Abstract
Alternative splicing is generally controlled by proteins that bind directly to regulatory sequence elements and either activate or repress splicing of adjacent splice sites in a target pre-mRNA. Here, we have combined RNAi and mRNA-seq to identify exons that are regulated by Pasilla (PS), the Drosophila melanogaster ortholog of mammalian NOVA1 and NOVA2. We identified 405 splicing events in 323 genes that are significantly affected upon depletion of ps, many of which were annotated as being constitutively spliced. The sequence regions upstream and within PS-repressed exons and downstream from PS-activated exons are enriched for YCAY repeats, and these are consistent with the location of these motifs near NOVA-regulated exons in mammals. Thus, the RNA regulatory map of PS and NOVA1/2 is highly conserved between insects and mammals despite the fact that the target gene orthologs regulated by PS and NOVA1/2 are almost entirely nonoverlapping. This observation suggests that the regulatory codes of individual RNA binding proteins may be nearly immutable, yet the regulatory modules controlled by these proteins are highly evolvable.
Alternative splicing is a process by which multiple messenger RNAs (mRNAs) can be generated by joining exons together in different combinations. This process is used to both increase protein diversity and to regulate gene expression (Nilsen and Graveley 2010). Approximately 95% of human genes contain introns and therefore have the potential to be alternatively spliced. Recent deep sequencing surveys of 10 human tissues found that nearly all (95%–98%) multi-exon human genes are alternatively spliced (Pan et al. 2008; Wang et al. 2008). Given the ubiquity of alternative splicing and the key roles it plays in the control of gene expression, it is important to develop a complete understanding of the mechanisms by which alternative splicing is regulated.
Alternative splicing is most commonly controlled by RNA binding proteins that bind to sequence elements called enhancers and silencers (Nilsen and Graveley 2010). Splicing regulators bound to these enhancers or silencers are thought to either recruit or inhibit assembly or activity of spliceosomal components at nearby splice sites. The best-characterized splicing regulator proteins are the SR and hnRNP protein families. SR proteins primarily bind to enhancer sequences in exons where they activate adjacent splice sites, while hnRNPs have mostly been shown to suppress splicing when bound to intronic silencers. In addition to SR and hnRNPs proteins, several other splicing regulators have been identified that function in a tissue specific manner (Chen and Manley 2009).
The mammalian proteins NOVA1 and NOVA2 (collectively named here as NOVA) are perhaps the best-characterized splicing regulators to date. NOVA1/2 encode RNA binding proteins with three KH-domains that recognize clusters of YCAY repeats. Over the past decade, several hundred splicing events have been shown to be regulated by NOVA1/2 (Ule et al. 2005, 2006; Licatalosi et al. 2008). A comparison of the locations of the NOVA1/2 binding sites with NOVA-regulated splicing events has revealed a stereotypical “RNA map” for NOVA1/2. Specifically, regions upstream of exons where NOVA inhibits splicing and regions downstream from exons where NOVA activates splicing were enriched with NOVA binding sites (Ule et al. 2006; Licatalosi et al. 2008). Similar “RNA maps” that link the position of binding sites to typical activities of the regulatory proteins have also been developed for mammalian FOX1/2 (Zhang et al. 2008; Yeo et al. 2009), PTB (Xue et al. 2009), and four D. melanogaster hnRNP proteins (Blanchette et al. 2009). Such maps, splicing expression data, and RNA sequence motifs have recently been used to predict regulated tissue-specific splicing changes in mouse, strongly supporting the existence of a splicing code (Wang and Burge 2008; Barash et al. 2010; Zhang et al. 2010), a decipherable sequence-based information system that dictates the splicing pattern of a given pre-mRNA under a specific condition. Though considerable progress has been made, interpreting this code remains a formidable task in the field. In particular, it is unclear how the mouse splicing code can be applied to different species, especially distantly related organisms such as Drosophila. Moreover, the extent to which the RNA maps of individual splicing regulators are static or plastic throughout evolution has been unknown.
We were interested in exploring the conservation of the splicing code between distantly related organisms. As a first step in this process, we sought to generate an RNA map of Pasilla (PS) (Seshaiah et al. 2001), the D. melanogaster ortholog of NOVA1/2. To identify PS-regulated exons, we used RNA-seq (Wold and Myers 2008) to identify splicing events that changed upon depletion of PS by RNAi. We conclude that the RNA map of PS and NOVA1/2 is highly conserved between mammals and insects.
Results
Transcriptome analysis of untreated and ps(RNAi) S2 cells
To identify regulatory targets of PS, S2-DRSC cells were cultured in biological quadruplicate with the presence of a 444 bp dsRNA fragment corresponding to the ps mRNA sequence. In parallel, untreated S2-DRSC were cultured in biological triplicate to serve as a control (Fig. 1A). After four days of treatment, total RNA was isolated. Semiquantitative RT-PCR was used to demonstrate that the ps mRNA levels were ∼60% lower in the ps(RNAi) cells than in the untreated S2 cells (Supplemental Fig. 1). The efficiency of RNAi was also confirmed by the RNA-seq data as the number of normalized reads (fragments per kilobase of exon model per million mapped reads [FPKM]; Trapnell et al. 2010) for ps mRNA FPKM were ∼4.7-fold lower in the ps(RNAi) cells than untreated S2 cells. No other genes containing a KH or RRM domain had a significant change in gene expression (see Supplemental Methods).
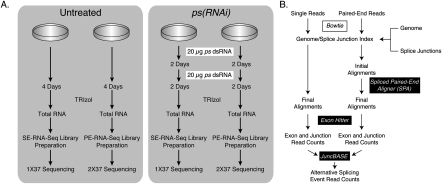
Figure 1.
Experimental and analytical approach. (A) S2-DRSC cells were either untreated or treated with two 20-μg doses of dsRNA. After a total of 4 d of incubation with the dsRNA, total RNA was isolated and used for preparing either single-end or paired-end RNA-seq libraries. The single-end libraries were sequenced using 37–45 cycles, while the paired-end libraries were sequenced using 37 cycles for each read. The single-end sequences were trimmed from the 3′ end to a total length of 37 nt prior to alignment. (B) Sequence analysis involved aligning all reads to a combined database of the genome and splice junctions using Bowtie (Langmead et al. 2009). The paired-end alignments were further analyzed using Spliced Paired-End Aligner (SPA) to identify mate pairs that map to the same chromosome, oriented toward one another and within 200 kb of one another. The aligned reads were then analyzed using exonhitter.pl (McManus et al. 2010) to count the number of reads that mapped to exons, splice junctions, and exon–intron boundaries. The read counts were then further analyzed using juncBASE to identify alternative splicing events that were significantly different between the Untreated and ps(RNAi) samples.
RNA-seq libraries were prepared from each biological replicate by performing two rounds of poly(A)+ enrichment, RNA fragmentation, random hexamer-primed cDNA synthesis, linker ligation, PCR enrichment, and size selection. The libraries were sequenced using both single-read and paired-end methodology with read lengths between 37 nt and 45 nt. Paired-end reads had an approximate insert size of 175 ± 50 bp (Supplemental Fig. 2). For consistency, all reads were trimmed to 37 nt from the 3′ end prior to alignment.
Our mapping strategy involved simultaneously aligning the reads to the genome sequence and splice junction sequences (Fig. 1B). We first performed a series of experiments to determine the alignment parameters that maximized the number of reads that reliably aligned to the splice junctions, as they provide the most information regarding splicing events. To do this, we used Bowtie (Langmead et al. 2009) to align the reads against the D. melanogaster genome sequence, 58,212 annotated splice junction sequences, and 221,388 predicted splice junction sequences. The predicted splice junctions contained all possible in-order junctions formed from annotated splice sites from the same gene as well as junctions formed by annotated splice sites from different genes, but within 2 kb away. Our reads were 37 nt; therefore, our splice junction sequences were 62 nt long (31 nt on either side of the splice junction) to ensure a ≥6-nt overhang of the read mapping from one side of the junction onto the other. This yielded an even coverage of alignment positions across all splice junctions (Supplemental Fig. 3A). There is a decrease in coverage for overhang lengths ≤5 nt as a result of reads no longer uniquely aligning to a junction. To test the effect of trimming the reads, we determined the number of reads that aligned to both the genome and splice junctions when we altered the number of bases trimmed from the 5′, 3′, or both ends of the read and allowed for up to two mismatches (Supplemental Fig. 4). These analyses revealed that the yield of splice junctions was maximized by not trimming the reads at either end—trimming from either end increased the number of reads that could be uniquely aligned to the genome, but reduced the number of reads that were uniquely aligned to the splice junctions (Supplemental Fig. 4). Using our alignment parameters, we observed a high correlation between biological (r2 = 0.88–0.89) and technical (r2 = 0.92–1.0) replicates of our samples when comparing the number of reads that aligned to splice junctions (Supplemental Fig. 5).
As our splice junction data set contains nearly four times as many predicted junctions as annotated junctions, we assessed the criteria that could be used to distinguish between true splice junctions and false-positive splice junctions. To do this, we generated a set of 5,409,600 splice junctions in which annotated exons from different chromosomes were randomly selected and spliced together in silico. Alignments to these junctions are considered false-positives, as such junctions are thought to rarely exist when compared to annotated junctions. Comparison of the alignment results of one lane of data containing 6.2 million paired-end reads to the genome and either the annotated or random splice junctions revealed that the false-positive rate could be greatly reduced (0.006% false-positive) by requiring at least three different start positions (offsets) for reads spanning the junction (Supplemental Fig. 3B). Thus, we have instituted a cutoff of at least three distinct offsets to consider a predicted splice junction as a confident splice junction. This filtering resulted in a final junction data set of 28,926 confident junctions from the ps(RNAi) and control samples.
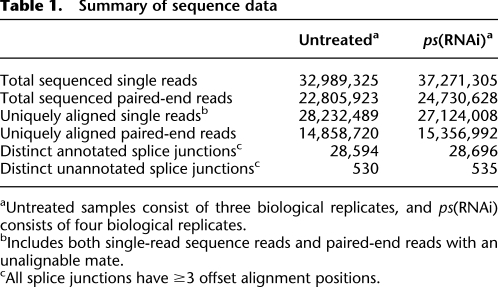
We aligned all single reads to the combined genome and splice junctions using the parameters outlined above. For our paired-end alignments, we used our spliced paired-end aligner (SPA), which aims to uniquely map mate pairs to ensure consistency between mappable reads, particularly when one or both reads align to splice junctions, and to uniquely place the pairs in instances where the individual reads could not be mapped uniquely. SPA treats each read of the mate pair as a single read and aligns them using Bowtie to the combined genome and splice junction sequences. All mapping positions for reads that can be mapped up to 10 possible locations are reported. SPA then processes the Bowtie output to identify mate pair combinations in which both reads map to the same chromosome, to opposite strands, are oriented toward one another, and are <200 kb apart (the size of the largest annotated D. melanogaster intron). Out of 47.5 million paired reads, 30.2 million (64%) were uniquely aligned given all three criteria. Importantly, the distribution of the distance between mate pairs was consistent with the insert size selected during the library preparation (Supplemental Fig. 2). The remaining reads were further analyzed to include cases where one read could be uniquely aligned, but the other read had no valid alignment. These are most likely instances where the unalignable read had a high error rate. This step “rescued” an additional 9.4 million (20%) reads from the paired-end sequence data that were treated as single reads. In summary, our alignment strategy yielded 115.8 million uniquely mapped 37-nt read sequences from the untreated and ps(RNAi) samples, of which ∼5% (5.76/115.8 million) map to splice junctions (Table 1).
Table 1.
Summary of sequence data
aUntreated samples consist of three biological replicates, and ps(RNAi) consists of four biological replicates.
bIncludes both single-read sequence reads and paired-end reads with an unalignable mate.
cAll splice junctions have ≥3 offset alignment positions.
Identification of PS-affected splicing events
To identify changes in splicing upon depletion of PS, we used our JuncBASE (junction based analysis of splicing events), which takes as input genome coordinates of all annotated exons and all confidently identified splice junctions (including annotated and novel junctions) to find sets of exons and junctions that can be classified as one of eight types of alternative splicing events: cassette exons, alternative 5′ splice sites, alternative 3′ splice sites, mutually exclusive exons, coordinate cassette exons, alternative first exons, alternative last exons, and retained introns (see Methods; Fig. 2). After identifying each splicing event, JuncBASE calculates the sum of single reads, mate pairs, and splice junction reads that align to either the inclusion or exclusion isoforms in both the untreated and ps(RNAi) samples, to determine if a shift in splicing has occurred upon depletion of PS. Mate pairs aligning to an isoform are treated as one event giving evidence for that isoform, instead of considering each mate as an independent read. A Fisher's exact test was performed on 2 × 2 contingency tables comprised of these read counts [inclusion vs. exclusion, untreated vs. ps(RNAi)]. This recapitulates key aspects of the approach described in Wang et al. (2008) with several distinctions. First, to identify retained intron events, we found that counting read alignments throughout the entire intron (as evidence for the inclusion of the intron) was confounded in cases where additional splice sites reside within the intron; therefore, only reads spanning the exon–intron boundaries of the 5′ and 3′ splice sites were used as evidence for retention of the intron (red bars with black lines in Fig. 2). Moreover, each end of the intron was tested separately for intron retention and then P-values were combined for a final P-value associated with the retained intron event (Supplemental Fig. 6). We also extended the Wang et al. (2008) method to identify coordinate cassette exons—two or more exons skipped or included as a group. Finally, we did not examine tandem 3′ untranslated region (UTR) events (alternative polyadenylation), seeing that there were few reads that gave direct evidence of a poly(A) site and that there are no exon reads or junction reads that are specific to the exclusion isoform (Supplemental Methods).
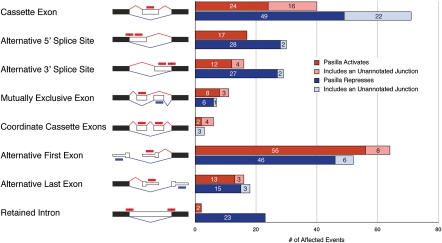
Figure 2.
405 PS-regulated pre-mRNA processing events. (Black boxes) Constitutive regions; (white boxes) alternative regions. (Red lines) Splice junctions for the inclusion isoform; (blue lines) junctions for the exclusion isoform. (Red bars) Exonic reads that support the inclusion isoforms; (blue bars) exonic reads that support the exclusion isoforms; (red bars with a black line) reads that support the inclusion isoform, but have a shared portion with the exclusion isoform. Thinner portions of the boxes in alternative first exons and alternative last exons correspond to UTRs.
From the 2 × 2 tables of counts constructed for each splicing event, we were able to classify 494 splicing events from 323 genes that changed significantly in the ps(RNAi) sample (Benjamini-Hochberg adjusted P-value ≤ 0.05; Supplemental Fig. 7; Supplemental Data Sets 1–3). Within each of the eight types of alternative splicing, we identified a nonredundant set of splicing events with no overlapping introns. If two events had an overlapping intron, the event with the lowest P-value was kept. From our nonredundant set, we identified 405 total splicing events affected by ps(RNAi) (Fig. 2). Semiquantitative RT-PCR experiments validated all 16 tested splicing events we identified by RNA-seq (Fig. 3; Supplemental Fig. 8). We did not observe a general decrease in gene expression upon RNAi, as noted in previous studies (Supplemental Fig. 5E,F; Blanchette et al. 2005), and moreover, our approach to identify changes in splicing accounts for any change in overall expression (row and column sums of the 2 × 2 contingency tables).
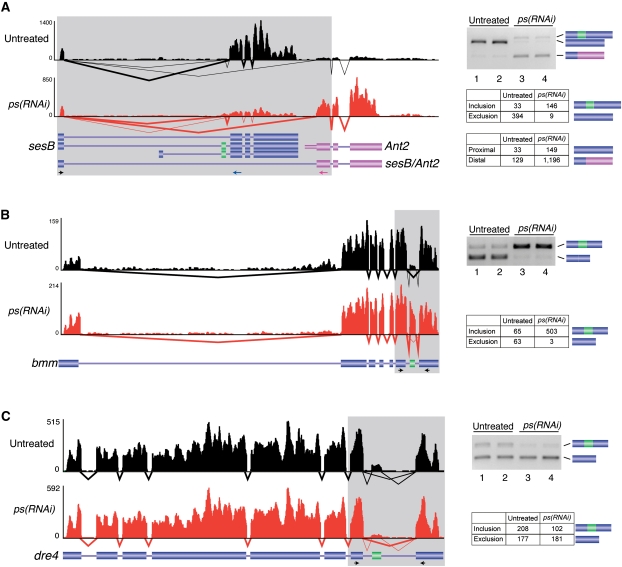
Figure 3.
Examples of PS-regulated splicing events. Alternative splicing events in the sesB/Ant2 (A), bmm (B), and dre4 (C) genes were identified from the RNA-seq data and validated by RT-PCR. In each case, the RNA-seq coverage and splicing patterns for both the untreated (black) and ps(RNAi) (red) samples are shown along with the annotated transcript models. The RT-PCR validation assays were performed in biological duplicates for both the untreated (lanes 1,2) and ps(RNAi) samples (lanes 3,4). The number of read counts supporting each splicing event in each sample is indicated in the tables on the right.
Figure 3 highlights three examples of PS-regulated splicing events we identified and validated. The first example involves the adjacent sesB and Ant2 genes (Fig. 3A). In untreated cells, the first exon of sesB is most frequently spliced to the downstream constitutive sesB exon (top isoform). However, in ps(RNAi) cells, splicing is strongly switched to favor the expression of two different isoforms. The first involves splicing of the first exon of sesB to the first exon of Ant2 (bottom isoform). The second isoform involves an increased inclusion of an alternative cassette exon (green) in sesB. A second example of a PS-regulated splicing event involves a cassette exon in the bmm gene (Fig. 3B). In this case, the exon is included in ∼50% of the transcripts in untreated cells as determined by RNA-seq, but nearly constitutively included in the ps(RNAi) cells. The final example involves the dre4 gene where a cassette exon is activated by PS and therefore skipped more frequently in the ps(RNAi) cells than in the untreated cells (Fig. 3C).
Our analysis method is quite sensitive, as we were able to identify PS-affected splicing events even when an isoform is normally expressed at a low level. For example, in untreated cells there were 865 reads supporting the inclusion of a validated PS-affected cassette exon in the cg gene, but only nine reads supporting the exclusion of the exon (Supplemental Fig. 8B). However, in ps(RNAi) cells, the exon is constitutively included as there are 831 reads supporting inclusion of the exons and no reads supporting exclusion. These results indicate that this exon is normally repressed by PS at a very low level. The change in inclusion of this exon in the ps(RNAi) cells is observed in the RT-PCR validation experiment (Supplemental Fig. 8B). While splicing events such as this can be detected when measuring both exon and splice-junction reads, they can be missed if only exon reads are considered.
Interestingly, while most (327 of 405) of the PS-affected splicing events contained entirely annotated junctions, 19% (77 of 405) of the affected splicing events involved unannotated splice junctions. Strikingly, 90% (69 of 77) of these affected, unannotated splice junctions were expressed in untreated S2-DRSC cells while only 10% (8 of 77) of the unannotated splice junctions are exclusively expressed in the ps(RNAi) cells (Fig. 2). A particularly striking example involving these unannotated splice junctions is found in the trol gene in which a group of nine contiguous exons, which are annotated as being constitutive, are coordinately skipped in untreated cells (with 52 reads spanning the skipping junction and only 136 reads total for all nine exons and inclusion junctions connecting these exons) but coordinately included in the ps(RNAi) cells (where no reads support skipping of these exons but 3573 reads support inclusion of the nine exons) (Supplemental Fig. 8A).
We next analyzed several general features of the set of PS-affected splicing events. We find that PS predominantly functions as a splicing repressor, as a majority (60%) of the affected splicing events we identified was repressed by PS. Of the splicing events that only involve splicing [and not differential promoter or poly(A) site use], PS regulated the greatest number of cassette exons (111), roughly equal numbers of alternative 5′ or 3′ splice sites (47 and 45, respectively), and relatively few intron retention events (25), mutually exclusive exons (18), and coordinately regulated cassette exons (nine) (Supplemental Fig. 9). However, when considering what fraction of expressed alternative splicing events are affected by PS, the greatest fraction of mutually exclusive splicing events (62%) were affected, and between 10% and 20% of cassette, alternative 5′ and 3′ splice sites, and only 1.5% of intron retention events (Supplemental Fig. 9). Perhaps surprisingly, we find that PS affected a significant number (116) of alternative first exon events and a smaller number (34) of alternative last exon events. However, it is unclear if these events are changing due to a direct or secondary effect of PS on the coordination of splicing with either transcription or polyadenylation.
A conserved Pasilla/NOVA-RNA map
Mammalian NOVA1/2 is known to bind directly to sequences that match the consensus motif YCAY and has the highest affinity for YCAY repeats (Jensen et al. 2000). The amino acids of NOVA1/2 that contact RNA in a sequence-specific manner (Lewis et al. 1999, 2000) are conserved in PS, suggesting that PS also recognizes YCAY motifs. Consistent with this, biochemical experiments confirm that recombinant PS can bind to YCAY containing RNA (Supplemental Fig. 10). To investigate potential PS-binding sites in silico, we identified overrepresented hexamers in conserved sequences within and 150 nt upstream of and downstream from our set of affected cassette exons compared to a set of unaffected cassette exons (Benjamini-Hochberg adjusted P-value ≥ 0.95). The top five hexamers, in order, were CACCAC, CCACCA, CAACAA, AACAAC, and ACAACA (P-value < 2.0 × 10−4, not correcting for 4096 tested hexamers). Consistent with the known RNA binding preference of NOVA1/2 and PS, the top two hexamers contain a YCAY sequence.
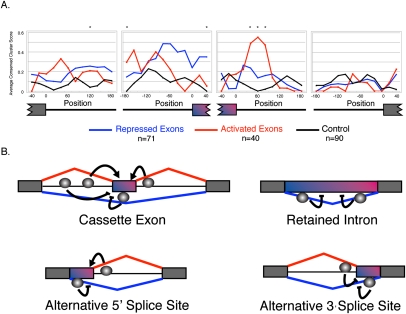
NOVA1/2 has previously been shown to preferentially repress downstream splice sites and activate upstream splice sites (Ule et al. 2006; Licatalosi et al. 2008). We were interested in determining whether a similar RNA map exists for PS. Using the binding model introduced for NOVA targets (Ule et al. 2006; Supplemental Methods), we tested for the enrichment of conserved clusters of YCAY motifs in 45-nt sliding windows across introns and exons of PS-activated and PS-repressed splicing events as well as unaffected splicing events to serve as a control (Fig. 4; Supplemental Fig. 11); which yielded 452 tests across cassette exons, alternative 5′ splice sites, alternative 3′ splice sites, alternative first exons, alternative last exons, and retained introns. We observed that positions upstream and within PS-repressed cassette exons had a higher average conserved YCAY cluster score than the control exons; however, only one position within the alternative exon had an overrepresentation of YCAY clusters with an uncorrected P-value < 0.01 for 452 tests. We observe that positions downstream from PS-activated cassette exons had an overrepresentation of conserved YCAY clusters, with a peak enrichment 60 nt downstream from the cassette exon (uncorrected P-value < 0.01). We also find conserved YCAY clusters further upstream of PS-activated exons and conserved YCAY clusters near upstream constitutive exons of PS-repressed exons (uncorrected P-value < 0.01). The locations of these enriched YCAY motifs near PS-regulated cassette exons are analogous to the locations of the YCAY motifs near NOVA-regulated exons (Ule et al. 2006).
Figure 4.
A Pasilla RNA-map. (A) Each position in the graph represents the average conserved YCAY cluster score, within a centered sequence window of 45 nt. The conserved YCAY cluster score was calculated for cassette exons that are activated by PS, repressed by PS, and unaffected cassette exons (Fisher's exact test, Benjamini-Hochberg adjusted P-value ≥ 0.95). Only regions adjacent to introns >400 nt were used for scoring. Positions with enriched YCAY cluster scores are indicated by an asterisk (Wilcoxon-rank sum test, uncorrected P-value < 0.01). (B) Positions near cassette exon events, alternative 5′ splice site events, alternative 3′ splice site events, and retained intron events with an enrichment of YCAY clusters. Gray spheres indicate the relative positions containing enriched binding sites. Detailed plots of average conserved YCAY cluster scores are shown in Supplemental Figure 9.
The conservation of the regulatory “map” is not due to the conservation of NOVA and PS target genes. Out of 47 mouse genes that were identified as targets of NOVA (Ule et al. 2006), 33 had at least one Drosophila ortholog (Li et al. 2006; Ruan et al. 2008). Of the 33 Drosophila orthologs, 23 were expressed in the S2 cell line. Amongst these 23 NOVA target genes with an S2-expressed Drosophila ortholog, only four were also a target of PS.
In addition to the regulatory map for cassette exons, we created maps for other alternative splicing events. There is an enrichment of YCAY motifs near PS-regulated alternative 5′ splice sites (Bonferroni-adjusted P-value < 0.05) and 3′ splice sites (uncorrected P-value < 0.01) and the enrichment near alternative 5′ splice sites is significant given multiple-testing correction for 452 tests (Fig. 4B; Supplemental Fig. 11A,B). The alternative 5′ and 3′ splice site map of PS is consistent with the asymmetric action of NOVA that was observed in the validated targets of NOVA (Ule et al. 2006).
We identified 23 introns that were retained significantly more often in the absence of PS. Positions adjacent to both the 5′ and 3′ splice sites of these introns had a significant enrichment of conserved YCAY clusters (Bonferroni-corrected P-value < 0.05). Therefore, the effect of PS on alternative splicing is not only dependent on its position with respect to a splice site, but also depends on the context of the type of alternative splicing event. However, a common pattern observed in cassette exon events, alternative 5′ splice sites, alternative 3′ splice sites, and retained intron events, was the presence of YCAY binding sites within the alternative exon (or portion of an exon) that is normally repressed in the presence of PS (Supplemental Fig. 11).
Finally, the NOVA RNA map contains an enrichment of YCAY motifs near the proximal poly(A) site in cases of alternative polyadenlyation events regulated by NOVA. While we observe an enrichment of YCAY motifs near PS-regulated alternative last exons in an analogous position, the enrichment is not statistically significant (Supplemental Fig. 11D).
Although the general RNA regulatory maps of NOVA and PS are quite similar, the scoring method used to predict NOVA exon targets was insufficient to distinguish PS-regulated exons from control exons or the direction of regulation (Supplemental Fig. 12). Perhaps, this is due to the lack of a significant silencer region directly upstream of the alternative exon (NISS2) and the lack of a significant enhancer region further downstream from the alternative exon (NISE3) that were identified near NOVA-regulated exons.
Discussion
We have identified 405 splicing events that are affected by RNAi depletion of PS. Interestingly, we found that 19% of the PS-regulated alternative splicing events contain an unannotated splice junction. The large proportion of unannotated splicing events in our affected set of exons demonstrates the benefits of using RNA-seq as a method to detect regulated alternative splicing events over other methods such as microarrays, which rely on predefined splicing events to probe. Previous studies using splice junction microarrays to identify alternative splicing events in Drosophila found that 29%–35% of their affected junctions were annotated as constitutive (Blanchette et al. 2005, 2009), which suggests that many of these junctions were participating in unannotated alternative splicing events. This work only looked at potential splice junctions formed between annotated splice sites; therefore, future work could identify even more cases of alternative splice junctions if unannotated splice sites are used.
The large number of unannotated splice junctions we identified also indicates that the D. melanogaster transcript annotations remain incomplete. More importantly, the fact that 90% of these unannotated splice junctions are observed in untreated S2-DRSC cells, and are not specific to the ps(RNAi) cells, indicates that these junctions are normally expressed and are not aberrant splicing events induced by depleting a splicing regulatory protein.
By calculating conserved YCAY cluster scores across affected cassette exons, alternative 5′ splice sites, and alternative 3′ splice sites that were activated or repressed by PS, we found that the PS RNA map recapitulates the major features of the NOVA RNA map. Both NOVA1/2 and the Drosophila ortholog, PS, appear to activate splicing of upstream exons and repress splicing of downstream exons or exons they directly bind to; however, further studies profiling transcriptome-wide PS binding are necessary to confirm the direct targets, as was confirmed with NOVA2 (Licatalosi et al. 2008). Ule et al. (2006) proposes a molecular mechanism for the action of NOVA; however, further work will need to be performed to determine if the same molecular mechanism is used by PS.
The PS RNA map is not similar to patterns of regulation identified for nonhomologous proteins such as PTB (Xue et al. 2009) and HNRNPC (König et al. 2010), further supporting an ancestrally conserved mechanism of splicing regulation for the orthologous proteins NOVA and PS. Although the PS and NOVA RNA maps are similar to the mammalian FOX-1/2 map, a recent study suggests that these maps are in part similar due to the proteins combinatorial affects on a subset of alternative splicing events (Zhang et al. 2010).
The four shared target genes of NOVA and PS (out of 23 NOVA target genes with at least one expressed Drosophila ortholog) are cac(CG1522), cora(CG11949), msn(CG16973), and caki(CG6703). According to their Gene Ontology (GO) annotations, cac has voltage-gated calcium channel activity and caki has calmodulin-dependent protein kinase activity and is also involved with cell adhesion. Both cac and caki have GO annotations involved with adult locomotory behavior, which suggests a possible ancestral role for a neurological function of PS and NOVA.
Although there are a few shared target genes, the RNA-maps generated from the targets of NOVA and from the targets of PS are based on almost entirely distinct sets of genes. This further supports the role of cis-acting binding sites in driving the evolution of alternative splicing, a result seen from a previous study examining orthologous NOVA-regulated exons in vertebrates (Jelen et al. 2007). These results suggest a general evolutionary model common to alternative splicing, transcription regulation, and miRNAs (Meireles-Filho and Stark 2009; Shomron et al. 2009). In general it appears that targeting molecules retain their sequence specificity over long periods of evolutionary time, and changes in the regulation, including large-scale changes in targets, occur through gains and losses of cis-acting binding sites.
While there was little overlap in NOVA and PS targets based on our studies in S2 cells, we cannot assume that NOVA and PS do not have shared targets in other contexts. Our study was performed in cell culture while the NOVA studies were performed in mouse brain. In the context of a different cell line or in ps mutant flies, we might observe an increase in overlapping genes; however, a previous study of ps mutants (Seshaiah et al. 2001) suggests that PS may have a different physiological function than NOVA. Homozygous ps mutants showed strong defects in salivary gland development, but neurological defects were not observed (Seshaiah et al. 2001). Nonetheless, a GO analysis of the set of PS-affected genes, using Funcassociate (Berriz et al. 2009), identified overrepresented terms corresponding to neuronal functions (regulation of neurogenesis, locomotion, regulation of axonogeneis, chemosensory behavior, etc.) as well as sexual reproduction (anatomical structure morphogenesis, gamete generation, oogenesis, etc.) and the cytoskeleton (cytoskeletal protein binding, actin binding, cytoskeletal protein binding, etc.) (Funcassociate adjusted P-value < 0.01) (Supplemental Data Set 4). While the expression pattern of PS and the ps mutant phenotype suggests that PS has an important role in salivary gland development, our results suggest that PS may also have additional roles not only in sexual reproduction and cytoskeleton dynamics, but also in neural function, like NOVA1/2. Thus, despite little overlap in the regulatory targets, the regulatory mechanisms and physiological functions of orthologous splicing regulators may be conserved.
In conclusion, we have identified and classified hundreds of alternative splicing events that are affected by one splicing regulator and have shown that the overall RNA map relating the position of binding sites for the factor to its affect on splicing is conserved from insects to mammals. Future studies that deplete other splicing regulators in Drosophila may also reveal regulatory maps that can perhaps be applied to mammalian splicing regulators as well.
Methods
RNA interference
RNA interference was performed essentially as described previously (Park et al. 2004; Park and Graveley 2005). A vector encoding double-stranded RNA for ps was generated as described previously (Park et al. 2004; Park and Graveley 2005). Briefly, cDNA fragments encoding for the specific dsRNA were amplified by RT-PCR with gene-specific primers (Supplemental Table 1) from total RNA isolated from S2-DRSC cells. The cDNA fragment was then cloned into the pCRII-TOPO vector (Invitrogen) and sequenced to verify the identity of the insert. DNA templates were amplified with M13 forward and M13 reverse primers. PCR products were used in individual in vitro transcription reactions with the Ampliscribe High Yield Transcription SP6 (Epicentre) kit and T7 kits (Epicentre) to generate the sense and antisense RNA strands. After DNase I digestion, the two single-stranded RNAs were annealed to generate double-stranded RNAs. Integrity of the PCR products, the single-stranded RNA transcripts, and dsRNAs were monitored by agarose gel electrophoresis.
S2-DRSC cells (obtained from the Drosophila Genomics Resource Center at Indiana University) were cultured with Schneider's medium (Sigma/Aldrich) plus 10% heat-inactivated fetal calf serum (FCS) (HyClone) at 27°C. One day prior to dsRNA treatment, cells were split into six-well culture dishes at a density of 1 × 106 cells/mL. Immediately prior to the addition of dsRNA, the culture medium was replaced with fresh Schneider's medium without FCS, followed by the addition of 20 μg of each dsRNA directly into the FCS-free medium and the cells incubated for 5 h at 27°C. After incubation with the dsRNA, 10% FCS was added back to cell culture. After 2 d, a second dose of 20 μg of dsRNA was added to each well in the same manner as described above and the cells incubated for two additional days after the re-addition of 10% FCS. After the dsRNA treatment, total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's directions. Parallel dsRNA treatments and total RNA preparations were performed independently for each replicate. Untreated S2-DRSC cells were used as a reference. To monitor the level of mRNA depletion, primer sets (Supplemental Table 2) that amplify regions of the targeted mRNAs outside of the dsRNA region were used for RT-PCR amplification, and compared with the results from the untreated cells (Supplemental Fig. 1).
Deep sequencing
All sequencing libraries were prepared with the mRNA-Seq Sample Prep Kits (Illumina) according to the manufacturer's instructions. Briefly, poly(A)+ RNA was purified from total RNA with oligo(dT) magnetic beads. The poly(A)+ RNA was fragmented using divalent cations under elevated temperature, followed by first and second strand cDNA synthesis primed with random hexamers. The cDNA fragments were end-repaired using T4 DNA polymerase and Klenow DNA polymerase, and phosphorylated at their 5′ ends with T4 polynucleotide kinase. After adding “A” bases to the 3′ end of the DNA fragments, Illumina adaptor oligonucleotides were ligated to the ends and ∼300-bp fragments were isolated from an agarose gel, enriched by PCR amplification, and gel-purified again. The samples were quantitated using a NanoDrop, loaded onto a flow-cell for cluster generation, and sequenced on an Illumina Genome Analyzer II using either single-read or paired-end protocols (Illumina).
Transcript annotations
Coding and noncoding transcript annotations were obtained from FlyBase r5.11 (Tweedie et al. 2009) and MB5 (http://www.modencode.org) and merged into a nonredundant set, allowing a 10-nt difference in the start and end coordinate of the first and last exon, respectively. Gene loci were inferred from a set of nonredundant transcripts by combining all transcripts with overlapping exons into a single gene locus (Supplemental Data Set 5).
Splice junction sequences
A database of 58,212 annotated and 221,388 unannotated (novel) splice junction sequences was created. 215,757 of the unannotated splice junctions were generated by joining every annotated exon with all possible downstream exons within the same gene. An additional set of 5631 novel junction sequences were created by joining every pair of exons from different gene loci that were ≤2 kb away. A separate database of 5,409,600 random splice junctions was created by joining each annotated 5′ splice site with 50 randomly drawn annotated 3′ splice sites located on a different chromosome and from each annotated 3′ splice site with 50 randomly drawn annotated 5′ splice sites from a different chromosome. All splice junctions contained 31 nt of exon sequence on either side of the junction in order to force an alignment overhang of at least 6 nt from one side of the splice junction to the other.
Single-read sequence alignments
Bowtie (Langmead et al. 2009) (with parameters: -m 1 -v 2 -best -y) was used to align the single-read sequences against a combined index containing both the D. melanogaster genome (dm3 assembly) (Adams et al. 2000; Celniker et al. 2002) and the splice junctions. All reads were first trimmed from the 3′ end to a total length of 37 nt. Reads that mapped uniquely with up to two mismatches were reported.
Paired-end sequence alignments
Paired-end alignments were conducted using Spliced Paired-End Aligner (SPA), which is a custom Perl script (spa.pl; Supplemental material) that uses Bowtie to independently align each read of a mate-pair and then parses the output files to identify the optimal alignment position for each read of each mate pair. Specifically, spa.pl calls Bowtie (version 0.9.9.2) to separately align each read of the mate pair to the combined genome and splice junction database, using the parameters: -v 2 -k 10 -m 10 -y -B 1, which reports all mapping locations for each read that maps with up to two mismatches to 10 or fewer locations using 1-based alignment coordinates. Next, spa.pl collects the genomic coordinates of the reads that map not only to the genome, but also the splice junctions, and then considers all possible combinations of the alignment positions of both reads of each mate pair (up to 100 possibilities, if both reads map to 10 locations). These possibilities are then filtered to identify mate-pair combinations in which both reads align to the same chromosome, the reads are oriented toward one another (i.e., there is both a “forward” and “reverse” read), and the reads are located within 200 kbp of one another on the genome. In cases where exactly one combination of mapping locations fulfills all three criteria, the mapping locations of the reads are reported. Optionally, but not used for this study, in cases where more than one combination of mapping locations fulfills all three criteria, the combination with the shortest genomic location is reported. In cases where one read of the mate pair can be mapped uniquely, but the other read cannot be mapped at all, the read is harvested as a uniquely aligned single read.
Associating mapped reads to annotation features
To reconcile the aligned reads with annotated features we used exonhitter.pl (McManus et al. 2010). Briefly, this involves intersecting alignment intervals with exons associated with known or predicted gene models. It is important to note that the assignment of reads to exons and introns is not always unambiguous, since there may exist distinct exons and introns that overlap when considering all isoforms of a given gene. Therefore, exonhitter.pl constructs such a mapping, noting when the mapping is unique and when it is ambiguous. Specifically, exonhitter.pl outputs a number of different output files containing read counts and feature coordinates, including: gene_hit_counts (counts of the number of reads aligning to each gene), unique_exon_hit_counts (counts of the number of reads aligning to unique exons), ambiguous_exon_hit_counts (counts of the number of reads aligning to ambiguous exons), junction_hit_counts (counts of the number reads that align to a given junction), unique_jctn_exons (lists the junction coordinates associated with reads that align to junction windows along with the exons that they map to for cases where both junction and exon assignments are unique), ambiguous_jctn_exons (same as the previous list except for cases where one or both of the exons are ambiguous), unique_mate_jctn (cases where the reads of a mate pair can be assigned uniquely to different exons allowing for the potential inferrence of a junction connecting the exons), ambiguous_mate_jctn (same as the previous list except for cases where one or both of the exons are ambiguous), unique_intron (read that can be assigned uniquely to an intron), ambiguous_intron (same as the previous list except for cases where the read cannot be uniquely assigned to a specific intron), and intergenic (list of reads whose alignments fall outside all annotated gene boundaries). In addition to the output files listed above, there are additional output files that consider boundary events: exon_intron_unique and exon_intron_ambiguous (lists the number of reads that straddle an exon:intron boundary and associate with a unique or ambiguous exon), exon_intron_jctn_hit_count (lists the number of reads that can be interpreted as straddling an exon:intron boundary specified by the exon boundary coordinate), and gene_boundary (lists the number of reads that straddle a gene boundary).
JuncBASE
JuncBASE (junction based analysis of splicing events) is a series of Python scripts that use the Bowtie, spa.pl, and exonhitter.pl output files to calculate exon exclusion and inclusion counts to splicing events and to identify statistically significant affected splicing events. It is important to note that all splice junctions used to identify significantly affected splicing events had at least three distinct offsets aligning to the junction given the pool of our alignments for all samples (based on criteria from our true- vs. false-positive analysis).
Figure 2 contains diagrams of exclusion and inclusion isoforms for the eight types of alternative splicing that were examined. Junctions and exons that are part of an exclusion isoform are depicted in blue and portions of inclusion isoforms are depicted in red. For both the untreated and ps(RNAi) samples, we counted the reads that aligned to the inclusion isoform or the exclusion isoform. For the paired-end alignments, if both ends of a read aligned to unique regions of an isoform (e.g., one read within a cassette exon and the other read to a junction that includes the cassette exon), the count was only incremented by one.
A more detailed explanation on identifying alternative splicing events and assigning read counts is described in Supplemental Methods. JuncBASE is available at http://compbio.berkeley.edu/proj/juncbase/.
Motif analysis
Overrepresented hexamers near affected cassette exons
Any overlapping phastCons conserved element within each affected cassette exon and 150 nt into the flanking introns were extracted from the UCSC Genome Browser MySQL database (http://genome.ucsc.edu/, April 2006 Assembly) (Chiaromonte et al. 2002; Kent et al. 2003; Schwartz et al. 2003; Blanchette et al. 2004; Siepel et al. 2005; Rhead et al. 2010). This was also done for the set of unaffected cassette exons (those with a Benjamini-Hochberg adjusted P-value ≥ 0.95). The proportion of hexamer sequences within the affected sequences and the control sequences were used to perform a Z-test for the difference in population proportions. No hexamer was significant given a Bonferroni-corrected P-value of 0.05 for 4096 tests. The top five scoring hexamers are reported; they have a raw P-value < 2.0 × 10−4.
Calculation of conserved YCAY motif clusters
Conserved YCAY motifs were searched near cassette exons, alternative 5′ splice sites, alternative 3′ splice sites, alternative first exons, alternative last exons, and retained intron events. For each AS event, a set of control exons were identified as those events with a Benjamini-Hochberg adjusted P-value > 0.95.
The average conserved YCAY cluster score was calculated in 45-nt windows with a step of 20 nt near each alternative splicing event, similar to what was described in (Ule et al. 2006). See Supplemental Methods for more details on the calculation of the YCAY conserved cluster score. If a cassette exon had multiple flanking introns, the longest intron was taken. For alternative 5′ and 3′ splice sites, the cluster scores were calculated near the constitutive splice site as well as near the alternative splice sites. If an alternative 5′ or 3′ splice site had multiple exclusion introns, the longest one was chosen. For alternative first exon events, the cluster scores are calculated near the constitutive splice site, near both alternative splice sites, and near the transcriptional start site; similarly for alternative last exon events, except near the poly(A) site. For retained intron events, cluster scores were calculated near both 5′ and 3′ splice sites. Plots of the average conserved cluster score were made only from introns that were ≥400 nt.
To identify specific windows that had a significant enrichment of YCAY motifs, we performed a Wilcoxon rank sum test on every window for every event, which yielded 452 tests. Positions with a raw P-value ≤ 0.01 are analogous to positions of conserved YCAY clusters near mouse NOVA target cassette exons. Moreover, we report positions that have a significant P-value given a more stringent Bonferroni correction of 0.05 upstream of alternative 5′ splice sites and within retained introns.
Validation
Alternative splicing events identified by analysis of the RNA-seq data were validated by RT-PCR. Briefly, PCR primers were designed to amplify multiple isoforms with different sizes. By comparing the splicing patterns between untreated cells and ps(RNAi) treated cells, the data obtained with Illumina sequencing were substantially confirmed for all genes tested by RT-PCR (Fig. 3; Supplemental Fig. 8) with gene specific primers (Supplemental Table 2).
Acknowledgments
We thank the UCHC Translational Genomics Core for use of the Illumina Genome Analyzer, M. Shatsky and L. Pozzi for helpful discussions during algorithm development, and all members of the modENCODE Drosophila Transcriptome Group for helpful discussions. This work was funded by an award from the National Human Genome Research Institute modENCODE Project (U01 HG004271) to S.E. Celniker under Department of Energy contract no. DE-AC02-05CH11231, R01-GM062516 to B.R.G., and R01-GM071655 to S.E.B. A.N.B. is also supported by a National Science Foundation Graduate Research Fellowship.
Author contributions: L.Y. and J.W.P. performed the experiments; L.Y. and B.R.G. designed the experiments; A.N.B., M.O.D., K.D.H., and B.R.G. performed the computational and statistical analysis; all authors designed the study and analyzed the data; A.N.B. and B.R.G. wrote the paper with input from all authors.
Footnotes
[Supplemental material is available for this article. The RNA-sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession nos. GSM461176–GSM461181.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.108662.110.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ 2010. Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP 2009. Next generation software for functional trend analysis. Bioinformatics 25: 3043–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Labourier E, Green RE, Brenner SE, Rio DC 2004. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol Cell 14: 775–786 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Green RE, Brenner SE, Rio DC 2005. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev 19: 1306–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Green RE, MacArthur S, Brooks AN, Brenner SE, Eisen MB, Rio DC 2009. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Mol Cell 33: 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al. 2002. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol 3: research0079 doi: 10.1186/gb-2002-3-12-research0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley J 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaromonte F, Yap VB, Miller W 2002. Scoring pairwise genomic sequence alignments. Pac Symp Biocomput 7: 115–126 [DOI] [PubMed] [Google Scholar]
- Jelen N, Ule J, Zivin M, Darnell RB 2007. Evolution of Nova-dependent splicing regulation in the brain. PLoS Genet 3: 1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB 2000. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc Natl Acad Sci 97: 5740–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D 2003. Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci 100: 11484–11489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Chen H, Edo C, Buckanovich RJ, Yang YY, Musunuru K, Zhong R, Darnell RB, Burley SK 1999. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure 7: 191–203 [DOI] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK 2000. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell 100: 323–332 [DOI] [PubMed] [Google Scholar]
- Li H, Coghlan A, Ruan J, Coin LJ, Hériché J-K, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, et al. 2006. TreeFam: A curated database of phylogenetic trees of animal gene families. Nucleic Acids Res 34: D572–D580 doi: 10.1093/nar/gkj118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ 2010. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res 20: 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles-Filho AC, Stark A 2009. Comparative genomics of gene regulation-conservation and divergence of cis-regulatory information. Curr Opin Genet Dev 19: 565–570 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Park JW, Graveley BR 2005. Use of RNA interference to dissect the roles of trans-acting factors in alternative pre-mRNA splicing. Methods 37: 341–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR 2004. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci 101: 15974–15979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res 38: D613–D619 doi: 10.1093/nar/gkp939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H, Chen Z, Coghlan A, Coin LJM, Guo Y, Hériché J-K, Hu Y, Kristiansen K, Li R, et al. 2008. TreeFam: 2008 Update. Nucleic Acids Res 36: D735–D740 doi: 10.1093/nar/gkm1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W 2003. Human-mouse alignments with BLASTZ. Genome Res 13: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshaiah P, Miller B, Myat MM, Andrew DJ 2001. Pasilla, the Drosophila homologue of the human Nova-1 and Nova-2 proteins, is required for normal secretion in the salivary gland. Dev Biol 239: 309–322 [DOI] [PubMed] [Google Scholar]
- Shomron N, Golan D, Hornstein E 2009. An evolutionary perspective of animal microRNAs and their targets. J Biomed Biotechnol 2009: 594738 doi: 10.1155/2009/594738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. 2009. FlyBase: Enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res 37: D555–D559 doi: 10.1093/nar/gkn788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, et al. 2005. Nova regulates brain-specific splicing to shape the synapse. Nat Genet 37: 844–852 [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586 [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold B, Myers RM 2008. Sequence census methods for functional genomics. Nat Methods 5: 19–21 [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. 2009. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell 36: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH 2009. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol 16: 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer A, Zhang M 2008. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev 22: 2550–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB 2010. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science 329: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]