Abstract
Regulation of gene expression by sequence-specific transcription factors is central to developmental programs and depends on the binding of transcription factors with target sites in the genome. To date, most such analyses in Caenorhabditis elegans have focused on the interactions between a single transcription factor with one or a few select target genes. As part of the modENCODE Consortium, we have used chromatin immunoprecipitation coupled with high-throughput DNA sequencing (ChIP-seq) to determine the genome-wide binding sites of 22 transcription factors (ALR-1, BLMP-1, CEH-14, CEH-30, EGL-27, EGL-5, ELT-3, EOR-1, GEI-11, HLH-1, LIN-11, LIN-13, LIN-15B, LIN-39, MAB-5, MDL-1, MEP-1, PES-1, PHA-4, PQM-1, SKN-1, and UNC-130) at diverse developmental stages. For each factor we determined candidate gene targets, both coding and non-coding. The typical binding sites of almost all factors are within a few hundred nucleotides of the transcript start site. Most factors target a mixture of coding and non-coding target genes, although one factor preferentially binds to non-coding RNA genes. We built a regulatory network among the 22 factors to determine their functional relationships to each other and found that some factors appear to act preferentially as regulators and others as target genes. Examination of the binding targets of three related HOX factors—LIN-39, MAB-5, and EGL-5—indicates that these factors regulate genes involved in cellular migration, neuronal function, and vulval differentiation, consistent with their known roles in these developmental processes. Ultimately, the comprehensive mapping of transcription factor binding sites will identify features of transcriptional networks that regulate C. elegans developmental processes.
Knowledge of the entire genome sequence of a multicellular animal provides an unprecedented opportunity to systematically decipher how this genetic information reliably produces a complex organism. As a first step, the genomes of organisms that serve as models for developmental biology should be interrogated to define a comprehensive list of the functional elements encoded within the genome (Celniker et al. 2009). Of particular importance are the regulatory elements bound by sequence-specific transcription factors (TFs), which drive proper spatial and temporal gene expression as the body plan unfolds from a single-celled embryo.
The nematode Caenorhabditis elegans provides one of the best model systems to study transcriptional regulatory networks during development (Okkema and Krause 2005). The developmental fate of each cell is invariant and traceable and provides a precise blueprint on which to map developmental regulatory networks. The shorter intergenic regions of the compact C. elegans genome simplify assignment of TF binding sites to candidate target genes, thus facilitating the process of constructing a potential regulatory network (Stein et al. 2003). Finally, the worm genome encodes many TFs that are highly conserved in both sequence and function with humans, making such studies in C. elegans broadly relevant (Reece-Hoyes et al. 2005). Despite these advantages, little systematic analysis of regulatory networks controlled by TFs in C. elegans has been performed to date, partly due to the relative paucity of reagents such as antibodies against native TFs. To sidestep this limitation and to systematically probe the relationships between many TFs and their candidate target genes, as part of the modENCODE Consortium we have developed methods to tag transcription factors with an epitope against which high-quality antibodies are available (Sarov et al. 2010). We then established an experimental pipeline to identify the genome-wide binding sites of these tagged C. elegans TFs using ChIP-seq, which we first applied to the FoxA factor PHA-4 (Zhong et al. 2010).
We have since used this pipeline to identify the binding sites genome-wide for another 21 sequence-specific TFs as well as at additional developmental stages for PHA-4. These factors represent a variety of different classes of TFs, and most have known, important roles in developmental processes. Here, we first describe general properties of these data sets to show basic principles of how TFs interact with the genome. We then build a regulatory network focusing on these TFs to begin to visualize potential regulatory hierarchies of TFs during development. Finally, we focus on comparing the binding sites of the three HOX transcription factors LIN-39, MAB-5, and EGL-5. Despite the critical roles these three factors play in specifying cell fates along the anterior–posterior axis in C. elegans, detailed knowledge of their in vivo targets was lacking. Our results demonstrate that the HOX factors have a subset of common target genes, but many of their specific functions in different cell types in different body regions are likely due to unique sets of target genes. In sum, the identification of binding sites for transcription factors can shed light on the mechanism of gene regulation by transcription factors and provide insight into how they direct diverse responses to developmental and environmental cues. Our data build a foundation to develop a deeper understanding of the transcriptional regulatory mechanisms directing metazoan development and should be applicable to regulatory networks in more complex organisms.
Results
Identification of genome-wide binding sites for 22 transcription factors
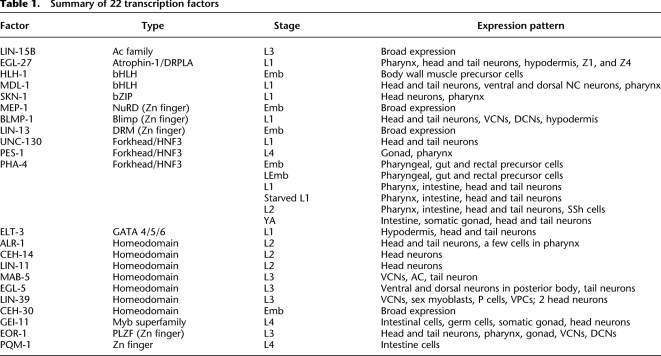
We used the established modENCODE pipeline (Sarov et al. 2010; Zhong et al. 2010) to identify in vivo binding sites for 22 sequence-specific transcription factors (TFs) in C. elegans (Table 1). These factors belong to diverse TF families, representing bHLH, homeobox, FOX, GATA, and C2H2 zinc finger DNA binding domains. Most of these TFs have known roles in C. elegans development and/or homeostasis.
Table 1.
Summary of 22 transcription factors
Detailed analysis of the transgenic lines expressing these tagged TFs, including transgene copy number, assessment of production of full-length tagged proteins, and comparison of transgenic and endogenous expression patterns, is presented elsewhere (Sarov et al. 2010). Additionally, five lines have been tested for rescue of mutant phenotypes, and all five show substantial rescue. For the purposes of ChIP-seq, we visualized the in vivo expression of each GFP-tagged transgenic factor using fluorescence microscopy and briefly described the specific tissues in which the TFs are expressed, including neurons, muscle, gut, and hypodermis (Supplemental Fig. S1; Table 1). In many cases, the expression pattern of a TF changes over time. For instance, we found that the expression pattern of PHA-4 varies over development: At embryonic, L1, and L2 stages it is highly expressed in pharynx, head, and tail neurons, with weaker intestinal expression; while in adults, PHA-4 becomes detectable in the somatic gonad, weaker in the pharynx, and stronger in intestinal cells; moreover, in some cells, PHA-4 is not nuclear at this stage. PHA-4 is known to have distinct tissue-specific functions at different stages of development, with a primary role in pharynx development in embryos (Mango et al. 1994; Ao et al. 2004) and somatic gonad development in larvae (Azzaria et al. 1996; Updike and Mango 2007), and a role in regulating environmental responses in later stages (Panowski et al. 2007; Zhong et al. 2010). Based on the expression patterns and phenotypic information from WormBase and relevant publications, we selected a developmental stage at which each tagged factor shows peak expression and/or is most strongly linked to a phenotypic outcome for ChIP-seq analysis. While most factors were assayed at a single developmental stage, we examined PHA-4 at multiple stages (late embryo, fed L1, L2, and young adult), in addition to the two previously analyzed (embryo and starved L1) (Zhong et al. 2010).
For each factor, at least two independent cultures of worms expressing the GFP-tagged transcription factor were grown to the desired stage and briefly re-examined for nuclear GFP expression prior to fixation and lysis. We then performed chromatin immunoprecipitation (ChIP) with an anti-GFP antibody to enrich for DNA fragments bound by each TF, followed by direct high-throughput sequencing with the Illumina Genome Analyzer platform (Zhong et al. 2010). The immunoprecipitated chromatin for each replicate was sequenced to a depth of at least 1 million mapped reads, along with input DNA as a control (Supplemental File S1). We then used the program PeakSeq to define binding peaks for each factor, and calculated the q-value of each peak from the pooled reads of two replicates (Supplemental Fig. S2; Rozowsky et al. 2009). Only peaks that spanned >50 bp and were reproduced in both replicates with a q-value <0.001 were included in further analyses (Supplemental Figs. S3, S4). All the raw and processed data are available for each factor at the GEO and the modENCODE databases (http://intermine.modencode.org/) and summarized in Supplemental File S2.
A total of 16,700 binding sites were identified for the 22 factors. Of these, 7811 (∼46.8%) were exclusively occupied by only one factor, while the rest were occupied by two or more factors (Supplemental Fig. S5). A subset of 304 sites bound by 15 or more factors have been defined as Highly Occupied Target (HOT) regions and are described in detail elsewhere (Gerstein et al. 2010; Supplemental File S3). Because these HOT regions do not appear to correspond to sequence-specific gene regulation, we removed them from all subsequent analyses described here.
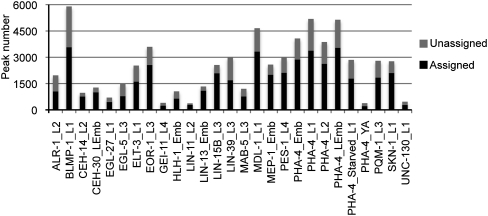
Different factors bind to varying numbers of sites genome-wide (Fig. 1; Supplemental File S2). In particular, BLMP-1, MDL-1, and most PHA-4 samples (mixed and late embryos, starved and fed L1s, and L2s) have the most binding sites (more than 4000), while EGL-27, UNC-130, LIN-11, GEI-11, and young adult PHA-4 bind to the fewest number of sites (less than 1000).
Figure 1.
Binding sites for 22 transcription factors. Chart of the total number of binding sites genome-wide, for each TF listed in alphabetical order. These binding sites are divided into those that ultimately were assigned to specific gene targets, and those that remained unassigned.
Most binding sites sit in close proximity to the transcript start sites of candidate gene targets
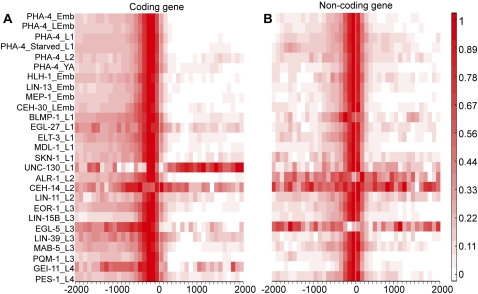
We next sought to associate the binding sites of each factor with candidate gene targets, including those predicted to function as RNA (non-coding) as well as those that function as protein (coding). Only binding sites reproduced in both replicates (q ≤0.001) and located within 2 kb upstream and 300 bp downstream of the transcript start site (TSS) of one or more annotated genes were assigned (Methods; Supplemental Figs. S6, S7; Supplemental Files S4, S5). The majority of C. elegans transcripts receive a trans-spliced leader RNA at the 5′ end, masking the precise start of transcription, so we used the 5′-most end of the trans-spliced transcript instead to estimate the transcript start site. Based on histone modifications that mark promoters, the typical transcription start site is ∼250 bp upstream of the trans-splice site (Kolasinska-Zwierz et al. 2009), close to our estimates. Binding sites further than 2 kb upstream of the TSS or 300 bp downstream from any annotated gene model remained unassigned (Fig. 1). Despite the compact genome of C. elegans, we found that the majority of binding peaks were assigned to a single gene target with this window size. The mean distribution of binding over all targets for a given factor demonstrates that the majority of binding sites for coding genes lie within 500 bp upstream of the TSS (Fig. 2A). For non-coding genes, the binding sites are even closer to the TSS, as they sit between 300 bp upstream and 200 bp downstream of the TSS (Fig. 2B; Supplemental Fig. S7B).
Figure 2.
Analysis of gene targets for 22 transcription factors. (A,B) Heat map showing the distribution of binding relative to the TSS (“0”) for each factor for coding (A) and non-coding (B) gene targets.
A few factors do not correspond to this trend. UNC-130 and CEH-14 exhibit a different pattern for coding targets, and CEH-14, ALR-1, and EGL-5 show a different pattern for non-coding target genes (Fig. 2). These factors tend to have relatively few binding sites in the genome, which reduces the effectiveness of the normalization procedure and results in a high uniform signal across the interval. However, UNC-130 appears to preferentially bind downstream from the start site. Closer investigation of individual gene targets for UNC-130 shows that, although UNC-130 does not bind to introns more frequently than other TFs, it binds introns with particularly strong signal intensity. In sum, this analysis suggests that a large amount of regulatory information can potentially reside in the relatively short regions immediately upstream of the 5′ end of genes in the compact C. elegans genome.
Non-coding RNA genes are also candidate targets of TFs
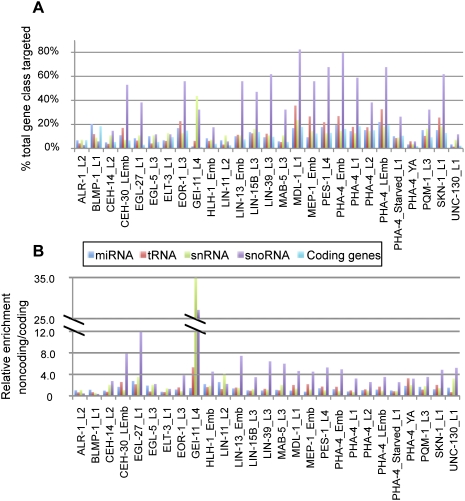
Although the majority of candidate targets correspond to protein-coding genes, we found that a significant fraction of known non-coding RNAs (ncRNAs) are also candidate target genes for most TFs, suggesting that there is substantial spatial and temporal regulation of miRNAs, tRNAs, snRNAs, and snoRNAs. For this analysis, we did not count ncRNA genes located in introns and transcribed in the same direction as the parent gene as candidate targets, because such embedded genes are typically spliced from the parent transcript and are not independently regulated. However, such organization is less frequent in C. elegans than in mammals, and most ncRNAs appear to be separate transcription units (Deng et al. 2006). We calculated the percentages of each type of ncRNA bound by each factor. In general, snoRNAs are the most enriched non-coding RNAs bound by these factors. For example, MDL-1 occupied ∼82% of the promoters of predicted snoRNAs (Fig. 3A), and most TFs show twofold or greater enrichment for binding to snoRNAs compared to coding genes. Eleven factors (PHA-4, LIN-13, MEP-1, CEH-30, BLMP-1, EOR-1, LIN-15B, LIN-39, PQM-1, GEI-11, PES-1) each bind to >10% of all annotated ncRNA genes (Supplemental File S6).
Figure 3.
Non-coding RNA genes are frequent targets of TFs. (A) Chart showing the fraction of each gene class (miRNA, snoRNA, snRNA, tRNA, and coding) bound by each TF. (B) Chart showing the relative enrichment of the fraction of each non-coding class bound by each TF relative to the fraction of coding genes bound by that TF.
Out of all factors examined, only GEI-11 is predicted to preferentially regulate non-coding RNAs. GEI-11 is the ortholog of the DNA-binding SNAP190 component of the SNAPc complex, a well-characterized transcriptional regulator of snRNA genes (Wong et al. 1998). Indeed, as expected based on this homology, GEI-11 binds to a higher fraction of snRNA genes than any other kind of ncRNA and is by far the most strongly associated with ncRNAs relative to coding genes, compared to all other factors (Fig. 3B). Taken together, our analysis indicates that multiple TFs might regulate a wide variety of non-coding RNAs. Potentially, tissue-specific regulation of different types of important structural RNAs might influence the efficiency of major cellular processes such as splicing and translation.
Gene Ontology (GO) enrichment analysis demonstrates binding site specificity
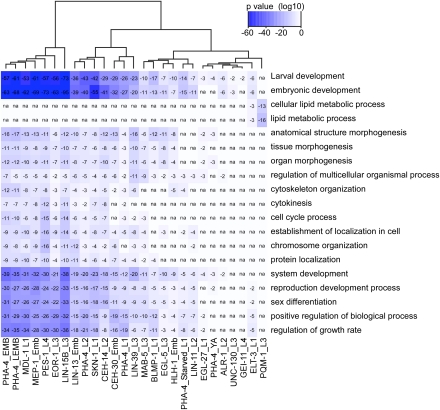
To classify functions for the targeted coding genes bound by each of the 22 TFs, we performed Gene Ontology (GO) enrichment analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) (Huang da et al. 2009). For each factor, DAVID identified various biological processes that are over-represented in the list of bound genes. Many broad GO terms related to developmental processes were enriched among gene targets bound by multiple factors, as expected given the known importance of many of the TFs in C. elegans development (Fig. 4; Supplemental File S7). Factors that bound to the greatest number of sites showed the greatest similarity in GO categories. However, we also noted instances in which particular factors shared more specific GO categories, such as ELT-3 and PQM-1, which both bind to genes involved in lipid metabolism, while most other factors do not. These observations are consistent with the possibility that many of the sites discovered in our ChIP-seq experiments reflect functionally relevant TF binding events.
Figure 4.
Correlation of GO categories between factors. The similarity between GO categories for each TF was calculated and organized into a heat map, which was clustered by TF based on this similarity.
A regulatory network of C. elegans transcription factors
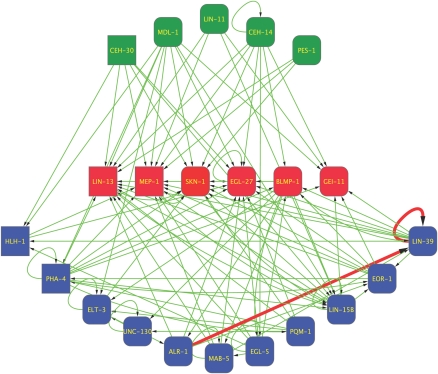
With these data, we can begin for the first time to get a sense of the regulatory relationships that might exist between a relatively large number of transcription factors. To identify potential interactions between transcription factors as both regulators and targets, we constructed a network among these 22 TFs (Fig. 5). If a TF bound to the regulatory region of a gene encoding one of the other 21 TFs, we drew an edge between these two factors, originating at the TF protein and pointing toward the bound gene. The resulting network shows a surprisingly large number of potential regulatory interactions. Consistent with previous studies, we can detect an autoregulatory relationship for TFs containing a HOX domain such as LIN-39 (Wagmaister et al. 2006). Additionally, we found that ALR-1 binds to the lin-39 regulatory region, which is consistent with data from a yeast one-hybrid interaction assay (W. Liu and D. Eisenmann, pers. comm.). Notably, grouped at the top level are factors that regulate multiple TF genes but are not targets themselves. Conversely, the factors in the middle level of the network are primarily targets and not regulators, whereas the bottom level of factors exhibits both characteristics. This stratification is not explained by the number of binding sites bound by these factors, as factors in the middle level bind to as many sites as those that are in the top level (see Fig. 1). Possibly, the TFs in the middle layer might regulate fewer TFs because all six have fundamental roles in global regulation of transcription, chromatin formation, and splicing, rather than regulating specific spatial or temporal events. As such, they might be common endpoints for regulation once tissues have been specified, to set the basic metabolism of the cells in that tissue.
Figure 5.
A C. elegans TF regulatory network. The relationships between TF genes as proximal targets and TF proteins as regulators. Edges represent a binding event, with the arrow pointing to the target gene. (Thick red edges) Highlight interactions discussed in the text; (green squares) TFs that bind other TF genes but are not targets themselves; (red squares) TF genes that are primarily targets and less frequently regulators; (blue squares) factors that are both targets and regulators; (squared corners) factors analyzed in embryos; (rounded corners) factors analyzed in larvae.
Related factors have correlated binding expression profiles
TFs often work together or form a complex with other transcriptional regulators in order to properly regulate expression of downstream targets. We were therefore interested in the overall similarities in binding profiles between TFs and wished to determine which factors shared the strongest overlap in target genes. We examined the binding profile correlation between each possible pair of TFs (Methods).
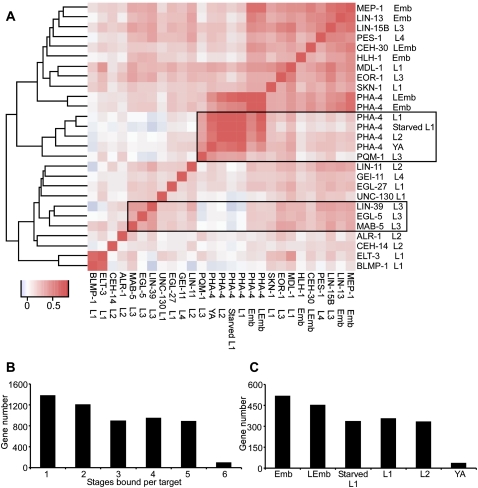
We uncovered multiple clusters with particularly close binding correlations (Fig. 6A). One cluster is composed of diverse types of TFs at different stages, in the upper-right corner of the clustergram. The formation of this group is likely driven by binding to many sites that are promiscuously bound by unrelated factors. Although the approximately 300 HOT sites that are bound by the vast majority of factors (Gerstein et al. 2010) were not included in our correlation analysis, binding sites that did not quite meet the criteria for HOT sites were included, and likely influence this cluster. Indeed, many of the factors in this cluster have the strongest association with HOT sites (Gerstein et al. 2010).
Figure 6.
Correlation of target gene binding by the 22 TFs. (A) Peak regions of all 22 factors (without HOT sites) were collected and binned into 100-bp windows. The signal in each bin for each factor was used in pairwise correlations as shown in the heat map. The heat map reflects the co-bound frequency (correlation coefficient) of each pair of TFs, with red indicating more co-bound regions and blue indicating less. TFs have been clustered along both axes based on similarity in their bound regions. (B) A histogram showing the number of genes targeted by PHA-4 at one, two, or more stages of development. (C) A histogram showing the number of unique PHA-4 binding sites at each stage.
In contrast, two other clusters appear to be based on TF function (boxed in Fig. 6A). One cluster consists of PHA-4 binding at post-embryonic stages, while embryonic stages are present in the non-specific cluster described in the previous paragraph. This observation suggests that PHA-4 has a major shift in targets from embryos to larval stages, as described before (Zhong et al. 2010), which mirrors the shift of PHA-4 from developmental regulator to an environmental sensor. The intriguing presence of the stress-responsive factor PQM-1 (Shapira et al. 2006) in the larval cluster suggests that PQM-1 and PHA-4 might share common targets involved in reacting to environmental challenges.
Because we analyzed PHA-4 binding at multiple stages, we assessed whether the addition of more stages substantially contributes new information about binding sites (Figure 6B). We found that more than 1300 sites are bound by PHA-4 at only a single stage, while 885 binding sites are bound at five of six stages (excluding young adult, at which PHA-4 has diminished binding genome-wide). Moreover, most individual developmental stages contribute many new binding sites (Fig. 6C). Thus, monitoring TFs at multiple stages is worthwhile, especially for factors such as PHA-4 that are known to have diverse roles during development.
Another notable cluster consists of three HOX TFs: LIN-39, MAB-5, and EGL-5, all of which have significant roles in establishing the larval body plan (Fig. 6A). Four other homeobox-containing factors included in the analysis—ALR-1, LIN-11, CEH-14, and CEH-30—do not share particularly close correlation and are present in different groups in the clustergram (Fig. 6A). This finding indicates that LIN-39, MAB-5, and EGL-5 likely share a functionally relevant core set of targets, while other homeobox factors have more diverse targets.
The target genes of LIN-39, MAB-5, and EGL-5 are consistent with their known developmental functions
LIN-39, MAB-5, and EGl-5 are expressed in overlapping regions of the animal, with LIN-39 in the mid-body, MAB-5 slightly more posterior, and EGL-5 in the tail (Wang et al. 1993). They are involved in specifying cell fates along the anterior–posterior axis in C. elegans and show complex relationships with each other that differ at distinct times and tissues, and between sexes (for review, see Kenyon et al. 1997). For instance, in the hermaphrodite epithelium, LIN-39 promotes vulval induction in concert with Ras signaling in the central vulval precursor cells P3.p–P8.p (Maloof and Kenyon 1998), while MAB-5 is required to repress vulval fate in the more posterior P7.p and P8.p cells (Clandinin et al. 1997). EGL-5 is required in P12.p for its specification and brief posterior migration to the hindgut, a process that again can be hindered by MAB-5 activity (Li et al. 2009). Additionally, LIN-39 and MAB-5 are required for migration of the Q neuroblasts and their daughters, but LIN-39 promotes anterior migration of QR descendants, while MAB-5 promotes posterior migration of QL descendants (Salser and Kenyon 1992). EGL-5 is also involved in neuronal migration, but it is required for the anterior migration of a different neural cell, the HSN (Chisholm 1991). Given the overlap and complex relationships in diverse tissues between these three TFs, the high correlation of their target genes is of interest.
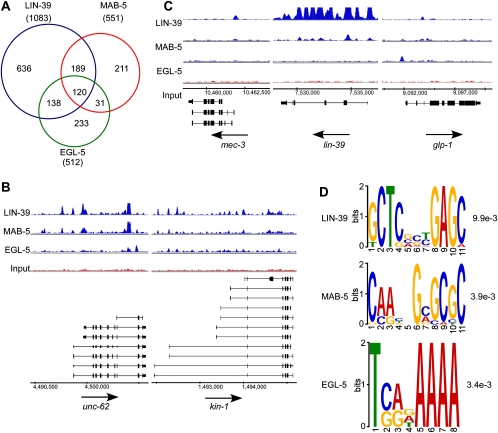
First, we looked at the precise overlap in target genes (Fig. 7A). A total of 120 target genes are bound by all three factors (common targets), with substantial fractions of the total targets bound only by a given factor (unique targets). To independently confirm a subset of these interactions, we repeated the chromatin immunoprecipitation of each factor and performed qPCR over selected loci (Supplemental Fig. S8). All loci we examined showed significantly enriched binding by the tested factor, relative to a control locus.
Figure 7.
Common and unique targets of LIN-39, MAB-5, and EGL-5. (A) Venn diagram overlap of gene targets for LIN-39, MAB-5, and EGL-5. (B) Example of common binding sites in the targets unc-62 and kin-1. (C) Example of unique target genes for each factor. Note that the lin-39 locus exhibits increased reads across the gene in the LIN-39 and input tracks because of the extra copies of the LIN-39:GFP transgene in the genome, and therefore does not necessarily represent binding. (D) Enriched consensus DNA motifs for each set of unique target genes, with e-value.
The common set of LIN-39/MAB-5/EGL-5 target genes comprise a wide diversity of potential functions, with significantly over-represented GO categories generally associated with embryonic and larval development, organ and tissue morphogenesis, and various metabolic processes (Supplemental File S8). Many of the common target genes are known to have functions in neuronal development as well, such as rgef-1, pqbp-1.2, cab-1, cmk-1, ncs-2, ric-3, snt-2, kin-1, and unc-76. One intriguing target is unc-62 (Fig. 7B), which encodes a co-factor of HOX proteins (Yang et al. 2005) but has not been previously suggested as a target of any HOX TF. These common targets potentially underlie the potential antagonistic and complementary relationships of these three TFs.
In addition to the common targets, each HOX TF has unique targets that are likely to be important for the specialized, distinct functions of LIN-39, MAB-5, and EGL-5. For instance, LIN-39 binds to multiple mec genes, including mec-3, mec-6, mec-7, and mec-12 (Fig. 7C). To our knowledge, no role for LIN-39 in mechanosensation has been documented to date, but this observation suggests that LIN-39 could be involved in that process or that LIN-39 could be repressing expression of these genes in other cells besides mechanosensory neurons. One MAB-5 unique target is lin-39 (Fig. 7C), consistent with the known role of MAB-5 in preventing vulval development in Pn.7p and Pn.8p (Kenyon 1986; Clandinin et al. 1997), and suggesting that MAB-5 might directly inhibit LIN-39 expression to repress vulval development. Candidate targets unique to EGL-5 correspond to several genes with known roles in cell migration, such as vab-8, unc-49, and mig-17. Intriguingly, we found that EGL-5 also targets two components of the Notch signaling pathway, glp-1 and emb-4 (Fig. 7C). A recent report has shown that Notch signaling cooperates with EGL-5 to influence differentiation of the epithelial Y cell (Jarriault et al. 2008).
Consistent with the diverse functions and unique targets of these factors, we found that the binding sites within these targets were associated with different sequence motifs for each factor (Fig. 7D). Each sequence motif is found in more than one-third of its corresponding set of unique gene targets, indicating that a reasonable fraction of the targets might be regulated through this motif (Supplemental File S9). These motifs do not contain an obvious core HOX motif (TAAT), so whether they are bound by the HOX factors directly or by a cofactor remains to be determined.
In sum, classification of both common and unique targets of these HOX factors support previous findings and point to new avenues for investigating the molecular mechanisms that underlie their important function in structuring the C. elegans body plan. This analysis provides an excellent example of the utility of global analysis of TF binding sites in a developmental model organism.
Discussion
Transcription factors play key roles in diverse aspects of development and physiology, including sex determination, early pattern formation, organogenesis, and response to environmental cues. Identifying the in vivo DNA binding sites of these factors and their candidate target genes is therefore a critical first step toward understanding how an organism develops and functions. The many experimental and computational advantages of the compact and relatively well-annotated genome of C. elegans are highly conducive to the in vivo analysis of TF binding sites within a precisely defined, highly reproducible developmental context. Therefore, having established an experimental ChIP-seq pipeline, we have comprehensively documented and analyzed the binding profiles of 22 C. elegans TFs to date.
Mapping the binding sites for these 22 TFs to both coding and non-coding transcripts demonstrated that a surprisingly large number of candidate gene targets can be assigned to most of the factors, in some cases encompassing almost 20% of annotated genes. Why do TFs exhibit such extensive binding, and how many of these sites are of functional consequence? The binding profile of a given factor reflects an amalgamation of binding sites from the many different cell types that are sampled simultaneously from the whole animal. Indeed, the TFs that are expressed in the most tissues tended to have the most binding sites. Many of these binding sites might reflect rarely used instances of gene-specific regulation that only occur under unusual circumstances, such as an environmental challenge that is not typically encountered in the laboratory. For such cases, the TF might constitutively occupy the site but would not impart any differential regulation of the target. Finally, given the compact nature of the C. elegans genome, some binding sites might sit close to a gene but are not involved in its regulation. Such sites might mediate higher-order genomic interactions and influence nuclear organization, instead. To tease apart the diverse effects that a binding event might have will require many future studies, both genome-wide and at the level of single-gene analysis.
The binding of many TFs to non-coding RNA genes suggests unexpected regulation of what are often considered constitutively expressed genes. For instance, in mammals, snoRNAs are not typically individually regulated, but tend to be located in introns of coding genes and are processed out as the parent mRNA undergoes splicing. However, in C. elegans, many snoRNAs are not found in introns and appear to have independent, developmentally variable regulation (Deng et al. 2006). Our results indicate that these snoRNAs, along with other non-coding RNA genes, are indeed potential targets of TFs, suggesting that their regulation is diverse and subject to developmental or environmental influences. Alternatively, the association of these TFs with highly transcribed ncRNAs such as tRNAs, snoRNAs, and snRNAs might reflect open chromatin favorable for TF binding, so some of these sites might be opportunistic rather than regulatory. Deciphering this issue will require a means to analyze the transcript levels of these RNAs in a tissue-specific manner.
With our data, we have been able to construct a first-generation regulatory network for these profiled factors. This network clearly shows that different TFs have different behaviors. A subset including CEH-30, CEH-14, LIN-11, MDL-1, and PES-1 all regulate genes encoding other TFs, but are not themselves targets of regulation. This finding suggests that these factors have relatively fewer inputs from other factors and might act at an upstream point in a regulatory pathway to initiate a cascade of TF regulation. Alternatively, they could be bound by other TFs within a limited number of cells (e.g., a few neurons), so that the binding event could not be detected in our whole-animal preps. The majority of factors are both targets and regulators of other TFs, and the network indicates that there are multiple reciprocal interactions, suggesting complex relationships. Importantly, the mechanism of regulation (activation or repression) cannot be determined from this analysis. Possibly, a factor might repress its candidate TF gene, so that the two factors might not exhibit similar phenotypes or expression patterns. Lineage-specific analyses in transcription factor mutant backgrounds, such as those used in Murray et al. (2008), must be employed to determine this level of regulation. Ultimately, having additional TF binding profiles will permit the expansion of this network, broadening its utility, and will also permit us to construct more focused networks in a given tissue or at given time in development. Larger networks will be increasingly biologically informative regarding transcriptional hierarchies.
The HOM-C class of HOX factors, which includes LIN-39, MAB-5, and EGL-5, is responsible for directing particular aspects of the C. elegans body plan. The presence of more than one of these factors (e.g., both LIN-39 and MAB-5) in a cell can produce a different fate than if that cell expresses only one (or neither) of the factors. Thus, these factors exhibit complex relationships with each other, and these relationships, in turn, are influenced by cellular context and the sex of the organism. Possibly, these factors interact at the promoters of common target genes to differentially influence gene expression and can have opposing or synergistic effects on these targets depending on cell type. Alternatively, each factor might act on a different set of gene targets whose products, in turn, interact in diverse ways to influence cell fate. Because these factors are expressed in multiple cell types, it is possible that some targets that appear in common are actually bound only by a single factor in a given subset of cells. Further investigation of the roles of individual targets should shed light on how these factors influence the execution of cell fate and location. Finally, all of our analyses have been carried out in hermaphrodites to date, but these factors, especially MAB-5, have important roles in male-specific cell fate decisions as well. Ultimately, performing ChIP-seq analysis of the binding sites of all three factors in males, although not technically straightforward, would be very interesting for comparison to their binding profiles in the hermaphrodite.
In summary, the genome-wide ChIP-seq data for these 22 transcription factors provide an unprecedented view, both in breadth and resolution, of the developmental transcriptional network in C. elegans. Moreover, these data expand the current knowledge of how Hox genes function as master regulators of A/P pattern formation. Notably, these 22 factors represent only the initial output of this aspect of the modENCODE project. Continued global mapping of transcription factor binding sites for more factors and at more developmental stages will build on this foundation for further evaluation of transcriptional regulatory mechanisms during C. elegans development and will ultimately give general insight into similar mechanisms in other organisms as well.
Methods
Strains and growth conditions
Strain information is listed in Supplemental Table 1. Each transgenic strain carries a 30–40-kb fosmid containing the entire transcription factor locus along with flanking regions. The TF has been tagged in frame with a dual GFP:3xFLAG tag at its C terminus (Sarov et al. 2010; Zhong et al. 2010). For factors represented by more than one strain, we selected the strain that appeared healthiest and had the most robust expression, although in most cases, independent strains for each factor were remarkably similar. Strains were maintained in liquid culture as described (Zhong et al. 2010), with the exception of OP109, OP120, OP177, OP178, OP179, OP184, and OP201, which were grown on plates. Worms were staged by bleaching and L1 starvation and then grown to the desired stage by direct visual examination of standard developmental milestones (Brenner 1974).
ChIP-seq analysis
ChIP assays were carried out as described previously (Zhong et al. 2010). Briefly, worms were collected at desired stages and cross-linked with 2% formaldehyde for 30 min at room temperature. Formaldehyde was then quenched by addition of 1 M Tris (pH 7.5). After sonication, cell extracts containing DNA fragments with an expected range between 200 and 800 bp were immunoprecipitated using anti-GFP antibodies. The enriched DNA fragments and input DNA (genomic DNA from the same prep) were used to prepare libraries for sequencing by the Illumina GA platform. In order to run four samples in one flow cell, sequencing libraries were multiplexed as described in Lefrancois et al. (2009).
ChIP-qPCR
Primers are listed in Supplemental Table S3. A 15-μL PCR reaction with each primer set was run in a Roche LightCycler 480 machine using the SYBR Green I Master kit (Roche 04 707 516 001) according to the manufacturer's instructions. The PCR program was as follows: Step 1: 95°C for 5 min; Step 2: 95°C for 30 sec; Step 3: 55°C for 30 sec, Step 4: 72°C for 45 sec. Repeat Steps 2–4 44 times; Step 5: 72°C for 5 min; Step 6: 4°C. The enrichment was estimated by subtracting the enrichment of a given gene in the ChIP sample from the enrichment of the same gene in the IgG control sample. The negative control gene (fer-1) did not show significant differences in enrichment between the ChIP sample and IgG control sample.
Calling binding sites/peaks from ChIP-seq data
In total, 27 ChIP-seq data sets from 22 transcription factors were included in the present study (Table 1). For each data set, we first pooled raw signals from two biological replicates and normalized with corresponding input reads to remove background signals. We then used PeakSeq (Rozowsky et al. 2009) to find peak regions of each factor from the pooled reads as well as for each replicate. We used two methods to find the correlation between two biological replicates. First, for each replicate, we took the binding peaks called by PeakSeq from pooled reads (q-value cutoff of 0.001), and binned them into 100-nt windows. Then we counted the raw reads at each window from both replicates and calculated the correlation between replicates. The correlation coefficients between two replicates are shown in Supplemental Figure S2A. Second, we only compared the top 40% peaks of two biological replicates; the percentages of overlapped peaks (≥1 nt overlap) are shown in Supplemental Figure S2B.
Subsequently, in order to have a set of binding peaks with good quality for further analysis, we used a stringent method to filter out unqualified peaks. We first collected the peaks called by PeakSeq from pooled reads. These pooled peaks were overlapped with the peaks called from each replicate. We only kept peaks from the pooled reads that overlapped >50% of each replicate's peaks and had a minimum length of 50 nt (Supplemental Fig. S3). We then calculated the total genome coverage of 27 data sets using pooled peaks and overlapped peaks (Supplemental Fig. S4). The coverage changes dramatically using different PeakSeq q-value cutoffs for all pooled peaks, while it is quite stable using overlapped peaks. Therefore, we decided to use the overlapped peaks from pooled reads with q-value ≤0.001 as qualified peaks. All the analyses in this paper were based on these overlapped/qualified peaks.
Calling target coding and non-coding genes
We calculated the signal intensity within the window 2 kb upstream to 2 kb downstream of TSSs of both coding and non-coding predicted gene targets for each factor (Fig. 2A,B; Supplemental Fig. S6). The signals over all targets were normalized and averaged to represent the overall distribution of the binding pattern for all targets of a given factor (Methods; Kidder and Palmer 2010). To assign a binding site to an annotated gene, we first collected coding and non-coding gene annotations from WormBase 170 and 200, respectively, while miRNAs were collected from miRBase v14 (Gerstein et al. 2010). We associated genes to TF binding sites using two steps: First, a binding peak is assigned to “proximal” genes when the middle point of the peak is located within 500 bp upstream and 300 downstream of the TSS (transcription start site) of a transcript. Only if the peak cannot be assigned to a proximal gene, is it assigned to a “distal” gene (upstream from 500–2000 bp). The fractions of proximal and distal genes associated with each factor are shown in Supplemental Figure S7. The majority of genes, especially ncRNAs, are proximal genes. An aggregation plot of binding signals (from upstream 5000 bp to downstream 5000 bp of the TSS), shows almost no binding signal beyond 2000 bp upstream, consistent with prior analysis (Zhong et al. 2010). When we plotted the average raw signal around all targeted coding and non-coding transcripts, the raw reads were scaled from 0 to 1 for each factor. The HOT regions (Gerstein et al. 2010) and genes targeted by HOT regions were removed from the final list. We also summarized the numbers of genes associated by individual peaks (Supplemental Table S2). Most peaks are only associated with one or two genes.
To identify high-stringency unique targets of LIN-39, MAB-5, and EGL-5, we added additional criteria to our peak calling analysis before we assigned a binding site to annotated genes. We first used the same method described above to define overlapped/qualified peaks of a given factor, then removed the overlapped/qualified peaks that appeared in all peaks found by PeakSeq from pooled reads of any of the other two factors. For instance, a unique peak of LIN-39 has to be an overlapped/qualified peak of LIN-39 that does not appear in peaks found by PeakSeq from pooled reads of either EGL-5 or MAB-5. In the end, we defined high-stringency unique peaks for that given factor and assigned them to annotated genes. Such gene targets were considered to be truly unique targets of a given factor and were used for GO analysis and motif analysis.
Gene Ontology (GO) analysis
The Gene Ontology of targeted coding genes by each factor (minus HOT regions) were analyzed using DAVID (Huang da et al. 2009; Supplemental File S4). Both proximal and distal gene targets were used for the GO analysis. GO level 3 (biological processes) was used in Figure 4 and Supplemental Files S7 and S8. To evaluate the closeness of GO enrichments, we pooled all enriched GO terms of 22 factors, ranked them by P-value, and kept only the top 30 enriched GO categories. We then manually removed the redundant categories and kept 19 terms per factor for the heat map in Figure 4. We used the average linkage cluster method to cluster different factors based on the P-value (log 10) of enriched GO terms.
Pairwise analysis of binding sites of 22 factors
We combined the binding site data for all factors into a large set of regions, binned those regions into 100-bp windows, and calculated the binding signal normalized to the input signal within each window to create an “average” or model binding distribution for each factor. We correlated the distributions between each pair of factors and performed hierarchical clustering of the correlation coefficients of the pairs. We took all the overlapped/qualified peaks of 27 data sets and merged them into a large set of TF binding sites. We then binned these binding regions into 100-nt windows, and the raw signals (normalized over each input) of each window were calculated from each factor. The Pearson correlation coefficient of each pair of factors was calculated and plotted in Figure 6A. We computed the hierarchical clustering of the correlation coefficients and reordered the factors into different clusters. The hierarchical dendrogram was produced using the average linkage cluster method with a correlation metric distance.
Motif analysis
We searched for enriched motifs from the common and high-stringency unique binding sites of LIN-39, MAB-5, and EGL-5 as follows: The central 200-bp sequence under the top 200 peaks for each factor (ranked by PeakSeq q-value) was extracted. This was the window size we optimized to find the known motifs from PHA-4 data sets in a previous paper (Zhong et al. 2010). We also performed a localization test for each motif we found for all the TFs in the Consortium paper (Gerstein et al. 2010) and found that the majority of the enriched motifs were located within the 200-bp range. We used these sequences in a motif search using MEME (Bailey and Elkan 1994). We generated the position weight matrices (PWMs) for the enriched motifs for the unique binding sites of three factors. Some enriched motifs are simply tandem repeats that were removed, leaving the complex motifs highlighted. The e-value, which is the estimation of expected number of motifs found from shuffled input sequences, was also calculated using MEME, and is listed along with each motif.
Acknowledgments
We thank Hannah Monahan, Debasish Raha, Phil Lacroute, and Ghia Euskirchen for Illumina sequencing; and Mike Wilson and Guoneng Zhong for data scoring and pipeline tracking. We thank Xiuqiong Zhou for making growth medium and Beijing Wu for two GFP-TF images. We also thank Marc Perry and Nicole L. Washington for publishing the data sets to the DCC website. This work is funded by NIH HG004267.
Footnotes
[Supplemental material is available for this article. The ChIP-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession nos. GSE14545, GSE15625, GSE17454, GSE17456, GSE25779, GSE25782–GSE25785, GSE25787, GSE25795–GSE25796, GSE25798–GSE25805, GSE25808–GSE25811, and GSE26152–GSE26153.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.114587.110.
References
- Ao W, Gaudet J, Kent WJ, Muttumu S, Mango SE 2004. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science 305: 1743–1746 [DOI] [PubMed] [Google Scholar]
- Azzaria M, Goszczynski B, Chung MA, Kalb JM, McGhee JD 1996. A fork head/HNF-3 homolog expressed in the pharynx and intestine of the Caenorhabditis elegans embryo. Dev Biol 178: 289–303 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm A 1991. Control of cell fate in the tail region of C. elegans by the gene egl-5. Development 111: 921–932 [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Katz WS, Sternberg PW 1997. Caenorhabditis elegans HOM-C genes regulate the response of vulval precursor cells to inductive signal. Dev Biol 182: 150–161 [DOI] [PubMed] [Google Scholar]
- Deng W, Zhu X, Skogerbo G, Zhao Y, Fu Z, Wang Y, He H, Cai L, Sun H, Liu C, et al. 2006. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression. Genome Res 16: 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE Project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jarriault S, Schwab Y, Greenwald I 2008. A Caenorhabditis elegans model for epithelial–neuronal transdifferentiation. Proc Natl Acad Sci 105: 3790–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C 1986. A gene involved in the development of the posterior body region of C. elegans. Cell 46: 477–487 [DOI] [PubMed] [Google Scholar]
- Kenyon CJ, Austin J, Costa M, Cowing DW, Harris JM, Honigberg L, Hunter CP, Maloof JN, Muller-Immergluck MM, Salser SJ, et al. 1997. The dance of the Hox genes: patterning the anteroposterior body axis of Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol 62: 293–305 [PubMed] [Google Scholar]
- Kidder BL, Palmer S 2010. Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res 20: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J 2009. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 41: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M 2009. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics 10: 37 doi: 10.1186/1471-2164-10-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kulkarni RP, Hill RJ, Chamberlin HM 2009. HOM-C genes, Wnt signaling and axial patterning in the C. elegans posterior ventral epidermis. Dev Biol 332: 156–165 [DOI] [PubMed] [Google Scholar]
- Maloof J, Kenyon C 1998. The Hox gene lin-39 is required during C. elegans vulval induction to select the outcome of Ras signaling. Development 125: 181–190 [DOI] [PubMed] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J 1994. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development 120: 3019–3031 [DOI] [PubMed] [Google Scholar]
- Murray JI, Bao Z, Boyle TJ, Boeck ME, Mericle BL, Nicholas TJ, Zhao Z, Sandel MJ, Waterston RH 2008. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods 5: 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema PG, Krause M 2005. Transcriptional regulation. WormBook 2005: 1–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A 2007. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447: 550–555 [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Deplancke B, Shingles J, Grove CA, Hope IA, Walhout AJ 2005. A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol 6: R110 doi: 10.1186/gb-2005-6-13-r110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB 2009. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol 27: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser SJ, Kenyon C 1992. Activation of a C. elegans Antennapedia homologue in migrating cells controls their direction of migration. Nature 355: 255–258 [DOI] [PubMed] [Google Scholar]
- Sarov M, Murray J, Schanze K, Pozniakovski A, Niu W, Angermann K, Preston E, Zinke A, Enst S, Janette J, et al. 2010. The TransgeneOme of C. elegans: A platform for in vivo analysis of protein function. (in press). [Google Scholar]
- Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, Tan MW 2006. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci 103: 14086–14091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol 1: E45 doi: 10.1371/journal.pbio.0000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Mango SE 2007. Genetic suppressors of Caenorhabditis elegans pha-4/FoxA identify the predicted AAA helicase ruvb-1/RuvB. Genetics 177: 819–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagmaister JA, Miley GR, Morris CA, Gleason JE, Miller LM, Kornfeld K, Eisenmann DM 2006. Identification of cis-regulatory elements from the C. elegans Hox gene lin-39 required for embryonic expression and for regulation by the transcription factors LIN-1, LIN-31 and LIN-39. Dev Biol 297: 550–565 [DOI] [PubMed] [Google Scholar]
- Wang BB, Muller-Immergluck MM, Austin J, Robinson NT, Chisholm A, Kenyon C 1993. A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell 74: 29–42 [DOI] [PubMed] [Google Scholar]
- Wong MW, Henry RW, Ma B, Kobayashi R, Klages N, Matthias P, Strubin M, Hernandez N 1998. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol Cell Biol 18: 368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Sym M, Kenyon C 2005. The roles of two C. elegans HOX co-factor orthologs in cell migration and vulva development. Development 132: 1413–1428 [DOI] [PubMed] [Google Scholar]
- Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HY, Preston E, et al. 2010. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet 6: e1000848 doi: 10.1371/journal.pgen.1000848 [DOI] [PMC free article] [PubMed] [Google Scholar]