Catalytic | 27.0 | 64.1 | 9.0 | 23.0 | 19.3 | 57.7 | 601 | 1.0 |
| CASTp | 32.1 | 26.4 | 41.5 | 26.7 | 19.3 | 54.0 | 5625 | 9.1 |
| ConSurfDB9 | 28.2 | 24.4 | 47.4 | 27.4 | 23.1 | 49.5 | 7651 | 12.8 |
| FOD | 32.4 | 40.5 | 27.2 | 29.0 | 30.5 | 40.6 | 4076 | 6.6 |
| PASS | 32.3 | 31.2 | 36.5 | 28.6 | 17.5 | 53.9 | 3024 | 4.9 |
| Pocket-Finder | 31.4 | 25.3 | 43.3 | 28.3 | 19.3 | 52.4 | 4841 | 8.3 |
| Q-SiteFinder | 30.9 | 26.6 | 42.5 | 26.7 | 19.8 | 53.5 | 3956 | 6.4 |
| SuMo | 38.6 | 43.5 | 17.9 | 22.6 | 18.9 | 58.5 | 1222 | 2.0 |
| WebFEATURE100 | 38.3 | 61.7 | 0 | 36.2 | 19.1 | 44.7 | 47 | 0.1 |
| WebFEATURE99 | 33.7 | 66.3 | 0 | 30.9 | 19.0 | 50.2 | 2161 | 3.5 |
| WebFEATURE95 | 29.6 | 73.1 | 0 | 30.3 | 17.9 | 51.8 | 4530 | 7.4 |
Residue types concerning polarity (polar, charged, hydrophobic) and secondary structure (helix, β-strand, coil) are taken into account. The total number of predicted residues and the percentage (in reference to the residues of chains for which a method gave any results) are given. Statistically significant similarity to the catalytic set is given in bold (proportion test, α = 0.05)
The size of predicted sites was expressed as a radius (R) of a sphere containing all its Cβ atoms (Cα for glycine). In respect of the median of R, denoted as μ 1/2(R), all the methods except WebFEATURE100 predicted bigger sites than catalytic ones which have μ 1/2(R) equal to 6.2 Å (Table 3). Moreover ConSurf, WebFEATURE99 and 95 have even μ 1/2(R) over 20 Å. These values are high, taking into account that μ 1/2(R) calculated for the whole chains is equal to 32 Å. Therefore such high values may indicate that these programs produce large sites or a prediction relates to more than one site. The argument for the latter is that ConSurf and WebFEATURE, contrary to other programs, indicate residues not necessarily corresponding to one site and may be distributed across whole protein structure. Indeed, the maximum of R for WebFEATURE99 or 95 and ConSurf is over 80 and 50 Å, respectively, while the maximum of R taking into account all residues is below 90 Å. Concurrently, relatively high maximum of R in the case of CASTp, shows that this program identifies large pockets (only the biggest pocket was considered).
Table 3.
Minimum, median and maximum radii of spheres that contain the catalytic residues and the predicted by the eight methods
| Radius, R (Å) | |||
|---|---|---|---|
| Minimum | Median | Maximum | |
| CSA | 3.0 | 6.2 | 12.57 |
| CASTp | 4.64 | 13.01 | 46.07 |
| ConSurfDB9 | 10.12 | 24.15 | 59.01 |
| FOD | 3.67 | 14.13 | 27.88 |
| PASS | 5.70 | 11.06 | 30.17 |
| Pocket-Finder | 4.53 | 12.27 | 35.99 |
| Q-SiteFinder | 6.68 | 11.45 | 20.43 |
| SuMo | 3.00 | 7.43 | 31.16 |
| WebFEATURE100 | 3.00 | 3.00 | 22.87 |
| WebFEATURE99 | 3.00 | 25.74 | 86.16 |
| WebFEATURE95 | 3.00 | 23.11 | 83.70 |
The analysis of the minimum of R reveals that WebFEATURE (99 or 95) may produce very small sites (single residue), while the smallest site predicted by ConSurfDB9 is bigger than the average catalytic site from the test set.
Polarity, secondary structure and solvent accessibility
In order to assess whether a prediction is likely to be correct the distributions of polarity, secondary structure as well as solvent accessibility within the sets of predicted residues were examined and the results were compared with relevant distributions within the set of catalytic residues. Table 2 contains marked in bold fractions that are similar to those of catalytic set according to the proportion test (α = 0.05). It is clearly seen that only ConsurfDB9, Q-SiteFinder and WebFEATURE (100 and 95) manifest the fraction of polar residues at similar level as in CSA. Only WebFEATURE (100 and 99) showed accordance with the fraction of charged residues. Therefore none of the methods predicted a set of residues that reproduces the distribution of polarity observed for the catalytic residues.
Subsequently, secondary structure fractions within the catalytic and predicted sets were compared. The resulting significance of the proportion test marked in Table 2 shows that only predictions of CASTp, Q-SiteFinder, SuMo and WebFEATURE100 are in accordance with the fractions of all three secondary structure states within the catalytic set. Other methods excluding ConSurfDB9 and FOD show correspondence with two (β-strand, coil) or one state (β-strand). The discordances are caused by the over-representation of residues forming helical structures or under-representation of coiled structures. The latter are very often involved in catalysis [96]. The highest agreement is observed for β-strand. Nonetheless the frequency of residues forming β structures does not show significant difference between the catalytic and non-catalytic sets (χ2 decomposition), and this feature is not informative. Hence, CASTp, Q-SiteFinder, SuMo andWebFEATURE100 appear as the best methods reproducing the secondary structure distribution of the catalytic residues.
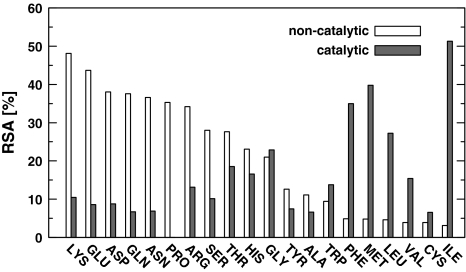
Figure 1 shows the median of relative solvent accessibilities (RSAs) of the 20 amino acids, taking into account catalytic and non-catalytic residues separately. Within the set of non-catalytic residues, polar ones tend to have high RSA compared to hydrophobic ones and their exposure to solvent is in accordance with their hydrophobicity. However, RSA analysis of the catalytic residues reveals quite opposite tendency. The majority of hydrophobic residues (I, L, M, F) have higher median RSA than polar or charged, which are catalytic (Fig. 1). Additionally the majority of polar residues performing catalysis (R, N, D, Q, E, K, S) have substantially lower median RSA compared with their non-catalytic equivalents. Consequently, it may be expected that methods for active site identification should detect polar residues more buried than typically and hydrophobic residues more exposed to solvent.
Fig. 1.
Relative solvent accessibilities (RSAs) of the 20 amino acid residues for catalytic and non-catalytic residues separately
The catalytic residues have the median of RSA equal to 10.3%, whereas the median for non-catalytic residues equals 20.9%. According to Mann–Whitney-Wilcoxon (MWW) test these two values are statistically different (p = 0.0000). The majority of methods give the median of RSA for predicted residues above the median of RSA for catalytic ones (MWW test, Table 4). The exceptions are ConSurf, FOD and WebFEATURE100. ConSurf produces the biggest set of residues and almost half of them are hydrophobic. However CASTp, PASS, Pocket-Finder also predict substantial portion of hydrophobic residues but higher median of RSA indicates that these residues form cavities or clefts. In turn, WebFEATURE100 produces small sets of residues which are mainly charged and additionally buried within a protein. FOD is in the middle of ConSurf and WebFEATURE100, but similarly to the former, hydrophobic residues predominate in its set of predicted residues. According to MWW test only the median of RSA for SuMo and WebFEATURE99 are statistically equal to the one for catalytic residues (p equals 0.834 and 0.096 respectively). The high maxima of RSA that even exceed 100%, indicate that all the methods are able to point residues highly exposed to solvent (data not shown).
Table 4.
Medians of the relative solvent accessibilities (RSAs) of the catalytic and non-catalytic residues as well as the predicted and non-predicted by the eight methods
| Median RSA (%) | ||
|---|---|---|
| Catalytic | Non-catalytic | |
| CSA | 10.3 | 20.9 |
| CASTp | 18.4 | 21 |
| ConSurf 9 | 4.9 | 24 |
| ConSurfDB9 | 4.1 | 24.4 |
| FOD | 9.3 | 21.7 |
| PASS | 22.7 | 22.4 |
| Pocket-Finder | 13.7 | 21.7 |
| Q-SiteFinder | 14.7 | 21.3 |
| SuMo | 10.5 | 21.1 |
| WebFEATURE100 | 4.1 | 18.8 |
| WebFEATURE99 | 14.5 | 21 |
| WebFEATURE95 | 18.2 | 21 |
Performance
The quality of predictions was measured using parameters commonly applicable to assessment of binary classifications. The predicted sets of residues were compared with the catalytic ones. The accuracy was measured assuming two perspectives that differ in definition of true positive (TP). First perspective regards only correctly indicated residues as TPs (AA perspective), while the second is more liberal and residues within catalytic sphere are counted as TPs (SO perspective).
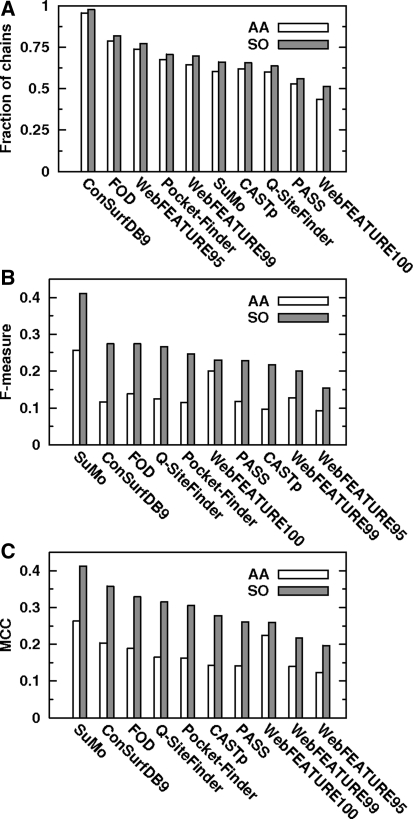
Firstly, in order to assess reliability of the methods, fraction of chains for which a method produced at least one true positive (TP) was calculated. Figure 2A shows that regardless of the assumed perspective (AA/SO) the most infallible is ConSurfDB9, then FOD, WebFEATURE95, Pocket-Finder and WebFEATURE99. The poorest yields are produced by PASS and WebFEATURE100. Moreover, there is no method producing at least one TP for all chains. Noteworthy is the increase in number of chains with at least one TP after transition from AA to SO perspective. The increase is the highest for SuMo and WebFEATURE100. This result proves that some predictions which at first are regarded as failures, may be valuable, because they are in the vicinity of a catalytic site.
Fig. 2.
Fractions of chains for which a method produced at least one true positive (A), F-measure (B) and MCC (C) parameters for both perspectives (AA and SO)
Figures 2B, C show F-measure and MCC calculated for each method and concerning two perspectives. According to this chart both parameters have maximum value near 0.25 (AA perspective) and 0.4 (SO perspective). With regard to AA perspective, SuMo and WebFEATURE100 are on the top of the ranking, while WebFEATURE95 is at the end. When SO perspective is considered, the higher agreement between F-measure and MCC is observed. Accordingly SuMo, ConSurfDB9, FOD, Q-SiteFinder and Pocket-Finder are among five the best methods, while WebFEATURE95 gives the worst outcome. More detailed analysis of these parameters reveals that only WebFEATURE descends in classification after change from AA to SO perspective, while other methods ascend or remain unchanged depending on the parameter considered. It is due to the small number of residues produced byWebFEATURE, which results in lower susceptibility to improvement of the outcome after softening of the criterion for TPs. With reference to SO perspective almost linear decrease in MCC is observed (Fig. 2C), indicating slight differences between subsequent methods. Contrary to that, F-measure shows outstanding high value for SuMo in comparison to other methods (Fig. 2B), and thus strengthens the position of a leader.
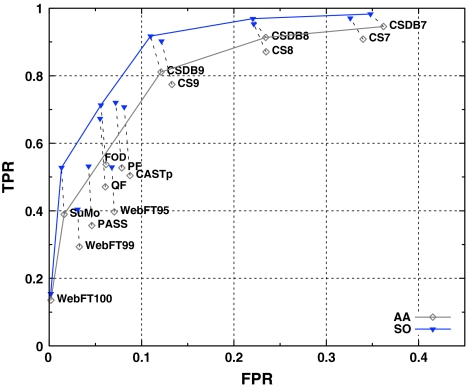
Even though F-measure as well as MCC are based on the four parameters (TP, TN, FP, FN), they do not provide complete information about the system [97]. Accordingly, mutual performances of the methods were visualized using points in the ROC space (Fig. 3). The x axis indicates false positive rate (FPR), while the y axis represents true positive rate (TPR). Figure 3 shows that softening of the criterion for TPs moves points in the ROC space towards upper-left corner. The least significant movement is in the case of WebFEATURE100. The overall relative arrangement of the points in the ROC space does not change substantially.
Fig. 3.
Points in the ROC space representing following results: CASTp, ConSurf—CS, ConSurfDB—CSDB, FOD, PASS, Pocket-Finder—PF, Q-SiteFinder—QF, SuMo, WebFEATURE—WebFT. ROC convex hulls are denoted by solid lines, while points related to the same method but different perspectives are connected by dashed-lines
The optimal methods were established through determination of the ROC Convex Hull (ROCCH). Regardless of the assumed perspective convex hull contains points representing ConSurfDB7, ConSurfDB8, ConSurfDB9, SuMo, WebFEATURE100. Additionally ROCCH related to SO perspective has an extra point corresponding to FOD. Majority of the optimal methods found in this way belong to the group of knowledge-based approaches.
Further examination of the ROC graphs revealed that, excluding ConSurf (ConSurfDB), all the methods have FPR below 10%. Therefore the main drawback of ConSurf is high number of over-predictions. The highest TPR with FPR below 10% have FOD and Pocket-Finder for AA and SO perspectives respectively. More detailed analysis of the points related to SO perspective revealed, however that Pocket-Finder has slightly higher TPR than FOD, but the latter is on the ROCCH, and therefore should be regarded as optimal. PASS has the lowest FPR among geometrical approaches, but also the lowest TPR. On the other hand Pocket-Finder has the highest TPR and lower FPR than CASTp. Additionally, the latter has slightly lower TPR than Pocket-Finder and therefore is unarguably not optimal in comparison to Pocket-Finder. Considering methods based on the physicochemical approaches, FOD has higher TPR than Q-SiteFinder, and comparable FPR, which demonstrates better performance of the former. Choosing the best method among the knowledge-based is not so obvious. ConSurf is able to generate different results depending on parameters that control the program or are used to interpret the results. Generally ConSurfDB gives optimal results compared to ConSurf with default parameters. Moreover ConSurfDB9 is the best (the closest to the point (0,1)) option in these group. Unfortunately it produces unsatisfactory high FPR. In contrast, WebFEATURE100 has the lowest FPR but also the lowest TPR. The value of the latter which is less than 0.2 is indisputably not satisfactory. SuMo is better than WebFEATURE95 and 99. Even though the latter has slightly higher TPR, SuMo has much lower FPR and is on the ROCCH.
Therefore the best representative of each approach are Pocket-Finder, FOD and SuMo. FOD outperforms Pocket-Finder in terms of TPR and FPR. Clear statement whether SuMo or FOD is better, depends on assumed costs of FP and FN errors. This reasoning is based on lemma claiming that for any set of cost and class distribution there is a point on the ROCCH with minimum expected cost [88]. Minimum expected cost (m mec) is defined as the product of cost ratio and the reciprocal of the class ratio, and is used to determine whether one classification model is better than another. Moreover it may be easily transformed into the so-called iso-performance line such as fragment of the ROCCH [98]. Therefore slopes of the ROCCH define range of minimum expected cost related to each point on the ROCCH. According to that SuMo corresponds to such a set of operating conditions that m mec = (4.0, 17.8) and m mec = (4.3, 32.0) considering AA and SO perspectives, respectively. FOD in turn is optimal when m mec = (3.8, 4.3) regarding SO perspective. Hence the choice of optimal method depends on classifier conditions. Because in our case the approximate probabilities of negative and positive classes are known, the only parameter which should be carefully considered is the cost ratio. When proportion of non-catalytic residues to catalytic ones is equal to 100:1 and cost of false positives is 10 times as expensive as false negatives, then m mec = 10, what perfectly fits the SuMo’s optimal range. However change of cost ratio from 10 to 25 causes that FOD is optimal (SO perspective).
Considering AA perspective, SuMo is the optimal method. It has the highest TPR with FPR below 10% among methods on the ROCCH. The situation is slightly different when SO perspective is examined. Then FOD as well as SuMo should be considered because the assessment of the method depends on classifier conditions and the choice is not obvious.
Comparison of the chains with different success rate
In order to create characteristics of chains differing in success rate of active site prediction, one representative set of results was chosen for each method. Therefore, among methods having a few variants, ConSurfDB9 and WebFEATURE95 were selected as exemplars. Subsequently, instances of failures, defined as no TP, were aggregated for each chain separately. Hence the maximum number of failures (NF) may be equal to 8, as such a number of methods is analysed. The chains have been divided into three groups depending on the number of failures. As a result there are hard (NF > 4), medium (4 > NF > 0) and easy (NF = 0) chains.
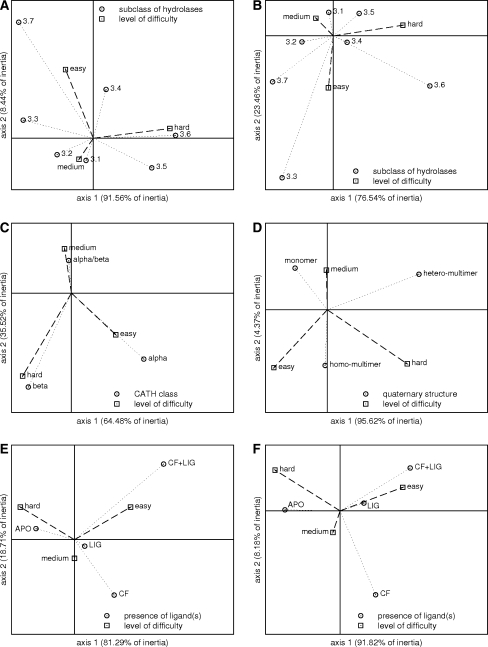
Figure 4 presents the results of correspondence analysis (CA). It visualizes relationship between a chain difficulty and features such as a subclass of hydrolases (panels A, B), a class of catalytic domain in CATH classification (panel C), a quaternary structure according to PQS (panel D) and a presence of different types of ligands (panels E, F). The vertical and horizontal axes intersect at point (0,0). The vertical axis is the one with the highest inertia. The CA was made by means of the statistical package STATISTICA. Column and row standardization was used to plot the points on the maps.
Fig. 4.
Correspondence analysis presenting relation between chain difficulty and subclass of hydrolases (A, B), CATH class (C), quaternary structure (D) and presence of ligands (E, F). The two perspectives are analyzed: AA (A, E) and SO (B, C, D, F)
EC subclasses
Correspondence analysis allowed to explore relationship between subclasses of hydrolases and the level of difficulty of active site prediction. The enzymes belonging to subclasses 3.8 and 3.11 according to the EC classification were excluded from the calculations (small number of representatives). Figure 4A presents the relationship between a hydrolase subclass and a group of difficulty (hard, medium, easy) within AA perspective. Dimension 1 is the most reliable indicator of an associations (inertia of 91.6%). It distinguishes between easy or medium and hard chains, and subclasses 3.4, 3.5, 3.6 and 3.1, 3.2, 3.3, 3.7. The strongest association is observed for hard chains and hydrolases acting on acid anhydrides (EC 3.6.-.-) and hydrolases acting on carbon–nitrogen bonds other than peptide bonds (EC 3.5.-.-). Moreover easy or medium chains are associated with subclasses 3.1, 3.2, 3.3 and 3.7. When dimension 2, accounting for 8.4% of the variation in the data is taken into account, easy chains are separated from the medium. Then medium chains are related to esterases (EC 3.1.-.-) and glycosydases (EC 3.2.-.-), two of the most abundant subclasses in the data set, while easy chains are associated with hydrolases acting on ether bonds (EC 3.3.-.-) and hydrolases acting on carbon–carbon bonds (EC 3.7.-.-). The latter association is however not highly reliable, because these two subclasses are poorly represented in the data set. Similar inferences can be drawn from the results obtained for SO perspective, even though the coordinates of points in the CA map are different (Fig. 4B). Noteworthy is the point related to peptidases (EC 3.4.-.-), which is close to the origin of the coordinate system. It clearly denotes that this subclass of hydrolases contains miscellaneous chains regarding difficulty of active site prediction, with tendency to contain hard and easy chains.
CATH classes
Relation between a structure of catalytic domains regarding class level of CATH classification and the rate of success was determined by means of CA as well. Domains that belong to class 4 were excluded from the analysis (small number of representatives). Figure 4C summarizes the final results for SO perspective. It presents explicit division of the points into three clusters. The chains containing catalytic domains that represent the mainly beta class appear as hard for active site prediction, while the enzymes assigned to the mainly alpha class are rather easy for the methods. The medium results are generally observed for the α/β class. Aforementioned conclusion can be drawn based on the results obtained for AA perspective (data not shown).
Quaternary structure
The influence of a quaternary structure on the rate of success was examined and Fig. 4D contains the resulting CA map for the SO perspective. As can be seen from the graph, hydrolases that are monomers are associated with the group of medium chains. Multimers in turn, composed of identical subunits are related to two groups: easy and medium chains, while the one build of different subunits is associated with hard chains. Similar map has been obtained for AA perspective (data not shown).
Ligands
The presence of ligands is another factor that may influence the quality of results. 124 chains contain heterogeneous molecules. The majority of them are ligands bound with a protein molecule (distance from a protein below 6 Å) but they are not required for biological activity. Another group of ligands are cofactors, which are mainly in the form of a metal ion (Zn, Ca, Mg, Co, Mn), but there is also a pyridoxal 5′-phosphate (kyruneninase, 1QZ9) and a nicotinamide adenine dinucleotide (NAD, adenosylhomocysteinase, 1B3R). Therefore the chains were divided into four groups. The first contains chains with no ligands (APO), the second comprises structures with cofactors (CF), the third contains chains that have cofactors as well other ligands (CF + LIG), and the fourth encompasses chains with ligands that are not cofactors (LIG). The correspondence analysis was applied to our data set regarding a chain difficulty and a presence of different types of ligands (APO, CF, CF + LIG, LIG,). Figures 4E, F present the results for AA and SO perspectives, respectively. Regardless of the assumed perspective there is a clear separation between hard, medium and easy chains. Unquestionably apo structures are associated with hard chains, and structures having bound a cofactor and a ligand (CF + LIG) are related to easy chains. In turn, medium chains are associated with structures having bound cofactors or other ligands (AA perspective). However a change from AA to SO perspective, causes that structures with ligands are more related to easy chains.
Secondary structure, polarity and solvent accessibility
The relation between secondary structure elements of catalytic residues and the level of difficulty was confirmed (χ2 statistics, p = 0.0280 and 0.0183 for AA and SO perspectives, respectively). With reference to AA perspective, the fraction of catalytic residues forming helices does not differ significantly among three groups of difficulty (χ2 decomposition, p = 0.0766). In turn when SO perspective is assumed such situation is in the case of β-strands (p = 0.5664). The common conclusion is that regardless of the assumed perspective, the difference always lies in coils. The most distinctive is the set of easy chains, which has the smallest fraction of catalytic residues located in coils (p = 0.0075 and p = 0.0041 for AA and SO perspectives respectively). In contrast to secondary structure, polarity distributions are similar among hard, medium and easy chains (p = 0.2216 and p = 0.4442 for AA and SO perspectives, respectively).
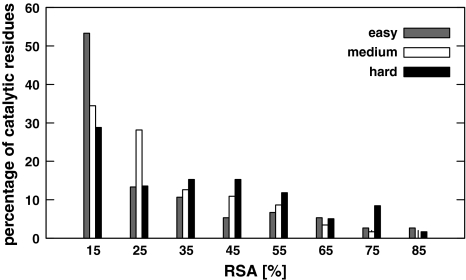
Figure 5 presents histograms of RSA for catalytic residues of three sets of chains: hard, medium and easy, distinguished according to SO perspective. Similar histograms are for AA perspective (not shown). The median of RSA for the catalytic residues of hard, medium and easy chains are equal to 16.6, 10.6 and 7.65%, while the median of RSA for non-catalytic residues is above 19%. Therefore the median of RSA of catalytic residues increases with difficulty, however it does not exceed the median RSA of non-catalytic ones.
Fig. 5.
Distribution of RSA for catalytic residues of easy, medium and hard chains
Examples of the most difficult chains
There is no chain for which all the methods failed, however there are 13 up to 17 chains (depending on the assumed perspective) posing a problem for six or seven methods. The most infallible methods for these hard cases are ConSurfDB9 and WebFEATURE95. Table 5 contains a list of the hardest chains (NF = 7) and their short characteristics.
Table 5.
List of the hardest chains and their characteristics including name, length of polypeptide chain, EC number, CATH ids, quaternary structure, catalytic residues and function
| PDB ID | Name | EC | Lengtha | CATHb | PQS | Acitve sited | Functione |
|---|---|---|---|---|---|---|---|
| 1CFR:A | Type-2 restriction enzyme Cfr10l | 3.1.21.4 | 285 (283) | 3.40 | Homo-tetramer | 190K | DNA binding, magnesium ion binding, type II specific deoxyribonuclease activity |
| 1RDD:A | Ribonuclease H | 3.1.26.4 | 155 | 3.30 | Monomer | 124H | Magnesium ion binding, nucleic acid binding, ribonuclease H activity |
| 1V0E:A | Endo-alpha-sialidase | 3.2.1.129 | 666 | 2.40 | Homo-trimer | 581E | Endo-alpha-sialidase activity |
| 2.120 | 596R | ||||||
| 3.30 | 647R | ||||||
| 4.10 | |||||||
| 1CVR:A | Arg-gingipain | 3.4.22.37 | 435 (432) | 2.60 | Monomerc | 152G | Calcium ion binding, cysteine-type endo-peptidase activity |
| 3.40 | 211H | ||||||
| 3.40 | 212G | ||||||
| 244C |
aNumber in parenthesis denotes number of residues in pdb file
bOnly first two levels are given, catalytic domains are in bold
cEven though PQS states it is a heterotrimer; two additional chains are short peptides and therefore should be treated as ligands
dCatalytic residues according to CSA
eAccording to UniProtKB
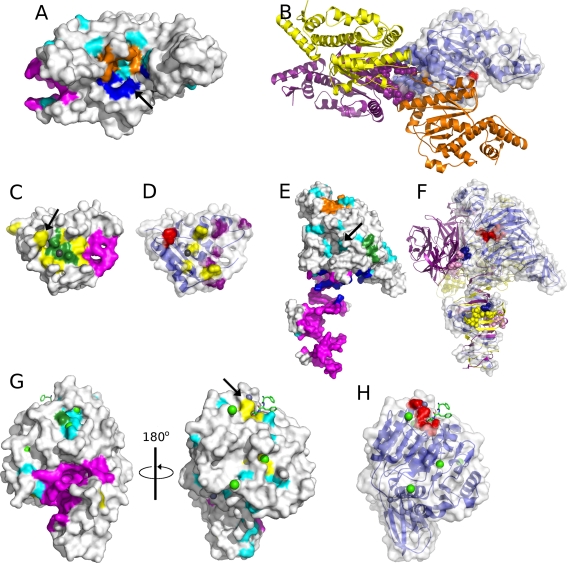
Type-2 restriction enzyme Cfr10I (1CFR:A) is the structure for which only FOD pointed at a catalytic residue. Catalytic site of this hydrolase (indicated by an arrow in Fig. 6A or red spheres in Fig. 6B) is in the form of a shallow pocket which together with second monomer serves to DNA binding [99, 100]. Most of the methods (CASTp, Q-SiteFinder, SuMo, PASS) indicate other pocket which is much deeper (magenta in Fig. 6A). Interestingly this pocket according to PQS database is involved in protein–protein contacts (Fig. 6B), and thus has functional significance [99]. Pocket-Finder in turn finds the pocket close to the catalytic residue, but even assuming the SO perspective it still fails to point at it. Similarly WebFEATURE95’s predictions are near the correct result (cyan in Fig. 6A). Unfortunately even though there was a catalytic residue among those pointed out, the answer where the active site is located would be equivocal, because the residues predicted by WebFEATURE are scattered across the whole structure.
Fig. 6.
Surface representations of structures found as the hardest chains. Colouring scheme applied to opaque surface: ConSurfDB9-yellow, CASTp-magenta, FOD-blue, Q-SiteFinder-orange, SuMo-green, WebFEATURE95-cyan. Catalytic residues are red spheres or pointed by an arrow. (A) Surface of type-2 restriction enzyme Cfr10I (1CFR:A) with depicted predictions of CASTp, FOD, Pocket-Finder and WebFEATURE95. (B) Ribbon model of quaternary structure of type-2 restriction enzyme Cfr10I with one chain as transparent surface. The four monomers are blue, purple, orange and yellow, two chains have residues indicated by CASTp as spheres. (C) Surface of ribonuclease H (1RDD:A) with depicted predictions of CASTp, ConSurfDB9, SuMo and magnesium ion as sphere. (D) Transparent surface of ribonuclease H with underlying ribbon model. RNA and DNA binding sites [99] are shown as yellow and purple spheres, respectively. (E) Surface of endo-alpha-sialidase (1V0E:A) with depicted predictions of CASTp, FOD, SuMo, Q-SiteFinder and WebFEATURE95. (F) Ribbon model of quaternary structure of endo-alpha-sialidase and transparent surface of one chain. The three monomers are blue, purple and yellow. Residues involved in sialic acid binding are shown as spheres. (G) Surface of Arg-gingipain (1CVR:A) with depicted predictions of CASTp, SuMo, WebFEATURE95 and ConSurfDB9. Ligand molecule in sticks, calcium ions in green spheres and zinc ions in grey spheres. (H) Transparent surface of Arg-gingipain with underlying ribbon model
Another interesting enzyme is ribonuclease H (1RDD:A). The predictions for this hydrolase are generally related to two sites on the protein’s surface. First site which is ‘spongy’ because it contains visible holes (C-terminus and loop) is indicated by CASTp, PASS, Pocket-Finder and Q-SiteFinder (magenta in Fig. 6C) and has no particular function ascribed, even though it is in close proximity to DNA-binding sites [101] (purple spheres in Fig. 6D). Second site is formed by the predictions of SuMo, ConSurf, FOD, WebFEATURE95 and additionally it has bound magnesium ion (grey sphere in Fig. 6C), [102]. Alternatively this site may bind two manganese ions [103] and generally it constitutes a metal-binding site [101]. Nonetheless only one method (ConSurf) pointed at the catalytic residue which is annotated in CSA and described in [104] (indicated by an arrow in Fig. 6C or red spheres in Fig. 6D). Therefore the common failure in prediction of the active site is due to the high solvent accessibility of its residue.
Endo-alpha-sialidase (1V0E:A) is an example of a homotrimeric enzyme (Fig. 6F). Its single chain caused a problem for all the methods except forWebFEATURE95. The structure exhibits mushroom-like shape (Fig. 6E) and is built of four distinct domains [105]. Generally six sites were identified by the methods (Fig. 6E): CASTp and Pocket-Finder in the tail-spike domain (magenta), FOD between the β-propeller domain and the tail-spike domain (blue), SuMo on the surface of the β-propeller (green), Q-SiteFinder between the N-terminal and the β-propeller domain (orange), WebFEATURE95 in the cavity of β-propeller (cyan) and PASS in the opposite cavity of the β-propeller (not shown). Since the sites indicated by CASTp, Pocket-Finder and FOD form contacts between monomers, residues predicted by Q-SiteFinder, SuMo and PASS have no particular function. Only among the residues indicated byWebFEATURE95 are those forming the active site cleft. Additionally the hydrolase contains two sialic acid-binding sites which are assemblies of residues from adjacent monomers (Fig. 6F). First is between the tail-spike domains indicated by CASTp and Pocket-Finder and the second between the β-barrel and the β-propeller. The second site is identified by none of the methods, even though together with the active site it forms a long pocket with a ridge.
Arg-gingipain (1CVR:A) in turn has almost flat active site within a domain composed of six-stranded β-sheets sandwiched by α helices (Fig. 6G, H) [106]. Nevertheless the methods reveal four sites (Fig. 6G). First, indicated by CASTp, FOD, PASS, Pocket-Finder and Q-SiteFinder is in the form of a tunnel like pocket, penetrating the structure (magenta), which is filled with water and does not have assigned any particular function [106]. The second and third are small and shallow. They are predicted by SuMo, WebFEATURE95 and ConSurf, WebFEATURE95 respectively. Indeed they bind calcium ions. However only the set of residues indicated by ConSurf (fourth site) contains the catalytic residues. Consequently presence of a definite cavity turned out to be misleading, while real active site as usual in such an open β-sheet enzymes is in a crevice outside the carboxyl end of the β-sheet [106].
Discussions
Demand for a method allowing accurate identification of active sites is still not satisfied. Currently very popular is utilizing machine learning approaches such as neural networks [51–53], support vector machines [54–56] or Naive Bayes classifications [57] in order to face this challenge. Here, however we validated simple approaches that use geometric criteria (CASTp, Pocket-Finder, PASS), physicochemical features (FOD, Q-SiteFinder) or knowledge-based patterns (ConSurf, SuMo, WebFEATURE). Relatively high number of over-predictions compared to the number of true positives is an obvious drawback of analysed methods. It is manifested in the number of pointed residues by a method, and in the radii of predicted sites. The most similar sites, regarding their size to those documented in CSA were produced by SuMo. Polarity analysis showed another shortcoming of tested methods. Correct reproduction of polarity distribution of catalytic residues appeared very challenging, concurrently denoting that these feature is highly informative. In turn a few methods yielded good accordance of secondary structure elements distribution with the set of catalytic residues, showing that these feature is easier to reproduce than polarity. Relative solvent accessibility of amino acid residues alone as well as in conjunction with information about polarity is a good indicator of correctness of catalytic site prediction. Only two methods: SuMo andWebFEATURE99 succeeded in reproduction of RSA characteristics related to the catalytic residues. In spite of that, a glance at polarity, secondary structure, RSA provides useful information about correctness of predicted sites. More accurate assessment of the predictions was carried out by means of calculation of MCC and F-measure parameters and plotting points in the ROC space. The best method among analysed turned out to be SuMo, which is a representative of the knowledge-based approaches. Second place takes FOD, classified as the physicochemical approach. It exhibits acceptable level of FPR and higher TPR than SuMo. In turn Pocket-Finder appears as the best method among geometric approaches. Unfortunately none of the methods exceeds MCC values obtained by the neural network approach using sequence and structure information [52]. MCC calculated for the AA perspective that equals to 0.26 for SuMo is not the highest that have ever been achieved. The obtained results, however are not below expectations as the analysed methods are more suitable for ligand-binding site prediction rather than catalytic residues. The adopted strategy for the assessment is due to well characterisation of active sites in enzymes and that the information is gathered consistently in one place (CSA). Therefore the obtained results referring to MCC, ROC analysis define minimum expectations for the methods for active site prediction. The summary of the evaluation that lists size, polarity, secondary structure, solvent accessibility and the overall performance based on MCC, F-measure and points in the ROC space is presented in Table 6. The more accurate method is denoted by the higher number of asterisks.
Table 6.
Summary of the evaluation regarding size, polarity, secondary structure, solvent accessibility of the predicted sites in comparison to the catalytic ones, as well as TPR, FPR and the overall performance of the methods based on MCC, F-measure and the ROC analysis
| Method | Size | Polarity | Secondary structure | Solvent accessibility | TPR | FPR | Performance |
|---|---|---|---|---|---|---|---|
| CASTp | ** | * | *** | * | ** | ** | ** |
| ConSurf | * | ** | * | ** | *** | * | ** |
| FOD | ** | * | * | ** | ** | ** | *** |
| PASS | ** | * | ** | * | ** | ** | * |
| Pocket-Finder | ** | * | ** | * | ** | ** | ** |
| Q-SiteFinder | ** | ** | *** | * | ** | ** | ** |
| SuMo | *** | * | *** | *** | ** | ** | *** |
| WebFEATURE | ** | ** | ** | ** | * | *** | * |
The more accurate results are denoted by the higher number of asterisks
Correspondence analysis revealed that esterases (EC 3.1.-.-) and glycosydases (EC 3.2.-.-) form structures of moderate difficulty, while hydrolases acting on acid anhydrides (EC 3.6.-.-) and on carbon–nitrogen bonds other than peptide bonds (EC 3.5.-.-) usually are hard. There is no clear association between any of hydrolase subclasses and the easy chains. In turn association between CATH class and difficulty is clear. Accordingly the α-proteins are easy, the α/β-medium and the β-hard. The relation between quaternary structure and difficulty of prediction is equivocal. Monomers are of moderate difficulty, while multimers—moderate or hard. Knowledge of quaternary structure may be crucial for the success rate in binding site prediction in cases when it is located in the pocket on the edge of subunits. Hence if there is no information about quaternary structure it is advised to perform such an analysis as some methods are sensitive to it, especially the ones based on the geometrical approaches. The presence of a ligand appeared another factor affecting the results. Higher success rate was observed for structures with bound ligand, cofactor or both than for ‘unbound’ structures. Similar observations were reported elsewhere [22], hence conformational flexibility should be taken into account in the future. There is no difference between polarity distribution within catalytic residues of different groups of difficulty. On the other hand in the hardest group there is a particularly high frequency of catalytic residues forming loops in comparison to other groups. Regarding RSA markedly hard are structures with the catalytic residues exhibiting RSA similar to the RSA of non-catalytic residues.
General observations concerning factors determining difficulty of catalytic site prediction have the confirmation in the examples of the hardest structures. Definite pockets turned out misleading in the case of type-2 restriction enzyme Cfr10I and Arg-gingipain. This problem was observed previously [107] but in the case of the former protein this could be negligible if the quaternary structure was concerned. Similarly this information could be helpful in the case of endo-alpha-sialidase. In turn ribonuclease H, has the catalytic site highly exposed to solvent.
Conclusions
In conclusion, the diversity of protein structures and functions requires multiple approaches in order to correctly predict their active sites. Even though the tested methods are more suitable for prediction of ligand-binding sites than active sites it was shown that they may be also valuable in the latter. The main challenge is the determination of the most suitable one for a particular protein structure. The alternative solution may be a protocol providing for many methods, not only designed for binding-site prediction. Especially useful would be calculations yielding information about quaternary structure as well as dynamic nature of proteins. Simultaneously intensive studies devoted to characterization of binding sites are necessary in order to guide improvement of in silico approaches for function prediction.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Katarzyna Prymula, Email: prymula@chemia.uj.edu.pl.
Tomasz Jadczyk, Email: jadczyk@agh.edu.pl.
Irena Roterman, Email: myroterm@cyf-kr.edu.pl.
References
- 1.Brenner SE. A tour of structural genomics. Nat Rev Genet. 2001;2:801–809. doi: 10.1038/35093574. [DOI] [PubMed] [Google Scholar]
- 2.Chandonia J-M, Brenner SE. The impact of structural genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 3.Grabowski M, Joachimiak A, Otwinowski Z, Minor W. Structural genomics: keeping up with expanding knowledge of the protein universe. Curr Opin Struct Biol. 2007;17:347–353. doi: 10.1016/j.sbi.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gileadi O, Knapp S, Lee WH, Marsden BD, Müller S, Niesen FH, Kavanagh KL, Ball LJ, von Delft F, Doyle DA, Oppermann UCT, Sundström M. The scientific impact of the structural genomics consortium: a protein family and ligand-centered approach to medically-relevant human proteins. J Struct Funct Genomics. 2007;8:107–119. doi: 10.1007/s10969-007-9027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hajduk PJ, Huth JR, Tse C. Predicting protein druggability. Drug Discov Today. 2005;10:1675–1682. doi: 10.1016/S1359-6446(05)03624-X. [DOI] [PubMed] [Google Scholar]
- 6.Hajduk PJ, Huth JR, Fesik SW. Druggability indices for protein targets derived from NMR-base screening data. J Med Chem. 2005;48:2518–2525. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 7.Vajda S, Guarnieri F. Characterization of protein-ligand interaction sites using experimental and computational methods. Curr Opin Drug Discov Devel. 2006;9:354–362. [PubMed] [Google Scholar]
- 8.Weigelt J, McBroom-Cerajewski LDB, Schapira M, Zhao Y, Arrowmsmith CH. Structural genomics and drug discovery: all in the family. Curr Opin Chem Biol. 2008;12:32–39. doi: 10.1016/j.cbpa.2008.01.045. [DOI] [PubMed] [Google Scholar]
- 9.Levitt DG, Banaszak LJ. POCKET: a computer graphics method for identifying and displaying protein cavities and their surrounding amino acids. J Mol Graph. 1992;10:229–234. doi: 10.1016/0263-7855(92)80074-n. [DOI] [PubMed] [Google Scholar]
- 10.Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J Mol Graph. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 11.Peters KP, Fauck J, Frömmel C. The automatic search for ligand binding sites in proteins of known three-dimensional structure using only geometric criteria. J Mol Biol. 1996;256:201–213. doi: 10.1006/jmbi.1996.0077. [DOI] [PubMed] [Google Scholar]
- 12.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: easurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binkowski TA, Naghibzadeh S, Liang J. CASTp: computed atlas of surface topography of proteins. Nucleic Acids Res. 2003;31:3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendlich M, Rippmann F, Barnickel G. LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J Mol Graph Model. 1997;15:359–363. doi: 10.1016/s1093-3263(98)00002-3. [DOI] [PubMed] [Google Scholar]
- 15.Brady GP, Stouten PF. Fast prediction and visualization of protein binding pockets with PASS. J Comput Aided Mol Des. 2000;14:383–401. doi: 10.1023/a:1008124202956. [DOI] [PubMed] [Google Scholar]
- 16.Ondrechen MJ, Clifton JG, Ringe D. THEMATICS: a simple computational predictor of enzyme function from structure. Proc Natl Acad Sci USA. 2001;98:12473–12478. doi: 10.1073/pnas.211436698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elcock AH. Prediction of functionally important residues based solely on the computed energetics of protein structure. J Mol Biol. 2001;312:885–896. doi: 10.1006/jmbi.2001.5009. [DOI] [PubMed] [Google Scholar]
- 18.Kortvelyesi T, Silberstein M, Dennis S, Vajda S. Improved mapping of protein binding sites. J Comput Aided Mol Des. 2003;17:173–186. doi: 10.1023/a:1025369923311. [DOI] [PubMed] [Google Scholar]
- 19.Landon MR, Lancia DR, Yu J, Thiel SC, Vajda S. Identification of hot spots within druggable binding regions by computational solvent mapping of proteins. J Med Chem. 2007;50:1231–1240. doi: 10.1021/jm061134b. [DOI] [PubMed] [Google Scholar]
- 20.An J, Totrov M, Abagyan R. Comprehensive identification of druggable protein ligand binding sites. Genome Inform. 2004;15:31–41. [PubMed] [Google Scholar]
- 21.An J, Totrov M, Abagyan R. Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol Cell Proteomics. 2005;4:752–761. doi: 10.1074/mcp.M400159-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Laurie ATR, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 23.Bryliński M, Prymula K, Jurkowski W, Kochanczyk M, Stawowczyk E, Konieczny L, Roterman I. Prediction of functional sites based on the fuzzy oil drop model. PLoS Comput Biol. 2007;3:e94. doi: 10.1371/journal.pcbi.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 25.Zhang B, Rychlewski L, Pawowski K, Fetrow JS, Skolnick J, Godzik A. From fold predictions to function predictions: automation of functional site conservation analysis for functional genome predictions. Protein Sci. 1999;8:1104–1115. doi: 10.1110/ps.8.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichtarge O, Sowa ME. Evolutionary predictions of binding surfaces and interactions. Curr Opin Struct Biol. 2002;12:21–27. doi: 10.1016/s0959-440x(02)00284-1. [DOI] [PubMed] [Google Scholar]
- 27.Madabushi S, Yao H, Marsh M, Kristensen DM, Philippi A, Sowa ME, Lichtarge O. Structural clusters of evolutionary trace residues are statistically significant and common in proteins. J Mol Biol. 2002;316:139–154. doi: 10.1006/jmbi.2001.5327. [DOI] [PubMed] [Google Scholar]
- 28.Aloy P, Querol E, Aviles FX, Sternberg MJ. Automated structure-based prediction of functional sites in proteins: applications to assessing the validity of inheriting protein function from homology in genome annotation and to protein docking. J Mol Biol. 2001;311:395–408. doi: 10.1006/jmbi.2001.4870. [DOI] [PubMed] [Google Scholar]
- 29.Landgraf R, Xenarios I, Eisenberg D. Three-dimensional cluster analysis identifies interfaces and functional residue clusters in proteins. J Mol Biol. 2001;307:1487–1502. doi: 10.1006/jmbi.2001.4540. [DOI] [PubMed] [Google Scholar]
- 30.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(1):S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 31.Armon A, Graur D, Ben-Tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 32.Nimrod G, Glaser F, Steinberg D, Ben-Tal N, Pupko T. In silico identification of functional regions in proteins. Bioinformatics. 2005;21(1):i328–i337. doi: 10.1093/bioinformatics/bti1023. [DOI] [PubMed] [Google Scholar]
- 33.Dou Y, Zheng X, Wang J. Prediction of catalytic residues using the variation of stereochemical properties. Protein J. 2009;28:29–33. doi: 10.1007/s10930-008-9161-0. [DOI] [PubMed] [Google Scholar]
- 34.Wallace AC, Borkakoti N, Thornton JM. TESS: a geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. Application to enzyme active sites. Protein Sci. 1997;6:2308–2323. doi: 10.1002/pro.5560061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleywegt GJ. Recognition of spatial motifs in protein structures. J Mol Biol. 1999;285:1887–1897. doi: 10.1006/jmbi.1998.2393. [DOI] [PubMed] [Google Scholar]
- 36.Kinoshita K, Furui J, Nakamura H. Identification of protein functions from a molecular surface database, eF-site. J Struct Funct Genomics. 2002;2:9–22. doi: 10.1023/a:1011318527094. [DOI] [PubMed] [Google Scholar]
- 37.Wangikar PP, Tendulkar AV, Ramya S, Mali DN, Sarawagi S. Functional sites in protein families uncovered via an objective and automated graph theoretic approach. J Mol Biol. 2003;326:955–978. doi: 10.1016/s0022-2836(02)01384-0. [DOI] [PubMed] [Google Scholar]
- 38.Barker JA, Thornton JM. An algorithm for constraint-based structural template matching: application to 3D templates with statistical analysis. Bioinformatics. 2003;19:1644–1649. doi: 10.1093/bioinformatics/btg226. [DOI] [PubMed] [Google Scholar]
- 39.Spriggs RV, Artymiuk PJ, Willett P. Searching for patterns of amino acids in 3D protein structures. J Chem Inf Comput Sci. 2003;43:412–421. doi: 10.1021/ci0255984. [DOI] [PubMed] [Google Scholar]
- 40.Yao H, Kristensen DM, Mihalek I, Sowa ME, Shaw C, Kimmel M, Kavraki L, Lichtarge O. An accurate, sensitive, and scalable method to identify functional sites in protein structures. J Mol Biol. 2003;326:255–261. doi: 10.1016/s0022-2836(02)01336-0. [DOI] [PubMed] [Google Scholar]
- 41.Stark A, Russell RB. Annotation in three dimensions. PINTS: patterns in non-homologous tertiary structures. Nucleic Acids Res. 2003;32:3341–3344. doi: 10.1093/nar/gkg506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stark A, Sunyaev S, Russell RB. A model for statistical significance of local similarities in structure. J Mol Biol. 2003;326:1307–1316. doi: 10.1016/s0022-2836(03)00045-7. [DOI] [PubMed] [Google Scholar]
- 43.Stark A, Shkumatov A, Russell RB. Finding functional sites in structural genomics proteins. Structure. 2004;12:1405–1412. doi: 10.1016/j.str.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Shulman-Peleg A, Nussinov R, Wolfson HJ. Recognition of functional sites in protein structures. J Mol Biol. 2004;339:607–633. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jambon M, Imberty A, Deléage G, Geourjon G. A new bioinformatic approach to detect common 3D sites in protein structures. Proteins. 2003;52:137–145. doi: 10.1002/prot.10339. [DOI] [PubMed] [Google Scholar]
- 46.Jambon M, Andrieu O, Combet C, Deléage G, Delfaud F, Geourjon C. The SuMo server: 3D search for protein functional sites. Bioinformatics. 2005;21:3929–3930. doi: 10.1093/bioinformatics/bti645. [DOI] [PubMed] [Google Scholar]
- 47.Laskowski RA, Watson JD, Thornton JM. Protein function prediction using local 3D templates. J Mol Biol. 2005;351:614–626. doi: 10.1016/j.jmb.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei L, Altman RB (1998) Recognizing protein binding sites using statistical descriptions of their 3D environments. Pac Symp Biocomput 497–508 [PubMed]
- 50.Taroni C, Jones S, Thornton JM. Analysis and prediction of carbohydrate binding sites. Protein Eng. 2000;13:89–98. doi: 10.1093/protein/13.2.89. [DOI] [PubMed] [Google Scholar]
- 51.Stahl M, Taroni C, Schneider G. Mapping of protein surface cavities and prediction of enzyme class by a self-organizing neural network. Protein Eng. 2000;13:83–88. doi: 10.1093/protein/13.2.83. [DOI] [PubMed] [Google Scholar]
- 52.Gutteridge A, Bartlett GJ, Thornton JM. Using a neural network and spatial clustering to predict the location of active sites in enzymes. J Mol Biol. 2003;330:719–734. doi: 10.1016/s0022-2836(03)00515-1. [DOI] [PubMed] [Google Scholar]
- 53.Tang Y-R, Sheng H-Y, Chen Y-Z, Zhang Z. An improved prediction of catalytic residues in enzyme structures. Protein Eng Des Sel. 2008;21:295–302. doi: 10.1093/protein/gzn003. [DOI] [PubMed] [Google Scholar]
- 54.Youn E, Peters B, Radivojac P, Mooney SD. Evaluation of features for catalytic residue prediction in novel folds. Protein Sci. 2007;16:216–226. doi: 10.1110/ps.062523907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang T, Zhang H, Chen K, Shen S, Ruan J, Kurgan L. Accurate sequence-based prediction of catalytic residues. Bioinformatics. 2008;24:2329–2338. doi: 10.1093/bioinformatics/btn433. [DOI] [PubMed] [Google Scholar]
- 56.Pugalenthi G, Kumar KK, Suganthan PN, Gangal R. Identification of catalytic residues from protein structure using support vector machine with sequence and structural features. Biochem Biophys Res Commun. 2008;367:630–634. doi: 10.1016/j.bbrc.2008.01.038. [DOI] [PubMed] [Google Scholar]
- 57.Zhang K, Xu Y, Chen G. PECB: prediction of enzyme catalytic residues based on Naive Bayes classification. Int J Bioinform Res Appl. 2008;4:295–305. doi: 10.1504/IJBRA.2008.019576. [DOI] [PubMed] [Google Scholar]
- 58.Bhinge A, Chakrabarti P, Uthanumallian K, Bajaj K, Chakraborty K, Varadarajan R. Accurate detection of protein: ligand binding sites using molecular dynamics simulations. Structure. 2004;12:1989–1999. doi: 10.1016/j.str.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 59.Bliznyuk AA, Gready JE. Identification and energetic ranking of possible docking sites for pterin on dihydrofolate reductase. J Comput Aided Mol Des. 1998;12:325–333. doi: 10.1023/a:1008039000355. [DOI] [PubMed] [Google Scholar]
- 60.Kurowski MA, Sasin JM, Feder M, Debski J, Bujnicki JM. Characterization of the cofactor-binding site in the SPOUT-fold methyltransferases by computational docking of S-adenosylmethionine to three crystal structures. BMC Bioinformatics. 2003;4:9. doi: 10.1186/1471-2105-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang DT-H, Oyang Y-J, Lin J-H. MEDock: a web server for efficient prediction of ligand binding sites based on a novel optimization algorithm. Nucleic Acids Res. 2005;33:W233–W238. doi: 10.1093/nar/gki586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laurie ATR, Jackson RM. Methods for the prediction of protein-ligand binding sites for structure-based drug design and virtual ligand screening. Curr Protein Pept Sci. 2006;7:395–406. doi: 10.2174/138920306778559386. [DOI] [PubMed] [Google Scholar]
- 63.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei L, Altman RB, Chang JT (1997) Using the radial distributions of physical features to compare amino acid environments and align amino acid sequences. Pac Symp Biocomput 465–476 [PubMed]
- 66.Liang MP, Banatao DR, Klein TE, Brutlag DL, Altman RB. WebFEATURE: An interactive web tool for identifying and visualizing functional sites on macromolecular structures. Nucleic Acids Res. 2003;31:3324–3327. doi: 10.1093/nar/gkg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stuart AC, Ilyin VA, Sali A. LigBase: a database of families of aligned ligand binding sites in known protein sequences and structures. Bioinformatics. 2002;18:200–201. doi: 10.1093/bioinformatics/18.1.200. [DOI] [PubMed] [Google Scholar]
- 68.Chalk AJ, Worth CL, Overington JP, Chan AWE. PDBLIG: classification of small molecular protein binding in the protein data bank. J Med Chem. 2004;47:3807–3816. doi: 10.1021/jm040804f. [DOI] [PubMed] [Google Scholar]
- 69.Kinoshita K, Nakamura H. eF-site and PDBjViewer: database and viewer for protein functional sites. Bioinformatics. 2004;20:1329–1330. doi: 10.1093/bioinformatics/bth073. [DOI] [PubMed] [Google Scholar]
- 70.Kellenberger E, Muller P, Schalon C, Bret G, Foata N, Rognan D. sc-PDB: an annotated database of druggable binding sites from the protein data bank. J Chem Inf Model. 2006;46:717–727. doi: 10.1021/ci050372x. [DOI] [PubMed] [Google Scholar]
- 71.Snyder KA, Feldman HJ, Dumontier M, Salama JJ, Hogue CWV. Domain-based small molecule binding site annotation. BMC Bioinformatics. 2006;7:152. doi: 10.1186/1471-2105-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teyra J, Paszkowski-Rogacz M, Anders G, Pisabarro MT. SCOWLP classification: structural comparison and analysis of protein binding regions. BMC Bioinformatics. 2008;9:9. doi: 10.1186/1471-2105-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gomis-Rüth FX. Structure and mechanism of metallocarboxypeptidases. Crit Rev Biochem Mol Biol. 2008;43:319–345. doi: 10.1080/10409230802376375. [DOI] [PubMed] [Google Scholar]
- 74.Vocadlo DJ, Davies GJ. Mechanistic insights into glycosidase chemistry. Curr Opin Chem Biol. 2008;12:539–555. doi: 10.1016/j.cbpa.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 75.Holliday GL, Almonacid DE, Bartlett GJ, O’Boyle NM, Torrance JW, Murray-Rust P, Mitchell JBO, Thornton JM. MACiE (mechanism, annotation and classification in enzymes): novel tools for searching catalytic mechanisms. Nucleic Acids Res. 2007;35:D515–D520. doi: 10.1093/nar/gkl774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holliday GL, Mitchell JBO, Thornton JM. Understanding the functional roles of amino acid residues in enzyme catalysis. J Mol Biol. 2009;390:560–577. doi: 10.1016/j.jmb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 77.Reis P, Holmberg K, Watzke H, Leser ME, Miller R. Lipases at interfaces: a review. Adv Colloid Interface Sci. 2009;147–148:237–250. doi: 10.1016/j.cis.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 78.Yon JM, Perahia D, Ghélis C. Conformational dynamics and enzyme activity. Biochimie. 1998;80:33–42. doi: 10.1016/s0300-9084(98)80054-0. [DOI] [PubMed] [Google Scholar]
- 79.Hammes GG. Multiple conformational changes in enzyme catalysis. Biochemistry. 2002;41:8221–8228. doi: 10.1021/bi0260839. [DOI] [PubMed] [Google Scholar]
- 80.Pantoja-Uceda D, Arolas JL, García P, López-Hernández E, Padró D, Aviles FX, Blanco FJ. The NMR structure and dynamics of the two-domain tick carboxypeptidase inhibitor reveal flexibility in its free form and stiffness upon binding to human carboxypeptidase B. Biochemistry. 2008;47:7066–7078. doi: 10.1021/bi800403m. [DOI] [PubMed] [Google Scholar]
- 81.Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 82.Botos I, Wlodawer A. The expanding diversity of serine hydrolases. Curr Opin Struct Biol. 2007;17:683–690. doi: 10.1016/j.sbi.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Porter CT, Bartlett GJ, Thornton JM. The catalytic site atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.UniProt Consortium, The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.BLASTClust: version 2.2.18, ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html
- 87.Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009;37:D323–D327. doi: 10.1093/nar/gkn822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Provost F, Fawcett T (2000) Robust classification for imprecise environments. Mach Learn 203–231, Kluwer Academic Publishers
- 89.Bartlett GJ, Porter CT, Borkakoti N, Thornton JM. Analysis of catalytic residues in enzyme active sites. J Mol Biol. 2002;324:105–121. doi: 10.1016/s0022-2836(02)01036-7. [DOI] [PubMed] [Google Scholar]
- 90.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 91.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 92.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. The protein data bank. Acta Crystallogr D Biol Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 93.IUBMB: Nomenclature Committee of the International Union of Biochemistry and Molecular Biology, http://www.chem.qmul.ac.uk/iubmb/
- 94.Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH-a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 95.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. J Mol Biol. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 96.Kallenbach N. Breaking open a protein barrel. Proc Natl Acad Sci USA. 2001;98:2958–2960. doi: 10.1073/pnas.071051798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 98.Fawcett T. An introduction to ROC analysis. Pattern Recogn Lett. 2006;27:861–874. [Google Scholar]
- 99.Siksnys V, Skirgaila R, Sasnauskas G, Urbanke C, Cherny D, Grazulis S, Huber R. he Cfr10I restriction enzyme is functional as a tetramer. J Mol Biol. 1999;291:1105–1118. doi: 10.1006/jmbi.1999.2977. [DOI] [PubMed] [Google Scholar]
- 100.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 101.Tadokoro T, Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 102.Katayanagi K, Okumura M, Morikawa K. Crystal structure of Escherichia coli RNase HI in complex with Mg2 + at 2.8 A resolution: proof for a single Mg(2+)-binding site. Proteins. 1993;17:337–346. doi: 10.1002/prot.340170402. [DOI] [PubMed] [Google Scholar]
- 103.Tsunaka Y, Takano K, Matsumura H, Yamagata Y, Kanaya S. Identification of single Mn(2+) binding sites required for activation of the mutant proteins of E.coli RNase HI at Glu48 and/or Asp134 by X-ray crystallography. J Mol Biol. 2005;345:1171–1183. doi: 10.1016/j.jmb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 104.Oda Y, Yoshida M, Kanaya S. Role of histidine 124 in the catalytic function of ribonuclease HI from Escherichia coli. J Biol Chem. 1993;268:88–92. [PubMed] [Google Scholar]
- 105.Stummeyer K, Dickmanns A, Mühlenhoff M, Gerardy-Schahn R, Ficner R. Crystal structure of the polysialic acid-degrading ndosialidase of bacteriophage K1F. Nat Struct Mol Biol. 2005;12:90–96. doi: 10.1038/nsmb874. [DOI] [PubMed] [Google Scholar]
- 106.Eichinger A, Beisel HG, Jacob U, Huber R, Medrano FJ, Banbula A, Potempa J, Travis J, Bode W. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 1999;18:5453–5462. doi: 10.1093/emboj/18.20.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Levitt M. A simplified representation of protein conformations for rapid simulation of protein folding. J Mol Biol. 1976;104:59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.