Abstract
RNA-seq was used to generate an extensive map of the Drosophila melanogaster transcriptome by broad sampling of 10 developmental stages. In total, 142.2 million uniquely mapped 64–100-bp paired-end reads were generated on the Illumina GA II yielding 356× sequencing coverage. More than 95% of FlyBase genes and 90% of splicing junctions were observed. Modifications to 30% of FlyBase gene models were made by extension of untranslated regions, inclusion of novel exons, and identification of novel splicing events. A total of 319 novel transcripts were identified, representing a 2% increase over the current annotation. Alternate splicing was observed in 31% of D. melanogaster genes, a 38% increase over previous estimations, but significantly less than that observed in higher organisms. Much of this splicing is subtle such as tandem alternate splice sites.
High-throughput sequencing of cDNA libraries (RNA-seq) is an effective and accurate method for transcriptome profiling. RNA-seq has been applied in many organisms including humans (Campbell et al. 2008; Cloonan and Grimmond 2008; Cloonan et al. 2008; Marioni et al. 2008; Mortazavi et al. 2008; Mudge et al. 2008; Nagalakshmi et al. 2008; Pan et al. 2008; Shendure 2008; Sultan et al. 2008; Wang et al. 2008; Filichkin et al. 2009; Levin et al. 2009; Perkins et al. 2009; Wurtzel et al. 2009). RNA-seq is not dependent on prior knowledge and requires no design work, thus reducing labor and enabling de novo discovery. Several additional features of RNA-seq make it likely to supplant microarrays as the key technology for transcriptome profiling, including accurate and sensitive measuring of expression level, identification of alternate splicing isoforms, and direct discovery of novel transcripts (Graveley 2008; Shendure 2008).
Highly accurate digital measurement of gene expression by RNA-seq can be obtained by counting the number of sequencing reads which map to annotated transcripts from appropriately prepared libraries (Mortazavi et al. 2008; Wang et al. 2009). In this way, RNA-seq offers increased sensitivity and larger dynamic range over microarray, effectively detecting rare transcripts with greater sensitivity and abundant transcripts with superior discrimination (Marioni et al. 2008). Furthermore, RNA-seq generates reads which straddle exon–exon junctions (junction reads). When junction reads are properly aligned to a reference genome the associated splice event can be inferred. Thus, direct discovery of alternate splicing isoforms is enabled without dependence on prior knowledge. While splicing variants can also be detected with custom microarrays, probes must be specifically designed to interrogate exon–exon junctions and are thus limited in scope to known or predicted junctions. Finally, direct annotation of gene models and novel transcript discovery are possible with RNA-seq (Yassour et al. 2009). This can be accomplished by alignment of reads to a reference genome and inference of transcript models from coverage and splicing data. While genome-tiling arrays can also be used to identify novel transcriptional units, RNA-seq is not subject to the inherent limitations of hybridization platforms. Due to the several advantages of the RNA-seq approach we have applied this technology to characterize the transcriptome of D. melanogaster.
D. melanogaster is an important model organism and has contributed significantly to our understanding of gene expression and development. The transcriptome of D. melanogaster is well annotated and functionally characterized. Analysis of transcript expression using spotted arrays has yielded insight into the developmental expression patterns of annotated D. melanogaster transcripts; however, this platform is limited to a subset of annotated genes (Arbeitman et al. 2002). Additionally, tilling arrays have been used to demonstrate that >29% of unnanotated introns and intergenic regions are transcribed within the first 24 h of development at specific time points in development (Manak et al. 2006). Tilling arrays, however, are limited in their resolution by the size of probes used. Expressed sequence tags (ESTs) provide the primary evidence for annotation efforts. Although more than 800,000 D. melanogaster ESTs have been sequenced, not all predicted or annotated genes are supported by ESTs (Stapleton et al. 2002a,b). Furthermore, these EST efforts have focused primarily on embryo and adult libraries, specifically adult head and gonad tissues, leaving intermediate stages of development underrepresented. Continuation of Sanger-based EST sequencing efforts is not an ideal solution for improvement of gene models because of its higher cost in comparison to next-generation sequencing platforms. More recently, high throughput sequencing has been recognized as an ideal solution for transcriptome profiling with base pair resolution. In D. melanogaster the 5′-end SAGE method has been combined with Illumina sequencing to generate a detailed profile of transcription start sites throughout development (Ahsan et al. 2009). RNA-seq will enable interrogation of all transcripts, including potentially unannotated transcripts without the need for further EST characterization (Torres et al. 2008). We therefore propose that characterization of the D. melanogaster transcriptome by RNA-seq technology will greatly facilitate ongoing annotation efforts.
Results
Extensive transcriptome profiling of D. melanogaster by RNA-seq
In order to gain a broad sampling of the D. melanogaster transcriptome, RNA-seq experiments were performed at all stages of the D. melanogaster life cycle. Poly(A)+ transcripts from 10 distinct stages during the live cycle of D. melanogaster were isolated to generated cDNA libraries which were sequenced on the Illumina GA II instrument. As shown in Table 1, 12 independent cDNA libraries were generated, including embryonic, larval, pupal, and adult. Some libraries were staged as specific windows: 2–4-h embryo, 14–16-h embryo, third instar larva, 3-d pupa, and 17-d adult. Additional libraries were derived from broadly staged mixed samples: embryo, larvae, and pupa. Three-day-old male and female adults were sequenced separately for discovery of sex-specific variation. Finally, one library of mixed-age pupal RNA was sequenced in replicate as a validation of the technology. These libraries were selected to represent a broad diversity of RNA molecules (Arbeitman et al. 2002). A total of 272 million paired-end reads of 64–100 bp in length were obtained.
Table 1.
RNA-collection and sequencing yields 690× coverage of the D. melanogaster transcriptome
Sequenced reads were aligned to the D. melanogaster genome (UCSC dm3/FlyBase r5.23) using BLAT (Kent 2002). In total, 180.5 million reads were aligned to the genome with 142.2 million mapped to unique locations, representing >356× sequence coverage of the 30Mb D. melanogaster transcriptome. The current FlyBase annotation (v5.23) was used to calculate digital counts of gene expression for each of the annotated 14,797 FlyBase genes. The FlyBase annotation is often considered the “gold standard” of D. melanogaster gene models because it is the most comprehensive set of curated models. Other annotations based on experimental and computational analysis are also available; a prominent example is the gene models (MB7) produced by the modENCODE consortium (http://www.modENCODE.org/). For robustness we used both of these annotations as references throughout our analysis of RNA-seq data.
Read alignment with BLAT enabled the direct detection of junction reads. These reads are essential to the identification of exon–exon junctions and alternate splicing. To maximize sensitivity for detecting junction reads we used two independent computational approaches (see Supplemental materials). Approximately 6% of all uniquely aligned reads were junction reads, of which, 94% are consistent with annotated FlyBase events, suggesting high concordance between experimental evidence and existing annotations. In total, 90.6% of annotated junctions (46,820) were observed with 93.6% consistency between the two approaches (see Supplemental Table 1; Supplemental Fig. 1). Additionally, junction reads supported ∼120,000 candidate novel junctions. Analytical methods were used to reduce these to 7776 well-supported candidates (see Supplemental materials), of which 2012 (25.9%) are reported in the modENCODE MB7 gene models.
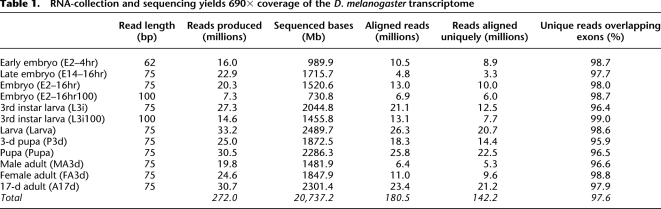
To determine the technical robustness of RNA-seq we considered the positive predictive value (PPV) and sensitivity of the platform. The PPV of RNA-seq for expressed sequences is defined as the percent of uniquely aligned reads which overlap with annotated transcripts. Across all samples, 97.6% of uniquely aligned reads overlap with annotated sequences, suggesting that RNA-seq is highly specific to expressed sequences (Table 1). The sensitivity of RNA-seq is calculated as the percentage of annotated bases covered by sequenced reads for the entire transcriptome and for each gene individually. With all pooled reads considered, 93.4% and 91.9% of FlyBase and modENCODE annotated bases were observed respectively, suggesting a high degree of sensitivity (Fig. 1A). Random sampling of pooled reads from all samples was used to simulate transcriptome coverage at various read depths. The curve of these plots is asymptotic, suggesting that deeper sequencing would offer low returns at the base-pair level. The majority of annotated genes are well covered by RNA-seq reads. For example, >70% of annotated genes are covered for >95% of their length (Fig. 1B). We conclude that RNA-seq is a robust technology for transcriptome profiling including the characterization of gene models.
Figure 1.
Deep RNA-seq covers 93% of the annotated D. melanogaster transcriptome. (A) 93.4% and 91.9% of the FlyBase and modENCODE annotated transcriptome is observed by pooled RNA-seq reads. Simulated read densities indicate that the current depth of sequencing approaches saturation. (B) Annotated transcripts are well covered by RNA-seq reads. More than 70% of annotated genes are covered for >95% of their length. (C) Most unobserved genes can be categorized into classes which are not expected to be observed due to size, genomic duplication, or lack of polyadenylation. (D) A novel exon is identified in wupA, which is supported by two novel junctions with 487 and 428 reads, respectively. (E) RT-PCR designed to validate the novel junction successfully amplifies an appropriately sized product from cDNA but not from genomic DNA.
We observed, however, that not all annotated genes are well represented by RNA-seq reads. In total, 1133 genes (7.7%) are covered at <25% by unique reads. To determine the limitations of RNA-seq it is important to understand the nature of these underrepresented genes. The lack of gene coverage may be due to two primary causes: either reads were generated but could not be mapped uniquely to the reference sequence, or no reads originated from the gene in question. The majority of underrepresented genes fall into classes which are not expected to be observed due to sequence features and limitations of the chosen methodology (Fig. 1C). For example, tRNAs, histones, 5SrRNAs, and Ste/Su(Ste) are highly similar and occur in gene clusters. Reads from these genes are not likely to map uniquely to the reference genome. Furthermore, unpolyadenylated transcripts such as miRNA and snoRNA are not likely to be observed because of the poly(A) selection used in the RNA purification strategy (see Methods). Finally, 128 gustatory, odorant, and ionotropic receptors (small molecules whose expression is tightly restricted to specific neuronal cells) were not observed in our data potentially due to their restricted expression pattern although size selection may also play a role (see Methods). We conclude that underrepresented genes are predominantly due to explainable limitations of the library preparation and read mapping methodologies employed.
RNA-seq data provides evidence for modifying 30% of gene annotations
The depth of our RNA-seq data enables us to evaluate not only the accuracy of current gene model annotations but also their completeness. While RNA-seq and current gene models are largely consistent, a significant portion of current models are incomplete. Evidence from junction reads and coverage was used to extend annotated gene models (see Methods). Even under stringent criteria, 30% of genes (4418) underwent some level of modification during this process (see Supplemental Table 2). The most significant modification was the addition of exon sequences, which affected 25% of gene models (3692). For example, a novel exon identified in wupA is depicted (Fig. 1D). RT-PCR experiments were designed to confirm the expression of this novel exon. Subsequent isolation and sequencing of an appropriately sized PCR product confirmed the prediction. Additionally, the untranslated regions (UTRs) of >8% of genes (1274) were extended by RNA-seq coverage in genes with previously unannotated UTRs. The ability of RNA-seq to accurately define 5′ and 3′ UTRs is limited due to biases in sequencing; therefore, we restricted these analysis to genes whose UTRs were previously unannotated.
RNA-seq detects 319 novel transcripts
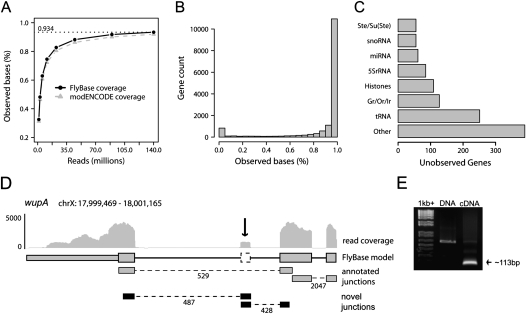
In addition to modifying current gene models, RNA-seq enables the identification of novel transcripts. Evidence from junction reads and read coverage was combined to identify 319 novel transcripts mapping completely outside of FlyBase gene annotations (see Methods). These novel genes are distributed across chromosomes roughly as expected by chromosome size and between strands in nearly equal proportions (Fig. 2A). Several features stand out for these novel genes compared to annotated genes. First, these novel genes are on average smaller than annotated genes. The FlyBase median gene size is 1560 bp, while the median size of these novel transcripts is 315 bp (Fig. 2B). Second, the majority of novel transcripts contain only two exons (Fig. 2C). Third, these novel genes are often expressed in stage and/or sex-specific patterns (Fig. 2D). For example, many novel transcripts are expressed in male but not female adults, although they are observed in mixed larva and pupa samples (Fig. 2D, “Male”). Other clusters express most abundantly in a specific stage (Fig. 2D, “Larva”).
Figure 2.
RNA-seq detects 319 novel transcripts. (A) Novel transcripts detected by RNA-seq are identified on all chromosomes distributed as expected by chromosome size. (B) Novel transcripts (median size 315 bp) are much smaller than annotated FlyBase transcripts (median size 1560 bp). (C) The majority of novel transcripts identified in this study have two exons. (D) Clustering identifies many novel transcripts expressed at specific time points or in sex-specific patterns. For example, many novel transcripts are expressed in male but not female adults, although they are observed in mixed larva and pupa samples (Male). Other clusters express most abundantly in a specific stage (Larva). (E) A novel transcript is depicted, and the associated junction is supported by 50 sequenced reads. Experimental validation by RT-PCR was obtained in pupae cDNA (see Supplemental Fig. 2).
For validation of these results, novel transcripts derived from RNA-seq were compared to two recently described data sets. One study using comparative genomic techniques published a set of 146 novel introns confirmed by ESTs or RT-PCR (Hiller et al. 2009). RNA-seq identified 55% of these introns (80 of 146). Additionally, RNA-seq identifies 61% predicted coding introns (57 of 94) and 18% of predicted introns of messenger-like-noncoding-RNAs (mlncRNAs) from the same study (23 of 129). These results are consistent with the authors' own experimental validation rate at ∼60%, which was lowest for the mlncRNAs due primarily to low and stage-specific expression. Thus we conclude that our current depth and breadth of sequencing is adequate for identification of some novel transcripts. Second, significant overlap has been observed between our data set and the novel genes identified by modENCODE. Of the novel transcripts identified, 45% (144 out of 319) are supported by the modENCODE annotation. Finally, by RT-PCR the junctions supporting nine of these novel transcripts were validated. An example novel transcript is illustrated in Figure 2E; this junction, which was supported by only 50 sequenced reads, was validated by RT-PCR in the pupae cDNA. The experimental evidence for these validations is included in the Supplemental material (see Supplemental Fig. 2).
RNA-seq provides an extensive digital expression profile of D. melanogaster development
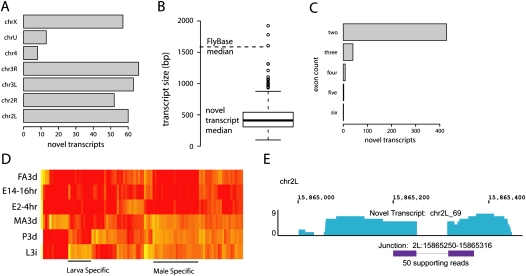
RNA-seq provides “digital” reading of gene expression levels (Mortazavi et al. 2008). To assess whether RNA-seq is consistent with previous technology in measuring gene expression we analyzed RNA expression in parallel on the microarray platform. A single RNA library generated previously and analyzed on the Affymetrix microarray was reanalyzed by RNA-seq. Expression levels were evaluated and normalized appropriately for each platform and the correlation coefficient calculated as R = 0.76 (Spearman), indicating a strong positive correlation between the platforms (see Supplemental Fig. 3). The largest differences between the two platforms occur for genes which were detected at very low levels on the array but were identified in a range of expression by RNA-seq. RNA-seq robustness was also observed in technical replicates. Total RNA extracted from pupa-staged flies was split to derive two separate sequencing libraries. These libraries were sequenced and analyzed in parallel, and the correlation coefficient of these replicates was calculated as R = 0.99 (Fig. 3D). Finally, we found that by random sub-sampling of RNA-seq reads at various depths, gene expression levels are highly robust and R = 0.99 correlation could be obtained with as few as 100,000 reads (see Supplemental Fig. 3).
Figure 3.
Sex-biased expression occurs in one-third of genes. (A) On average, 9995 genes are observed in each stage, 7214 genes are shared between all stages, and 12,490 genes are expressed in one or more stages. (B) 85.7% of genes exhibit greater than fourfold difference in expression between maximum and minimum expression time points. (C) The distribution of coefficient of variation calculated for each gene identifies genes whose expression is highly consistent across development. (D) Technical replicates of pupa RNA-seq exhibit nearly perfect correlation (R = 0.99). (E) Females and early embryos exhibit a high degree of correlation (R = 0.80). (F) 5251 exhibit fourfold difference in expression level between males and females: 1088 genes up-regulated in females are consistent with their expression in early embryos (red), suggesting their importance in embryonic development, 3486 genes are up-regulated in males (blue). (G) Genes without a known ortholog are abundantly expressed in males, many of which have known male-specific functions including seminal fluid proteins and male-specific transcripts. (H) Many genes on unassembled heterochromatin (chrU) exhibit male-specific expression. (I) Genomic PCR results suggest CG40968, CG40583, CG40992, and CG41561 are linked to chromosome Y.
To determine which genes are expressed in each developmental stage, the read counts and normalized expression levels were calculated for each annotated feature (see Methods). Libraries were grouped according to the major stages as appropriate to consider the total number of genes observed in each. In total, 84.4% of FlyBase genes (12,490) are expressed twofold above background, calculated as the average coverage of intergenic regions, in at least one sample. Furthermore, 48.8% of FlyBase genes (7214) were expressed twofold above background in all stages, suggesting that this group of genes is broadly expressed across the D. melanogaster lifecycle (Fig. 3A). On average, 67.5% of genes (9995) are observed within each stage. Since the majority of D. melanogaster genes are developmentally regulated (Arbeitman et al. 2002), we sought to identify genes which are differentially regulated during development and those which are invariant in their expression. Developmentally regulated genes were identified by considering the difference between maximum and minimum expression levels across all samples. The majority of genes expressed above background (85.7%; 10,702 of 12,490) exhibit fourfold or greater differences in expression level between their highest and lowest stages of expression, suggesting developmental regulation (Fig. 3B). Expressed genes which exhibit consistent expression were identified by calculating the coefficient of variation (Fig. 3C). Gene Ontology (GO) analysis was performed on the 3% of genes exhibiting the most consistent expression (Carmona-Saez et al. 2007; Nogales-Cadenas et al. 2009). Many of these genes are involved with basic cellular processes (246 genes, P = 1.47 × 10−16) and functions, such as translation (14 genes, P = 1.39 × 10−6).
It has been reported that a significant number of genes are differentially expressed between males and females (Arbeitman et al. 2002). We observed 5251 genes (35.5%) that exhibited fourfold difference in their expression levels between 3-d-old male and female adults (Fig. 3E,F). Of these, 1088 genes are up-regulated in female over male but consistent between female and embryonic expression (Fig. 3E,F). GO analysis of these genes identified functions related to embryonic development, including 45 genes involved in oogenesis (P = 5.99 × 10−13), 25 genes involved with DNA replication (P = 4.5 × 10−16), and 17 genes involved with rRNA processing (P = 1.08 × 10−9). In total, 3486 genes are up-regulated in males over females (Fig. 3F). GO analysis identified many expected functional groups, including 27 genes involved with sensory perception of chemical stimulus (P = 2.297 × 10−6), eight genes involved with post-mating behavior (P = 4.64 × 10−6), 38 genes involved with microtubule-based movement (P = 8.06 × 10−6), and 32 genes involved with oxidation reduction (P = 1.16 × 10−5).
Another class of genes up-regulated in males consists of genes without an identified ortholog among the 12 Drosophila Genomes Project (Fig. 3G; Clark et al. 2007). Of 1043 genes without an annotated Drosophila ortholog, nearly 200 exhibited male-specific expression patterns; in contrast, less than 20 genes exhibited a female-specific expression pattern. It is conceivable that these nonconserved genes are more rapidly evolving and that their male-specific expression can be related to the forces of male-driven evolution. Many known male-specific transcripts and sperm factors do not have an identifiable ortholog among Drosophila species, supporting this hypothesis; however, several genes do not have described molecular functions and it is reasonable to postulate that some of these may also be linked to male reproduction (see Supplemental Table 3).
Identification of novel D. melanogaster Y–linked genes
We observed that many genes with male-specific patterns of expression are located on chrU (Fig. 3H). The D. melanogaster chrU consists of unassembled contigs which are largely heterochromatic and have not been assigned to physical genomic locations due to difficulty in assembly. Chromosome Y is wholly heterochromatic; therefore, much of its sequence is currently unassembled and assigned as chromosome U. Only eight genes are annotated on chromosome Y according to the current FlyBase annotation. We hypothesized that genes with male-specific expression annotated on chrU might potentially be linked to chromosome Y. To test for Y-linkage, PCR primers were designed for eight genes exhibiting male-specific expression. Amplification of genomic DNA in males but not in females is indicative of Y-linkage. PCR products were obtained only in males for four of the eight tested genes: CG41561, CG40968, CG40583, CG40992 (Fig. 3I). Evidence linking three of these genes to the Y chromosome has been reported previously (Carvalho et al. 2000, 2001, 2003; Vibranovski et al. 2008; Krsticevic et al. 2009). Assignment of CG41561 to chromosome Y is believed to be a novel discovery as no FlyBase or GenBank record links this gene to chromosome Y.
Alternate splicing is observed in 31% of D. melanogaster genes
A key feature of eukaryotic gene expression is alternate splicing. In humans, as many as 95% of transcripts are believed to be alternately spliced; in contrast, in the current FlyBase annotation only 26% of genes have multiple splice isoforms. Our analysis estimates that alternate splicing occurs in at least 4618 genes (31%) of the D. melanogaster genome, a 38% increase over FlyBase estimates. The detection of junction reads in RNA-seq data enables the discovery of the alternate splicing events summarized in Table 2. We profiled the extent of defined alternate splicing events: skipped exons (SE), retained introns (RI), alternate donor splice site (ADSS), alternate acceptor splice site (AASS), and alternate last exon (ALE) (see Supplemental materials). Alternate first exons (AFE), a transcriptionally regulated event detectable by RNA-seq, were also profiled. Each of these splicing events has the capacity to generate transcript diversity by modifying the coding and regulatory sequences of a transcript (Wang et al. 2008).
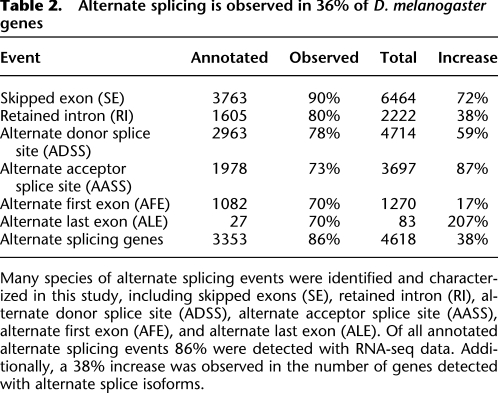
Table 2.
Alternate splicing is observed in 36% of D. melanogaster genes
Many species of alternate splicing events were identified and characterized in this study, including skipped exons (SE), retained intron (RI), alternate donor splice site (ADSS), alternate acceptor splice site (AASS), alternate first exon (AFE), and alternate last exon (ALE). Of all annotated alternate splicing events 86% were detected with RNA-seq data. Additionally, a 38% increase was observed in the number of genes detected with alternate splice isoforms.
We observed 80% of annotated alternate splicing events (9181) across all categories of splicing events examined. The most common annotated event is the SE, of which we observed 90% of events (3390). The SE was also the most common novel event observed, of which we observed 3074 new events. The other classes were also broadly observed (Table 2). Of 3353 genes with annotated splicing events profiled in this study (23% of all genes) we detected annotated alternate splicing events in 86% (2899 genes). Additionally, we observed many novel alternate splicing events. The largest increase over annotation is the ALE event of which we detected 64 novel events, a twofold increase.
Alternate splicing is an essential feature of sexual determination and differentiation in D. melanogaster, and it has been estimated that as many as 22% of multi-transcript genes exhibit sex-biased splicing patterns (Telonis-Scott et al. 2009). In our data set 3-d-old males and females were sequenced separately to enable assessment of sex-specific alternate splicing. Of alternately spliced genes, 2308 were expressed in both males and females at a sufficient level to enable examination of alternate splicing. Of genes with annotated alternate splice isoforms, differential splicing was observed in 23.5%. In a recent report, interrogation of 417 genes with exon-microarrays led to identification of 33 genes shown to have sex-biased splicing. Among these candidate genes, we confirmed 14 (Telonis-Scott et al. 2009), most unconfirmed genes were expressed below our threshold in one or the other sex (see Supplemental Table 4). Therefore, our analysis is quite sensitive, identified hundreds of specific transcripts, and confirms that widespread sex-specific regulation of alternate splicing occurs in D. melanogaster.
Tandem alternate splicing in the D. melanogaster transcriptome
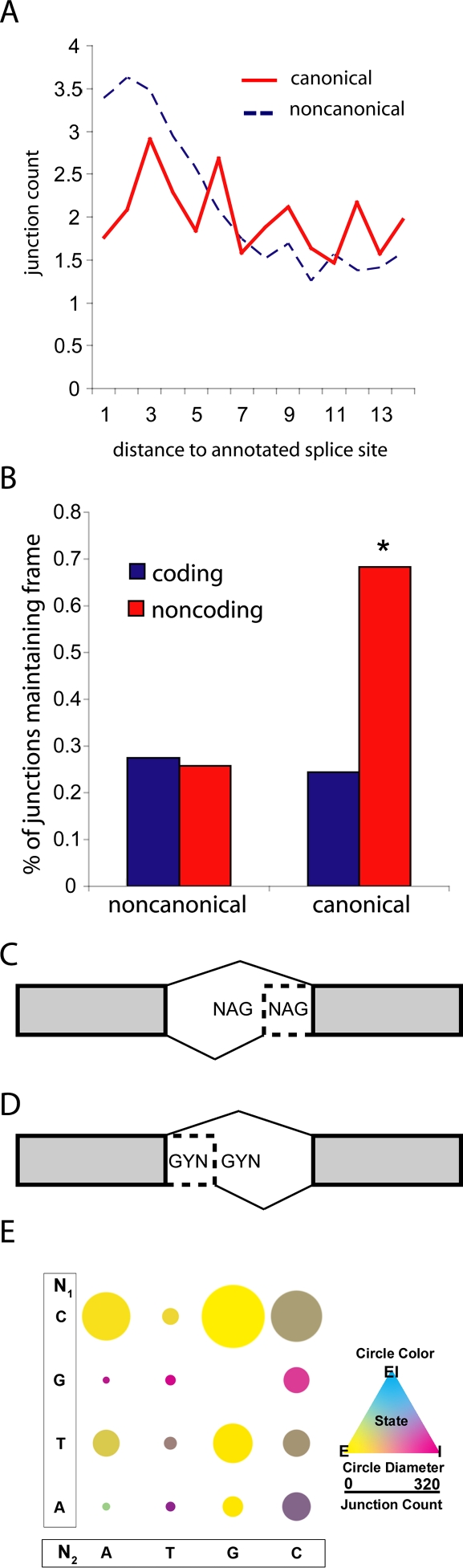
A striking observation regarding alternate splicing is that many of the 120,000 candidate novel junctions occur near annotated junctions, exhibiting a 3-bp periodicity in their distance from annotated junctions (Fig. 4A). In fact, when these are further partitioned into coding and noncoding junctions, this periodicity is strongly observed only in coding regions (Fig. 4B). These observations led us to consider mechanisms which might result in the observed spacing. One explanation for this observation is the existence of tandem alternate splice sites (TASSs) in the D. melanogaster genome. Subtly different alternate splice isoforms can be observed when multiple potential splicing sites are located in close physical proximity. We profiled two well-described classes of TASS: the GYNGYN donor site and the NAGNAG alternate acceptor site (Hiller et al. 2004, 2006, 2007). Though many thousand TASS sites exist within the transcriptome of D. melanogaster only a small fraction are known to splice alternately. Furthermore, splicing at NAGNAGs is far more common than at GYNGYNs, with 135 and seven annotated sites, respectively. From the 7776 novel junctions identified, tandem alternate splicing was observed at 90% of annotated NAGNAG sites and 29% of GYNGYN sites. In addition to annotated sites, we observe alternate splicing at 242 novel NAGNAG splicing sites and 22 novel GYNGYN sites (see Supplemental Table 5).
Figure 4.
RNA-seq detects abundant subtle alternate splice isoforms. (A) Distribution of novel splicing junctions near annotated exon–exon junctions approximates the frequency of read indel events. The distance between novel splice sites and annotated splice sites is plotted separately for canonical and noncanonical novel junctions. (B) Canonical novel junctions in coding regions conserve frame. The portion of novel junctions which conserve frame is calculated for all candidate junctions across two partions: canonical/noncanonical and coding/noncoding. Only canonical novel junctions within coding regions conserve frame more than expected by chance (Binomial test n = 34,060, P-value = 2.2 × 10−16). (C) Constitutively splicing NAGNAGs can be exonic (E) and intronic (I), named for where the second NAG is incorporated. Alternate splicing NANAGs result in both E and I states. (D) Constitutively splicing GYNGYNs can be of two forms: exonic (e) or intronic (i), named for where the first GYN is incorporated. Alternate splicing GYNGYNs result in both e and i states. Diagram adapted from Hiller et al. (2006). (E) For each possible NAGNAG splice site N1 (A, T, G, or C) and N2 (A, T, G, or C), the genomic count (circular diameter) and the proportion of exonic (E), intronic (I), and alternative (EI) splicing (circle color) was calculated.
Consistent with previous reports, we observed that the most important characteristic of determining the dominant splice site at a NAGNAG are the bases N1 and N2 (Fig. 4E; Sinha et al. 2009). For example, CAG is a strong acceptor site while GAG is a weak acceptor site. When weak sites or strong sites are paired, alternate splicing frequently occurs. Conversely, when a weak and a strong site are paired, constitutive splicing is the norm. The most abundant alternate splice pair CAGCAG demonstrated alternate splicing at 53.2% of observed sites, suggesting that while N1N2 are highly informative for splice site strength, other factors (cis or trans) likely contribute to determine alternate splicing. These observations suggest that TASSs are a common mechanism for alternate splicing in D. melanogaster.
Discussion
We have generated the first map of Drosophila transcription based on paired-end RNA-seq. The extensive nature of these data is illustrated by the broad range of 10 distinct developmental time points sampled. Additionally, from the 142.2 million uniquely mapped sequencing reads, 95% of annotated genes and 90% of annotated junctions were observed in these data.
RNA-seq data provides direct experimental evidence for validating and modifying current gene models and identifying novel genes. In our data sets, a total of 97.5% of uniquely mapped reads overlapped with annotated models suggesting a high level of concordance between experimental evidence and current annotations. Even using stringent criteria, ∼30% of FlyBase genes underwent some level of modification when the coverage and junction reads data were combined with annotated gene models. These changes include extensive modification to coding and noncoding exons in 25% of gene models and the extension of previously unannotated UTRs for 8% of gene models. Furthermore, we identified 319 novel transcripts across the genome. It remains to be determined whether these transcripts have phenotypic consequences or represent noisy transcription. It seems likely that narrower developmental time points and tissue-specific samples may be necessary to identify the full range of transcripts in the D. melanogaster transcriptome. This is based on our observation that novel transcripts are usually small, of low abundance, and exhibit stage and sex-specific expression patterns. From these observations we conclude that, while existing models are largely complete, RNA-seq data sets are valuable resources for the continued refinement of gene models and identification of novel transcripts.
Our data provide interesting insights into the extent of alternate splicing in D. melanogaster and highlight mechanisms which contribute to the subtle variety of splice isoforms. We identified 7776 potential novel splice events with strong experimental support. Many of these events are well supported by RNA-seq reads but have not been validated by additional experimental methodologies. On a gene-by-gene basis, interested researchers may want to validate alternate splicing events which occur in their particular gene of interest. We estimate that alternate splicing occurs in 5014 genes (36%) of the D. melanogaster genome, a 61% increase over FlyBase estimates. In comparison to mammals, where alternate splicing is estimated to occur in 95% of genes, this number is relatively low. Interestingly, however, we observed extensive amounts of subtle alternate splicing, including 264 novel TASS sites identified.
We observed that sex-specific differences are extensive in D. melanogaster at the gene regulatory level and at the level of alternate splicing. Of genes with annotated alternate splice isoforms, differential splicing was observed in 23.5%. This suggests that extensive sex-specific regulation occurs at the level of alternate splicing. Furthermore, while the majority of genes (85.7%) appear to be developmentally regulated, there is a significant degree of sex-specific regulation in gene expression; more than one-third of genes are differentially expressed between males and females. From this we conclude that both gene regulation and alternate splicing contribute significantly to the differences between males and females in D. melanogaster.
Several directions remain to further utilize these data. For example, it is reasonable that evidence for RNA editing can be extracted from our reads (Stapleton et al. 2006). Additionally, recent studies have demonstrated that de novo assembly of transcripts from RNA-seq data is a potential avenue for gene annotation (Yassour et al. 2009). Finally, for simplicity we have ignored all novel noncanonical splicing events detected in our data despite some events having very high support, and examination of these may shed additional light on the variation introduced by alternate splicing in the D. melanogaster genome.
Methods
Stocks and staging
CS flies were reared at room temperature (23°C–24°C) for collection. Flies were staged to generate 12 separate collections. For early embryo (E2–4hr), adults were set for 2 h of egg laying on grape juice plates. Embryos were then collected and allowed to age for an additional 2 h, resulting in embryos from 2 to 4 h in age. Late embryos (E14–16hr) were collected as above, but allowed to age for 14 h. The broadly staged embryos (E2–16hr and E2–16hr100) were set for 14 h of egg laying on grape juice plates, followed by 2 h of aging resulting in embryos from 2 to 16 h in age. For third instar larva (L3i and L3i100), ∼30 wandering larvae were collected. For pupa collections, third instar wandering larvae were collected into new vials and approximately 30 pupae were collected at 24, 48, 72, 96, and 108 h. Mixed pupa sample (P) consisted of equimolar amounts of total RNA from each of these collections, whereas 3-d pupa (P3d) consisted of only the 72-h collection. Three-day-old adults (MA3d and FA3d) were collected when emerging and separated as males and virgin females into new vials, followed by 48 h of aging. Seventeen-day-old adults were collected separately, allowed to age for 17 d, and total RNA from each was combined in equimolar amounts.
RNA isolation
Whole-body samples were homogenized in 1 mL of TRIzol (Invitrogen) per 50–100 mg of tissue using a glass-Teflon homogenizer. RNA was purified according to the suggested manufacturer's protocol (Invitrogen).
Library preparation
For each library, mRNA was purified from 20 μg of total RNA using an mRNA purification kit (Invitrogen). Double-stranded cDNAs were made from mRNA using the Superscript Double-Stranded cDNA Synthesis kit (Invitrogen) and random hexamer primers (Invitrogen, 3 μg/μL). On a 2% agrose gel, 200–400-bp cDNAs were selected and used as the DNA template for the Illumina library construction. The quality and quantity of the resulting double-stranded cDNAs was assessed using the Nanodrop 7500 spectrophotometer (Nanodrop).
DNA sequencing libraries were generated using the size-selected cDNAs according to the manufacturer's protocol. Cluster generation and sequencing was performed on the Illumina cluster station and Illumina GA II following manufacturer's instructions.
Read alignment
Several alignment programs written specifically for mapping next-generation sequencing data were considered for this study, including SOAP, SOAP2, Bowtie/Tophat, and BWA. BLAT was chosen for two important reasons: First, it outperformed the other alignment algorithms in the number of reads aligned and in our assessment of alignment quality. Short-read alignment programs, while efficient, offer their substantial increases in speed at the expense of sensitivity and accuracy. Second, BLAT natively supports alignments of intron-sized gaps and the direct discovery of junction reads. Small indels of two to three base pairs were permitted as these might correspond to polymorphism or sequencing error; larger indels were filtered out. Alignments with gaps >39 bp were additionally processed to determine whether they corresponded to introns. While BLAT does not make use of paired-end information, custom scripts were developed to disambiguate read pairs with multiple mapping locations if a unique concordant alignment could be identified. Read pairs mapping to separate chromosomes were discarded, as these are likely artifacts of library preparation or alignment errors (McManus et al. 2010).
Modifying gene models
A reference-guided assembly approach which joined overlapping reads to form blocks of coverage was used similar to approaches previously described (Denoeud et al. 2008). Novel junctions were incorporated into annotated gene models if they shared a common splice site with the model. Novel junctions were also incorporated if their associated block of read coverage overlapped with the annotated exons of that locus and they were derived from the appropriate strand. Additionally, 5′ and 3′ UTRs were extended based on composite coverage only if the UTR was previously annotated as 10 bp or smaller.
Identifying novel transcripts
A reference-guided assembly approach which joined overlapping reads to form blocks of coverage was used. Reads mapping outside of annotated FlyBase genes which could not be connected to gene models were considered for the detection of novel transcripts. Novel transcripts were required to contain at least one canonical intron and be covered by 20 or more reads.
Gene expression counts
The number of reads uniquely aligning to transcribed regions of each gene was calculated for all genes in the annotated transcriptome. The gene read count was calculated as the number of unique reads which aligned to the genome within the exons of each gene. The expression level of each gene was calculated in reads per kilobase per million reads (RPKM) according to the formula:
where C is the read count, N is the sum of aligned reads in the experiment, and L is the length of the transcript (Mortazavi et al. 2008).
GO analysis
GO analysis was performed using the GeneCoDis2.0 web server (Carmona-Saez et al. 2007; Nogales-Cadenas et al. 2009). The D. melanogaster organism is supported on this server, and GO categories “Biological Process,” “Molecular Function,” and “Cellular Component” were selected.
Sex-specific splicing
We counted the number of reads occurring within each exon of alternate splicing genes and tested whether the male to female ratio met the expected ratio based on gene expression counts, excluding genes which were not observed in both sexes. Our criteria required that first an exon expression level exhibit greater than twofold between male and female, and second that the χ2 P-value exceed 1 × 10−4.5. This estimate excludes those genes whose alternate splicing might lead to down-regulation of the transcripts.
Acknowledgments
We thank Graeme Mardon for critical reading of the manuscript. We thank Ming Cao for contributing to formatting of the manuscript and technical support of the analyses. We thank the staff of the Human Genome Sequencing Center who performed the sequencing of genomic libraries. B.D. is supported by training grant T32 EYO7102-16. H.W. is supported by postdoctoral fellowship EY19430-01. This work was partially supported by NHGRI/NIH grant 5U54HG003273 (R.G.) and Retinal Research Foundation and the NEI/NIH grant R01EY016853 (R.C.).
Footnotes
[Supplemental material is available for this article. The sequencing data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA012173 and to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE24324.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.107854.110.
References
- Ahsan B, Saito TL, Hashimoto S, Muramatsu K, Tsuda M, Sasaki A, Matsushima K, Aigaki T, Morishita S 2009. MachiBase: A Drosophila melanogaster 5′-end mRNA transcription database. Nucleic Acids Res 37: D49–D53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270–2275 [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. 2008. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A 2007. GENECODIS: A web-based tool for finding significant concurrent annotations in gene lists. Genome Biol 8: R3 doi: 10.1186/gb-2007-8-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Lazzaro BP, Clark AG 2000. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci 97: 13239–13244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Dobo BA, Vibranovski MD, Clark AG 2001. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci 98: 13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Vibranovski MD, Carlson JW, Celniker SE, Hoskins RA, Rubin GM, Sutton GG, Adams MD, Myers EW, Clark AG 2003. Y chromosome and other heterochromatic sequences of the Drosophila melanogaster genome: How far can we go? Genetica 117: 227–237 [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 [DOI] [PubMed] [Google Scholar]
- Cloonan N, Grimmond SM 2008. Transcriptome content and dynamics at single-nucleotide resolution. Genome Biol 9: 234 doi: 10.1186/gb-2008-9-9-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5: 613–619 [DOI] [PubMed] [Google Scholar]
- Denoeud F, Aury JM, Da Silva C, Noel B, Rogier O, Delledonne M, Morgante M, Valle G, Wincker P, Scarpelli C, et al. 2008. Annotating genomes with massive-scale RNA sequencing. Genome Biol 9: R175 doi: 10.1186/gb-2008-9-12-r175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC 2009. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR 2008. Molecular biology: Power sequencing. Nature 453: 1197–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M 2004. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat Genet 36: 1255–1257 [DOI] [PubMed] [Google Scholar]
- Hiller M, Huse K, Szafranski K, Rosenstiel P, Schreiber S, Backofen R, Platzer M 2006. Phylogenetically widespread alternative splicing at unusual GYNGYN donors. Genome Biol 7: R65 doi: 10.1186/gb-2006-7-7-r65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Nikolajewa S, Huse K, Szafranski K, Rosenstiel P, Schuster S, Backofen R, Platzer M 2007. TassDB: A database of alternative tandem splice sites. Nucleic Acids Res 35: D188–D192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Findeiss S, Lein S, Marz M, Nickel C, Rose D, Schulz C, Backofen R, Prohaska SJ, Reuter G, et al. 2009. Conserved introns reveal novel transcripts in Drosophila melanogaster. Genome Res 19: 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krsticevic FJ, Santos HL, Januario S, Schrago CG, Carvalho AB 2009. Functional copies of the Mst77F gene on the Y chromosome of Drosophila melanogaster. Genetics 184: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JZ, Berger MF, Adiconis X, Rogov P, Melnikov A, Fennell T, Nusbaum C, Garraway LA, Gnirke A 2009. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol 10: R115 doi: 10.1186/gb-2009-10-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak JR, Dike S, Sementchenko V, Kapranov P, Biemar F, Long J, Cheng J, Bell I, Ghosh S, Piccolboni A, et al. 2006. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet 38: 1151–1158 [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y 2008. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Duff MO, Eipper-Mains J, Graveley BR 2010. Global analysis of trans-splicing in Drosophila. Proc Natl Acad Sci 107: 12975–12979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, Huntley JJ, Luo S, Zhang L, van Velkinburgh JC, Farmer AD, et al. 2008. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PLoS ONE 3: e3625 doi: 10.1371/journal.pone.0003625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A 2009. GeneCodis: Interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res 37: W317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Perkins TT, Kingsley RA, Fookes MC, Gardner PP, James KD, Yu L, Assefa SA, He M, Croucher NJ, Pickard DJ, et al. 2009. A strand-specific RNA-Seq analysis of the transcriptome of the typhoid bacillus Salmonella typhi. PLoS Genet 5: e1000569 doi: 10.1371/journal.pgen.1000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J 2008. The beginning of the end for microarrays? Nat Methods 5: 585–587 [DOI] [PubMed] [Google Scholar]
- Sinha R, Nikolajewa S, Szafranski K, Hiller M, Jahn N, Huse K, Platzer M, Backofen R 2009. Accurate prediction of NAGNAG alternative splicing. Nucleic Acids Res 37: 3569–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M, Carlson J, Brokstein P, Yu C, Champe M, George R, Guarin H, Kronmiller B, Pacleb J, Park S et al. 2002a. A Drosophila full-length cDNA resource. Genome Biol 3: RESEARCH0080 doi: 10.1186/gb-2002-3-12-research0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M, Liao G, Brokstein P, Hong L, Carninci P, Shiraki T, Hayashizaki Y, Champe M, Pacleb J, Wan K, et al. 2002b. The Drosophila gene collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res 12: 1294–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M, Carlson JW, Celniker SE 2006. RNA editing in Drosophila melanogaster: New targets and functional consequences. RNA 12: 1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. 2008. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960 [DOI] [PubMed] [Google Scholar]
- Telonis-Scott M, Kopp A, Wayne ML, Nuzhdin SV, McIntyre LM 2009. Sex-specific splicing in Drosophila: Widespread occurrence, tissue specificity and evolutionary conservation. Genetics 181: 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres TT, Metta M, Ottenwalder B, Schlotterer C 2008. Gene expression profiling by massively parallel sequencing. Genome Res 18: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Koerich LB, Carvalho AB 2008. Two new Y-linked genes in Drosophila melanogaster. Genetics 179: 2325–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R 2009. A single-base resolution map of an archaeal transcriptome. Genome Res 20: 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Kaplan T, Fraser HB, Levin JZ, Pfiffner J, Adiconis X, Schroth G, Luo S, Khrebtukova I, Gnirke A, et al. 2009. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc Natl Acad Sci 106: 3264–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]