Abstract
We have used a combination of three high-throughput RNA capture and sequencing methods to refine and augment the transcriptome map of a well-studied genetic model, Caenorhabditis elegans. The three methods include a standard (non-directional) library preparation protocol relying on cDNA priming and foldback that has been used in several previous studies for transcriptome characterization in this species, and two directional protocols, one involving direct capture of single-stranded RNA fragments and one involving circular-template PCR (CircLigase). We find that each RNA-seq approach shows specific limitations and biases, with the application of multiple methods providing a more complete map than was obtained from any single method. Of particular note in the analysis were substantial advantages of CircLigase-based and ssRNA-based capture for defining sequences and structures of the precise 5′ ends (which were lost using the double-strand cDNA capture method). Of the three methods, ssRNA capture was most effective in defining sequences to the poly(A) junction. Using data sets from a spectrum of C. elegans strains and stages and the UCSC Genome Browser, we provide a series of tools, which facilitate rapid visualization and assignment of gene structures.
Constructing a comprehensive transcriptome is a critical step in establishing a genomic framework for an organism. Caenorhabditis elegans is a well-characterized organism that provides a powerful system for genetic research. The organism is transparent, has a short life cycle, and its cell lineage is fully characterized. The C. elegans genome is relatively small in size (∼100 Mb) and completely sequenced (The C. elegans Sequencing Consortium 1998), which makes C. elegans a straightforward model system for transcriptome analysis.
Several technologies have been developed to study the transcriptome of an organism, including microarrays (Schena et al. 1998; Kim et al. 2001), expressed sequence tag (EST) libraries (Waterston et al. 1992; McCombie et al. 1992), and tag-based methods such as serial analysis of gene expression (SAGE) (Velculescu et al. 1995; Jones et al. 2001). While these methods are high-throughput, they are used to analyze only a portion of the full transcriptome and have limited ability to identify new transcripts, provide accurate annotation of genes, and present a full picture of a transcriptome.
Genome tiling arrays are another method that was recently developed for measuring expression levels and have the advantage of being able to discover new genes and changes in gene models. However, this method has sensitivity limitations, requires a large amount of input RNA, and depends on prior knowledge of the genome sequence (Wang et al. 2009).
Characterizing the transcriptome of an organism at high resolution has been recently facilitated by advances in RNA high-throughput sequencing (RNA-seq) (Wang et al. 2009). Among the organisms that RNA-seq has been applied to are Saccharomyces cerevisiae, Schizosaccharomyces pombe, Arabidopsis, human, and mouse cells (Cloonan et al. 2008; Lister et al. 2008; Marioni et al. 2008; Morin et al. 2008; Mortazavi et al. 2008; Nagalakshmi et al. 2008; Sultan et al. 2008; Wilhelm et al. 2008). RNA-seq has been used by several groups to map and assess transcriptome expression in C. elegans at various developmental stages and mutant backgrounds (Shin et al. 2008; Hillier et al. 2009; Ramani et al. 2009; Gent et al. 2010). This approach enables a broader look at the pattern of gene expression with single-base-pair resolution by high-throughput sequencing of mRNA. Another important advantage of this approach is the ability to quantify expression levels of low-abundance transcripts (Mortazavi et al. 2008).
Although RNA-seq methods have been extremely helpful in transcriptome annotation, there are challenges inherent in achieving a well-defined transcriptome, especially when current methods for RNA-seq have biases and limitations. In providing data to define the C. elegans transcriptome, we tried to address several important needs: (1) Strandedness information for each transcribed region. (2) Extending coverage to regions that may be missed in individual RNA capture schemes. (3) Continuing a tradition of examining expression in a diversity of cell populations and stages. (4) Rapid tools for visualizing and evaluating gene annotation. (5) Information defining translational activity of the transcriptome.
Results and Discussion
Refining C. elegans transcriptome annotations by multiple RNA-seq methods
To improve the annotation of the C. elegans transcriptome and study the efficacy of RNA-seq methods, we used three different methods of capturing isolated poly(A)+ RNA:
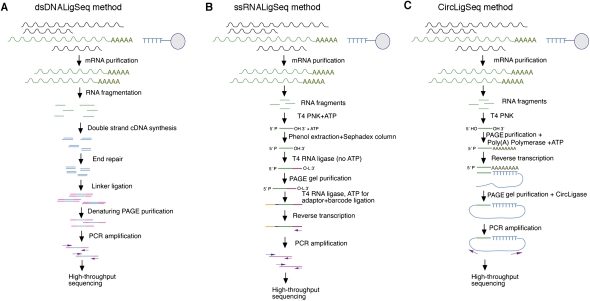
A strand-symmetric method (dsDNALigSeq) (Fig. 1A), in which cDNA is synthesized from mRNA fragments by first-strand synthesis using random primers and second-strand synthesis by hairpin priming of the first strand. Sequencing adapters are subsequently added to the cDNA fragments, and the fragments are then amplified by PCR. A number of previous transcriptome annotation studies for C. elegans have also used a similar method involving short dsDNA cDNA segments produced by fragmentation of longer double-stranded cDNA (Shin et al. 2008; Hillier et al. 2009; Ramani et al. 2009).
A strand-specific method (ssRNALigSeq) (Fig. 1B), in which mRNA fragments are ligated at their 3′ end with a 5′-adenylated adapter using T4 RNA ligase. The fragments are then ligated at their 5′ end to a sequencing adapter, reverse transcribed, and amplified.
A circularized strand-specific method (CircLigSeq) (Fig. 1C), in which mRNA fragments are polyadenylated and then reverse-transcribed with an anchored oligo(dT) primer that includes adapter sequences. The ssDNA product is then circularized using CircLigase (an enzyme that circularizes single-stranded DNA circles) (e.g., Ingolia et al. 2009) and is subjected to PCR amplification.
Figure 1.
Flowcharts describing the RNA-seq methods. Flowcharts describing the protocols to construct mRNA sequencing libraries using the dsDNALigSeq RNA-seq method (A), ssRNALigSeq RNA-seq method (B), or CircLigSeq RNA-seq method (C).
Both strand-specific methods (ssRNALigSeq and CircLigSeq) preserve the directionality of the sequence tag, while the strand-symmetric method (dsDNALigSeq) does not. We used the three methods to construct 17 libraries for RNA-seq from animals with several different genetic backgrounds and at several life stages (described in Table 1). In C. elegans, germ cells account for substantial diversity in gene expression (Reinke et al. 2004). To have substantial material from diverse germline and somatic cell types, libraries were prepared from populations of hermaphrodites, males, somatically hermaphrodite animals defective in sperm-to-oocyte switch [fem-3(q20)], and somatically hermaphrodite animals deficient in spermatogenesis [fem-1(hc17)] (Table 1).
Table 1.
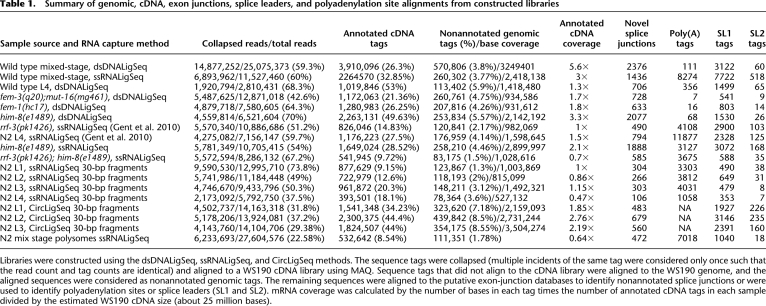
Summary of genomic, cDNA, exon junctions, splice leaders, and polyadenylation site alignments from constructed libraries
Libraries were constructed using the dsDNALigSeq, ssRNALigSeq, and CircLigSeq methods. The sequence tags were collapsed (multiple incidents of the same tag were considered only once such that the read count and tag counts are identical) and aligned to a WS190 cDNA library using MAQ. Sequence tags that did not align to the cDNA library were aligned to the WS190 genome, and the aligned sequences were considered as nonannotated genomic tags. The remaining sequences were aligned to the putative exon-junction databases to identify nonannotated splice junctions or were used to identify polyadenylation sites or splice leaders (SL1 and SL2). mRNA coverage was calculated by the number of bases in each tag times the number of annotated cDNA tags in each sample divided by the estimated WS190 cDNA size (about 25 million bases).
The libraries were sequenced using the Illumina Genome Analyzer system and were analyzed using MAQ and BLAT software. We sorted and trimmed the barcodes (present in all reads that were obtained with the ssRNALigSeq capture method), and trimmed homopolymer A tails for reads that were obtained using the CircLigSeq method. We “collapsed” the set of RNA-seq tags from each library such that repeated incidents of identical sequences were counted only once (to avoid PCR “jackpots”) and aligned the unitarized sequence set to the WS190 cDNA reference database (Table 1; WS190 was used, as the most current version represented in the UCSC Genome Browser). Tags that failed to align to the cDNA data set were next aligned to the C. elegans genome and splice junction databases (see below).
All methods include several steps that can be a source for biases, including 5′-end phosphorylation, 3′-cyclic phosphate removal, ligation steps, gel fractionation, PCR, cluster growth, and sequencing. In order to detect biases in the RNA-seq methods, we looked at both gene coverage and nucleotide preference. Conceptually we would expect differences at the termini of transcripts; in particular, the extreme capped 5′ ends of mRNAs might be expected to be lost in the ssRNALigSeq method (due to failure of ligation at the cap) and should inevitably be truncated in the dsDNALigSeq method due to the need for hairpin priming and opening. No conceptual barrier exists for capture of the 5′ ends with the CircLigSeq method. In contrast, identification of the 3′-poly(A) addition site [mRNA/poly(A) junction] would be difficult with CircLigSeq [due to the poly(A) tailing step incorporated into the protocol].
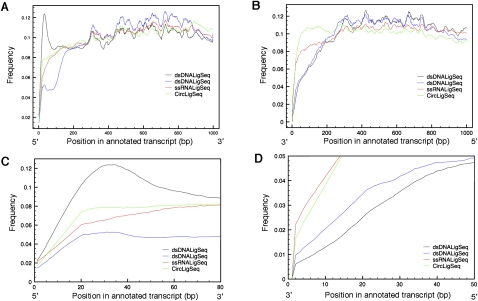
In examining the experimental coverage by the three methods, we found evidence consistent with these and other method-specific biases. Figure 2 shows coverage as a function of relative position within genes (distance from 5′ and 3′ ends of annotated RNA sequences). Few C. elegans mRNAs have been characterized precisely, particularly at the 5′ ends, so that such annotation-based analysis was by nature rather rough, yielding an indication of regional balance but not of recovery for extreme termini. When using the dsDNALigSeq method, there is apparent over-representation of the 5′ regions of genes (Fig. 2A,C), with a decrease at the 3′ end (Fig. 2B,D). With the ssRNALigSeq method, we saw a slight decrease in gene coverage toward the 5′ end of genes (Fig. 2C). We observed the most uniform coverage from the CircLigSeq method. Coverage differences at the end of genes using a similar dsDNALigSeq method were noted previously (Hillier et al. 2009).
Figure 2.
Transcript coverage by position by variety of RNA-seq methods. Transcript coverage was determined by comparing transcript position of RNA-seq tags generated by the dsDNALigSeq, ssRNALigSeq, and CircLigSeq methods. The sequence tags were mapped using BLAT software. Only transcripts that are longer then 1000 bp were considered in the analysis. The plots depict transcript coverage from the start of the transcript (A,C) or from the end of the transcript (B,D). C and D are magnified representations of A and B, respectively. For clarity, only RNA-seqs from N2 mixed stage constructed by the ssRNALigSeq method (red), fem-1(hc17) constructed by dsDNALigSeq method (black), N2 at L4 larval stage constructed by dsDNALigSeq method (blue), or N2 at L1 larval stage constructed by CircLigSeq method (green) are shown. The somewhat uneven coverage along the length of a canonical gene appears partly due to disproportionate contributions by a fraction of highly expressed genes.
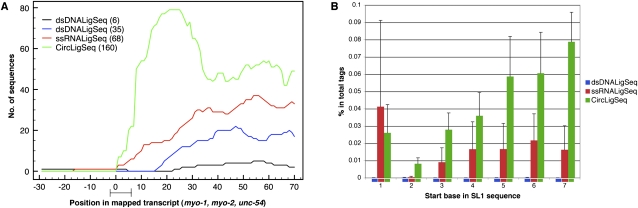
To further investigate the coverage at the extreme 5′ ends of transcripts, we examined three abundantly expressed muscle myosin genes with well-mapped transcription start sites (myo-1, myo-2, and unc-54 [Dibb et al. 1989; Okkema et al. 1993]; note (1) that these three genes lack the common trans-spliced leaders present on most C. elegans transcripts [Blumenthal 2004]) and (2) that myo-1 is referred to in some annotations as synonymous with let-75 (McKim et al. 1992). As shown in Figure 3A, we observed the highest coverage at the 5′ ends with the CircLigSeq method, with virtually no coverage at the extreme 5′ ends with the dsDNALigSeq method.
Figure 3.
Coverage at the 5′ region of genes changes significantly among the different RNA-seq methods. (A) Transcript coverage on the 5′ region was determined by mapping sequences generated by the three different RNA-seq methods to the well-annotated start sites of the myo-1, myo-2, and unc-54 genes (Dibb et al. 1989; Okkema et al. 1993). The sequence tags were mapped using BLAT software. The plots depict transcript coverage from 30 bases before the annotated start sites to 70 bases after. The bar on the x-axis presents the 1–5 base ambiguity and variation in natural start sites (Dibb et al. 1989; Okkema et al. 1993). For clarity, only RNA-seqs from N2 mixed stage constructed by the ssRNALigSeq method (red), fem-1(hc17) constructed by dsDNALigSeq method (black), N2 at L4 larval stage constructed by the dsDNALigSeq method (blue), or N2 at L1 larval stage constructed by the CircLigSeq method (green) are shown. The overall number of sequences that aligned to the assayed region are indicated in parentheses. A lack of coverage at the extreme 5′ ends was also observed using data that were derived from a similar dsDNALigSeq method by Hillier et al. (2009) (Supplemental Fig. 7S). (B) Frequency of sequence reads starting in SL1 splice leader sequences. A start frequency at each of the first seven bases of SL1 was calculated for the three RNA-seq methods by counting the portion of the sequence tags that start with the relevant 16 SL1 bases. None of these 16-mers is found in the C. elegans transcriptome outside of SL1. (Blue) dsDNALigSeq method; (red) ssRNALigSeq method; (green) CircLigSeq method.
Approximately 70% of C. elegans mRNAs are trans-spliced to 22-nt spliced leaders, SL1 or SL2, between the 5′ cap and the first 3′ splice site (Blumenthal 2004). To further examine the coverage of the different methods at the start site, we calculated the frequency of sequence tags that started with at least 16 bp from the SL1 sequence. We found very few tags from the dsDNALigSeq method that started with the first bases of the SL1 sequence (Fig. 3B), consistent with the 5′-end truncation of cDNAs as part of the dsDNALigSeq method. The CircLigSeq and ssRNALigSeq methods both yielded tags that start with the first bases of SL1, with a somewhat higher fraction with CircLigSeq (Fig. 3B). The recovery of ssRNALigSeq reads with the precise 5′ terminus of SL1 indicates either an alkali lability of the alpha–beta bond in the 5′ cap or a population of uncapped 5′ phosphorylated transcripts. Interestingly, the frequency of tags that started with the second base of SL1 was substantially lower than at the first base or other bases of SL1, with similar results observed for SL2 (Fig. 3B; data not shown). This would be consistent with a modification affecting the ability of alkali to cleave between the first and second encoded base of the splice leader. We found that alkali treatment for longer periods of times caused higher frequency of tags starting at the second base of SL1 (data not shown). One possible modification between the first and second bases of an mRNA would be a 2′-O-methylation of the sugar attached to the first templated base of the splice leader. This modification has been observed in other systems (e.g., Furuichi et al. 1975) and would certainly be expected to influence the rate of alkali cleavage.
We further noted differences between the methods in nucleotide preference in tag ends and flanking sequences (Supplemental Fig. 1S). Sequence tags that were generated using the dsDNALigSeq method have a significant preference toward C or G at the first position of the alignment (Supplemental Fig. 1SA), while other positions in the alignment have a slight preference against T. Sequences that were generated using the CircLigSeq method had significant preference toward A and T at the first position of the alignment and C and T at the second position of the alignment (Supplemental Fig. 1SC). Given the discrepancy between methods, the initial nucleotide preferences are likely to be at least in part an outcome of the technical processes. These biases were probably not caused as a result of the alkaline mRNA fragmentation step, because the RNA was fragmented in the same way in all methods. One possible source for differential tag recovery would be a sequence bias of the different ligases used in the RNA-seq methods (Ingolia et al. 2009). Despite the localized variations between methods within genes, we note that the nucleotide preferences do not seem to affect gene coverage (Fig. 2).
Refining and expanding models of the C. elegans transcriptome
The diversity of starting material (genetic backgrounds and developmental stages) and of methodology used to construct the RNA-seq libraries has the potential to extend analysis of the transcriptome. We first used the RNA-seq data to revise a commonly used set of transcript models by examining RNA-seq tags that fail to align to a recent mRNA reference data set (UCSC May 2008/WS190 cDNA) (Table 1). Of such sequences, 5.5% could be aligned to the WS190 C. elegans genome, defining additional regions in the transcriptome not annotated in WS190.
Extending the collection of defined C. elegans exon junctions
High-throughput sequencing of the transcriptome provides a valuable opportunity to discover and refine splicing patterns. Identification of novel splicing events from RNA-seq tags is challenging and has been tackled by several groups (Sultan et al. 2008; Hillier et al. 2009; Trapnell et al. 2009; Blekhman et al. 2010; Filichkin et al. 2010; Wang et al. 2010). To evaluate our ability to expand the inventory of splice junctions, we used a “hypothesis-testing” approach. Two lists of potential exon–exon junctions (one for each strand) were constructed by scanning the WS190 genome for splicing acceptor or donor sites that meet the splicing consensus sequences requirements (Blumenthal and Steward 1997; for details, see Methods). To minimize backgrounds of spurious matches, we chose relatively stringent constraints on both intron length (32 bp–2 kb) and splicing consensus sites. Each potential splice product in our lists represents a hypothesis that could be supported by finding RNA-seq reads that span the predicted junction. In our analysis, we discarded any alignment where there was more than one mismatch between the RNA-seq read, or where the RNA-seq fragment was aligned with <9 bp (out of 30 or 32 total) on each side of the splice junction.
To evaluate false discovery rates, we generated a number of “sham” lists of potential splice junctions in which putative splice donor and acceptor sequences were combined either at random across the genome, or were combined in localized regions but in configurations not consistent with canonical gene structures (e.g., with the acceptor upstream of the donor [instead of downstream]). Additional tests using a composition-controlled but randomized genome (Lowe and Eddy 1997) are described in Methods.
The RNA-seq data from this study provide tentative experimental support for at least 6447 exon junctions (Table 1) not annotated in WS190, of which 98% are unique in the potential exon–exon junction lists. In parallel with this analysis, false discovery rate upper bounds from 2% to 4.5% were calculated by aligning to the four different sets of “sham” exon–exon junction databases as described above. We note that the higher (4.5%) upper bound in this case represents the “sham” database consisting of splice acceptors upstream of (but close to) proximal splice donors. Since such conditions can generate real products (circularized exons) (e.g., Nigro et al. 1991), it is possible that the increased matched fraction in this set of exon–exon models may reflect a real presence of some of these “circularizing” junctions in the transcriptome.
Although these data strongly support the inferred higher complexity of the C. elegans transcriptome, it is critical to consider each potential junction as “provisional” depending on other available data and validation. As an additional test of the provisional splice junctions in aggregate, the start positions of the RNA-seq tags in the aligned putative exon junctions were examined. For spurious matches, we would expect a strong preference for “edge” alignments with a minimal number of matched bases on one side of the junction. Instead, the alignments were distributed evenly (Supplemental Fig. 2S). These observations suggest the authenticity of the substantial majority of assigned junctions from the analysis.
Of the putative non-WS190 junctions identified in this analysis, 60% (3884) were also supported by alignments to the putative junctions with reads from Hillier et al. (2009). The remaining 2563 are not seen in the Hillier data, potentially reflecting different populations of animals used to construct libraries, different sequencing approaches, and/or simple representation limits for relatively rare mRNAs in the two large (but certainly not exhaustive) data sets. We note in particular a group of 852 junction sequences observed in our male populations that were absent in WS190 and in the hermaphrodite RNA-seq data from our analysis and from Hillier et al. (2009). Given the slightly different approaches to identification of potential exon junctions used by Hillier et al. (2009) and in this study, it was also of interest to examine the total number of non-WS190 potential junctions present in the combined data sets. This value (11,215) exemplifies the remarkable complexity of the C. elegans transcriptome.
In addition to the evaluation of potential false discovery, it is critical to stress that not all potential exons will be identified by this method. In particular, the relatively stringent criteria on intron length and match to consensus eliminate a fraction of well-characterized introns. Relaxing these criteria substantially is of at best limited value for the approach used here, as this will increase the number of potential exon junctions. Thus the potential exon-junction list that was assembled computationally was by necessity a partial list. Aligning the WS190 cDNA reference database, we detect that 73% of the annotated splice junctions were predicted by our method, while the remaining 27% would include misannotations and true exon–exon junctions that do not meet our length or consensus splice-site criteria. These considerations, combined with the likelihood of additional junctions not captured in these data sets due to presence in rare mRNAs or rare cell types, suggest an even larger pool of mRNA junctions that remain to be characterized.
Identifying polyadenylation sites
Apparent extensions to annotated 3′ regions of transcripts are particularly common (Supplemental Fig. 4S). Looking for polyadenylation events in the RNA-seq data can give a better picture of the 3′ regions of transcripts. As noted above, the CircLigSeq method was not used to identify such events because a poly(A) tail is added in that method as part of library preparation (Fig. 1C). We thus used the dsDNALigSeq and ssRNALigSeq for poly(A) mapping. We searched for tags that have at least 18 bases that match the genome followed by a non-genome-encoded tail of at least seven A's. We found 43,823 such polyadenylated tags with a false discovery rate of 4.6%. Sixty-four percent of the poly(A) tags aligned to annotated gene regions in the genome. Interestingly, only 558 (1.3%) poly(A) tags were found in libraries that were constructed by the dsDNALigSeq method, from a 33% contribution of the dsDNALigSeq to the data sets, which suggests that the dsDNALigSeq method is very inefficient for recovering poly(A) tags. The low coverage at the 3′ end of genes when using the dsDNALigSeq method (Fig. 2) might explain the lack of poly(A) tags in the sequence tags.
Identifying trans-splicing events
Splice leader sequences in the RNA-seq tags can suggest trans-splicing sites and provide useful information about the beginning of transcripts. To detect trans-splicing sites, we searched for tags from the three different RNA-seq methods that have at least 10 bases that match the end of the splice leaders followed by at least 16 bp that match the genome. We found 35,080 tags with SL1 sites and 1846 tags with SL2 sites with a false discovery rate of 0.05%. Twenty-three percent of the tags containing SL1 sites and 9.5% of the tags containing SL2 sites were found in the libraries that were constructed by the dsDNALigSeq (from a 33% contribution to the datasets). This observation is in line with our finding that coverage at genes start sites is low when using the dsDNALigSeq method (see above).
Identification of sequences in a polysomal fraction
Polysome profiles can be of considerable value in revealing the distribution of mRNAs engaged by the translation machinery (Melamed and Arava 2007). We examined the sequence composition of a polysomal fraction obtained by sedimentation of disrupted mixed stage C. elegans on a sucrose gradient. RNA from the polysome fractions was poly(A) selected and sequenced using the ssRNALigSeq method.
We found some of the nonannotated transcribed regions to be sedimented in polysomal fractions, consistent with the possibility that these RNAs could be translated (Supplemental Figs. 5SA, 3S). Of the genes, 73.8% (12,074) had at least one polysome-sedimenting tag, compared to genes with RNA-seq tags from N2 mix-staged libraries when using the ssRNALigSeq method. The polysome-sedimenting tags primarily aligned to the 3′ ends of transcripts (data not shown), probably as a result of partial degradation of the RNA prior to poly(A) selection of the mRNA.
Examples of display-based transcriptome refinement by RNA-seq methods
To facilitate the global viewing of transcript structure, we have constructed an interface in which our data sets can be viewed in Genome Browser display (Fig. 4). For viewing and analysis, we used the UCSC Genome Browser with the WS190 version of the C. elegans genome. The displayed tracks include genome-matched tags that are missing in the current data set of predicted mRNA models (WS190 in this case), potential exon–exon junctions, polysome density map, polyadenylation sites, and trans-spliced sites. To provide a further evaluation of each potential exon–exon junction, each of these is shaded based on a score derived from the frequency and quality of matches from the RNA-seq data sets. Potential splices are shown with an orientation: Even with strand-symmetric data, we can still deduce the strand specificity of the splice site based on the intrinsic orientation of splice junctions (Supplemental Fig. 3S).
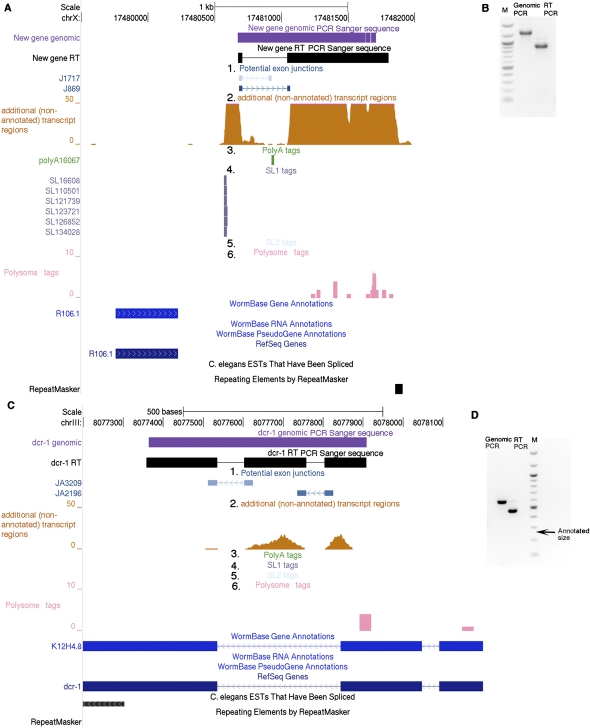
Figure 4.
Browser-based refinement of the transcriptome by RNA-seq. We used the UCSC Genome Browser custom tracks with the WS190 version of the C. elegans genome for viewing the following data sets: (1) “Potential exon-junctions” track (blue), which displays potential nonannotated exon–exon junctions that are supported by RNA-seq reads by two bars, each for the 23 bp of the adjacent exons, with a connecting arrow that indicates the exon-junction directionality. The bar shade indicates a strength score, calculated from the number of aligned tags to the exon junction and number of bases from each junction that are included in the sequence tag, with darker shades representing higher scores (score = 100 × number of alignments × coverage score). The coverage score equals 1 when the smallest base coverage of the exon is 9 or 10 bases; 1.2 when the smallest base coverage of exon is 11, 12, or 13; or 1.5 when the smallest base coverage of exon is 14, 15, or 16. (Supplemental Fig. S2). (2) RNA sequences from regions with no existing gene predictions [“additional (nonannotated) transcript regions,” orange]. In this custom track the bar height represents the number of sequences that align to each position. (3) A poly(A) tags track (green) that displays polyadenylation junctions identified by the RNA-seq. The arrow in each bar points to the start position of the putative poly(A) tail. (4) SL1 tags track (purple) and (5) SL2 tags track (blue-gray) that display trans-splice leader sites identified by the RNA-seq. The arrow in each bar indicates splice leader directionality. (6) A polysome tags track (pink) that displays observed tags from a polysome-enriched RNA pool. The browser shots exemplify the discovery of nonannotated transcribed regions from RNA-seq data. For chrX:17480500–17842000 (A) nonannotated genomic tags with a darkly shaded splice junction suggest a transcript. This transcript and splice were validated by RT-PCR and sequencing (B; PCR Sanger-sequence data not shown). The SL1 tags suggest the presence of a different transcript from the proximate R106.1 transcript. Polysome sedimentation (pink track in A) suggests that the transcript is present in polysome fractions. The light-shade exon junction and the poly(A) site do not have significant nonannotated genomic tag coverage at the same position, so this junction could be considered “provisional.” dcr-1 is an example of a gene that is studied by many research groups (e.g., Knight and Bass 2001; Duchaine et al. 2006; Pavelec et al. 2009), which we found to contain a predicted nonannotated exon. The exon is 195 bp long and is in-frame with the adjacent exon. This exon appears uniformly incorporated into the transcript; we see no evidence for differential splicing (C). The exon existence was confirmed by PCR, RT-PCR, and Sanger sequencing (D). The arrow in D indicates the size of the expected RT-PCR band from the WS190-annotated dcr-1. EST additions to GenBank (June 2010) further support the structures shown in this figure and Supplemental Figure 4SA.
We have used these data sets for refinement of a number of genes currently of interest for specific analyses (Fig. 4; Supplemental Figs. 4S, 5S). Confirming the refined models, several features of the revised transcription maps have appeared in WormBase and modENCODE during the course of this work (http://www.wormbase.org/db/gb2/gbrowse/c_elegans/), while other features [including polysome association, poly(A) sites, and numerous splice sites (cis and trans)] have not yet appeared (as of July 2010) in these resources.
We also found that RNA-seq tags at microRNA loci can help identify microRNA precursors (Supplemental Fig. 6S). microRNA precursors (pri-miRNA) are polyadenylated before they are processed to become mature microRNA, and therefore would be included as part of the RNA-seq libraries. Identification of microRNA polyadenylation sites is also possible from the RNA-seq tags (Supplemental Fig. 6S). The RNA-seq data and small RNA sequencing data can be used for comparing the levels of mature microRNAs and microRNA precursors and might shed light on microRNA function. Interestingly, changes in mature microRNA levels without noticeable changes in the levels of the microRNA precursors are seen in microRNA processing regulation (Ambros et al. 2003).
Data set integration
Taken together, a putative splice-junction data set, nonannotated genomic tags, polyadenylation sites, splice leader sites, polysome tags, and strand-specificity information from the strand-specific RNA-seq libraries provide a diverse and multifaceted image of transcript structure. The combination of multi-method RNA-seq and rich display should be of substantial utility both for characterized model systems (such as C. elegans) and emerging models where a mixture of RNA-seq and genome assembly will be a primary mode of functional genome characterization.
Methods
C. elegans strains
The following strains were used in this study: Bristol N2 (Brenner 1974), NL2099 rrf-3(pk1426) II (Sijen et al. 2001), BA17 fem-1(hc17)IV (Nelson et al. 1978), JK816 fem-3(q20)IV (Barton et al. 1987), PD3331 him-8(e1489)IV, and PD3330 rrf-3(pk1426) II; him-8(e1489) IV. The JK816 fem-3(q20) strain carries an additional background mutation upstream of mut-16 gene (Gent et al. 2010).
C. elegans population synchronization and harvesting
All strains were cultured as described in Brenner (1974). N2 were grown at 16°C and harvested as mixed stage or at larvae stage of development after synchronization. Synchronization was obtained by feeding animals following treatment with sodium hypochlorite solution to kill all stages except embryos. The germline mutant strains fem-3(q20) and fem-1(hc17) were raised at 25°C and harvested as young adults [equivalent stage to the him-8(e1489) males]. him-8(e1489) males were isolated as described in Gent et al. (2009). rrf-3(pk1426) and N2 at the fourth larval stage that were used for ssRNALigSeq sequencing are described by Gent et al. (2010).
ssRNALigSeq mRNA sequencing library preparation
Library samples for ssRNALigSeq mRNA sequencing were prepared using a similar scheme to that used commonly for small RNA sequencing library preparation (Lui et al. 2007; the protocol scheme is in Fig. 1B). mRNA was extracted from the collected animals and alkali-fragmented using reagents from Ambion [MicroPoly(A) purist and RNA fragmentation]. For longer fragments the mRNA was incubated with the fragmentation buffer for 5 min at 70°C, and for shorter fragments for 45 min at 98°C. mRNA fragments were then treated with T4 polynucleotide kinase (New England Biolabs) and ATP to form uniform species with 5′-monophosphorylated and 3′-hydroxylated ends. We then removed the ATP with an Illustra microspin G-25 column (GE Healthcare) and ligated the 3′ adapter (IDT Linker-1) using T4 RNA ligase 1 (New England Biolabs). The mRNA fragments were then excised and purified from a 6% acrylamide gel between 125 bp and 200 bp and ligated to the 5′ adapter with T4 RNA ligase 1 (New England Biolabs). Subsequent extension and amplification (PCR) were as described (Lui et al. 2007). With these samples we also used 3-nt or 4-nt barcodes in the 5′ adapter to allow pooling of samples and as a guard against contamination. The primers and 5′ adapter that were used are described in Lui et al. (2007).
dsDNALigSeq mRNA-sequencing library preparation
dsDNALigSeq mRNA-sequencing libraries (except N2 L4 sample) were prepared using reagents from Illumina (RS-100-0801). Total RNA was extracted from frozen tissue samples with mirVana (Ambion), and 10 μg was used for initial mRNA purification. The N2 L4 library was prepared using a slightly different protocol (Fig. 1A). mRNA was extracted from frozen tissue and fragmented as above for the ssRNALigSeq libraries. First-strand cDNA was synthesized from the mRNA fractions using random hexamer primers and Superscript II Reverse Transcriptase (Invitrogen). The second strand was synthesized using DNA polymerase I (Invitrogen). The cDNA was then treated with T4 polynucleotide kinase (New England Biolabs) and T4 DNA polymerase (New England Biolabs) to generate double-stranded cDNA fragments with blunt ends. We then ligated the cDNA fragments to an oligo duplex (SG-133, AGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG; SG-134, CCCTACACGACGCTCTTCCGATCT) with T4 DNA ligase (New England Biolabs). After ligation the cDNA fragments where excised between 175 and 300 bp and purified from a 6% acrylamide gel and then subjected to PCR amplification with primers SG-135, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT and SG-137, CAAGCAGAAGACGGCATACGAGCT.
CircLigSeq mRNA sequencing library preparation
mRNA sequencing library samples were prepared as described in Ingolia et al. (2009) (the protocol scheme is in Fig. 1C). mRNA was extracted and fragmented as above (alkaline hydrolysis: 45 min at 98°C) and treated with T4 polynucleotide kinase (New England Biolabs) without ATP to form species with 5′- and 3′-hydroxylated ends. The mRNA fragments were then excised and purified from a 12% acrylamide gel with an average size of 30 nt. To add a poly(A) tail, we incubated the fragments for 15 min at 37°C with 0.25 U of Escherichia coli poly(A) polymerase (NEB) and 1 mM ATP. Reverse transcription was then carried out using the modified DNA primer oNT1223 (Ingolia et al. 2009; synthesized by IDT). The reverse-transcription products were separated on a 12% acrylamide gel and circularized using CircLigase (Epicentre). The circularized fragments were subjected to PCR amplification with oNT1230 and oNT1231 primers (Ingolia et al. 2009). mRNA fragments were 30 bp long when using the CircLigSeq method (as optimized by Ingolia et al. 2009). Some of the libraries prepared with the ssRNALigSeq capture method were selected at 30 nt for comparisons (Table 1).
Polysome sedimentation and library preparation
Total polyribosomes were isolated from C. elegans N2 mix-staged animals and sedimented on a 10%–50% sucrose gradient as previously described (Davies and Abe 1995). The gradient was fractionated, and polyribosome-containing fractions were identified from UV absorbance measurements. Total RNA from these fractions was incubated at 65°C with proteinase K and extracted with acid-phenol:chloroform and ethanol precipitation. The isolated RNA was prepared for high-throughput sequencing using the ssRNALigSeq method (see above).
Sequence processing and alignment
The Illumina GAII system was used to obtain sequence reads. A read length of 36 bp was used for all samples, with the exception of the L4 dsDNALigSeq sample (33 bp). Reads that were obtained with the ssRNALigseq cloning method had barcodes, and reads that were obtained using the CircLigSeq method had homopolymer A tails. Before alignment all reads were processed by trimming the barcodes and the entire homopolymer A tail as needed. Next, identical sequences were collapsed in each sample to avoid PCR “jackpots.” All sequences were aligned to a cDNA reference set (version WS190) using MAQ software (http://maq.sourceforge.net) (Li et al. 2008) using the first 30 nt of each read (28 nt for short fragments) and allowing one mismatch. Sequences that did not align to the cDNA reference set were aligned to the C. elegans genome (version WS190). We will refer to these aligned sequences as the nonannotated genomic tags (Table 1). We further aligned the remaining unaligned sequences to the putative exon-junction database to find nonannotated exon junctions. To confirm the results, we aligned the sequences to both the cDNA set and the C. elegans genome (version WS190) using BLAT alignment software, with default parameters except that tileSize and stepSize were set to 11 and 5, respectively (Kent 2002). The false discovery rate (<0.01%) was calculated by aligning the sequences to a composition-matched randomized genome (Lowe and Eddy 1997; Fire et al. 2006). Polysome tags were aligned to the C. elegans genome using MAQ. The cDNA data set, derived from the C. elegans WS190 genome assembly, was downloaded from http://www.wormbase.org/biomart/martview. The source reads are available at the NCBI Gene Expression Omnibus (accession numbers GSE22410 and GSE19414). As an additional test for usability of the UCSC Genome Browser interface, we randomly chose 20 previously characterized genes from the WS190 annotation and examined their refinement using the browser tracks described above. For 15 of these, the present data provided corrections or additions to the inferred gene model. For seven genes we found extensions to the 5′ or 3′ gene regions. For nine genes polyadenylation sites were identified. For eight genes splice leader sites were identified. Five genes had nonannotated exon junctions. In one gene we observed a previously nonannotated internal coding exon.
Nucleotide preference analysis
The nucleotide abundance at each position along the length of the mRNA was calculated after normalizing to the mRNA nucleotide content, considering the number of sequences aligning to each cDNA in the WS190 cDNA data set in a certain sample. The cDNA template was used for the calculations rather then the sequence tag to allow extension of the analysis beyond the sequence ends and to avoid counting sequencing errors.
Construction of putative exon-junction databases
Two putative exon–exon junction databases (plus and minus strand) were created by predicting introns in C. elegans genomic sequence. The criteria used to identify putative introns were: (1) Presence of the invariant 5′-GT and 3′-AG sequences at the beginning and end of the intron; (2) length between 32 and 2000 bases; and (3) a sequence-match cutoff criterion based on a survey of acceptor and donor sites in the C. elegans genome (Staden 1984; P-value cutoff 0.03% for each splice junction; Blumenthal and Steward 1997). The resulting Potential Exon Junction-Sense database (PEJS) contained 16,652,231 putative exon junctions, with the antisense database (PEJA) containing 16,492,340. Each sequence in the database contains 46 bases (42 bases for the short RNA-seq fragments), 23 bases from each exon adjacent to the splice junction. We then eliminated previously annotated exon junctions from our putative exon-junction databases by alignment of our databases to the cDNA reference database. Experimental support (reads that matched with at most one mismatch in a segment with at least 9 bp on each side of the splice junction) was obtained for 0.02% of the resulting list of potential junctions (47% supported by a single read and 53% by multiple reads).
Four different sets of “irrational” exon–exon junction databases were created to estimate a “false-positive” rate at which spurious reads might align to our databases. Each of the four sham databases starts with the same set of potential donor and acceptor sites used to generate the PEJS and PEJA data sets, but joins donors and acceptors in combinations not expected to occur in bone fide mRNAs.
(1) Acceptor upstream of donor: Instead of canonical exon–exon junctions that connect 5′ donor sites to 3′ acceptor sites, aberrant exon–exon junctions were created by joining donor sites to upstream acceptor sites. The genomic length between the acceptor and donor positions was kept between 32 and 2000 bases. This database was subdivided into two databases for separate analyses according to the genomic length between the donor and acceptor sites; one where genomic length was a multiple of three (in-frame) and containing the out-of-frame remainder. Included in these sets would be both spurious matches of RNA sequence to the “irrational” exon–exon junction data set, cases where donor splicing to upstream acceptors occurs in natural mRNA synthesis (potentially generating circular exon RNAs) (e.g., Nigro et al. 1991), matches to genomic regions in which adjacent regions had been misplaced in genome assembly, and cases in which DNA had become rearranged to produce a donor-upstream configuration, so alignment rates represent an upper bound on false positives. Alignment to the RNA-seq data sets in this paper yielded matches to 0.0009% and 0.0007% of the acceptor-upstream (irrational) junctions (in-frame and out-of-frame, respectively). These values are >20-fold lower than those for donor-upstream (rational) junctions.
(2) Opposite strand, donor upstream: The donor and acceptor sites in this “irrational” database were required to sit on different strands of the same chromosome, with the acceptor site located 32–2000 bases downstream from the donor site. Matches to this data set would include spurious sequence matches as well as instances where a segment of the genome in one or more cells was inverted with respect to the published genome sequence. Of these junctions, 0.00057% yielded alignment to the RNA-seq data sets.
(3) Opposite strand, acceptor upstream: This set is similar to (2) except that the putative acceptor site is located 32–2000 bases upstream of the putative donor site, on the opposite strand. Of these junctions, 0.00052% yielded alignment to the RNA-seq data sets.
(4) Mixed chromosomes: Putative donor and acceptor sites on different chromosomes were used. Of these junctions, 0.0004% yielded alignment to the RNA-seq data sets.
An additional value (lower bound) for false discovery rate (0.015%) was calculated by aligning the putative exon-junction databases to a composition-matched randomized genome (Lowe and Eddy 1997; Fire et al. 2006).
Identification of polyadenylation sites and splice leaders
To produce a list of poly(A) tags, we searched for tags that did not match the genome in our initial screening and had at least seven consecutive A's at the end of the tag in the ssRNALigSeq sequences, or at least seven A's at the end of the tag or at least seven T's at the beginning of the tag in the dsDNALigSeq sequences. At least 18 bases from the remaining bases in the tag had to match uniquely the C. elegans genome for the tag to be considered in the database. The false discovery rate (4.6%) was calculated by aligning the identified polyadenylation sites to the C. elegans genome allowing two mismatches. Sequence tags that were produced by the CircLigSeq method were not used for this analysis because of their poly(A) tail.
To produce a list of tags that contain splice leaders, we searched for tags that did not match the genome in our initial screening and had at least 10 bases from SL1 or SL2 at the beginning of the tag, or at least 10 bases from the inverted SL1 or SL2 at the end of the tag in the dsDNALigSeq sequences. At least 17 bases from the remaining bases in the tag had to match uniquely the C. elegans genome for the tag to be considered in the database. The false discovery rate (0.05%) was calculated by aligning the identified splice leader sites to the C. elegans genome allowing two mismatches.
Acknowledgments
We thank Sam Gu for flow cell preparation; Cheryl Smith, Ziming Weng, Phil Lacroute, Anton Valouev, and Arend Sidow for flow cell preparation and sequencing; Jason Merker and Julia Pak for comments; Gary Schroth and Shujun Luo from Illumina for helping with the dsDNALigSeq method; and the Caenorhabditis Genetics Center for strains. This work was supported by NIH (R01GM37706 [A.Z.F.], T32HG00044 [J.I.G.]), National Science Foundation Graduate Research Fellowship and Stanford Graduate Fellowship program (M.R.S.), A*STAR (Agency for Science, Technology and Research) Singapore (H.Z.), and Stanford Dean's Fellowship and Machiah Foundation (A.T.L.).
Footnotes
[Supplemental material is available for this article. The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE22410.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.108845.110.
References
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13: 807–818 [DOI] [PubMed] [Google Scholar]
- Barton MK, Schedl TB, Kimble J 1987. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y 2010. Sex-specific and lineage-specific alternative splicing in primates. Genome Res 20: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T 2004. Operons in eukaryotes. Brief Funct Genomics Proteomics 3: 199–211 [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Steward K 1997. RNA processing and gene structure.In C. elegans II (ed. Riddle TL et al. ), pp. 117–145 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012–2018 [DOI] [PubMed] [Google Scholar]
- Cloonan N, Forrest ARR, Kolle G, Gardiner BBA, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5: 613–619 [DOI] [PubMed] [Google Scholar]
- Davies E, Abe S 1995. Methods for isolation and analysis of polyribosomes. Methods Cell Biol 50: 209–222 [DOI] [PubMed] [Google Scholar]
- Dibb NJ, Maruyama IN, Krause M, Karn J 1989. Sequence analysis of the complete Caenorhabditis elegans myosin heavy chain gene family. J Mol Biol 205: 603–613 [DOI] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354 [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong W-K, Mockler TC 2010. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Alcazar R, Tan F 2006. Unusual DNA structures associated with germline genetic activity in Caenorhabditis elegans. Genetics 173: 1259–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, Morgan M, Shatkin AJ, Jelinek W, Salditt-Georgieff M, Darnell JE 1975. Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci 72: 1904–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Schvarzstein M, Villeneuve AM, Gu SG, Jantsch V, Fire AZ, Baudrimont A 2009. A Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNAi. Genetics 183: 1297–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ 2010. Distinct Phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell 37: 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, Reinke V, Green P, Hirst M, Marra MA, Waterston RH 2009. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res 19: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SJ, Riddle DL, Pouzyrev AT, Velculescu VE, Hillier L, Eddy SR, Stricklin SL, Baillie DL, Waterston R, Marra MA 2001. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res 11: 1346–1352 [DOI] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS 2001. A gene expression map for Caenorhabditis elegans. Science 293: 2087–2092 [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui W-O, Pourmand N, Patterson BK, Fire A 2007. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res 67: 6031–6043 [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y 2008. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCombie WR, Adams MD, Kelley JM, FitzGerald MG, Utterback TR, Khan M, Dubnick M, Kerlavage AR, Venter JC, Fields C 1992. Caenorhabditis elegans expressed sequence tags identify gene families and potential disease gene homologues. Nat Genet 1: 124–131 [DOI] [PubMed] [Google Scholar]
- McKim KS, Starr T, Rose AM 1992. Genetic and molecular analysis of the dpy-14 region in Caenorhabditis elegans. Mol Gen Genet 233: 241–251 [DOI] [PubMed] [Google Scholar]
- Melamed D, Arava Y 2007. Genome-wide analysis of mRNA polysomal profiles with spotted DNA microarrays. Methods Enzymol 431: 177–201 [DOI] [PubMed] [Google Scholar]
- Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, McDonald H, Varhol R, Jones S, Marra M 2008. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques 45: 81–94 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson GA, Lew KK, Ward S 1978. Intersex, a temperature-sensitive mutant of the nematode Caenorhabditis elegans. Dev Biol 66: 386–409 [DOI] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B 1991. Scrambled exons. Cell 64: 607–613 [DOI] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S 2009. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics 183: 1283–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Nelson AC, Kapranov P, Bell I, Gingeras TR, Fraser AG 2009. High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol 10: R101 doi: 10.1186/gb-2009-10-9-r101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323 [DOI] [PubMed] [Google Scholar]
- Schena M, Heller RA, Theriault TP, Konrad K, Lachenmeier E, Davis RW 1998. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol 16: 301–306 [DOI] [PubMed] [Google Scholar]
- Shin H, Hirst M, Bainbridge MN, Magrini V, Mardis E, Moerman DG, Marra MA, Baillie DL, Jones SJM 2008. Transcriptome analysis for Caenorhabditis elegans based on novel expressed sequence tags. BMC Biol 6: 30 doi: 10.1186/1741-7007-6-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476 [DOI] [PubMed] [Google Scholar]
- Staden R 1984. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res 12: 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. 2008. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW 1995. Serial analysis of gene expression. Science 270: 484–487 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xi Y, Yu J, Dong L, Yen L, Li W 2010. A statistical method for the detection of alternative splicing using RNA-seq. PLoS ONE 5: e8529 doi: 10.1371/journal.pone.0008529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R, Martin C, Craxton M, Huynh C, Coulson A, Hillier L, Durbin R, Green P, Shownkeen R, Halloran N 1992. A survey of expressed genes in Caenorhabditis elegans. Nat Genet 1: 114–123 [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243 [DOI] [PubMed] [Google Scholar]