Abstract
Mirtrons are intronic hairpin substrates of the dicing machinery that generate functional microRNAs. In this study, we describe experimental assays that defined the essential requirements for entry of introns into the mirtron pathway. These data informed a bioinformatic screen that effectively identified functional mirtrons from the Drosophila melanogaster transcriptome. These included 17 known and six confident novel mirtrons among the top 51 candidates, and additional candidates had limited read evidence in available small RNA data. Our computational model also proved effective on Caenorhabditis elegans, for which the identification of 14 cloned mirtrons among the top 22 candidates more than tripled the number of validated mirtrons in this species. A few low-scoring introns generated mirtron-like read patterns from atypical RNA structures, but their paucity suggests that relatively few such loci were not captured by our model. Unexpectedly, we uncovered examples of clustered mirtrons in both fly and worm genomes, including a <8-kb region in C. elegans harboring eight distinct mirtrons. Altogether, we demonstrate that discovery of functional mirtrons, unlike canonical miRNAs, is amenable to computational methods independent of evolutionary constraint.
Canonical microRNAs (miRNAs) are ∼22-nucleotide (nt) regulatory RNAs derived from inverted repeat transcripts whose biogenesis involves a defined series of processing events (Kim et al. 2009). Primary-miRNA (pri-miRNA) transcripts are first cleaved by the nuclear RNase III enzyme Drosha (also known as RNASEN) to yield pre-miRNA hairpins. Following their cytoplasmic export via exportin 5, pre-miRNAs are cleaved on their terminal loop side by a Dicer-class RNase III enzyme to release miRNA/miRNA* duplexes. One side of the duplex, designated the mature miRNA, is preferentially transferred into an Argonaute protein and guides it to regulate target transcripts. Its partner miRNA* strand is inferred to be preferentially degraded on account of its lower steady-state accumulation, although miRNA* species may still be transferred into Argonaute proteins and have regulatory activities. Since RNase III enzymes typically cleave substrates leaving signature 2-nt 3′ overhangs, an appropriate geometry of cloned small RNA duplex ends provides evidence for their transit via a Drosha–Dicer pathway (Ambros et al. 2003; Friedlander et al. 2008; Berezikov et al. 2010; Chiang et al. 2010).
Since thousands of miRNAs are now known (Griffiths-Jones et al. 2008), one might presume that sufficient information exists to segregate bona fide miRNA genes from bulk genomic hairpins. Although bioinformatic strategies can enrich for genuine miRNA genes, the number of plausible pri-miRNA hairpins in a typical animal genome exceeds the number of confirmed miRNA hairpins by several orders of magnitude. Consequently, the most successful methods for computational miRNA gene finding rely upon evolutionary conservation of miRNA candidates (Grad et al. 2003; Lai et al. 2003; Lim et al. 2003a,b). In particular, conserved hairpins that diverge more quickly in their terminal loops relative to the hairpin stems are likely to be genuine miRNAs (Lai 2003; Lai et al. 2003; Berezikov et al. 2005).
The specificity of the comparative approach increases with the burgeoning amount of genome sequence now available (Rhead et al. 2010), and substantial computational efforts identified miRNAs that are well-conserved in particular animal clades, such as Drosophila (Ruby et al. 2007b; Sandmann and Cohen 2007; Stark et al. 2007b) or vertebrates (Hertel and Stadler 2006; Yousef et al. 2006; Huang et al. 2007; Sheng et al. 2007; Terai et al. 2007). Still, this approach leaves open the question of how many species-restricted miRNA genes exist. Machine learning approaches were implemented toward identifying generic structural features of miRNAs (Bentwich et al. 2005; Nam et al. 2005; Xue et al. 2005; Miranda et al. 2006; Brameier and Wiuf 2007; Helvik et al. 2007; Jiang et al. 2007; Ng and Mishra 2007; Ritchie et al. 2008; Batuwita and Palade 2009; Kadri et al. 2009; van der Burgt et al. 2009). Nevertheless, 10,000s to 100,000s of candidates are identified genome-wide at cutoffs that permit reasonable sensitivity for recovery of known miRNAs. Therefore, it is currently not possible to predict confidently, in silico, whether an arbitrary hairpin is competent for processing by the miRNA generating machinery in vivo.
Instead, the identification of newly evolved miRNAs has depended on small RNA sequencing, an approach revolutionized by recent technological advances. Many species-restricted genes have emerged via these methods (Morin et al. 2008; Goff et al. 2009; Berezikov et al. 2010; Chiang et al. 2010), and such studies affirm that miRNAs with strong evidence for Drosha–Dicer processing are not obviously structurally distinguished from many other genomic hairpins predicted in the genome. Indeed, it is frequently difficult to distinguish short RNAs that derive specifically from the miRNA biogenesis machinery, as opposed to fortuitous degradation fragments generated by other ribonucleolytic processes. As newly evolved miRNAs are generally expressed at lower levels than well-conserved miRNAs, it is conceivable that many genuine species-specific miRNAs eluded currently available small RNA datasets. Therefore, the number of miRNA genes in any given species remains a topic of debate.
In an alternative pathway for miRNA biogenesis, short hairpin introns termed mirtrons are spliced and debranched to generate pre-miRNA hairpin mimics (Berezikov et al. 2007; Okamura et al. 2007; Ruby et al. 2007a). These are then cleaved by Dicer and incorporated into typical miRNA silencing complexes. By searching for conserved hairpin introns, for which the terminal loop diverged more quickly than the duplexed stem, we identified a limited set of conserved mammalian mirtron candidates, of which three out of 13 were validated by small RNA cloning (Berezikov et al. 2007). However, additional newly evolved mirtrons were validated by mapping small RNA reads to introns, indicating that reliance upon evolutionary conservation recovered only a subset of mirtron loci (Berezikov et al. 2007). Indeed, novel mirtrons in mammals and avians were subsequently reported (Babiarz et al. 2008; Glazov et al. 2008).
The biogenesis of mirtrons at known mRNA splice sites provides evidence for an initial step of nuclear processing, whereas no comparable external reference data exists for the endogenous processing of substrate RNAs by Drosha. We therefore hypothesized that mirtron gene finding, absent input from comparative genomics, might be more feasible than for canonical miRNAs. In this report, we generated empirical evidence for structural and sequence parameters that are critical for mirtron processing. Together with the original set of 14 published mirtrons from Drosophila melanogaster (Okamura et al. 2007; Ruby et al. 2007a), these data enabled a computational model that achieved high sensitivity and specificity for genuine mirtrons. We also validated a number of novel mirtrons among the highest-scoring candidates. The computational mirtron model was also effective on C. elegans and substantially increased the number of validated mirtrons in this species. These efforts demonstrate substantially effective prediction of a class of endogenous Dicer substrates in these invertebrate genomes.
Results
Experimental definition of parameters of mirtron functionality
We performed structure-function assays to define critical features of productive mirtrons. S2 cells express a low level of the mirtron miR-1003 endogenously, whose levels increase substantially upon transfection of a mir-1003 expression plasmid (Okamura et al. 2007; Ruby et al. 2007a). We generated a panel of mir-1003 variants and monitored their processing into pre-miRNAs and mature miRNAs using Northern blotting. The constructs tested and their processing capacities are summarized in Figure 1, with representative primary data shown in Figure 2. Band quantification supported our conclusion that only certain variant constructs of appropriate size and hairpin overhangs yielded processed miR-1003 above endogenous expression (Figs. 1, 2). In addition, we performed RT-PCR tests using exonic primers flanking the mirtron cassettes and observed efficient and accurate splicing of all constructs (Fig. 2). We also sequenced the junctions of the spliced products and found them to exhibit nucleotide accuracy in the utilized splice junctions, as expected (data not shown). The following sections describe the mirtron variants and their biogenesis capacities in greater detail.
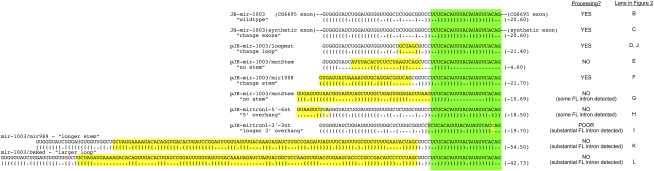
Figure 1.
Constructs used for structural analysis of mirtron biogenesis. Shown are sequence variants of the mir-1003 mirtron used for functional tests. (Green) The mature miRNA sequence; (yellow) the nucleotides differing from mir-1003. Their relative abilities to be processed in S2 cells are indicated (see also Fig. 2).
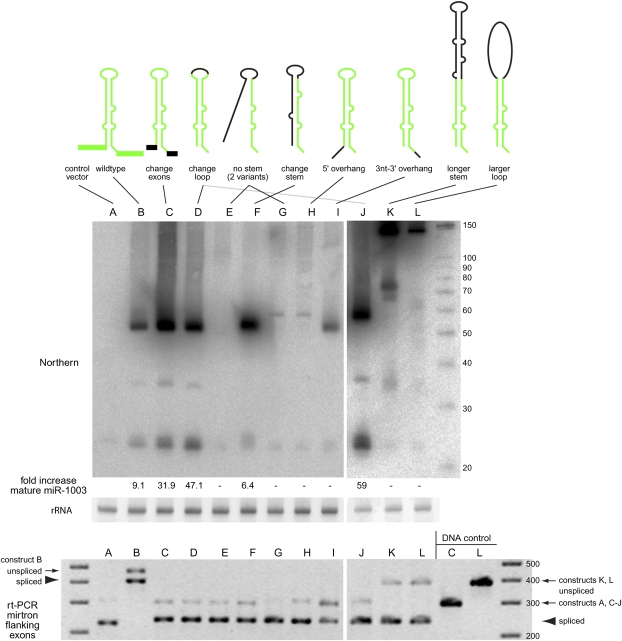
Figure 2.
Structure-function analysis of mirtron biogenesis. (Top) S2 cells were transfected with UAS-mirtron and ub-Gal4 plasmids and RNA was isolated and subjected to Northern blot using an LNA probe antisense to miR-1003. Ethidium bromide staining of 5S rRNA is shown as a loading control. The fold increase in mature miR-1003 above control transfections is indicated below; (−) No substantial increase in miR-1003 level (>2 folds) was detected. (A) Control transfection using empty expression vector shows that S2 cells express a low level of the mirtron-derived miRNA miR-1003. (B) Introduction of mir-1003 expression plasmid, which includes portions of its endogenous flanking exons, yields strongly elevated pre-mir-1003 and mature miR-1003. Neither substitution of mir-1003 exonic context (C), nor replacement of its terminal loop (D), interferes with its biogenesis. Extensive mutation of its miRNA* arm abolishes production of miR-1003 (E,G), although a small amount of pre-miRNA is detected in the later case. However, extensive mutation while maintaining hairpin structure supports efficient mirtron biogenesis (F). (H) Introduction of a 5′ hairpin overhang abolishes small RNA production. (I) Extension of the 3′ hairpin overhang strongly impairs mirtron processing, although pre-miRNA accumulated. (J–L) Starting with a terminal loop mutant of mir-1003 (J, see also lane D), structured (K), and unstructured (L) hairpin extensions were introduced. Both constructs yielded substantial amounts of ∼150 nt pre-miRNA product, with higher levels of the fully duplexed intron (K); however, neither supported accumulation of mature miRNA. A ∼75-nt product corresponding to approximately half of the long hairpin intron accumulated; its biogenesis is not known. (Bottom) The same RNA samples used for Northern blotting at top were subjected to RT-PCR analysis to verify splicing accuracy of the mirtron variants. We observed weaker bands for the unspliced products and stronger bands for the spliced products; the DNA template controls at the right provide a size marker to gauge the unspliced amplification products. Note that the wild-type mir-1003 construct in its native CG6995 context includes more exon sequence than the other constructs, leading to the larger sizes of its RT-PCR products (“B”).
Flanking exon and terminal loop contexts
We previously observed that introns can autonomously dictate their entry into the mirtron biogenesis pathway (Okamura et al. 2007). As shown in Figure 2C, substitution of flanking exonic sequence from its host transcript CG6695 with artificial sequence did not impede its processing into ∼22-nt miRNAs. We also generated a construct in which the loop of mir-1003 was substituted (Fig. 2D), and this was also effectively processed. Therefore, flanking exons and terminal loops do not seem to provide essential context for mirtron processing.
Hairpin structure
As with canonical miRNAs, we presumed that mirtrons must adopt some minimum pairing between the prospective miRNA/miRNA* duplex of the intronic stem. We disrupted the 5′ end of the intron in two different constructs, while maintaining the mature miR-1003 sequence (Fig. 1). Their ability to generate pre-miRNAs and mature miRNAs was essentially abolished (Fig. 2E,G), although the construct with greater predicted secondary structure accumulated a small amount of spliced intron (Fig. 2G). On the other hand, complete substitution of the 5′ arm so as to introduce a distinct hairpin, modeled on the general structure of mir-1008, now rescued the production of mature miR-1003 (Fig. 2F). Such flexibility in the miRNA* sequence affirmed that the overall degree of hairpin structure is a major determinant of mirtron functionality, although diverse patterns of hairpin imperfections are accommodated.
Hairpin overhangs
Canonical Drosophila mirtron hairpins often contain precise 2-nt 3′ overhangs, in which the “GU” splice donor is paired with the two nucleotides preceding the “AG” splice acceptor (Okamura et al. 2007; Ruby et al. 2007a). Such a configuration resembles a Drosha-cleaved pre-miRNA hairpin, and the 2-nt 3′ (i.e., 0:2) overhang is also optimal for recognition by the exportin 5 complex for trafficking into the cytoplasm (Yi et al. 2003; Lund et al. 2004; Okada et al. 2009). We tested whether other end-configurations were compatible with efficient mirtron biogenesis. We modified the mir-1003 hairpin to include a 5′ overhang of 6 nt (Fig. 1). This construct failed to be processed, although a small amount of spliced intron accumulated (Fig. 2H). We also made two variants in which the 3′ overhang was lengthened to 3 nt. Although this very minor alteration did not affect overall mirtron secondary structure, the construct with a 3-nt 3′ overhang accumulated a substantial amount of pre-miRNA hairpin, but not of mature miRNA (Fig. 2I). These results support the notion that short 3′ overhangs are optimal for mirtron biogenesis.
Hairpin length
It is not uncommon for plant pre-miRNA hairpins to be several hundred nucleotides in length (Llave et al. 2002; Park et al. 2002; Reinhart et al. 2002). In contrast, few animal pre-miRNA hairpins are greater than 80 nt in length, and almost none are greater than 100 nt in length. Therefore, most computational strategies for animal miRNAs involved folding candidate hairpins 100–120 nt in length (Lai et al. 2003; Lim et al. 2003a; Bentwich et al. 2005; Berezikov et al. 2006). This proved to be convenient for the analysis of RNA structures, as the complexity of confounding alternative structures increases strongly with increased transcript length. However, our deep-sequence analysis of cloned Drosophila small RNAs revealed several pre-miRNA hairpins 100–150 nt in length (Ruby et al. 2007b). Therefore, it was relevant to test the effect of increased loop length on mirtron processing.
In one construct, we extended the stem of mir-1003 by inserting 86 nt from the “long” miRNA, mir-989 (Fig. 1). This construct accumulated a full-length spliced intron near the expected size of ∼146 nt, and gave rise to some intermediately sized bands hybridizing to miR-1003 probe. However, these were not processed into mature ∼22-nt miRNAs (Fig. 2K). In a second construct, we introduced ∼100 nt of sequence from DsRed into the terminal loop of mir-1003, yielding a long unstructured region. As with the mir-1003/mir-989 hybrid construct, the prospective miRNA/miRNA* regions of mir-1003 were unchanged in this construct. This construct accumulated a band presumably reflecting the spliced intron, but also did not accumulate mature miR-1003 (Fig. 2L).
The accumulation of intronic RNAs from these extended mirtrons differentiates them from typical introns that are rapidly degraded following splicing. This might be due to their association with components of the mirtron pathway, or perhaps with other cellular machineries. Nevertheless, their failure to efficiently generate mature miRNAs demonstrated that intron length affects the entry of substrates into the mirtron biogenesis pathway. In particular, strong pairing of the mirtron hairpin base was not compatible with efficient processing of longer introns, even when these exhibit continuous duplex.
Unusual structural features of known Drosophila mirtrons
Extensive cloning of canonical miRNA genes suggests that preferred substrates of RNase III enzymes have extensive duplex structure and a tendency for smaller, symmetric internal loops as opposed to larger, asymmetric internal loops and bulges. These features were incorporated into scoring rubrics for canonical miRNAs that award continuous duplex regions and progressively penalize unpaired regions (Lai et al. 2003; Lim et al. 2003a,b; Ruby et al. 2007b; Stark et al. 2007a). CG6695_in5/mir-1003 exemplifies a presumably optimal mirtron that exhibits these features and is highly expressed (Fig. 3A). Curiously, several well-conserved mirtrons with high endogenous expression exhibit large internal loops of ≥4 nt on a side. For example, CG31163_in17/mir-1010 has a 1 + 4-nt internal loop and VHA-SFD_in3/mir-1006 has a 5 + 2-nt internal loop (Fig. 3B; Supplemental Fig. 1). In both cases, the most abundant reads form a duplex with a typical 2-nt 3′ overhang on the end closest to the terminal loop, indicating their precise Dicer-1 cleavage despite their imperfections. These unpaired nucleotides are apparently looped out in the hairpin structure, resulting in atypically long Dicer products such as the 26-nt miR-1006* (Fig. 3B).
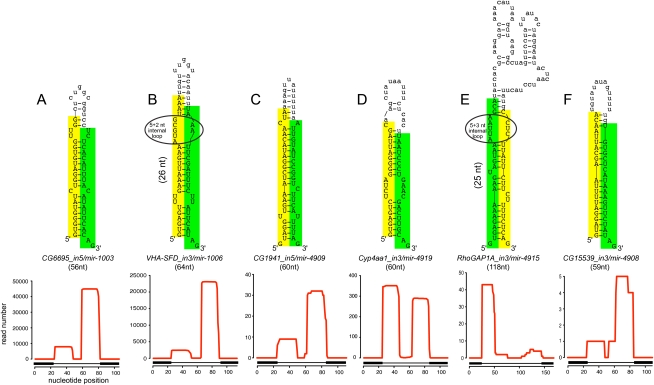
Figure 3.
Examples of known and novel mirtrons in D. melanogaster. The abundant small RNAs derived from each hairpin are highlighted, green for the miRNA and yellow for the miRNA*. Below the secondary structures are plots that show the abundance of cloned small RNAs across the aggregate D. melanogaster small RNA data. The small RNA density is highest at either end of each intron, with typically one side accumulating to a higher level; often this is the 3′ arm, but occasionally it is the 5′ arm. The black boxes below the graph indicate the exon–intron boundaries. (A) CG6695_in5/mir-1003 is an example of a conserved, abundantly expressed mirtron with optimal features, including a straight short intronic hairpin with a 2-nt 3′ overhang. (B) Vha-SFD_in3/mir-1006 is an example of a conserved, abundant-expressed mirtron with a large asymmetric internal loop (5 + 2 nt). CG1941_in5 (C) and Cyp4aa1_in3 (D) are novel mirtrons with typical straight hairpins and compatible overhangs. (E) RhoGAP1A_in3 is an expressed mirtron with an unusually large, unstructured terminal loop, a large asymmetric internal loop (5 + 3 nt) and single nucleotide overhangs at its 5′ and 3′ ends. (F) CG15539_in3 exhibits convincing mirtron features, but is on the borderline of confident cloning evidence; nevertheless, its reads exhibit a characteristic 2-nt 3′ overhang on the Dicer-1-cleaved end.
Although these sizes of internal loops are not unprecedented among miRNAs, their frequency in the small set of known mirtrons is much higher than with canonical Drosophila miRNA hairpins (Ruby et al. 2007b; Stark et al. 2007a). In miRbase Release 14, only nine of 142 canonical miRNAs have ≥4-nt internal loops in D. melanogaster, but two of 14 mirtrons have ≥4-nt internal loops (Supplemental Fig. 1). These observations suggested that a strongly progressive penalty on internal loops and bulges of increasing size, which we previously found useful for evaluating canonical miRNA hairpins in Drosophila (Lai et al. 2003), might not be appropriate for assessing mirtrons.
We also inspected mirtron hairpin end-structures. The crystal structure of the exportin 5/RanGTP/pre-miRNA hairpin complex provides physical evidence for its preference for a 2-nt 3′ overhang (Okada et al. 2009), and this was supported by our experimental tests (Figs. 1, 2). This was reflected in the expression of previously described Drosophila mirtrons: 12 of 14 initially cloned mirtrons (Ruby et al. 2007a) exhibit a 2-nt 3′ overhang (Supplemental Fig. 1). However, alternative hairpin end structures are also possible, since CG31772_in12/mir-1004 and CG3860/mir-1009 have 2-nt 5′ and 3-nt 3′ (i.e., 2:3) overhangs, while opa1-like_in6/mir-1016 has a 1:1 overhang. Although these loci have abundant expression for mirtrons (∼6000, ∼9000, and ∼2000 reads in the aggregate small RNA data, respectively), the five known Drosophila mirtrons with >10,000 reads all have 2-nt 3′ overhangs (CG6695_in5/mir-1003, Lerp_in6/mir-1012, CG18004_in2/mir1008, CG31163_in17/mir-1010, and VhaSFD_in3/mir-1006) (Supplemental Fig. 1), suggesting that a 0:2 overhang is preferred for optimal biogenesis. Finally, even though our experimental manipulation of mir-1003 showed that a 0:3 overhang is strongly detrimental, such a feature does not necessarily abolish processing completely, since the recently reported mirtron CG17560_in3/mir-2494 (Berezikov et al. 2010) has a 0:3 overhang and an aggregate count of ∼400 reads (Supplemental Fig. 1).
Altogether, we infer that a slightly broader range of endogenous pre-miRNA structures can be achieved by the mirtron pathway compared with Drosha cleavage, although there is likely a 5p:3p hierarchy of 0:2 > 2:3 > 1:1 > 0:3 hairpin overhangs with respect to efficacy for miRNA biogenesis.
Computational identification of mirtrons in Drosophila
Our functional assays indicated that mirtrons do not require particular exonic context of terminal loop sequences. Beyond the minimal requirements needed for successful splicing, structural characteristics and intron length play the predominant roles in determining entry into the mirtron pathway. In addition, mirtron biogenesis appeared more tolerant of relatively large internal loops within the miRNA/miRNA* duplex than anticipated from studies of canonical miRNAs. Finally, experimental tests showed that subtle alteration of hairpin overhangs strongly impeded mirtron maturation, indicating that the adoption of a restricted set of hairpin overhangs in known mirtrons actively reflected a key feature of efficient mirtron biogenesis.
These features contrast in many respects with canonical miRNAs and highlighted that mirtrons are not simply short miRNA hairpins, nor are they specifically defined solely by searching for intronic hairpins. For comparison, we checked the ability of the miRscan III algorithm (Ruby et al. 2007b) to classify mirtrons from bulk short introns. As expected, although it prioritized introns with hairpins, the strong majority of high-ranking candidates were not plausible mirtrons, since they lacked hairpin overhangs compatible with export or dicing and/or did not exhibit appropriate duplex between the intron termini (data not shown). We therefore sought to develop an alternative approach for mirtron gene finding. Notably, we hypothesized that the hairpin overhang feature could be uniquely exploited for mirtron prediction relative to canonical miRNA prediction, owing to the precision with which their hairpin ends could be inferred via splicing.
We utilized a machine learning approach to predict whether a candidate short intronic structure might form a functional mirtron using a positive training set of the 14 original validated D. melanogaster mirtrons (mir-1003→mir-1016, Supplemental Fig. 1) and a negative training set of candidate structures of 500 non-mirtron introns randomly selected from the collection of 50- to 120-nt introns lacking small RNA reads. We used UNAFold (Markham and Zuker 2008) to fold intronic sequences, keeping alternative predicted structures. We used three sets of features to represent different aspects of the structures in our SVM models: (1) a binary vector representation of the overhang configuration; (2) a set of structural descriptors motivated by our experimental data on relevant determinants of mirtron biogenesis; and (3) a set of structural similarity scores, based on pairwise comparison of structures using the relaxed base-pair score (RBP) (Agius et al. 2010). We combined the three feature sets using a standard linear kernel combination approach and trained an SVM model using LIBSVM, then ran the classifier on the 27,620 D. melanogaster introns 50–120 nt in length. A detailed description of the model is provided in the Methods section.
In evaluating the performance of the model, we note that this approach was intended to capture canonical mirtrons, for which both ends of the pre-miRNA are defined by the splicing event. Recently, we found that a subset of mirtrons with substantial 3′ overhangs are targets of exosome-mediated 3′–5′ trimming, permitting functional pre-miRNA mimics to be generated from so-called “tailed” mirtrons (Flynt et al. 2010). On this basis, we recently reclassified CG7927_in2/mir-2501 (Berezikov et al. 2010) as a tailed mirtron. This locus and other tailed loci score poorly as canonical mirtrons, as well they should (http://cbio.mskcc.org/leslielab/mirtrons); loci not generated by the canonical mirtron pathway will require distinct algorithms for their identification.
With this caveat in mind, we were encouraged by the strong recall of previously published mirtrons among the highest candidates. There were 16 annotated mirtrons positioned within the top 26 candidates genome-wide, with another known mirtron at rank 45 (Fig. 4A; http://cbio.mskcc.org/leslielab/mirtrons). This performance was notable given that canonical miRBase miRNAs are not differentiated from 1000s to 10,000s of other hairpins in single-genome assessments. To provide additional evidence that we did not simply fit the model to known data, we hoped to validate novel mirtrons among high-scoring candidates. To do so, we compiled published datasets of D. melanogaster small RNAs along with additional data that we generated for the modENCODE project (Celniker et al. 2009) (see the Methods and Supplemental Table S1) and inspected these for evidence of mirtron-like cloning patterns.
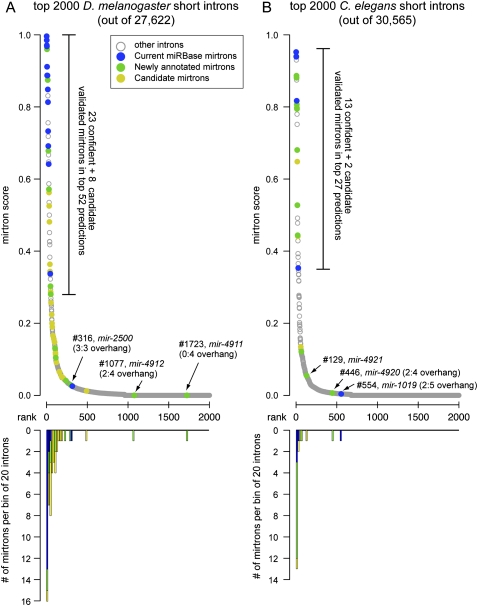
Figure 4.
Performance of the computational model for mirtron identification on the D. melanogaster and C. elegans genomes. (A) Performance of an SVM trained on the 14 original D. melanogaster mirtrons (mir-1003-mir-1016) and run across the fly genome. (B) Performance of the D. melanogaster model on C. elegans. In both cases we used as input the annotated short introns 50–120 nt in length; no evolutionary features were considered. The top graphs plot the scores of mirtron likelihood and illustrate that the scores quickly drop following the top predicted candidates. Highlighted in blue are mirtrons previously deposited in miRBase (note that the previously annotated C. elegans mir-2220 was reported earlier but not recognized as a mirtron; it is nonetheless included in the “blue” loci), novel mirtrons annotated in this study are in green, and candidate mirtrons are highlighted in gold. The bottom graphs utilize the same x-axis and plot the numbers of validated and candiate mirtrons in consecutive bins of 20 introns in the rank order. Note that a few validated mirtrons scored poorly, and most of these have atypical 3′ overhangs. The full rankings can be viewed in Supplemental Tables S3 and S4.
We considered highly confident evidence for transit via a splicing- and dicing-dependent pathway to be cases where small RNA reads of appropriate length (∼21–24 nt) were preferentially recovered from both ends of the intron and for which dominant reads exhibited 3′ overhangs on the duplex end closest to the terminal loop (Supplemental Fig. 2). Six novel mirtrons in the top 51 candidates had confident read evidence, including CG1941_in5 (ranked fifth), Cyp4aa1_in3 (ranked 22nd), and yl_in5 (ranked 48th) (Fig. 3C,D). Notably, several validated loci had relatively large internal loops (tex_in1 [29th]: 3:4 and 0:4 nt, Cyp4aa1_in3: 4:4 nt, and CG1718_in2 [51st]: 5:3 and 2:5 nt). The longest validated mirtron was RhoGAP1A_in3 (113th). It had a suite of seemingly suboptimal features underlying its lower score, such as long intron length, 1:1 hairpin overhang, and a 5:3 internal loop. Its relatively low number of reads was consistent with its suboptimal features; nevertheless, its cloning pattern provided evidence for a specifically processed mirtron (Fig. 3E). At the borderline of confident annotation was CG15539_in3 (ranked 14th). Although only five intron-terminal reads were recovered, these exhibited confident biogenesis patterns, since both miRNA and miRNA* were cloned, all reads extended to the intron termini, and these exhibited a 2-nt 3′ overhang on the Dicer-1-cleaved end (Fig. 3F).
Other high-scoring candidates had intron terminal reads, but otherwise had one or more confounding features to their evidence as genuine mirtrons. These included very few reads, atypical duplex end structure, lack of star strand reads, presence in only one library data set, and/or highly heterogeneous read length (Supplemental Fig. 3). To avoid inappropriate annotations, we highlighted a set of relatively high-scoring mirtron predictions with limited small RNA evidence as “candidates”; we expect that at least some of these may reach confident gene status following additional experimentation. For example, CG15160_in3 (#50) exhibits a typical mirtron structure with strong hairpin quality, and it was associated with single 5p and 3p reads that extend to the intron termini. These reads are arranged with a 2-nt overhang at either end, and two additional 3p reads fall just short of the splice acceptor site, but share their 5′ end with the “canonical” 3p read (Supplemental Fig. 3). It seems quite likely that this locus is processed by Dicer-1, but at a very low level in the available small RNA libraries. Other predictions lacking intron terminal reads, or exhibiting abundant reads from other regions of the intron incompatible with simple splicing/dicing history, were not considered as candidates. Altogether, the recovery of many high-scoring candidates with confident or candidate reads affirmed the efficacy of the computational model.
The D. melanogaster mirtron model is effective on C. elegans
Although we confidently validated the endogenous expression of many high-scoring mirtrons from our screen, most of the very highest-scoring candidates had been annotated previously (Ruby et al. 2007a). This potentially speaks to the efficacy of our computational approach; however, a concern arises whether the model was overfitted to the training set. C. elegans offered a potentially appropriate setting to conduct an entirely independent test of our model, since few mirtrons (four) had been previously identified in this species (Ruby et al. 2007a). If nematode mirtrons exhibited distinct properties from fruitfly mirtrons, as may be the case with mammalian mirtrons (Berezikov et al. 2007), then poor performance of the Drosophila model in C. elegans would not be interpretable. For example, two of the four known C. elegans mirtrons have overhangs that are atypical by fly standards, since mir-1019 has a 2:5 overhang and mir-1020 has a 2:4 overhang; mir-62 and mir-1018 have typical 0:2 overhangs (Supplemental Fig. 4). On this basis, it was unclear how the Drosophila model would perform in this species.
We ran our model on the 30,565 C. elegans introns 50–120 nt in length, and sought to validate the output using published C. elegans small RNA data (see Methods and Supplemental Table S2). We were pleased to observe that three of the four published mirtrons ranked in the top 27 candidates genome-wide (at ranks #1, #3, and #27). More importantly, we could confidently annotate nine novel mirtrons among the top 20 predictions, along with two other high-scoring loci with borderline evidence; a handful of additional novel and candidate mirtrons were ranked slightly lower (Fig. 4B; Supplemental Fig. 5). We note that three of the four previously annotated C. elegans mirtrons were called on the basis of only two reads each (mir-1018, mir-1019, and mir-1020) (Ruby et al. 2007a), but all of these currently have several hundred reads in the aggregate C. elegans small RNA data. Therefore, we expect some additional candidates may be validated in the future; this seems probable when considering that the majority of publicly available C. elegans small RNA data were generated from conditions aimed at depleting miRNA-class small RNAs (see Methods). Nevertheless, the strong performance of the D. melanogaster model on a completely independent species provided compelling validation of the notion that mirtrons can be effectively identified by a forward gene-finding approach. The core data from the genome-wide intron rankings and read evidence are summarized in Supplemental Tables S3 and S4 (D. melanogaster and C. elegans, respectively), and full observations are summarized at http://cbio.mskcc.org/leslielab/mirtrons/.
Notable cloned mirtrons with atypical features
Our previous experience demonstrated that certain unexpected hairpin loci generated small RNAs that permitted confident assessment of miRNA/miRNA* duplexes (Ruby et al. 2007b). We inspected mapped intronic reads from D. melanogaster and C. elegans in search of loci with characteristic small RNAs at both intron termini whose structures did not qualify them among the highest computational ranks. Such loci provided evidence of the existence of some atypical Dicer substrates (Supplemental Figs. 2, 5; http://cbio.mskcc.org/leslielab/mirtrons).
A mirtron derived from an alternatively spliced intron
All mirtrons described thus far derive from constitutively spliced introns. A mirtron overlapping the third intron of CG17560 has highly confident cloning features (Berezikov et al. 2010), but was missed by our pipeline because its miRNA-generating arm is not currently annotated as a splice isoform and, in fact, overlaps coding exon. The annotated intron is only 55 nt long, while the miRNA-generating intron is 72 nt long. Inspection of modENCODE mRNA-seq data generated by Graveley, Celniker, and colleagues provided complementary evidence that both splice sites are utilized in messenger RNAs (J Landolin, pers. comm.). Structurally, this mirtron is slightly unusual, as it has a 3-nt 3′ overhang; nevertheless, inclusion of the genuine mirtronic-intron into the starting pool showed that it ranked 13th overall genome-wide. We tallied ∼700 miRNA reads from CG17560_in3, a modest number that was 1–2 orders of magnitude less than many mirtrons with 0:2 overhangs (see http://cbio.mskcc.org/leslielab/mirtrons). This is consistent with our experimental data indicating that a hairpin with a 0:3 overhang is not efficiently processed (Fig. 2I), but indicates that some endogenous processing of such a substrate does occur.
The mRNA-associated splice site is absolutely conserved among the sequenced Drosophilids, slightly more so than is the splice site that generates the CG17560_in3/mir-2494 mirtron. Curiously, usage of the mirtronic splice site would put the remainder of the CG17560 mRNA out of frame. It remains to be seen whether the processing of this mirtron is part of a mechanism that regulates CG17560 translation, or whether its mirtron might derive from a distinct transcript that overlaps the same genomic space. In either case, we must bear in mind that our bioinformatics screen would not have recovered genuine mirtrons that derive from unannotated introns.
Mirtrons with unusual hairpin overhangs
As noted, CG17560_in3 has a 0:3 hairpin overhang (Fig. 5). If the 5′ base of such a mirtron hairpin was unpaired, it would exhibit a 1:4 hairpin overhang. We identified the validated mirtron yl_in5, ranked 48th genome-wide, as having such an overhang. Its total read counts were lower than with CG17560_in3 (∼400 vs. ∼700) (Supplemental Fig. 2), although differences in the transcription of their host genes may contribute to this disparity. ND23_in2 has a 0:4 overhang and was expressed lower still (<50 reads), yet its 3p reads extend to the 3′ end of the intron, indicating that it is not a tailed mirtron. Such hairpins with short, but noncanonical, 3′ overhangs are very likely disfavored relative to 0:2 or 2:3 overhangs, but appear to be specifically processed by the mirtron pathway in vivo at least to some extent.
Figure 5.
CG17560_in3 generates a mirtron from an alternatively spliced intron. Shown is a multiple sequence alignment and phastCons assessment of conservation (obtained from the UCSC Genome Browser). The splice acceptor used to generate the protein-coding transcript is highly conserved across the 12 sequenced Drosophilids; a different splice acceptor is used to generate the CG17560 mirtron. Small RNA mappings exhibit typical Dicer-1 cleavage patterns, including the generation of rare reads corresponding to the cleaved terminal loop. Other rare reads were not summarized in this schematic. Note the slightly atypical hairpin end of this mirtron, which terminates in a 3-nt 3′overhang. Usage of the mirtronic splice generates a frame-shift, since the typical splice site joins in the +2 coding frame, while the mirtron-spliced site joins in the +1 coding frame.
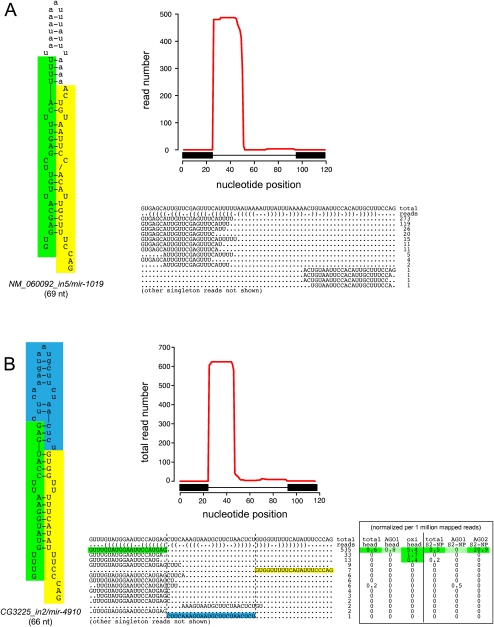
A few other cases of mirtrons with more unusual hairpin overhangs exist. For example, the atypical Drosophila mirtron CG3225_in2 exhibits a 4 + 7-nt overhang (Fig. 6) and appears structurally to be neither an effective exportin 5 cargo nor an effective Dicer substrate. Nevertheless, the recovery of a terminal loop read whose ends dovetail precisely with the miRNA/miRNA* duplex provides evidence for endogenous dicing. Although most mirtrons preferably generate mature small RNAs from their 3p arms (Ruby et al. 2007a), the dominant read from CG3225_in2 comes from its 5p arm (Fig. 6B), consistent with this end of the duplex being thermodynamically unstable (Khvorova et al. 2003; Schwarz et al. 2003). The same is true for C. elegans mir-1019, which has an unusual 2:5 hairpin overhang and whose 5p read is similarly dominant (Fig. 6A).
Figure 6.
Exceptional fly and worm mirtrons exhibit strongly unpaired hairpin termini. It is generally accepted that a defined short 3′ overhang is critical for nuclear export of pre-miRNA hairpins via exportin 5. Consequently, a strongly unpaired hairpin base is unfavorable for pre-miRNA maturation. (A) The exceptional C. elegans mirtron mir-1019 exhibits a 2 + 5 hairpin overhang, but still exhibits a typical pattern of mirtronic reads corresponding specifically to the ends of the intron. (B) Similarly, the atypical D. melanogaster mirtron CG3225_in2 exhibits strong evidence for Dicer-1 cleavage despite a 4 + 7 hairpin overhang, including a rare read corresponding to the cleaved terminal loop (highlighted in blue). Reads from this intron exhibit evidence for loading to the siRNA effector AGO2 instead of the miRNA effector, AGO1. Head data including AGO1-IP and oxidized RNA (which enriches for mature AGO2-loaded siRNAs) were reported by Ghildiyal et al. (2010) and S2 cell data from AGO1-IP and AGO2-IP were reported by Czech et al. (2008); to permit comparison between the total and IP levels, these read numbers were normalized per million mapped reads in each library. Note that these worm and fly mirtrons are further atypical in that their mature cloned species derive from their 5p arms; this correlates with the strong thermodynamic asymmetry associated with their unpaired hairpin bases. These mirtrons are exceptional, and few other introns with similarly unpaired bases were productively converted into short cloned RNAs.
CG3225_in2-derived small RNAs are also productively loaded into an Argonaute effector, although Argonaute-specific libraries generated by the Hannon and Zamore groups from S2 cells (Czech et al. 2008) and heads (Ghildiyal et al. 2010) showed preferred loading into AGO2 (the siRNA effector) relative to AGO1 (the miRNA effector). It was recently shown that other Dicer-1-generated small RNAs, in particular many miRNA* species, can be loaded into AGO2 (Czech et al. 2009; Okamura et al. 2009; Ghildiyal et al. 2010). A few other mirtrons with unusual overhangs but evidence for miRNA/miRNA* duplexes extending to the splice sites included ND23_in2 (0:4 overhang), CG2976_in2 (2:4 overhang), and CG32704_in1 (1:4 overhang) (Supplemental Fig. 3).
According to our current experimental knowledge, the capacity for miRNA production from these particular fly and worm loci is unexpected. Although most of these are quite lowly expressed, a few, such as CG3225_in2 and nematode mir-1019, achieved nontrivial levels (Fig. 6). We speculate that supplementary biogenesis factors may aid the processing of some atypical mirtrons, and thus we do not expect them to be identified by our model for canonical mirtrons. Nevertheless, inspection of vast quantities of fly and worm small RNAs indicated that these examples were not the norm, consistent with our experimental tests (Fig. 2) and the features of highly expressed mirtrons (Supplemental Figs. 1–5). Therefore, our computational model did not appear to be plagued by a substantial pool of false-negatives.
Individual messenger mRNAs that spawn multiple mirtrons
Canonical miRNA genes are often genomically clustered, reflecting their frequent organization into operons that generate multiple miRNAs from a common precursor transcript. Previous studies did not identify clustered mirtrons, but this might be expected given that their nuclear biogenesis differs fundamentally from that of canonical miRNAs. Moreover, as mirtrons comprise only 5%–10% the total pool of miRNA loci (<1 mirtron/5 megabases), their genomic clustering is statistically quite improbable.
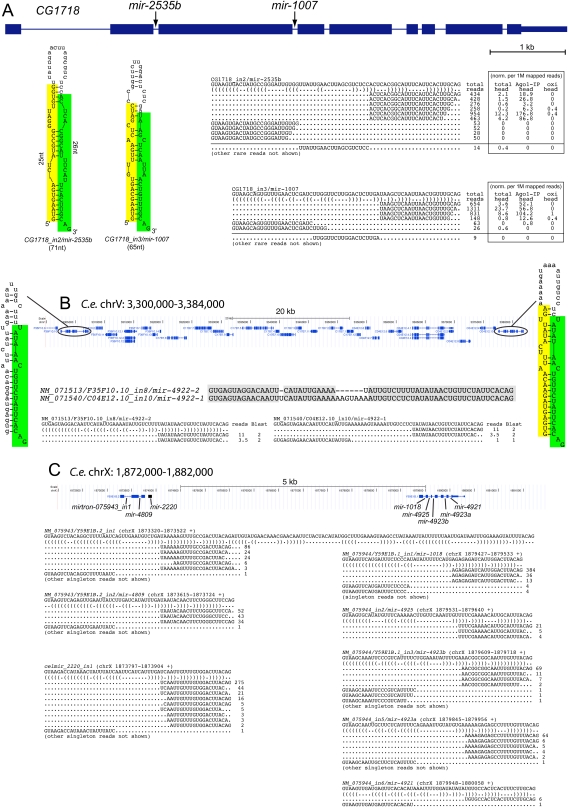
Unexpectedly then, we identified a mirtron in the second intron of CG1718, a gene previously annotated to harbor a mirtron in its third intron (mir-1007) (Fig. 7A). The hairpin in the second intron of CG1718 has two internal loops that are 5 nt on their longer sides, which might suggest it to be a mediocre Dicer substrate. These features indeed caused it to be scored lower than mir-1007 (ranked 10th), although it nevertheless ranked 51st genome-wide. Given their substantial structural differences, we were surprised to observe that the number of miRNA reads derived from these two mirtrons was similar, across the aggregate data set as well as in individual libraries. Examination of head libraries published by Zamore and colleagues (Ghildiyal et al. 2010) revealed that both mirtrons generated typical miRNAs that were enriched in AGO1-IP and depleted in oxidized samples, indicating that they bore free 3′ hydroxyl groups (Fig. 7A). We also observed rare terminal loop reads, whose ends precisely abutted the termini of the dominant miRNA and miRNA* species from both mirtrons, providing additional support for their endogenous cleavage by Dicer-1. Since the primary transcription across the second and third introns of CG1718 should be identical, we conclude that the maturation of these two mirtrons is unexpectedly comparable despite substantial differences in their structures.
Figure 7.
Clustered mirtrons in the D. melanogaster and C. elegans genomes. (A) Drosophila CG1718 generates mirtrons from both its second and third introns; CG1718_in2 was newly identified in this study. Curiously, while the hairpin structure of CG1718_in2 is seemingly suboptimal compared with the previously identified mir-1007, mature miRNAs accumulate to relatively similar levels from these mirtrons. Analysis of head libraries published by Ghildiyal et al. (2010) provided evidence that these mirtrons are expressed in the head and generate RNAs that populate AGO1, but not AGO2 complexes; this study used oxidation (oxi) of input samples to enrich for 2'O-methylated RNAs in mature AGO2 complexes. To permit comparison between the total and IP levels, these read numbers were normalized per million mapped reads in each library. Rarer reads were not shown, except for the informative cloned terminal loops that report on endogenous Dicer-1 processing; the full read patterns are available at http://cbio.mskcc.org/leslielab/mirtrons. (B) NM_071513 and NM_071540 are related genes that reside ∼70 kb apart on C. elegans chromosome V. Each gene bears a mirtron whose 3p arm is identical; thus, small RNA reads from this arm map to both mirtrons. We normalized the read numbers to assign half to each locus. On the basis of unique star arms, we can definitively annotate the expression of NM_071540. However, given that the hairpin of NM_071513 has only small symmetric loops, we infer that its processing should be equivalent, if not more efficient, to its paralog. (C) A supercluster of mirtron genes on C. elegans chromosome X. This <8-kb region was previously annotated to contain mir-1018 and mir-2220, of which mir-1018 was previously noted to be a mirtron (Ruby et al. 2007a). Although mir-2220 was earlier annotated as a canonical miRNA (Kato et al. 2009), we infer that it is similarly a mirtron, as its cloned RNAs begin and end with effective splice junctions. Here, we identify six additional mirtrons in this genomic region. Of these, NM_075943_in1 might appear to be a tailed mirtron based on the annotated splice junction; however, that its abundant 3p reads end with CAG suggests that it may be the product of alternative splicing, as seen for the Drosophila mirtron CG17560. Note that in all gene alignments only a subset of informative singleton reads, typically belonging to mirtron star species are shown.
We searched for other examples. CG6695 was previously reported to generate a mirtron from its fifth intron (mir-1003), which scored sixth overall in our computational assessment of the genome. We noticed that the first intron of this gene (which uses a highly conserved GCAAGT splice donor) ranked 49th overall in the genome and generated some short RNA reads (Supplemental Fig. 3). We did not annotate it as a mirtron at present since it generated only a small number or reads, some of which were of atypical size. Given its suboptimal hairpin, it is unlikely to be an effective substrate of the mirtron pathway compared with its partner intron mir-1003, which generated three orders of magnitude more reads. Nevertheless, it is tempting to speculate that minor nucleotide changes in the first intron of CG6695 might suffice to convert it into a more definitive mirtron substrate.
In C. elegans, we identified one case of a duplicated mirtronic gene, yielding mirtrons from NM_071513/F35F10.10_in8 (ranked 16th) and NM_071540/C04E12.10_in10 (ranked 20th). Although not adjacent, these genes have been retained within a 70-kb genomic interval (Fig. 7B). Their 3p miRNAs are identical, but these introns have diverged such that their 5p “star” regions are distinct. Evidence for independent expression based on star reads currently exists only for NM_071540/C04E12.10_in10. However, seeing how its hairpin has a 2-nt bulge, while its sister mirtron exhibits an optimal straight hairpin, it seems likely that both mirtrons are endogenously processed.
A straight case of mirtronic host gene duplication is perhaps unremarkable; however, the features of NM079544/Y59E1B.1 proved extraordinary. This transcript was previously shown to harbor the mirtron mir-1018 in its first intron (Ruby et al. 2007a), which indeed proved to rank first among worm introns. Our computational screen identified its second, third, and fifth introns as the ninth, 10th, and 14th highest mirtron candidates genome-wide, and small RNA cloning provided confident evidence for their endogenous expression as miRNAs (Fig. 7C). Further inspection revealed the sixth intron of this gene as the 129nd ranked candidate. This intron has a 4 + 4 internal loop that contributed to its lower score, and it generated far fewer reads than the other introns of NM079544. Nevertheless, miRNA and miRNA* were cloned, and precisely the same 3p read was recovered in five different libraries, indicating a high likelihood of a genuine processing event. Therefore, NM079544 encodes five distinct mirtrons.
Amazingly, the 65th highest-scoring candidate, which also generated a clear mirtronic pattern of small RNAs, derived from the second intron of the adjacent gene NM079543/Y59E1B.2. We independently identified the first intron of this gene in a survey for candidate tailed mirtrons (AS Flynt, EC Lai, unpubl.) akin to Drosophila mir-1017 (Ruby et al. 2007a; Flynt et al. 2010). It is conceivable that this is actually a case of an alternatively spliced intron analogous to D. melanogaster CG17560_in3/mir-2494 (Fig. 5), since abundant 3p reads from the hairpin terminate in the optimal splice acceptor CAG (Fig. 7C). In either case, two neighboring genes in C. elegans each generate multiple miRNAs via a splicing-dependent pathway.
This genomic region had a further surprise in store. Located between NM079543/Y59E1B.2 and NM079544/Y59E1B.1 lay the previously annotated mir-2220 (Kato et al. 2009). Interestingly, the 5p reads that define the mir-2220 pre-miRNA hairpin begin with GUAAGA (a functional intron donor and only 1 nt different from the optimal GUAAGU splice site), while its 3p reads end with the optimal splice acceptor CAG (Fig. 7C). We infer that mir-2220 is likely a mirtron derived from an unannotated noncoding RNA, or perhaps a longer isoform of NM079543/Y59E1B.2. When this presumptive intron was included in the starting pool, it ranked eighth genome-wide among mirtron candidates.
Unlike the case of the duplicated mirtrons on chromosome V, the seeds of the eight mirtrons in the chromosome X cluster are different, and thus presumably distinct in regulatory capacity. The NM079544/Y59E1B.1 (318/938 bp, 105 aa, six to seven introns) and NM079543/Y59E1B.2 (300/511 bp, 99 aa, two introns) mRNAs are themselves not obviously related in sequence, have different numbers of introns, and encode short predicted open reading frames with no similarity to each other or to the rest of the predicted C. elegans proteome. Therefore, it is conceivable that multiple genes in this region are putative noncoding RNAs that serve as host transcripts for mirtron operons.
Discussion
Challenges for computational identification of species-restricted miRNAs
The range of hairpin imperfections across known miRNA genes are comparable to those found in 100,000s of other predicted hairpins in typical animal genomes. The difficulty of identifying bona fide canonical miRNA genes is highlighted by the fact that certain single-base changes abolish Drosha or Dicer cleavage (Duan et al. 2007; Kawahara et al. 2007; Kotani et al. 2010), despite only subtle effects in overall hairpin quality. Consequently, the recent rise of next-generation sequencing technologies has made them the preferred method for miRNA discovery and has been indispensable for annotating recently evolved miRNAs. Even with cloned reads in hand, however, the evidence required to distinguish confident annotation of genuine miRNA hairpins processed by RNase III cleavage from degradation products incidentally mapped to inverted repeats remains a topic of debate. Therefore, direct computational identification of miRNA genes with reasonable specificity and sensitivity, as is possible with protein-coding genes, remains a desirable goal for the future.
High specificity of bioinformatic calls on miRNAs was reported possible, but this is necessarily accompanied by a tradeoff on sensitivity. Three years ago, the Microprocessor SVM strategy (Helvik et al. 2007) was benchmarked to be more sensitive and specific than other contemporary methods (Nam et al. 2005; Sewer et al. 2005; Xue et al. 2005). This approach selected 60% of human miRBase miRNAs at a score that excluded 95% of the initial hairpin set. However, while the initial hairpin set included 6.8 million loci, it was still insufficient to capture 2% of cloned miRNA loci. Such statistics indicate the daunting nature of whole-genome annotation, since there remained nearly half a million plausible good-scoring miRNA candidates.
The situation has not improved dramatically since then. For example, van Ham and colleagues reported last year that it was necessary to include 3.5 million hairpin candidates from the C. elegans genome to have 97.5% (128/132) sensitivity of known annotated miRNAs (van der Burgt et al. 2009). A restricted list of 3099 high-scoring candidates (“high L score”) exhibited higher specificity, but this retained only 34% of known C. elegans miRNAs. It must be kept in mind that from the hundreds of thousands of reasonable hairpin candidates in different animal species, only 150–800 have been cloned in any organism (Griffiths-Jones et al. 2008). Since most species-specific miRNAs are expressed at far lower levels than well-conserved ones, an absence of read evidence for high-scoring miRNA candidates does not necessarily invalidate them. The most recent studies from Drosophila (Berezikov et al. 2010) and mammals (Chiang et al. 2010) suggest a fairly limited repertoire of miRNAs in these animals, even accounting for very lowly expressed loci. Nevertheless, the tally of species-restricted canonical miRNA genes in any given genome remains controversial.
Effective computational prediction of mirtrons in flies and nematodes
In this study, we showed that the mirtron subclass of miRNA genes is amenable to effective computational discovery independently of evolutionary conservation. Indeed, the majority of mirtrons are relatively poorly conserved, and thus could not be identified using comparative genomics. Our model was predicated on features of known D. melanogaster mirtrons, but proved effective on C. elegans as an independent evaluation of performance. Curiously, at least some effective Dicer substrates exhibit features that might be expected to substantially inhibit their capacity for processing. For example, we identified several mirtrons with internal loops of 4–5 nt, structures that might have been expected to segregate them away from bulk-validated miRNA hairpins. While our experimental assays demonstrate that increased hairpin structure is clearly correlated with increased miRNA production, it is significant that a computational approach could still identify processed mirtrons with such atypical features.
On the basis of our computational and experimental efforts, we estimate that no more than a few 10s (∼30) of mirtrons in D. melanogaster are expressed at a level of 10 out of 400 million reads from a diverse set of stage- and tissue-specific libraries. A similar conclusion applies to C. elegans, although fuller support of this notion will come with the accumulation of more data from total RNA and ALG1/2-IP libraries. There may exist additional genuine mirtrons that are lowly expressed simply due to restricted expression of their host gene. However, our high precision and recall on our predictions suggests that most of the mirtrons remaining to be found likely have low expression due to compromised structural features, such as suboptimal hairpin structures, mirtron lengths, or hairpin overhangs.
Our efforts were aided by the restricted search space introduced by the nature of mirtron biogenesis, in which splicing substitutes for Drosha-mediated cropping. Current knowledge of how Drosha substrates are selected is scant beyond the notion that its partner Pasha (DGCR8 in mammals) identifies a junction between single-stranded and double-stranded RNA at the hairpin base to position Drosha cleavage approximately one helical turn into the hairpin stem (Han et al. 2006). Presumably the transcriptome of any animal cell contains many such junctions at hairpin bases that are not recognized as substrates. Although computational studies have examined the features of Drosha substrates (Han et al. 2006; Helvik et al. 2007; Ritchie et al. 2007, 2008), much greater understanding is clearly needed to yield effective predictions in genome-wide scans. A corollary inference from our studies is that if Drosha substrates and cleavage sites could be predicted effectively, it might be possible to identify canonical miRNA genes effectively.
Although our computational mirtron model has proven efficacy, there remains room for improvement for the future. Inclusion of mirtrons newly identified in this study may improve training of the model. We have documented the SVM scripts and made them available for download (http://cbio.mskcc.org/leslielab/mirtrons/), and various parameters can be modified as desired. In addition, our surveys were limited by their reliance on annotated splice junctions as input. It is conceivable that unannotated splice sites, either in known mRNAs (e.g., CG17560), unannotated mRNAs, or even ncRNAs (e.g., pri-mir-2220), might yield additional mirtrons. This may be addressed once newer transcriptome annotations based on ultrahigh-throughput RNA-seq evidence are available, which are currently under production by the modENCODE consortium (S Celniker, R Waterston, pers. comm.). However, as D. melanogaster and C. elegans currently stand as two of the best-annotated metazoans, refinement of genome annotations may not yield major revisions to mirtron predictions. It is also worth considering the set of apparent cloned mirtrons with poor structures and/or noncanonical overhangs (e.g., Fig. 6), which may, in principle, transit a distinct pathway. For example, Drosophila mir-1017 exhibits a 3′ terminal tail of ∼100 nt following splicing (Ruby et al. 2007a) and can only enter the mirtron pathway following additional processing by the RNA exosome (Flynt et al. 2010). Members of the “3'-tailed mirtron” class require separate bioinformatic criteria for classification, and it may be that certain intronic substrates only access the mirtron pathway in concert with other factors that remain to be identified. Nevertheless, the experimental data suggests that there are few such atypical mirtron-like loci that achieve even modest expression levels.
It is debatable whether poorly processed mirtrons from marginal hairpin structures ought to be considered as genuine mirtrons, even if associated with read patterns characteristic of Dicer processing. Indeed, efforts to annotate canonical miRNAs are only beginning to consider the efficiency of processing (Chiang et al. 2010). We suggest that this will be a critical parameter to assess with future deep-sequencing analyses. Detailed knowledge of specific structural or sequence features that compromise, but do not abolish, the processing of canonical miRNAs and mirtrons should be important for the rational assessment of newly evolved miRNAs, which may often harbor suboptimal features (Liu et al. 2008; Berezikov et al. 2010).
Methods
Mirtron structure-function tests
The wild-type UAS-DsRed-mir-1003 constructs, in its endogenous synthetic context, and the minimal mir-1003 mirtron cloned between DsRed and 2x-myc exons, were described earlier (Okamura et al. 2007). We cloned additional variants into UAS-DsRed-[AscI-mirtron-NotI]-2xmyc (pJH) using synthetic primers as listed in Figure 1, with overhang nucleotides that permitted direct cloning into the AscI and NotI sites of the parent vector. To generate the longer mirtron variants, we digested pJH-mir-1003/Nhe with NheI and inserted the designated oligonucleotides with compatible CTAG overhangs that destroyed the NheI sites. These inserts carried diagnostic BglII sites within their terminal loops (AGAUCU). We transfected 2 × 106 S2 cells with 0.25 μg of ub-Gal4 and 0.5 mg of UAS-DsRed-mirtron plasmids using Effectene (Qiagen) in 6-well plates, and extracted total RNA 2 d later. Northern analysis was performed as described (Okamura et al. 2007).
We determined the splicing accuracy and efficiency across the mirtron variant panel using RT-PCR. cDNA was prepared from DNA-free total RNA samples using random primer, and the fragments of the flanking exons were amplified with DsRedOut (5′-CCCACAACGAGGACTACAC-3′) and ReverseSplicing (5′-TTATGTCACACCACAGAAGTAAGGTTCC-3′) primers. The PCR products corresponding to the spliced fragments were purified from gels and sequenced, which confirmed accurate splicing of all the constructs.
Small RNA analysis
We downloaded published D. melanogaster small RNA datasets (Brennecke et al. 2008; Chung et al. 2008; Czech et al. 2008, 2009; Ghildiyal et al. 2008, 2010; Kawamura et al. 2008; Seitz et al. 2008; Hartig et al. 2009; Malone et al. 2009; Zhou et al. 2009) and C. elegans small RNA datasets (Ruby et al. 2006; Batista et al. 2008; Claycomb et al. 2009; de Wit et al. 2009; Gent et al. 2009, 2010; Kato et al. 2009; Stoeckius et al. 2009; van Wolfswinkel et al. 2009; Conine et al. 2010) from the NCBI Gene Expression Omnibus or Short Read Archive. We generated additional small RNA datasets for the modENCODE project, and these are available for download from the modENCODE DCC (http://www.modencode.org/). In addition, we also deposited all of the small RNA data in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/gds). We clipped the reads of their 3′ linkers and mapped them to the Dm5.23 and WS200/ce6 annotated genomes using Bowtie (Langmead et al. 2009). We required ≥18-nt perfect matching of the clipped insert, chromosome Uextra excluded, and all alignment locations were recorded. Supplemental Tables S1 and S2 summarize the library, accession numbers, and mapping information for the fly and nematode datasets, respectively.
We retrieved annotated introns from both species and included 25 nucleotides of flanking exon on both sides. We then visualized the mappings of short RNA reads mapping to the sense or antisense strands of these regions. Assessment of mirtron-like generating potential was judged by the enrichment of sense reads from both ends of the intron, relative to exonic reads or antisense reads. The read alignments (separated by library identity) and schematization of the spatial read density along the intron, were summarized in individual gene pages for D. melanogaster and C. elegans introns. The analyses are available at http://cbio.mskcc.org/leslielab/mirtrons/.
Computational prediction of mirtrons
We developed a machine learning approach to predict whether a candidate short intronic structure can form a functional mirtron, using a positive training set of the 14 original validated D. melanogaster mirtrons (Okamura et al. 2007; Ruby et al. 2007a) and a negative training set of 500 randomly selected nonmirtron introns (i.e., introns with no read evidence from among the other more than 27,600 D. melanogaster introns 50–120 nt in length). We used UNAFold (Markham and Zuker 2008) to fold intronic sequences, keeping alternate structures for analysis. Based on overhang constraints for effective substrate recognition by Dicer, we imposed three filters on intronic structures: (1) neither 5′ nor 3′ overhang exceeds 5 nt; (2) the 5′ overhang is less than or equal to the 3′ overhang; (3) counting from the first base pair of the stem, no more than 2 nt of the first 6 nt are unpaired on either the 5′ or the 3′ sides of the structure. These filters eliminated structures that are biochemically infeasible for biogenesis of conventional mirtrons, allowing the supervised learning algorithm (described below) to focus on likelier candidates. In particular, these filters culled some structures with large unpaired regions in the basal stem adjacent to a predicted 2-nt 3′ overhang, whose likely in vivo structure consists of large 5′ and 3′ overhangs.
For the 500 randomly selected nonmirtron introns, we first trained a preliminary support vector machine (SVM) using the positive set and all of the structures for each negative intron with the feature sets described below. Then, we picked out the highest-scoring structure for each negative intron so that the negative set was represented by the most mirtron-like structures, and retrained the SVM. We used three sets of features to represent different aspects of the structures in our SVM models: (1) a binary vector representation of the overhang configuration; (2) a set of structural descriptors, motivated by our experimental results on important determinants of mirtron function; and (3) a set of structural similarity scores, based on pairwise comparison of structures using the relaxed base-pair score (RBP). For 1, we used a binary vector to encode the paired 3′ and 5′ overhang lengths. For 2, we used the following list of 10 features, most of which were calculated on the hairpin substructure involving the last 25 nt of the intron (“ss25”): number of base pairs in ss25, number of bulges in ss25, number of nucleotides in ss25 bulges, number of AU base pairs in ss25, number of GU base pairs in ss25, number of GC base pairs in ss25, number of 5′ bulges in ss25, number of 3′ bulges in ss25, number of interior loops in ss25, the minimum free energy (mfe) of the full intron normalized by the intron length. For 3, we represented each ss25 substructure by its vector of distances to the analogous substructures in the training set using the RBP score to compare the RNA secondary structures. The RBP score generalizes the commonly used base-pair metric by counting differing base pairs up to a defined threshold, so that some base pairs that have similar but not necessarily identical indices in the two structures are considered as matches (Agius et al. 2010). Using a set of (dis)similarity scores between an example and the training set as a feature representation is often called an empirical kernel map (Schölkopf and Smola 2002).
We combined the three feature sets using a standard linear kernel combination approach and trained an SVM model using LIBSVM. We kept the RBP relaxation parameter fixed at 0.4, but we tuned the SVM cost parameter to optimize the ranks of eight (out of 14) mirtrons with the highest read counts (Supplemental Fig. 1), whose features we inferred to correlate with greater biogenesis efficiency. We then used the trained model to score all 27,620 D. melanogaster introns 50–120 nt in length annotated in Dm5.23; this data set was supplemented with the CG17560_in3 mirtronic sequence, which is not present in Dm5.23. For introns with multiple UNAFold structures that pass the overhang filters, we used the highest score among the candidate structures as the predicted score. While the SVM was trained on a relatively small subset of positive and negative examples, we obtained a model with high specificity and sensitivity in genome-wide analysis.
To address the possibility of overfitting to the D. melanogaster training data, we used the C. elegans genome as an independent test set. We scored the 30,565 short introns in the worm annotation WS200/ce6; this data set was supplemented with the inferred mir-2220 mirtronic intron (Fig. 7C), which is not currently annotated as an intron. Again, we obtained good detection of the validated worm mirtrons at the top of the ranked list of predictions (Fig. 4B), providing evidence that the model generalizes beyond the genome on which it was trained and optimized.
The computational analyses were integrated with the read mappings (http://cbio.mskcc.org/leslielab/mirtrons/), where one can also download the mirtron SVM script. The summary pages for D. melanogaster and C. elegans combine the top 1000 introns, ranked according to their mirtron-like features intersected with those introns containing more than five mapped reads. These groups are mostly overlapping, but a collection of low-scoring mirtrons generated substantial numbers of reads (although in most cases it is evident by inspection that these are not typically due to miRNA production). The fly and worm summary pages are sortable by various column headers, including by mirtron score or by read number. Each intron is linked to its genomic position in the UCSC Genome Browser (http://www.genome.ucsc.edu/) and to an individual gene page containing its optimal mirtron-like secondary structure, a schematic of the read density along the intron, and alignments of all of the small RNA reads mapped to the intron and/or to 25 nt of flanking exons (separated by individual library).
Acknowledgments
We thank the Hannon, Zamore, Forstemann, Siomi, Mello, Slack, Bartel, Ketting, Berezikov, and Fire labs, who enabled our mirtron validation efforts by depositing their published sets of small RNA sequences in public databases. We thank Steven Lianoglou for help with the HTML interfaces, Alex Flynt for providing advice on the splicing assay, and Jane Landolin for pointing out alternative splicing of CG17560 in mRNA-seq data. K.O. was supported by the Japan Society for the Promotion of Science. Work in E.C.L.'s group was supported by the Burroughs Wellcome Fund, the Alfred Bressler Scholars Fund, and the NIH (R01-GM083300 and U01-HG004261).
Footnotes
[Supplemental material is available for this article. Small RNA data have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). A full list of accession numbers can be found in Supplemental Table S1.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.113050.110.
References
- Agius P, Bennett KP, Zuker M 2010. Comparing RNA secondary structures using a relaxed base-pair score. RNA 16: 865–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. 2003. A uniform system for microRNA annotation. RNA 9: 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batuwita R, Palade V 2009. microPred: Effective classification of pre-miRNAs for human miRNA gene prediction. Bioinformatics 25: 989–995 [DOI] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37: 766–770 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120: 21–24 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Cuppen E, Plasterk RH 2006. Approaches to microRNA discovery. Nat Genet 38: S2–S7 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung W-J, Willis J, Cuppen E, Lai EC 2007. Mammalian mirtron genes. Mol Cell 28: 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Liu N, Flynt AS, Hodges E, Rooks M, Hannon GJ, Lai EC 2010. Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat Genet 42: 6–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameier M, Wiuf C 2007. Ab initio identification of human microRNAs based on structure motifs. BMC Bioinformatics 8: 478 doi: 10.1186/1471-2105-8-478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Okamura K, Martin R, Lai EC 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol 18: 795–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC 2010. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci 107: 3588–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel J, Sachidanandam R, et al. 2008. An endogenous siRNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ 2009. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell 36: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Linsen SE, Cuppen E, Berezikov E 2009. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res 19: 2064–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan R, Pak C, Jin P 2007. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet 16: 1124–1131 [DOI] [PubMed] [Google Scholar]
- Flynt AS, Chung WJ, Greimann JC, Lima CD, Lai EC 2010. microRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell 38: 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415 [DOI] [PubMed] [Google Scholar]
- Gent JI, Schvarzstein M, Villeneuve AM, Gu SG, Jantsch V, Fire AZ, Baudrimont A 2009. A Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNA interference. Genetics 183: 1297–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell 37: 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD 2010. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML 2008. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res 18: 957–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Davila J, Swerdel MR, Moore JC, Cohen RI, Wu H, Sun YE, Hart RP 2009. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS ONE 4: e7192 doi: 10.1371/journal.pone.0007192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad Y, Aach J, Hayes G, Reinhart BJ, Church G, Ruvkun G, Kim J 2003. Computational and experimental identification of C. elegans microRNAs. Mol Cell 11: 1253–1263 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Hartig JV, Esslinger S, Bottcher R, Saito K, Forstemann K 2009. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J 28: 2932–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helvik SA, Snove O Jr, Saetrom P 2007. Reliable prediction of Drosha processing sites improves microRNA gene prediction. Bioinformatics 23: 142–149 [DOI] [PubMed] [Google Scholar]
- Hertel J, Stadler PF 2006. Hairpins in a Haystack: Recognizing microRNA precursors in comparative genomics data. Bioinformatics 22: e197–e202 [DOI] [PubMed] [Google Scholar]
- Huang TH, Fan B, Rothschild MF, Hu ZL, Li K, Zhao SH 2007. MiRFinder: An improved approach and software implementation for genome-wide fast microRNA precursor scans. BMC Bioinformatics 8: 341 doi: 10.1186/1471-2105-9-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Wu H, Wang W, Ma W, Sun X, Lu Z 2007. MiPred: Classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res 35: W339–W344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadri S, Hinman V, Benos PV 2009. HHMMiR: Efficient de novo prediction of microRNAs using hierarchical hidden Markov models. BMC Bioinformatics 10: S35 doi: 10.1186/1471-2105-S1-S35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, de Lencastre A, Pincus Z, Slack FJ 2009. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol 10: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K 2007. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer–TRBP complex. EMBO Rep 8: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada T, Siomi MC, Siomi H 2008. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature 453: 793–797 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kotani A, Ha D, Schotte D, den Boer ML, Armstrong SA, Lodish HF 2010. A novel mutation in the miR-128b gene reduces miRNA processing and leads to glucocorticoid resistance of MLL-AF4 acute lymphocytic leukemia cells. Cell Cycle 9: 1037–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC 2003. microRNAs: Runts of the genome assert themselves. Curr Biol 13: R925–R936 [DOI] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, and Rubin GM 2003. Computational identification of Drosophila microRNA genes. Genome Biol 4: R42.41–R42.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP 2003a. Vertebrate microRNA genes. Science 299: 1540 doi: 10.1126/science.1080372 [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP 2003b. The microRNAs of Caenorhabditis elegans. Genes Dev 17: 991–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Okamura K, Tyler DM, Phillips MD, Chung WJ, Lai EC 2008. The evolution and functional diversification of animal microRNA genes. Cell Res 18: 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC 2002. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U 2004. Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham NR, Zuker M 2008. UNAFold: Software for nucleic acid folding and hybridization. Methods Mol Biol 453: 3–31 [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I 2006. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217 [DOI] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JW, Shin KR, Han J, Lee Y, Kim VN, Zhang BT 2005. Human microRNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acids Res 33: 3570–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KL, Mishra SK 2007. De novo SVM classification of precursor microRNAs from genomic pseudo hairpins using global and intrinsic folding measures. Bioinformatics 23: 1321–1330 [DOI] [PubMed] [Google Scholar]
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T 2009. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 326: 1275–1279 [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC 2009. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36: 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP 2002. MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie W, Legendre M, Gautheret D 2007. RNA stem-loops: To be or not to be cleaved by RNAse III. RNA 13: 457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie W, Theodule FX, Gautheret D 2008. Mireval: A web tool for simple microRNA prediction in genome sequences. Bioinformatics 24: 1394–1396 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP 2007a. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007b. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T, Cohen SM 2007. Identification of novel Drosophila melanogaster microRNAs. PLoS ONE 2: e1265 doi: 10.1371/journal.pone.0001265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schölkopf B, Smola A 2002. Learning with kernels. Massachusetts Institute of Technology Press, Cambridge, MA [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD 2008. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA strands in flies. Curr Biol 18: 147–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewer A, Paul N, Landgraf P, Aravin A, Pfeffer S, Brownstein MJ, Tuschl T, van Nimwegen E, Zavolan M 2005. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics 6: 267 doi: 10.1186/1471-2105-6-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Engstrom PG, Lenhard B 2007. Mammalian microRNA prediction through a support vector machine model of sequence and structure. PLoS ONE 2: e946 doi: 10.1371/journal.pone.0000946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M 2007a. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res 17: 1865–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. 2007b. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Maaskola J, Colombo T, Rahn HP, Friedlander MR, Li N, Chen W, Piano F, Rajewsky N 2009. Large-scale sorting of C. elegans embryos reveals the dynamics of small RNA expression. Nat Methods 6: 745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai G, Komori T, Asai K, Kin T 2007. miRRim: A novel system to find conserved miRNAs with high sensitivity and specificity. RNA 13: 2081–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burgt A, Fiers MW, Nap JP, van Ham RC 2009. In silico miRNA prediction in metazoan genomes: Balancing between sensitivity and specificity. BMC Genomics 10: 204 doi: 10.1186/1471-2164-10-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139: 135–148 [DOI] [PubMed] [Google Scholar]
- Xue C, Li F, He T, Liu GP, Li Y, Zhang X 2005. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinformatics 6: 310 doi: 10.1186/1471-2105-6-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef M, Nebozhyn M, Shatkay H, Kanterakis S, Showe LC, Showe MK 2006. Combining multi-species genomic data for microRNA identification using a Naive Bayes classifier. Bioinformatics 22: 1325–1334 [DOI] [PubMed] [Google Scholar]
- Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, Hannon GJ 2009. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA 15: 1886–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]