Abstract
The C. elegans genome has been completely sequenced, and the developmental anatomy of this model organism is described at single-cell resolution. Here we utilize strategies that exploit this precisely defined architecture to link gene expression to cell type. We obtained RNAs from specific cells and from each developmental stage using tissue-specific promoters to mark cells for isolation by FACS or for mRNA extraction by the mRNA-tagging method. We then generated gene expression profiles of more than 30 different cells and developmental stages using tiling arrays. Machine-learning–based analysis detected transcripts corresponding to established gene models and revealed novel transcriptionally active regions (TARs) in noncoding domains that comprise at least 10% of the total C. elegans genome. Our results show that about 75% of transcripts with detectable expression are differentially expressed among developmental stages and across cell types. Examination of known tissue- and cell-specific transcripts validates these data sets and suggests that newly identified TARs may exercise cell-specific functions. Additionally, we used self-organizing maps to define groups of coregulated transcripts and applied regulatory element analysis to identify known transcription factor– and miRNA-binding sites, as well as novel motifs that likely function to control subsets of these genes. By using cell-specific, whole-genome profiling strategies, we have detected a large number of novel transcripts and produced high-resolution gene expression maps that provide a basis for establishing the roles of individual genes in cellular differentiation.
The generation of specific cell types depends on spatial and temporal control of gene expression. The nematode Caenorhabditis elegans has been widely utilized to address this question because of its simple body plan and fully sequenced genome (Hillier et al. 2005). Although composed of fewer than 1000 somatic cells, the tissues of C. elegans adults include cell types characteristic of all metazoans such as muscle, nerve, intestine, and skin (Hall and Altun 2008). Moreover, the developmental origin of each of these cells is fully described in a complete map of cell divisions from fertilized zygote to sexually mature adult (Sulston and Horvitz 1977; Sulston et al. 1983). The C. elegans genome sequence is also precisely defined and, at ∼100 Mb, is about one-thirtieth the size of the human genome (Hillier et al. 2005). However, with 20,168 predicted genes (http://wiki.wormbase.org/index.php/WS200), the C. elegans protein-coding genome is only slightly smaller than that of humans (http://www.sanger.ac.uk/PostGenomics/encode/stats.html). Major classes of noncoding RNAs (ncRNAs) such as microRNAs (miRNAs) are also represented in C. elegans (Ruby et al. 2006; Kato et al. 2009). Thus, C. elegans provides a simple but representative model of development that depends on differential expression of a compact, well-described genome. Although C. elegans is completely sequenced, some predicted genes lack direct evidence of transcription, and other cryptic protein-coding genes and ncRNAs are likely to have been overlooked by gene prediction software (Hillier et al. 2009; Schweikert et al. 2009). In addition, the cell-specific expression patterns of the majority of C. elegans genes are unknown. Thus, the anatomy and development of the animal is defined at the resolution of the single cell, but a comparably precise atlas of gene expression is not currently available.
The goal of a comprehensive gene expression map has been achieved in part by analysis of promoter::GFP fusions for a broad array of protein coding genes (Dupuy et al. 2007; Hunt-Newbury et al. 2007; Murray et al. 2008; Liu et al. 2009). This methodology, however, is generally not quantitative and can be misleading if key regulatory elements are omitted from the reporter genes (Hunt-Newbury et al. 2007). We have adopted the alternative strategy of measuring native transcripts from a broad array of specific tissues and cell types. In addition, we used whole-genome tiling arrays in order to sample the entire nonrepetitive genome and therefore achieve an unbiased approach to transcript discovery. In addition to assigning gene expression to identified tissues and stages, our approach of analyzing different cell types and developmental periods also ensures detection of RNAs that may be selectively expressed during discrete temporal intervals or in limited numbers of cells. We accomplished this goal by utilizing recently developed methods for obtaining RNA from specific C. elegans cells (Roy et al. 2002; Zhang et al. 2002; Fox et al. 2005). Altogether, we sampled 13 embryonic cell types and 12 larval and adult tissues. We also produced tiling array data sets for whole-animal RNA isolated from seven different developmental stages. Additional profiling results were obtained from larval males and from the hermaphrodite gonad and soma. Thus, our data sets significantly enhance a growing body of tissue- and stage-specific gene expression for C. elegans (McKay et al. 2003; Pauli et al. 2006; Von Stetina et al. 2007; Meissner et al. 2009). Our results indicate that most protein coding genes (about 75%) are differentially expressed among the stages and cell types that we sampled. In addition to providing evidence of extensive gene regulation, these results should also greatly aid genetic analysis by suggesting cell types or developmental stages in which highly expressed transcripts are likely to function. For example, our results provide the first comprehensive description of gene expression in C. elegans primordial germ cells and led to the discovery that proteins encoded by a subset of these genes are expressed well before their established roles in meiosis and oogenesis. To identify novel transcripts, we utilized a recently developed computational method for recognizing transcribed regions irrespective of their annotation status (Laubinger et al. 2008; Zeller et al. 2008). This approach revealed a large number of previously unannotated transcripts encoded by at least 10% of the C. elegans genome. These novel transcripts show striking cell specificity that may be indicative of tissue-specific functions. To facilitate the use of these data for future studies of gene function, we provide online resources for visualizing transcribed regions in a genome browser and for estimating relative gene expression levels across tissue types and developmental stages.
Results
Strategies for profiling specific cell types and developmental stages
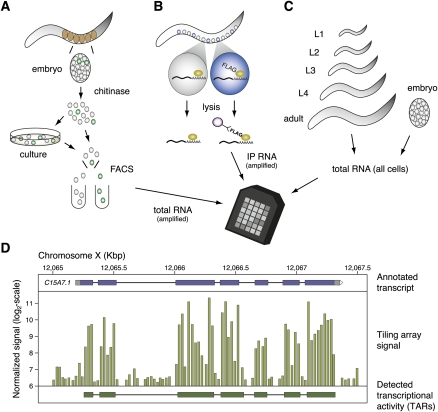
Cell-specific RNA was obtained from GFP-labeled embryonic cells isolated by FACS and from larval cells by use of the mRNA-tagging method (Fig. 1A,B, see also Supplemental Fig. S1). Altogether, we generated tiling array profiles from 25 different tissues, with each sample derived from one of five distinct developmental stages. The cell specificity of these data sets is reflected in the strong enrichment of native transcripts for the corresponding gene promoter used to mark each cell type for profiling (Supplemental Fig. S2). Corresponding reference data sets were collected from all cells for each of these developmental periods (Table 1; see Methods). We also generated an independent developmental series with total RNA isolated from whole animals at seven different ages (EE, LE, L1, L2, L3, L4, YA) (Table 1; Fig. 1C; Supplemental Table S1). An additional group of tiling array profiles was obtained from young adult hermaphrodite gonads, L4 hermaphrodite somatic cells, and L4 males. Because our samples were isolated by different methods, which could potentially result in biased representation, we used principal component analysis (PCA) to compare tiling array results (see below) obtained from specific cells and from whole animals. PCA shows that tiling array profiles obtained from whole embryos cluster with data sets generated from specific embryonic cells and that larval and adult profiles are grouped with data sets obtained from specific postembryonic and young adult tissues (Supplemental Fig. S3). Correlation analysis comparing cell type data sets with developmental series data sets also confirms that expression estimates derived from cell types generally correlate well with the corresponding developmental stage data set generated from total RNA (Supplemental Fig. S4). Thus, a global analysis of our tiling array results suggests that cell-specific profiling preserves overall patterns of temporally regulated gene expression.
Figure 1.
Strategies for generating tiling array data sets from specific C. elegans cells in embryos and larvae and from whole animals at defined developmental stages. (A) In the MAPCeL (Micro-array Profiling of C. elegans Cells) method, embryos are isolated from gravid adults and blastomeres released by treatment with chitinase. Dissociated embryonic cells are either sorted immediately or cultured for 24 hrs before FACS. Total RNA is amplified for tiling array analysis. (B) The mRNA-tagging strategy was used to isolate RNA from specific larval and adult cells. The epitope-tagged (FLAG) polyA-binding protein (PAB-1) is expressed under the control of cell-specific promoters. The PAB-1:RNA complex is immunoprecipitated and RNA is amplified for tiling array analysis. (C) Total RNA is isolated from synchronized populations of embryonic, larval and adult animals for tiling array analysis. (D) Tiling array data (middle) is shown in a region around the annotated transcript C15A7.1 (top). Each vertical bar corresponds to the signal of one probe feature. A transcript identified by mSTAD using only the tiling array signal is shown at the bottom.
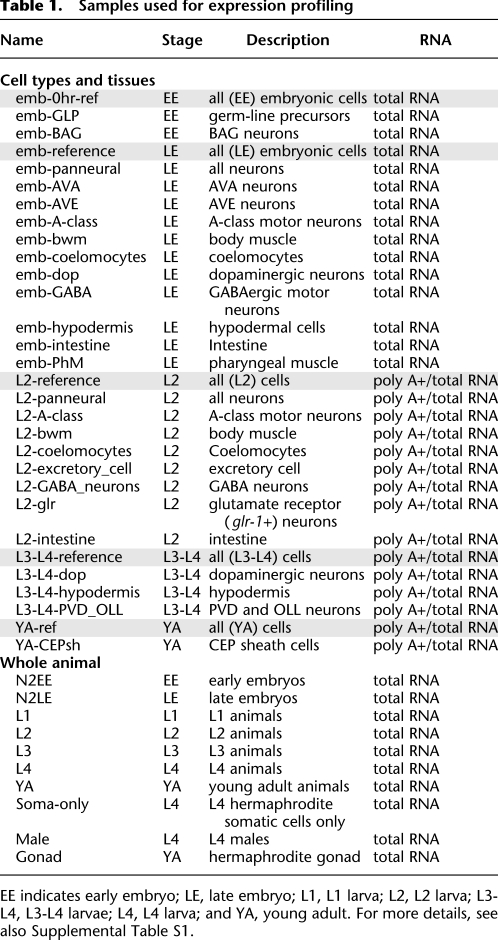
Table 1.
Samples used for expression profiling
EE indicates early embryo; LE, late embryo; L1, L1 larva; L2, L2 larva; L3-L4, L3-L4 larvae; L4, L4 larva; and YA, young adult. For more details, see also Supplemental Table S1.
Expression of annotated genes detected with tiling arrays
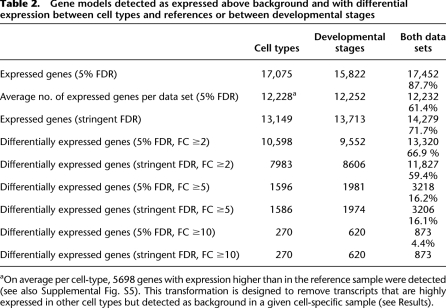
To evaluate expression of annotated protein-coding genes, we created probe sets corresponding to the constitutive exons of individual gene models annotated in WormBase and summarized the intensity values for each gene (see Methods). We then identified transcripts that are detectable above background in each sample with a statistical test (see Methods). The union of results derived from all cell types and stages detected a total of 17,452 genes (Table 2). Because these results were obtained from multiple independent comparisons, we conservatively adjusted the false discovery rates (FDRs) to limit the potential accumulation of false-positives (see Methods), which resulted in expression detected for 13,149 genes from the union of cell type–specific data sets and for 13,713 genes in at least one of the stage-specific data sets. When both groups of data sets were combined, we detected 14,279 expressed genes (Table 2).
Table 2.
Gene models detected as expressed above background and with differential expression between cell types and references or between developmental stages
aOn average per cell-type, 5698 genes with expression higher than in the reference sample were detected (see also Supplemental Fig. S5). This transformation is designed to remove transcripts that are highly expressed in other cell types but detected as background in a given cell-specific sample (see Results).
Our initial analysis identified an average of 12,228 transcripts in samples derived from a specific cell type or tissue (Table 2). To provide a more accurate estimate of genes expressed in each tissue type, we adopted a simple transformation designed to exclude transcripts likely to originate with the minor fraction (3%–20%) of unmarked cells isolated by FACS (Supplemental Table S1) or from nonspecific RNA generated by the mRNA-tagging protocol (Von Stetina et al. 2007). Transcripts that are highly expressed in a specific tissue might also be detectable at lower levels in profiles derived from other cell types due to this background. Thus, as a conservative measure, we restricted the set of expressed genes for each cell type to transcripts with a higher level of expression measured for a given cell type than for the corresponding reference (i.e., the “average” cell) at the same stage. This approach effectively excluded, for example, the myo-3 body muscle–specific transcript (Okkema et al. 1993) from data sets derived from nonmuscle cell types while generally retaining “housekeeping” genes such as ribosomal proteins that are likely to be widely expressed in all tissues (Von Stetina et al. 2007). This analysis detected between 4572 and 7199 expressed genes in each of the 25 cell types profiled (5698 genes on average) (see Supplemental Fig. S5).
Identification of transcriptionally active regions
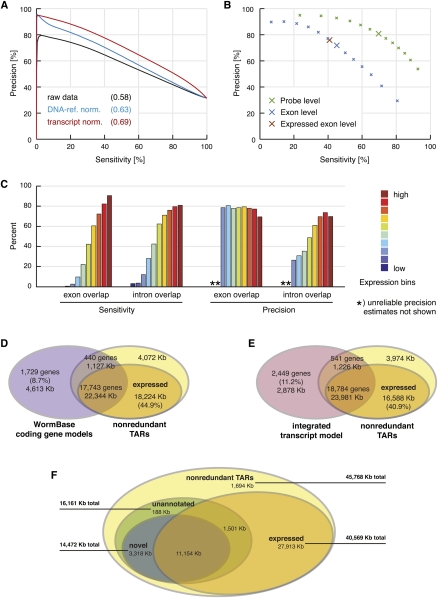
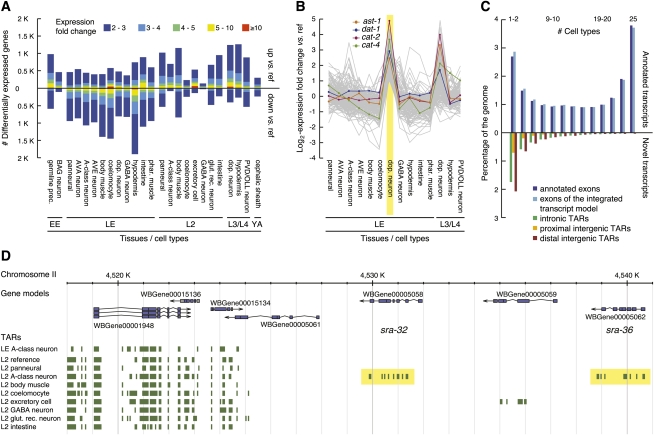
The high probe density (on average every 25 bp) of the tiling array made the nonrepetitive portion (about 85%) of the genome accessible to de novo identification of transcripts in a way that is not biased by potentially incomplete annotations (for an illustration, see Fig. 1D). However, this comprehensive representation of the genome does not allow for optimized probe sequences and consequently results in large variability in hybridization affinity. Our pivotal normalization step thus aimed at reducing probe sequence bias. This correction also improved the signal-to-noise ratio (exon intensity over background) to an even larger extent than observed for another method that additionally exploits reference hybridization to genomic DNA (Fig. 2A; for details, see Methods) (Huber et al. 2006). For the segmentation of hybridization signals into intergenic regions, exons, and introns, we used a method called mSTAD (Laubinger et al. 2008; Zeller et al. 2008). Although mSTAD is trained on hybridization signals corresponding to known (mostly protein-coding) genes (see Methods), it afterward predicts transcripts regardless of their annotation status. We first assessed the cross-validation accuracy of these predictions relative to annotated protein-coding gene models (Fig. 2B). Generally, the sensitivity of these predictions for annotated exons improved with the expression level of the corresponding genes, while high precision (about 80%) with respect to overlap with annotated exons was maintained across all expression levels (Fig. 2C). An additional evaluation of transcripts predicted by mSTAD in comparison to another tiling array-based method (Gerstein et al. 2010) further corroborated the conclusion that mSTAD accurately reconstructs expressed transcripts (Supplemental Figs. S6, S7; Supplemental Result SR4).
Figure 2.
De novo transcript identification with mSTAD and overlap of TARs with annotated and experimentally defined gene models. (A) Transcript normalization (red) improved exon probe recognition over raw data (black) and compared with normalization using genomic DNA hybridization as reference (blue). Sensitivity and precision were estimated after thresholding the intensity data with increasing cutoffs in a fivefold cross-validation. Sensitivity is defined as the percentage of exon probes with signal above the threshold among all annotated exon probes. Precision is defined as the percentage of annotated exon probes among those with signal above the threshold (see Methods). Values in parentheses indicate area under the curve. Based on data from LE-ref (see Table 1). (B) Cross-validation accuracy of mSTAD for probes (green), for exons (blue), and for exons with independently confirmed expression (brown). For exons, sensitivity is defined as the percentage of annotated exons for which all corresponding tiling probes were predicted as exonic by mSTAD. Precision is defined as the percentage of predicted exons for which all probes are annotated as such. Definitions for probes are as in A but with respect to predictions by mSTAD. Exon-level evaluation was repeated with the subset of predicted exons also detected as expressed by a statistical test (see Expressed Exon Level). Enlarged crosses correspond to predictions used for subsequent analysis. Based on data from LE-ref (Table 1). (C) Accuracy of exon and intron recognition increased with gene expression. Colored bars correspond to equally sized expression bins. Here exon overlap sensitivity equals the percentage of predicted exons, which overlap by at least 75% of their length with annotated exons. Exon overlap precision equals the percentage of exon predictions overlapping with annotated exons (by 75% or more) among all predicted exons (intron overlap sensitivity and precision are defined analogously with respect to predicted and annotated introns). Based on data from LE-ref (Table 1). (D) Overlap between nonredundant TARs (nrTARs), the portion detected as expressed and annotated coding gene models. About 45% of expressed nrTAR bases do not overlap with annotated coding gene models. (E) Overlap between TARs and the modENCODE integrated transcript model (Hillier et al. 2009; Gerstein et al. 2010). About 41% of expressed nrTAR bases do not overlap with the integrated transcript model. (F) Unannotated and novel TARs and their overlap with TARs expressed above array background. Unannotated TARs are defined as TARs without significant overlap (≥20 bp) with exons of annotated coding genes, pseudogenes, and ncRNAs. Novel TARs are defined as the subset of unannotated TARs without significant overlap (≥20 bp) with exons in the integrated transcript model (for details, see main text).
TARs map to protein-coding genes and to intergenic domains
From the transcriptionally active region (TAR) predictions of individual samples, we constructed nonredundant TARs (nrTARs) containing the union of nucleotides inclusive to a TAR in any of the samples analyzed (30 cell type and reference samples and seven developmental stages) (Table 1). In total, ∼45 Mb were covered by nrTARs, and the subset of expressed nrTARs (i.e., TARs that passed a statistical test for expression above background, see Methods) contained ∼40 Mb (Supplemental Table S2). We next compared on an individual-nucleotide basis the overlap between known transcripts and nrTARs predicted de novo from the tiling array data. In a comparison to protein-coding gene models annotated in WormBase (Rogers et al. 2008), about 84% of nucleotides (i.e., 22,344 kb ± 1127 kb) in annotated exons (from more than 90% of gene models) were also detected in nrTARs (Fig. 2D). The subset of “expressed” nrTARs covered about 80% of nucleotides in annotated exons from more than 90% of gene models (Supplemental Table S3) and additionally contained >18 Mb (about 45% of expressed nrTARs) outside of exons for annotated protein-coding genes (Fig. 2D). A similar comparison between nrTARs and the modENCODE integrated transcript model defined by transcriptome sequencing of polyadenylated RNA (Hillier et al. 2009) and EST evidence from WormBase (Gerstein et al. 2010) detected ∼25 Mb of overlapping exons or nrTARs corresponding to about 90% of nucleotides in exons of the integrated transcript model and about 57% of nucleotides in nrTARs (Fig. 2E). Nearly 41% of nucleotides within the expressed nrTARs were found outside of exons defined by the integrated transcript model (Fig. 2E). Taken together, most gene models (about 90%) were supported by nrTARs, whereas a substantial fraction of nrTARs could not simply be attributed to known transcripts.
Tiling array analysis detects 11 Mb of novel TARs from intergenic regions
We defined unannotated TARs as those that did not significantly overlap with exons of any coding gene, ncRNA, or pseudogene annotated in WormBase (Supplemental Fig. S8; Rogers et al. 2008). When we additionally required that TARs not overlap with any exons of the integrated transcript model, we obtained “novel TARs” (see Methods). In total, unannotated nrTARs covered ∼16 Mb of the genome; about 90% were also novel (Fig. 2F). Expression above background in any sample could be confirmed by a statistical test (see Methods) for ∼11 Mb of novel nrTARs (Fig. 2F; Supplemental Table S2; Supplemental Fig. S9A). These results suggest that our extensive profiling of cells and tissues as well as developmental stages revealed a significant fraction of the C. elegans transcriptome that went undetected by methods limited by analysis of polyadenylated transcripts or by sampling of fewer conditions (Rogers et al. 2008; Hillier et al. 2009). Our findings parallel results from a previous tiling array study that also detected abundant non-polyA+ transcription from intergenic regions (He et al. 2007). Although our transcript identification method was originally trained on annotated protein-coding genes, it is based purely on hybridization features (Laubinger et al. 2008; Zeller et al. 2008) and hence is expected to be capable of recognizing nonpolyadenylated as well as noncoding transcripts. We verified that TARs identified by mSTAD contain annotated ncRNAs, including snoRNAs, miRNAs, and pseudogenes (Supplemental Fig. S8; Supplemental Table S4). Moreover, on a per-nucleotide basis, almost 60% of the putative long ncRNAs (>2.7 Mb) predicted by Lu et al. (2011) was contained in the set of novel nrTARs described here (Supplemental Fig. S11). However, less than 20% of nucleotides from the novel nrTARs recognized by our approach were also predicted by Lu et al. (2011) (Supplemental Fig. S11). RT-PCR of a subset of these novel TARs confirmed expression (Fig. 3A,B).
Figure 3.
mSTAD detects TARs corresponding to protein-coding genes and to novel transcribed regions. (A) Novel TARs detected in larval L2 intestine. Enlarged region shows location of primers and predicted RT-PCR amplicon from two TARs, L2-int-1 and L2-int-2. (B) RT-PCR detects novel TARs expressed in specific cell types. TARs L2-int-1, 2, 3 are detected in RNA isolated from the larval L2 intestine (L2-int) but are not amplified from RNA in the absence of reverse transcriptase (L2-int-RT). (C) qPCR validates enrichment of novel TARs in specific cell types. Log2 ratio of enrichment in specific tissue versus corresponding reference samples (Table 1).
The majority of C. elegans genes are differentially expressed among cell types and developmental stages
We profiled a broad panel of tissues and developmental stages with the idea that this approach could reveal the prevalence of potential gene regulatory mechanisms that might modulate transcript abundance among different cell types or developmental periods. To detect changes in gene expression during development, we performed all pairwise comparisons (total, 21 comparisons) of the seven tiling array data sets obtained from staged embryos (EE, LE), larvae (L1, L2, L3, L4) and young adults (YA) (see Methods). To detect transcripts that are differentially expressed between cell types, we compared each of the 25 cell-specific data sets to its corresponding reference sample (total, 25 comparisons) (Table 1). In both cases, these comparisons were designed to detect transcripts that are either significantly depleted or enriched (twofold or more, FDR ≤ 0.05) (Fig. 4A). After correcting for multiple testing as above (Methods), this analysis produced conservative estimates of genes differentially expressed between stages (8606 on average) or between specific cell types and reference samples (7983) (Table 2; Supplemental Fig. S12). A combined, corrected threshold of FDR <0.11% for all 46 comparisons yielded 11,827 differentially expressed genes among all stages and cell types tested (Table 2). Although this stringent correction effectively reduced our estimate of the overall number of expressed transcripts from 88% to 72% of all C. elegans genes, the ratio of differentially expressed genes among detected genes in both cases is stable at about 75% (Table 2; Supplemental Fig. S13). On the basis of these results, we conclude that transcripts for a majority of C. elegans genes are regulated to achieve different levels of expression during development and between specific types of cells.
Figure 4.
Transcripts enriched or depleted in certain cell types. (A) Genes differentially expressed between a given cell type and the corresponding reference sample (FDR ≤0.05). Bars pointing up and down indicate the number of enriched and depleted genes, respectively, relative to reference. Expression fold change is color-coded (see key). (B) Log2-expression fold change relative to reference shown as gray lines for genes selectively enriched in LE dopaminergic neurons (highlighted in yellow). Four selectively enriched genes (ast-1, dat-1, cat-2, cat-4) with known function in these neurons are plotted in color (see key). (C) Coverage of the genome by expressed transcripts at base-pair resolution. Nucleotides in nonredundant TARs (nrTARs) (for 25 cell-type samples) (Table 1) were binned according to the number of samples for which a TAR was detected at the given position. Bars pointing upward correspond to expressed TARs overlapping with exons of annotated coding genes and those defined by the integrated transcript model. Bars pointing downward correspond to nucleotides in expressed novel TARs (for definition, see main text) organized into subgroups according to their location relative to annotated protein coding gene models (see key). Intergenic positions were classified as proximal if within 500 bp of any annotated gene and otherwise as distal. (D) 7TM genes are selectively expressed in a specific neuron. Two members (sra-32 and sra-36) of a tandem array (yellow highlights) of 7TM-encoding genes are selectively enriched in the A-type motor neuron data set derived from L2 larvae.
To validate the enrichment of genes in tissues and cell types as detected here, we compared a select number of our enriched gene lists to similar, independently derived data sets. Comparison of enriched gene sets from this study for the L2 excretory cell, embryonic and L2 intestine, L2 body wall muscle, and YA gonad with comparable tissue-specific profiles generated by other groups produced highly significant overlaps (1.8- to 8.1-fold overrepresentations, all hypergeometric P-values <1 × 10−15) (see Supplemental Results SR1). These comparisons reinforce the validity of each data set, particularly since the earlier profiles were generated with a variety of methods including GFP reporter imaging and serial analysis of gene expression (SAGE) (see Supplemental Results SR1) and also may differ from our samples in developmental age. Lists of cell or tissue-enriched transcripts are included as supplemental data (http://www.vanderbilt.edu/wormdoc/wormmap/) and should be useful for identifying genes with key roles in the corresponding cell type (see also Supplemental Results SR2). Examples of this application are provided for the embryonic germ cell precursors (see below) and for the larval excretory cell (Supplemental Result SR3).
Specific genes are selectively enriched in certain cell types or tissues
Among the genes that are enriched in a certain tissue, we further sought to distinguish genes that are selectively enriched in the given tissue relative to those with broadly elevated expression in many cell types. The information theoretic concept of Shannon entropy effectively allowed us to define this subset of selectively enriched genes by distinguishing patterns of broad and uniform expression (high entropy) from more restricted ones with a high degree of tissue specificity (low entropy) (see Methods; Schug et al. 2005). These lists of selectively enriched genes comprised about 20%–57% of all genes enriched in the corresponding tissue or cell type. Eighty-two percent of all genes selectively enriched in any cell type or tissue are specific to only one or two samples. Moreover, the overlap between tissues sampled at two time points is generally larger than between different tissues or cell types (Supplemental Fig. S14). For example, the set of genes selectively enriched in embryonic dopaminergic neurons also shows elevated expression in larval dopaminergic neurons and comprises known dopaminergic genes, including the ETS transcription factor, ast-1, and its downstream targets the dopamine transporter dat-1 and dopamine biosynthetic enzymes cat-2 and cat-4. (Fig. 4B; Supplemental Result SR2; Flames and Hobert 2009).
We further defined the set of genes selectively enriched in any of the 13 neuronal samples, but not enriched in non-neuronal tissue. Strikingly, this combined neuron-selective data set is most strongly enriched for putative 7-transmembrane (7TM) domain G-protein–coupled receptor (GPCR)-like proteins (FDR < 5.2 × 10−25) (Supplemental Fig. S15)
Our finding is consistent with earlier reports of selective expression of 7TM/GPCR genes in the C. elegans nervous system (Troemel et al. 1995; Chen et al. 2005). Cases of 7TM/GPCR genes that are expressed in specific neurons are also evident in our tiling array results. For example, sra-32 and sra-36 are uniquely detected in the L2 larval stage A-class neuron data set (Fig. 4D). Of the 1512 predicted members of the 7TM/GPCR family, 314 (about 21%) were not detected in any RNA-Seq–derived data set produced from whole animals (Hillier et al. 2009) for the modENCODE consortium (Gerstein et al. 2010). Among these are 66 family members that are detected in our tiling array assays (Supplemental File 1 7TM genes). Our findings provide an explanation for the relative lack of coverage of the 7TM/GPCR family in the RNA-Seq results and predict that transcripts encoded by other members of this large and diverse gene family could be detected by expanding our cell-specific profiling strategy to additional neuron types (Supplemental Fig. S16).
Novel TARs are differentially expressed and many are selectively detected in certain cell types
To quantitatively assess expression differences for TARs, in particular novel ones, probes contained within TARs from each cell-specific data set were compared to probes from the same region in the corresponding reference sample (see Table 1). This analysis revealed ∼5 Mb of TARs with significant expression changes between cell types and references or between developmental stages at an FDR ≤0.05 and twofold expression difference (see Methods; Supplemental Figs. S9B, S10). On average, 933 novel TARs are differentially expressed in a particular cell type in comparison to a reference sample of all cells at the corresponding developmental stage (Supplemental Figs. S9C, S12C). We used quantitative PCR (qPCR) to confirm that 10 of these differentially expressed novel TARs indeed show significant enrichment in the specific cell types initially identified in the comparison of tiling array results (Fig. 3C).
We next explored the extent to which novel TARs are selectively expressed with the goal of cataloging potentially rare transcripts that might be specifically detected in a limited subset of cells or in a discrete developmental period. To investigate the expression patterns of TARs on a per-nucleotide basis, we tabulated the frequency at which a given base was detected as transcribed across cell types and stages. Approximately 15% of bases covered by exons of gene models annotated in WormBase are detected in all 25 cell types profiled (Fig. 4C). A larger fraction of bases derived from gene models (about 75%), however, is expressed in at least one, but not all, of the cell types, and 11% is detected in no more than two cell types. For stages, we observed that 29% of bases from exons of annotated gene models is detected throughout development versus 8% expressed in no more than one developmental period (Supplemental Fig. S17). Bases corresponding to novel TARs that map to intergenic regions showed a stronger bias for cell- or stage-specific expression with more than 53% detected in either one or two cell types but less than 1% (∼34 kb) detected in all cell types (Fig. 4C). Bases located >500 nucleotides (nt) from a gene model (distal) comprised the majority (about 75%) of novel transcribed intergenic nucleotides uniquely detected in one or two cell types or in a single stage (Fig. 4C; Supplemental Fig. S17). Given the average intron length of 344 nt for C. elegans (Bradnam and Korf 2008), we suggest that these distal bases are more likely to correspond to new transcribed regions as opposed to exons belonging to existing gene models.
Online resources for visualization and data access
To facilitate further study of our tiling array-based expression data, we have made it accessible to the research community through two online visualization tools, both of which are linked from a project website (http://www.vanderbilt.edu/wormdoc/wormmap/). One of the utilities displays expression values across cell types and developmental stages for a user-defined subset of genes (http://fml.mpg.de/raetsch/wormviz/tileviz.jsp; see also Supplemental Fig. S18). Additionally, a customized genome browser (http://gbrowse.fml.mpg.de/cgi-bin/gbrowse/ce_WS199) visualizes TARs for all analyzed samples together with gene models and genomic features annotated in WormBase (see Supplemental Fig. S7; Rogers et al. 2008). Raw array data have been deposited at GEO (Barrett et al. 2009) (accession nos. GSE23245–GSE23271, GSE23278–GSE23287, GSE23769–GSE23770, and GSE25350–GSE25351), and supplemental data files are available for download from the project website (http://www.vanderbilt.edu/wormdoc/wormmap/) and from modMine (http://intermine.modencode.org).
Analysis of differentially expressed transcripts reveals cell-specific functions and clusters of co-regulated genes with candidate cis-acting motifs
Our quantitative analysis has identified transcripts that are differentially expressed across a broad array of cell types and developmental stages. We expect that these results will provide a useful resource for future studies of cell-specific gene function and for identifying the regulatory elements that define spatial and temporal patterns of gene expression. Below we feature examples of these approaches in order to illustrate potential applications of these data sets.
Cell-specific expression profiling of embryonic primordial germ cells reveals X-chromosome silencing and early activation of meiosis and oogenesis genes
Two primordial germ cells, Z2 and Z3, are born from the P4 blastomere within 2 h of fertilization and ultimately give rise to all germ cells in the adult (Sulston et al. 1983). Z2 and Z3 are mitotically dormant and largely transcriptionally quiescent throughout embryogenesis, even as somatic cells rapidly proliferate and actively transcribe genes. Although some genes are known to be expressed in Z2/Z3 in older embryos (e.g., nos-2 and pgl-1) (Kawasaki et al. 1998, 2004; Subramaniam and Seydoux 1999), the full complement of genes actively transcribed in Z2/Z3 during this period has not been previously defined. Comparison of the Z2/Z3 expression data set to a reference profile obtained from all embryonic cells (EE) (Table 1) identified 979 genes with significantly enriched (twofold or more, FDR ≤ 5%) expression in Z2/Z3 (Supplemental File Z2/Z3 #2). Because the germline reporter (Ppie-1::GFP::PGL-1) (Table 1) used for marking Z2/Z3 for FACS shows expression in hypodermal cells of older embryos (data not shown; see also Supplemental Fig. S14), we removed 335 genes from the Z2/Z3 list that were also enriched in an independent profile of embryonic hypodermal cells (Table 1, see Supplemental Protocol SP13). We refer to the resultant list of 644 genes as the “core Z2/Z3” data set (Supplemental File Z2/Z3 #2). This stringent treatment effectively excludes hypodermal genes from the core Z2/Z3 list but also eliminates other transcripts that are known to serve roles in germline development and are in fact expressed in Z2/Z3 (i.e., deps-1, ima-2, pgl-1, htp-3, cpg-2, daz-1) (Fig. 5, Supplemental Table S5).
Figure 5.
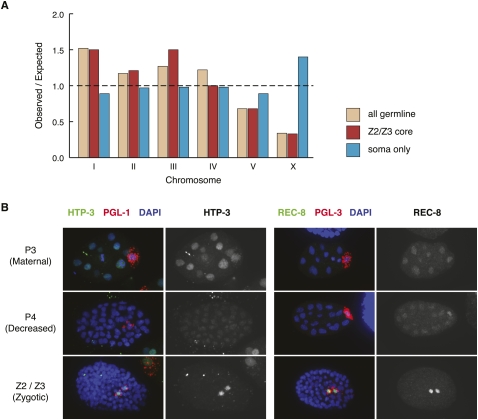
Global characteristics of Z2/Z3-enriched gene expression. (A) Z2/Z3-expressed genes are underrepresented on the X chromosome. The chromosomal locations of genes expressed in the adult germline (“all germline”) (Reinke et al. 2004), genes identified from tiling array profiles of Z2/Z3 minus hypodermis-expressed genes (“Z2/Z3 core”, see text), and genes identified from tiling array profiles of adult hermaphrodite somatic cells (“soma-only”) were determined. Observed/expected was calculated by comparing the number of genes per chromosome from each data set to the entire number of protein-coding genes on that chromosome. (B) Meiotic proteins are expressed in Z2/Z3. Antibodies to HTP-3 and REC-8 were used to stain mixed stage embryos, along with PGL-1 or PGL-3 to mark P granules in Z2/Z3. Stage of germline development is indicated on the left, with the earliest stage shown (P2/P3) likely showing persistence of maternal protein, the P4 stage showing decreased protein in the P4 cell, and the Z2/Z3 stage showing an increase in protein level in Z2 and Z3.
Two observations indicate that the core Z2/Z3 data set is strongly enriched for authentic Z2/Z3 transcripts. First, we detected significant overlap between the core Z2/Z3 data set and previously defined sets of transcripts produced in the adult germline (Reinke et al. 2000, 2004) and maternally loaded into embryos (397 of 644 Z2/Z3 genes, P < 8 × 10−72, hypergeometric test, see Methods) (Baugh et al. 2003). The core Z2/Z3 data set also significantly overlaps with a previously generated germline SAGE enriched gene data set (125 of 644 Z2/Z3 genes, P < 1.62 × 10−34) (Wang et al. 2009). Second, X-linked genes are as underrepresented in the core Z2/Z3 data set as they are in the adult germline (Fig. 5A; Supplemental File Z2/Z3 #2; Reinke et al. 2004). This result provides the first evidence that the X chromosomes are underexpressed in primordial germ cells, as has been previously documented in larval and adult germ cells. The specificity of this effect for germ cells is underscored by tiling array results obtained from adult hermaphrodite somatic cells (“soma only,” Fig. 5A), which instead show overrepresentation of X-linked transcripts.
Strikingly, many genes expressed in Z2/Z3 have known roles in germ cell proliferation, meiosis, and oocyte differentiation (Supplemental Table S5; Supplemental Fig. S19) events that do not take place in the germline until larval and adult stages (Kimble and Crittenden 2005). In contrast, genes expressed during spermatogenesis are underrepresented in the Z2/Z3 data set (Supplemental File Z2/Z3 #2). This observation suggests that the gene expression program for oocyte differentiation is activated in the primordial germ cells, whereas the program for spermatogenesis remains quiescent, even though spermatogenesis occurs prior to oogenesis in C. elegans hermaphrodites (Hubbard and Gerstein 2005; Shakes et al. 2009). Alternatively, many of the RNAs detected in this analysis could reflect the persistence of maternally provided transcripts in Z2 and Z3. To distinguish between maternal loading and zygotic transcription, we manually examined existing in situ hybridization images (NextDB; http://nematode.lab.nig.ac.jp) for genes in the core Z2/Z3 list (Supplemental Table S6). We found that 14 of 100 randomly selected genes in the core Z2/Z3 list show in situ signal in PGCs in embryos and/or L1s, compared with three of 100 randomly selected genes in the genome (P = 0.0047, Fisher's exact test). Of the 14 genes from the core Z2/Z3 list, 12 genes show new appearance of transcript signal in Z2/Z3 after a previously negative stage (i.e., P4 or an early stage of Z2/Z3 lacked signal) (prg-1, glh-1, ppw-2, lsl-1, pas-5, asb-1, iff-1, ucr-2.3, cpg-1, ife-3, rpl-11.1, hil-4). Of those 12 genes, 10 show signal in Z2/Z3 in newly hatched first stage larvae (L1s), and six show signal in embryos. Of the three genes from the control list, all three show signal in Z2/Z3 in L1s (top-2, cra-1, mel-46), and none show signal in Z2/Z3 in embryos. These observations confirm that at least a subset of transcripts detected in the core Z2/Z3 data set are products of zygotic transcription in the primordial germ cells of embryos.
Transcription in Z2/Z3 of genes with protein products known to act later in development raised the question of whether zygotically expressed transcripts are translated in Z2/Z3. We addressed this possibility by immunostaining embryos with antibodies against several meiotic proteins. Strikingly, immunostaining for both HTP-3 and REC-8 (Pasierbek et al. 2001; Goodyer et al. 2008) is easily detected in embryonic Z2/Z3. Both proteins appear to be maternally loaded in early embryos, to diminish significantly by the P4 stage, and then to turn on in Z2/Z3 (Fig. 5B). Thus, at least some of the zygotically expressed transcripts are indeed translated in the primordial germ cells of embryos. This finding confirms the validity of our Z2/Z3 transcription profiling and provides a clear example of how these data can lead to new discoveries about germ cell biology.
Self-organizing maps reveal cohorts of co-regulated genes during development and across specific cell types
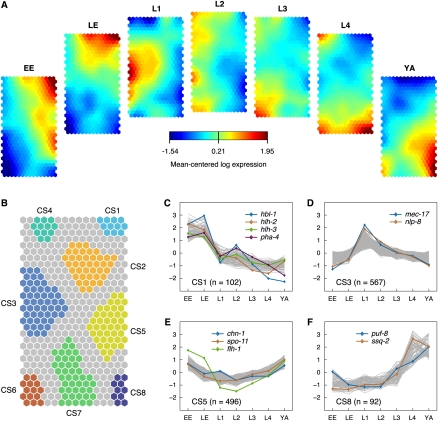
We used self-organizing maps (SOMs) to seek shared patterns of expression for transcripts derived from coding genes (see Methods). SOMs are a widely applied clustering technique that yields intuitive visualization of high-dimensional data sets, as, for example, generated with DNA microarrays (Jiang et al. 2001). SOMs are conceptually related to a technique previously proposed to construct a relational map of C. elegans gene expression (Kim et al. 2001). In the first instance, we fitted a SOM to the developmental stage data set (Fig. 6A) and identified eight regions that correspond to genes with shared patterns of either enrichment or depletion in specific developmental periods (Fig. 6B, see Methods). To demonstrate the variety of developmental expression patterns identified by this approach, we plotted the top 50% of best-fitting genes from each cluster (Fig. 6C–F; for additional clusters, see Supplemental Fig. S20). Cluster 1 (CS1) contains genes with elevated expression in the embryo (Fig. 6B,C). Notable examples from this group include the FoxA transcription factor, pha-4, the hunchback homolog, hbl-1, (Krause et al. 1997) and the helix-loop-helix transcription factors (bHLH) hlh-2 and hlh-3, for which independent studies have detected peak expression in the embryo (Azzaria et al. 1996; Krause et al. 1997; Fay et al. 1999). Cluster 5 (CS5) contains genes with elevated expression in embryonic stages and in the adult (Fig. 6B,E). Strikingly similar protein and transcript levels have been previously observed for a member of this group, the FLYWCH transcription factor flh-1, which blocks expression of specific miRNA genes during embryogenesis (Ow et al. 2008).
Figure 6.
Expression patterns during C. elegans development. (A) Component planes of a self-organizing map (SOM) fitted to the developmental stage data set. Each component plane visualizes mean-centered gene expression (log2-scale) in one stage as a color gradient from blue to red indicating low and high expression, respectively (see color bar): EE indicates early embryos; LE, late embryos; L1, larvae stage 1; L2, larvae stage 2; L3, larvae stage 3; L4, larvae stage 4; and YA, young adults. (B) Eight regions (CS1–CS8) of the SOM, which robustly clustered together, are color-coded (see main text for details). (C–F) Mean-centered log2-expression values of genes corresponding to four of the clusters in B are plotted for the 50% of best-fitting genes (additional clusters in Supplemental Fig. S20). Colored lines indicate the expression of a selected subset of genes (see key). mec-17 and nlp-8 encode neuron-enriched transcripts; chn-1 and spo-11 are highly expressed in the adult hermaphrodite gonad; puf-8 is highly expressed in embryonic and adult germline; and ssq-2 encodes a sperm-specific transcript. For other labeled genes, see Results.
We applied a similar SOM clustering procedure to the cell type–specific data sets in order to delineate genes that are coregulated in different tissues (Fig. 7A; Supplemental Fig. S21; see Methods). Because these cell types were sampled across a series of developmental stages, we also expected this approach to detect genes with temporally correlated expression. Figure 7A depicts the resultant regional clusters superimposed on the SOM. Clusters showing stage-specific expression include C1 (Fig. 7B), which features genes that are highly expressed in all postembryonic cell types and C7 (Fig. 7D), which is biased for genes expressed in late embryos and especially in neurons. C8 is dominated by genes that are highly expressed in neurons but are depleted or show weak expression in most other cell types (Fig. 7E). Examples of genes in this group include ric-4 (snap-25), a synaptic vesicle component that facilitates neurotransmitter release and is known to be exclusively expressed in neurons (Hwang and Lee 2003), and acy-1 (adenylate cyclase), a key regulator of neuron-dependent behavior (Reynolds et al. 2005). Several clusters detect highly expressed intestinal genes including C1 (post-embryonic cell types and larval intestine) (Fig. 7B) and C11 (embryonic and larval intestine) (Fig. 7F; for additional examples, see Supplemental Fig. S22).
Figure 7.
SOM clustering of tissue- and cell-type data. (A) Regions in the SOM for cell-type data as defined by k-means clustering. (B–F) Expression patterns of genes from selected clusters in the SOM. Cell types are indicated at bottom, and reference samples are shaded in gray. Box plots were generated from the mean-centered log2-expression values of prototypical genes for a given cell type with horizontal lines indicating the median, boxes delineating the interquartile range, and whiskers extending to the most extreme values within 1.5 times the median-quartile range; outliers are depicted as black crosses. Some of the SOM clusters correspond to peaked expression in a subset of cell types and/or developmental stages: (B) higher expression in larval stages and YA compared with embryo with a prominent peak for intestine, (C) elevated expression in L3/L4 reference, (D) high expression in LE neurons, (E) most neurons, and (F) intestine (for SOM component planes and additional clusters, see Supplemental Figs. S22 and S23).
DNA sequence motifs associated with cell-specific and developmentally regulated gene expression
Because each SOM cluster includes genes with similar patterns of expression, we searched for instances in which genes in a specific cluster share common DNA sequence motifs through which trans-acting factors might coordinate their expression. To explore this possibility, we applied the FIRE motif analysis program to the SOM clusters (Elemento et al. 2007). FIRE uses mutual information between the presence or absence of a short nucleotide sequence and the occurrence of a gene in a particular expression cluster to identify overrepresented motifs. FIRE produces optimized motifs and links the results to motifs that are available in public databases.
FIRE identified 20 upstream promoter motifs and nine overrepresented 3′ UTR sequences in genes contained in the SOM clusters derived from developmental stages (Figs. 6, 8A; Supplemental Figs. S23, S24). A canonical EBox and bHLH binding site is detected in cluster CS1, which, as noted above, includes transcription factors HLH-2 and HLH-3 (Thellmann et al. 2003). The overrepresentation of GATA-like transcription factor binding sites in four clusters (CS4, CS5, CS6, CS7) is likely indicative of the broad roles of GATA factors in multiple developmental pathways in C. elegans, including endodermal and hypodermal cell fate determination and differentiation, germline gene regulation, and aging (Koh and Rothman 2001; McGhee et al. 2007; Budovskaya et al. 2008; del Castillo-Olivares et al. 2009). The second highest-ranking motif corresponds to a GC-rich sequence that has been previously identified by computational analysis of germline expressed genes (Li et al. 2010). This motif is also similar to a putative transcriptional activation site for the E2F homolog, EFL-1, that promotes gene expression in the germline (Chi and Reinke 2006). Detection of these GATA and E2F sites in cluster CS5 is consistent with our finding that genes which contain these 5′ sites and which are enriched in germline precursor (GLP) cells (Supplemental File Z2/Z3 #2) are also overrepresented in this cluster (23 GLP genes with the GATA site are 1.6-fold overrepresented, P < 0.017; 40 GLP genes with the E2F site are 1.8-fold overrepresented, P < 2.74 × 10−4). These results validate our approach and suggest that other motifs revealed by this strategy may also correspond to binding sites for transcription factors that regulate developmental gene expression.
For SOM clusters derived from cell-specific profiles (Fig. 8B), FIRE identified 45 overrepresented sequences including 35 upstream motifs and 10 RNA sequences that map to 3′ UTR domains (Fig. 8B; Supplemental Figs. S23, S24). As noted above for the SOM clusters derived from developmental stages, the highest scoring motif matches a GATA transcription factor-binding site. In C. elegans, the elt-2 GATA transcription factor is known to interact with this sequence to drive expression of intestine-specific genes (McGhee et al. 2009). Our results also reflect this role; the GATA motif is overrepresented in cluster C11, which contains transcripts enriched in the embryonic and larval intestine profiles (Figs. 7F, 8B), and in C1 and C4, both of which show peak expression in larval intestine (Fig. 7B, 8B; Supplemental Fig. S22). The accurate identification of the GATA factor-binding site by the FIRE algorithm suggests that other motifs associated with specific SOM clusters may also correspond to specific transcription factor binding sites. An interesting example includes the sequence TTTCG[TC]AA[CT] (Fig. 8B), which is overrepresented in genes enriched in embryonic neurons in cluster C7 (Fig. 7D) and also reciprocally depleted in genes that are underexpressed in embryonic neurons in cluster C5 (Supplemental Fig. S22). This motif is bound by the vertebrate C/EBPalpha transcription factor (Grange et al. 1991), which has been shown to function with NeuroD to regulate neural gene expression (Sandelin et al. 2004; Calella et al. 2007). It will be interesting to determine whether C/EBP and NeuroD homologs exercise similar functions in C. elegans neural development.
Figure 8.
Selected regulatory elements discovered in stage and cell-type expression clusters. FIRE analysis identifies motifs over- and underrepresented in developmental profile clusters (A) and cell-type profile clusters (B). Complete results are shown in Supplemental Figure S23. A heat map indicates whether each motif is overrepresented (yellow) or underrepresented (blue) in each cluster. Motifs are arranged in rows and clusters in columns. Significant overrepresentation is indicated by red box outlines and underrepresentation is indicated by blue outlines (P ≤ 0.05, Bonferroni-corrected). The optimized motif logo, location of the motif (5′ upstream promoter or 3′ UTR), mutual information with the genes in the cluster, and matching transcription factors and miRNAs listed in public data bases for indicated motifs are shown alongside the heat map.
FIRE also identified 3′ UTR binding sites for two distinct groups of miRNA genes belonging to the mir-58 and mir-51 families (Fig. 8B; Supplemental Figs. S23, S24). Members of the mir-58 family (mir-58,-80,-81,-82) are abundantly expressed throughout development (Lim et al. 2003; Kato et al. 2009), but assays with promoter::GFP reporter genes have detected cell-specific patterns of expression (Martinez et al. 2008). For instance, mir-58 is expressed in a broad array of cell types, including the excretory canal, intestine, pharynx, and hypodermis, but is excluded from the nervous system (Isik et al. 2010). Emerging evidence indicates that transcript destabilization is the principle mechanism whereby miRNAs down-regulate gene expression (Bagga et al. 2005; Guo et al. 2010). Thus, the absence of mir-58 expression in the nervous system predicts that neuronal transcripts carrying the mir-58 recognition sequence should escape mir-58-induced degradation. And, in fact, our result showing that the mir-58 sequence is overrepresented in neuron-enriched transcripts (cluster C8) (Figs. 7E, 8B) is consistent with this model. The motif for the mir-51 family (mir-51,-52,-53,-54,-55,-56) is also overrepresented in C8 (Figs. 7E, 8B) and in SOM clusters C9 and C10 that are dominated by transcripts enriched in hypodermal cells and neurons (Supplemental Fig. S22). This pattern suggests that mir-51 family genes may have limited roles in regulating transcript levels in neurons and in the hypodermis. Conversely, the observation that the mir-58 and mir-51 motifs are significantly underrepresented in C2 and C3 is suggestive of strong regulation by these miRNAs in the tissues that contribute to this cluster. In considering this question, we noted that C2 and C3 include an expression peak for the L3/L4 reference sample (Fig. 7C). Because germline tissue is rapidly proliferating at this stage (Kimble 1981), we compared the genes in clusters C2 and C3 to separate tiling array profiles obtained from the adult hermaphrodite gonad and L4 males at L4 stage (see Supplemental Result SR5). These comparisons show a significant overlap, thereby indicating that C2 and C3 genes are largely expressed in the germline and specifically enriched for sperm expression. Thus, we speculate that members of the mir-58 and mir-51 gene families may have significant roles in modulating transcript levels in the germline. The mir-58 and mir-51 motifs were previously identified by FIRE analysis of an independent group of whole-animal microarray data sets from C. elegans (Elemento et al. 2007). Our results have now confirmed that each of these motifs is associated with a group of co-regulated genes and have also provided additional clues pointing to the specific cell types in which mir-51 and mir-58 might function.
Discussion
We have used whole-genome tiling arrays to profile RNA isolated from specific cells and developmental stages of C. elegans. Our strategy of sampling a variety of different cell types and developmental periods was designed to capture potentially rare or transiently expressed transcripts as well as to provide a detailed spatial and temporal map of gene expression.
To monitor expression of individual protein-coding genes, we derived intensity values from aggregated probe sequences corresponding to each annotated gene model. Our combined set of tiling array data from 25 different cell types and seven developmental stages (Fig. 1) detected about 90% of known protein coding genes (Table 2; Fig. 2; Supplemental Table S3).
In addition to detecting expressed genes, our analysis also revealed that about 75% of all detected genes show at least twofold, statistically significant differences in transcript levels between cell types or developmental stages (Table 2; Supplemental Fig. S13). To document this trend of widespread differential expression among cell types and throughout development, we tabulated the frequency of transcription of a given nucleotide across tissues and stages. This analysis revealed that whereas 15% of exonic sequence is detected in all of the cell types that we sampled, a larger fraction (60%) shows more limited transcription with about 11% in no more than one or two cell types (Fig. 4C). Our results also indicate that coding sequence is dynamically expressed during development with about 8% of bases from exons uniquely detected in only one embryonic, larval or adult stage (Supplemental Fig. S17). As we extensively sampled the C. elegans nervous system, we investigated the subset of genes selectively expressed in neuronal tissue. Among these genes, we noted striking enrichment of members of the 7TM-GPCR family, which is known for highly specific expression in the nervous system (Chen et al. 2005). The restricted expression of 7TM-GPCR genes potentially explains why many members of this family still lack experimental support (Hillier et al. 2009; Schweikert et al. 2009). Our results, however, suggest that profiles of more cell types should confirm expression of additional annotated gene models (see Supplemental Fig. S16) or genes newly predicted from the genome sequence. Overall, our finding of widespread differential gene expression underscores the conclusion that most C. elegans genes are extensively regulated and points to the key role of differential gene expression in the determination of cell fates and developmental progression. In practice, our data on genes that are selectively enriched in a particular cell type or developmental period should be especially useful for identifying genes with cell- or stage-specific functions (Zhang et al. 2002; Colosimo et al. 2004; Blacque et al. 2005; Cinar et al. 2005; Touroutine et al. 2005; Von Stetina et al. 2007; McGhee et al. 2009; Chatzigeorgiou et al. 2010; Smith et al. 2010; Hallem et al. 2010). For example, our analysis of the transcripts enriched in the primordial germ cells (Z2/Z3) revealed two new features of germline development. First, compared to the autosomes, relatively few X-linked genes are expressed in C. elegans primordial germ cells (Fig. 5A). This embryonic silencing of X-linked genes mirrors a similar effect in larval and adult germ cells (Reinke et al. 2004). Thus, except for a brief period of activation of X-linked genes in oocytes, the X chromosome is underexpressed throughout all stages of germ cell development. Second, Z2/Z3 express genes involved in meiosis and oogenesis, well in advance of when those processes actually occur. Moreover, at least some of those transcripts are translated into protein (Fig. 5B). This result raises the interesting question of whether those proteins serve additional nonmeiosis and nonoogenesis roles or instead whether primordial germ cells express many more of their repertoire of gene products than they need at that stage. Notably, the primordial germ cells of Drosophila and mice also express meiosis proteins, indicating that early expression of this protein class is conserved (Baltus et al. 2006; Mukai et al. 2006; Rogers et al. 2008). More detailed investigation of the individual genes expressed in Z2/Z3 should provide a wealth of new information about the processes occurring in primordial germ cells, including mitotic quiescence and chromatin regulation. As one example, cyclin B is important for mitotic quiescence of germ cells in other systems such as Drosophila (Deshpande et al. 1999). Of the several orthologs of cyclin B in the C. elegans genome, only cyb-2.2 is enriched in the Z2/Z3 data set, making it a candidate to mediate mitotic quiescence in Z2/Z3.
In addition to using our tiling array results to identify genes expressed in specific cell types or developmental periods, we also sought evidence for more complex patterns in which cohorts of genes might be similarly regulated across tissues or among different developmental stages. For this purpose, we used the unbiased strategy of SOMs to cluster coexpressed genes (Figs. 6, 7). This approach revealed, for example, a striking cluster with consistently elevated transcript levels in both embryonic and larval neurons that is largely composed of genes with established neuron-specific functions (Fig. 7E). Other clusters could reflect genes with common functions in a wide array of cell types during a particular developmental period (Fig. 7B,D). Thus, our approach has confirmed known groups of coregulated genes as well as suggested novel clusters that could point to previously unstudied biological roles for batteries of coexpressed genes. In addition to providing a direct read-out of cell-specific gene expression, our microarray data should also substantially enhance the accuracy of SVM-based strategies that rely on gold standard training sets for ab initio identification of cell-specific expression from whole-animal microarray data (Chikina et al. 2009). Motif analysis of the SOM results derived from our data sets identified highly overrepresented flanking sequences in genes belonging to specific clusters (Fig. 8; Supplemental Fig. S23). Each case could be indicative of a regulatory mechanism involving a shared trans-acting factor. For example, a consensus binding site for a GATA factor with a broad role in regulating intestine-specific genes in C. elegans (McGhee et al. 2009) was specifically overrepresented in SOM clusters defined by transcripts with high expression levels in tiling array data sets derived from intestinal cells (Fig. 8B). Overrepresented motifs in the 3′ UTR regions include recognition sites for two large and highly expressed groups of closely related miRNAs, the mir-58 and mir-51 families (Fig. 8B). Our analysis of these results points to potential roles for both mir-58 and mir-51 in regulating transcript abundance in the germline, a suggestion consistent with the recent observation that the Drosophila ortholog of the mir-58 family, bantam, is required for germline stem cell fate (Yang et al. 2009).
Our tiling array results confirm expression of the vast majority (about 90%) of C. elegans protein coding genes recently identified by RNA-Seq analysis (Supplemental Table S3; Hillier et al. 2009). Additionally, our machine-learning algorithm also identified a substantial number of TARs arising from intergenic regions (Fig. 2D–F). A conservative treatment of these data that uses a statistical test for expression above background leads to the estimate that ∼11 Mb of intergenic sequence, or about 10% of the C. elegans genome, encodes novel RNAs that have not been previously annotated in WormBase or detected by RNA-Seq (Supplemental Table S2). One explanation for this difference is that we assayed total RNA from embryonic cells and developmental stages and that the poly-A+ pull-downs that we used for sampling postembryonic cell types (Fig. 1B) also include a significant non-polyA+ fraction (Von Stetina et al. 2007). In contrast, recent RNA-Seq results for C. elegans were limited to purified poly-A+ RNA (Hillier et al. 2009). Because the known families of short ncRNAs were manually excluded from our list of intergenic RNAs, we propose that these transcripts define potentially new types of ncRNA. An independent analysis of C. elegans transcriptomics data that includes the tiling array results used in this work also reports a substantial number (∼4.6 Mb) of putative ncRNAs from intergenic regions with a large overlap (>2.5 Mb) to our ncRNA predictions (Supplemental Fig. S11; Lu et al. 2011). Our analysis indicates that transcription of these novel TARs shows an even stronger bias for cell-specific expression than transcripts derived from protein coding genes (Fig. 4C). In this respect, our findings are similar to an earlier report that a majority of unannotated human transcripts are expressed in only one of the 11 different cell lines sampled (Birney et al. 2007). Although the extent of intergenic transcription from the mammalian genome is controversial (van Bakel et al. 2010), mounting evidence points to multifaceted roles for long intergenic ncRNAs (lincRNAs), including transcriptional control, imprinting, dosage compensation and maintenance, and remodeling of chromatin structure (Rinn et al. 2007; Hirota et al. 2008; Wilusz et al. 2009; Tsai et al. 2010). Nevertheless, in every case, definitive tests are required to establish specific functions for candidate regulatory ncRNAs. The tissue-specific patterns of ncRNA expression (Fig. 4C; Supplemental Fig. S12C) that we have revealed for C. elegans should provide a valuable guide to the likely focus of mutant phenotypes that perturb expression of specific ncRNAs (Mercer et al. 2008). We note, for example, that the recent discovery of an in vivo role for the lincRNA, Evf2 (also known as Dlx6os1), in neuronal differentiation hinged on prior knowledge of Evf2 expression in a specific brain region (Bond et al. 2009).
Although the tiling array results reported here should provide a useful resource for defining the roles of specific genes in cell fate and development, RNA-Seq data derived from these cell specific RNA samples would offer a more accurate representation of gene structure and substantially greater dynamic range for measuring differential gene expression. With the recent development of effective methods for excluding ribosomal RNA from sequencing templates (Armour et al. 2009; Albrecht et al. 2010), it should now be feasible to use RNA-Seq for a direct test of the ncRNA transcripts predicted by our tiling array results (W.C. Spencer and D.M. Miller, unpubl.). The fact that cell-specific tiling arrays detected predicted coding genes that were not touched by RNA-Seq analysis of C. elegans transcripts derived from the whole animal, also suggests that deep sequencing of RNA isolated from individual cell types could reveal additional protein-coding genes (see Supplemental Fig. S16).
Methods
Sample production
Nematode culture
C. elegans strains were maintained as described (Brenner 1974). We used N2 as the wild-type strain. Other strains used in this study are listed in Supplemental Table S1.
Construction of cell-specific 3XFLAG::PAB-1 plasmids
To express 3XFLAG-tagged PAB-1 in specific cell types, promoters were amplified and cloned into the pSV41(Pgateway::3XFLAG::PAB-1 + unc-119 minigene) plasmid using the Gateway cloning system (Invitrogen). Transgenics were obtained by microparticle bombardment or by microinjection (see Supplemental Protocols SP1–SP4).
Isolation of cell-specific RNA by the mRNA-tagging method
Cell-specific RNA was isolated from transgenics expressing 3XFLAG-tagged PAB-1 using the mRNA-tagging strategy (Roy et al. 2002) described by Von Stetina et al. (2007; see Supplemental Protocol SP5).
Preparation and primary cell culture of embryonic cells and isolation of fluorescently labeled embryonic cells by FACS
Embryonic cells were isolated by FACS as previously described (Christensen et al. 2002; Fox et al. 2005, 2007). Cell types were sorted to a fractional purity ranging from 80%–97% (Supplemental Table S1). RNA was extracted from sorted cells in TRIzol LS, treated with DNase I, and purified using the DNA-free RNA kit from Zymo Research (see Supplemental Protocols SP6–SP8).
RNA amplification and microarray hybridization
RNA from sorted cells and mRNA-tagging lines was amplified and labeled using the WT-Ovation Pico, WT-Ovation Exon, and Encore Biotin kits from NuGEN for application to C. elegans tiling arrays (Affymetrix). Pearson correlation coefficients between replicates were determined to confirm consistent microarray data quality (see Supplemental Protocols SP9, SP10).
RT-PCR and qPCR to validate novel and differentially expressed transcripts
Primers were designed to produce short amplicons (75–150 bp) using Batch-Primer3 (You et al. 2008). RT-PCR was performed using the same cDNA produced for microarray analysis for template and GoTaq polymerase (Promega). qPCR was performed using the same cDNA produced for microarray analysis for template and Sso-Fast Eva green reaction mix on a CFX96 real-time PCR machine (Bio-Rad) (Fig. 3; see Supplemental Protocols SP11, SP12).
Immunochemistry for Z2/Z3 protein expression
Embryos were fixed using methanol/acetone (Strome and Wood 1983). Images were acquired with a Volocity spinning disk confocal system (Perkin-Elmer/Improvision) fitted on a Nikon Eclipse TE2000-E inverted microscope (Fig. 5B; for details, see Supplemental Protocol SP13).
Computational analyses of tiling array data
Array annotation
Tiling array features were mapped to the C. elegans genome and WormBase gene annotation (Rogers et al. 2008). Additionally repetitive tiling probes were flagged (see Supplemental Protocol SP14). Based on annotated protein-coding gene models, tiling probes were annotated into exonic, intronic, intergenic, and ambiguous categories.
Normalization and transcript identification
Raw tiling array data were normalized to correct for uneven background (Borevitz et al. 2003; Zeller et al. 2009), between-array variability with quantile normalization (Bolstad et al. 2003), and probe-sequence effects with transcript normalization (Zeller et al. 2008). We evaluated the extent to which normalizing for probe sequence effects improved subsequent transcript recognition in comparison to DNA reference normalization (Huber et al. 2006) on the basis of the above probe annotation (Fig. 2A). In this context, we defined sensitivity as the percentage of tiling probes with signal above a cutoff (true-positives, TP) among all annotated exon probes and undetected ones (false-negatives, FN): Sn = TP/(TP + FN). Precision was defined as the percentage of annotated exon probes (TP) among those with signal above the cutoff (including true- and false-positives, FP): Pr = TP/(TP + FP). Varying the threshold parameter across the whole range of measured array intensities resulted in curves showing different trade-offs between precision and sensitivity (Fig. 2A; see Supplemental Protocol SP15).
For de novo identification of TARs from tiling array data, we employed mSTAD (margin-based segmentation of tiling array data), a machine-learning–based method (Laubinger et al. 2008; Zeller et al. 2008). Its internal parameters were trained on hybridization patterns and tiling probe annotations in regions around experimentally confirmed genes. Genome-wide TAR predictions were generated in a two-fold cross-validation scheme. Cross-validation accuracy was assessed with respect to annotated genes confirmed by full-length cDNA sequences (Fig. 2B,C) as well as to the modENCODE integrated transcript model (Supplemental Fig. S6; Supplemental Protocols SP16, SP17; Gerstein et al. 2010). Additionally, accuracy was compared to modMine TARs (Supplemental Fig. S6; Supplemental Protocol SP17; Supplemental Result SR4).
Identification of new transcripts
“Unannotated” TARs were identified in comparison to coding and noncoding genes and pseudogenes annotated in WormBase as those with <20 nt overlap to annotated exons. If additionally a given TAR did not overlap by ≥20 nt with exons of the integrated transcript model, we called it a novel transcript (Fig. 2D–F; see Supplemental Protocol SP18). Taking the per-nucleotide union of TARs obtained in individual samples, we obtained nonredundant (nr) TARS (analogously for expressed nrTARs, differentially expressed nrTARs, unannotated nrTARs, and novel nrTARs) (Fig. 2F; see Supplemental Protocol SP18). For each position within expressed nrTARs, we counted the number of samples in which a TAR was detected to generate histograms of sample specificity (Fig. 4C; Supplemental Fig. S17; see Supplemental Protocol SP18).
Detection of expressed transcripts and significant expression differences
For each annotated protein-coding gene and predicted TAR, we constructed a probe set for expression summarization (see Supplemental Protocol SP19). Subsequently, transcript expression was estimated using a customized RMA pipeline (see Supplemental Protocol SP19; Bolstad et al. 2003; Irizarry et al. 2003; Gautier et al. 2004). A Mann-Whitney U test with an empirical background model and FDR correction for multiple testing was used to detect expressed transcripts (Benjamini and Hochberg 1995). Genes and TARs with an FDR ≤0.05 were reported as expressed above background (see Table 2; Supplemental Fig. S9A; see also Supplemental Protocol SP20). We detected differentially expressed transcripts using a method based on linear models (Smyth 2004). Genes and TARs were called differentially expressed if the FDR was ≤0.05 and the fold change (FC) ≥2.0 (Table 2; Fig. 4A; Supplemental Figs. S9B,C, S12; see Supplemental Protocol SP21). To more strictly correct for potential false-positives resulting from multiple sample comparisons, we divided individual FDR estimates by the number of samples or sample comparisons, respectively. This resulted in an adjusted FDR of 1.3 × 10−4 for expression above background and of 7.4 × 10−4 for differential expression (Table 2; Supplemental Fig. S12; Supplemental Protocol SP22). We called genes “selectively enriched” in a given tissue (see Results) if they met the following requirements: (1) enriched expression in a given tissue (FDR ≤0.05 and FC ≥2.0), (2) fold change versus reference among the upper 40% of the positive FC range observed for this gene across all tissues, and (3) fold-change entropy among the lower 40% of the distribution observed for all genes (see Supplemental Protocol SP23; Schug et al. 2005).
Self-organizing maps
We adopted SOMs (Kohonen 1982) as a means of discovering, clustering and visualizing gene expression similarity with respect to cell types or developmental stages. One SOM was fitted to mean-normalized log2-transformed gene expression estimates from the developmental stage data set (Fig. 6; Supplemental Fig. S20) and another one to those from cell type samples (Fig. 7; Supplemental Fig. S21; see Supplemental Protocol SP25). Regions in the SOM corresponding to characteristic and coherent expression patterns were afterward identified by k-means clustering of the SOM units (with k = 8 and k = 14 for the developmental and the cell type data set, respectively) (see Supplemental Protocol SP25). The top half of more coherent SOM units were identified by means of silhouette coefficients resulting in the clusterings shown (Figs. 6, 7; Supplemental Protocol SP25; (Rousseeuw 1987). Finally, we visualized prototypical gene expression patterns for each SOM region. Plotted are genes with a best-matching SOM unit within one of these regions and a quality error below the 50th and 20th percentile for developmental and cell type data sets, respectively (Figs. 6, 7; Supplemental Figs. S20, S22).
Motif discovery
Regulatory elements were identified using the FIRE algorithm (Elemento et al. 2007). Gene clusters produced from SOM analyses were submitted to the Integrated Genomics Exploration Tools (IGET) website (http://iget.princeton.edu) for analysis using FIRE.
GO/Protein domain enrichment analysis
Gene lists were tested for gene ontology (GO) or protein domain enrichment using the enrichment widgets on the modMINE website (http://intermine.modencode.org). Significance of enrichment was determined using hypergeometric tests, and P-values were corrected for multiple-testing using FDR (Benjamini and Hochberg 1995).
Acknowledgments
We thank E. Hallem, N. Ringstand, and P. Sternberg for Pgcy-33::GFP; B. Grant for Punc-122::RFP; O. Hobert for Prig-3::GFP; Harald Hutter for Pglr-1::dsRed; S. Kim for the Pges-1 mRNA tagging line; P. Roy for the Pmyo-3 mRNA tagging line; D. Marlee and A. George for help with building the Pclh-4 mRNA tagging line; S. Schultheiss for advice with the motif analysis; J. Tap for helpful discussions; D. Anastassiou for support; V. Varadan for advice; G. Seydoux for encouragement; D. Hall and Z. Altun for use of images from WormAtlas; N. Kurn, S. Wang, and J.D. Heath (NuGEN) for help with developing amplification and labeling methods for tiling array hybridization; the Vanderbilt Functional Genomics Shared Resource for microarray processing; and the Veterans Administration Medical Center Flow Cytometry Core and the Vanderbilt University Medical Center Flow Cytometry Core for FACS. Some of the strains used in this work were provided by the C. elegans Genetic Center, which is supported by NIH NCRR. Some GFP expression data were obtained from the Genome BC C. elegans Gene Expression Consortium, which is funded by Genome Canada and Genome British Columbia. This work was supported by the Max Planck Society and DFG RA1894/1-1 (G.R., S.R.H., G.Z.); an EMBL postdoctoral fellowship (G.Z.); NIH grants HG004263 (D.M.M., V.R.), NS49743 (J.D.W.), GM34059 (S.S.), GM83548 (L.P.), R01 NS064273 (S. Shaham), NS26115 (D.M.M.), MH077302 (D.M.M.), and P50 DK44757 (A. Fogo); and NIH grants to Vanderbilt University: P30 CA68485, P60 DK20593, P30 DK58404, HD15052, P30 EY08126, and PO1 HL6744.
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession nos. GSE23245–GSE23271, GSE23278–GSE23287, GSE23769–GSE23770, and GSE25350–GSE25351.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.114595.110.
References
- Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T 2010. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res 38: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, Shah JK, Dey J, Rohl CA, Johnson JM, et al. 2009. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods 6: 647–649 [DOI] [PubMed] [Google Scholar]
- Azzaria M, Goszczynski B, Chung MA, Kalb JM, McGhee JD 1996. A fork head/HNF-3 homolog expressed in the pharynx and intestine of the Caenorhabditis elegans embryo. Dev Biol 178: 289–303 [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563 [DOI] [PubMed] [Google Scholar]
- Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC 2006. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 38: 1430–1434 [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. 2009. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res 37: D885–D890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP 2003. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889–900 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, Warner A, Khattra J, Holt RA, Ou G, Mah AK, et al. 2005. Functional genomics of the cilium, a sensory organelle. Curr Biol 15: 935–941 [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD 2009. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci 12: 1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Liang D, Plouffe D, Chang H-s, Zhu T, Weigel D, Berry CC, Winzeler E, Chory J 2003. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res 13: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, Korf I 2008. Longer first introns are a general property of eukaryotic gene structure. PLoS ONE 3: e3093 doi: 10.1371/journal.pone.0003093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK 2008. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calella AM, Nerlov C, Lopez RG, Sciarretta C, von Bohlen und Halbach O, Bereshchenko O, Minichiello L 2007. Neurotrophin/Trk receptor signaling mediates C/EBPalpha, -beta and NeuroD recruitment to immediate-early gene promoters in neuronal cells and requires C/EBPs to induce immediate-early gene transcription. Neural Develop 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzigeorgiou M, Yoo S, Watson JD, Lee WH, Spencer WC, Kindt KS, Hwang SW, Miller DM 3rd, Treinin M, Driscoll M, et al. 2010. Specific roles for DEG/ENaC and TRP channels in touch and thermosensation in C. elegans nociceptors. Nat Neurosci 13: 861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Pai S, Zhao Z, Mah A, Newbury R, Johnsen RC, Altun Z, Moerman DG, Baillie DL, Stein LD 2005. Identification of a nematode chemosensory gene family. Proc Natl Acad Sci 102: 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Reinke V 2006. Promotion of oogenesis and embryogenesis in the C. elegans gonad by EFL-1/DPL-1 (E2F) does not require LIN-35 (pRB). Development 133: 3147–3157 [DOI] [PubMed] [Google Scholar]
- Chikina MD, Huttenhower C, Murphy CT, Troyanskaya OG 2009. Global prediction of tissue-specific gene expression and context-dependent gene networks in Caenorhabditis elegans. PLoS Comput Biol 5: e1000417 doi: 10.1371/journal.pcbi.1000417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M, Estevez A, Yin X, Fox R, Morrison R, McDonnell M, Gleason C, Miller DM 3rd, Strange K 2002. A primary culture system for functional analysis of C. elegans neurons and muscle cells. Neuron 33: 503–514 [DOI] [PubMed] [Google Scholar]
- Cinar H, Keles S, Jin Y 2005. Expression profiling of GABAergic motor neurons in Caenorhabditis elegans. Curr Biol 15: 340–346 [DOI] [PubMed] [Google Scholar]
- Colosimo ME, Brown A, Mukhopadhyay S, Gabel C, Lanjuin AE, Samuel AD, Sengupta P 2004. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr Biol 14: 2245–2251 [DOI] [PubMed] [Google Scholar]
- del Castillo-Olivares A, Kulkarni M, Smith HE 2009. Regulation of sperm gene expression by the GATA factor ELT-1. Dev Biol 333: 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Yanowitz JL, Schedl PD 1999. Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell 99: 271–281 [DOI] [PubMed] [Google Scholar]
- Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al. 2007. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol 25: 663–668 [DOI] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S 2007. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell 28: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Stanley HM, Han M, Wood WB 1999. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Dev Biol 205: 240–253 [DOI] [PubMed] [Google Scholar]
- Flames N, Hobert O 2009. Gene regulatory logic of dopamine neuron differentiation. Nature 458: 885–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox RM, Von Stetina SE, Barlow SJ, Shaffer C, Olszewski KL, Moore JH, Dupuy D, Vidal M, Miller DM III 2005. A gene expression fingerprint of C. elegans embryonic motor neurons. BMC Genomics 6: 42 doi: 10.1186/gb-2007-8-9-r188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox RM, Watson JD, Von Stetina SE, McDermott J, Brodigan TM, Fukushige T, Krause M, Miller DM III 2007. The embryonic muscle transcriptome of Caenorhabditis elegans. Genome Biol 8: R188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry Ra 2004. affy: Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE Project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, Zetka M 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell 14: 263–274 [DOI] [PubMed] [Google Scholar]
- Grange T, Roux J, Rigaud G, Pictet R 1991. Cell-type specific activity of two glucocorticoid responsive units of rat tyrosine aminotransferase gene is associated with multiple binding sites for C/EBP and a novel liver-specific nuclear factor. Nucleic Acids Res 19: 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Altun ZF 2008. C. elegans atlas. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Hallem EA, Spencer WC, McWhirter RD, Zeller G, Henz SR, Rätsch G, Miller DM III, Horvitz HR, Sternberg PW, Ringstad N 2010. Receptor-type guanylate cyclase is required for carbon dioxide sensation by Caenorhabditis elegans. Proc Natl Acad Sci 108: 254–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Wang J, Liu T, Liu XS, Li T, Wang Y, Qian Z, Zheng H, Zhu X, Wu T, et al. 2007. Mapping the C. elegans noncoding transcriptome with a whole-genome tiling microarray. Genome Res 17: 1471–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, Coulson A, Murray JI, Bao Z, Sulston JE, Waterston RH 2005. Genomics in C. elegans: So many genes, such a little worm. Genome Res 15: 1651–1660 [DOI] [PubMed] [Google Scholar]
- Hillier LW, Reinke V, Green P, Hirst M, Marra MA, Waterston RH 2009. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res 19: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K 2008. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456: 130–134 [DOI] [PubMed] [Google Scholar]
- Hubbard EJA, Greenstein D 2005. Introduction to the germ line. In WormBook (ed. The C. elegans Research Community, Wormbook) doi: 10.1895/wormbook.1.18.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Toedling J, Steinmetz LM 2006. Transcript mapping with high-density oligonucleotide tiling arrays. Bioinformatics 22: 1963–1970 [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, et al. 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol 5: e237 doi: 10.1371/journal.pbio.0050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SB, Lee J 2003. Neuron cell type-specific SNAP-25 expression driven by multiple regulatory elements in the nematode Caenorhabditis elegans. J Mol Biol 333: 237–247 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15 doi: 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isik M, Korswagen HC, Berezikov E 2010. Expression patterns of intronic microRNAs in Caenorhabditis elegans. Silence 1: 5 doi: 10.1186/1758-907X-1-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Ryu J, Kiraly M, Duke K, Reinke V, Kim SK 2001. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc Natl Acad Sci 98: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, de Lencastre A, Pincus Z, Slack FJ 2009. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol 10: R54 doi: 10.1186/gb-2009-10-5-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S 1998. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94: 635–645 [DOI] [PubMed] [Google Scholar]
- Kawasaki I, Amiri A, Fan Y, Meyer N, Dunkelbarger S, Motohashi T, Karashima T, Bossinger O, Strome S 2004. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics 167: 645–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS 2001. A gene expression map for Caenorhabditis elegans. Science 293: 2087–2092 [DOI] [PubMed] [Google Scholar]
- Kimble J 1981. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev Biol 87: 286–300 [DOI] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL 2005. Germline proliferation and its control. In WormBook (ed. The C. elegans Research Community, Wormbook) doi: 10.1895/wormbook.1.13.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Rothman JH 2001. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development 128: 2867–2880 [DOI] [PubMed] [Google Scholar]
- Kohonen T 1982. Self-organized formation of topologically correct feature maps. Biol Cybern 43: 59–69 [Google Scholar]
- Krause M, Park M, Zhang JM, Yuan J, Harfe B, Xu SQ, Greenwald I, Cole M, Paterson B, Fire A 1997. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development 124: 2179–2189 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Zeller G, Henz SR, Sachsenberg T, Widmer CK, Naouar N, Vuylsteke M, Scholkopf B, Rätsch G, Weigel D 2008. At-TAX: A whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol 9: R112 doi: 10.1186/gb-2008-9-7-r112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Panea C, Wiggins CH, Reinke V, Leslie C 2010. Learning “graph-mer” motifs that predict gene expression trajectories in development. PLoS Comput Biol 6: e1000761 doi: 10.1371/journal.pcbi.1000761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP 2003. The microRNAs of Caenorhabditis elegans. Genes Dev 17: 991–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Long F, Peng H, Aerni SJ, Jiang M, Sanchez-Blanco A, Murray JI, Preston E, Mericle B, Batzoglou S, et al. 2009. Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell 139: 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZJ, Yip KY, Wang G, Shou C, Hillier LW, Khurana E, Agarwal A, Auerbach R, Rozowsky J, Cheng C, et al. 2011. Prediction and characterization of non-coding RNAs in C. elegans by integrating conservation, secondary structure and high throughput sequencing and array data. Genome Res (this issue). doi: 10.1101/gr.110189.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, Walhout AJ 2008. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res 18: 2005–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, Goszczynski B, Tian H, Krich ND, Khattra J, Holt RA, et al. 2007. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev Biol 302: 627–645 [DOI] [PubMed] [Google Scholar]
- McGhee JD, Fukushige T, Krause MW, Minnema SE, Goszczynski B, Gaudet J, Kohara Y, Bossinger O, Zhao Y, Khattra J, et al. 2009. ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev Biol 327: 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, et al. 2003. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol 68: 159–169 [DOI] [PubMed] [Google Scholar]
- Meissner B, Warner A, Wong K, Dube N, Lorch A, McKay SJ, Khattra J, Rogalski T, Somasiri A, Chaudhry I, et al. 2009. An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet 5: e1000537 doi: 10.1371/journal.pgen.1000537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS 2008. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci 105: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai M, Kitadate Y, Arita K, Shigenobu S, Kobayashi S 2006. Expression of meiotic genes in the germline progenitors of Drosophila embryos. Gene Expr Patterns 6: 256–266 [DOI] [PubMed] [Google Scholar]
- Murray JI, Bao Z, Boyle TJ, Boeck ME, Mericle BL, Nicholas TJ, Zhao Z, Sandel MJ, Waterston RH 2008. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods 5: 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow MC, Martinez NJ, Olsen PH, Silverman HS, Barrasa MI, Conradt B, Walhout AJ, Ambros V 2008. The FLYWCH transcription factors FLH-1, FLH-2, and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes Dev 22: 2520–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev 15: 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli F, Liu Y, Kim YA, Chen PJ, Kim SK 2006. Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development 133: 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, et al. 2000. A global profile of germline gene expression in C. elegans. Mol Cell 6: 605–616 [DOI] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323 [DOI] [PubMed] [Google Scholar]
- Reynolds NK, Schade MA, Miller KG 2005. Convergent, RIC-8-dependent Galpha signaling pathways in the Caenorhabditis elegans synaptic signaling network. Genetics 169: 651–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A, Antoshechkin I, Bieri T, Blasiar D, Bastiani C, Canaran P, Chan J, Chen WJ, Davis P, Fernandes J, et al. 2008. WormBase 2007. Nucleic Acids Res 36: D612–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw P 1987. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20: 53–65 [Google Scholar]
- Roy PJ, Stuart JM, Lund J, Kim SK 2002. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418: 975–979 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B 2004. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32: D91–D94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ Jr 2005. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol 6: R33 doi: 10.1186/gb-2005-6-4-r33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweikert G, Zien A, Zeller G, Behr J, Dieterich C, Ong CS, Philips P, De Bona F, Hartmann L, Bohlen A, et al. 2009. mGene: Accurate SVM-based gene finding with an application to nematode genomes. Genome Res 19: 2133–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes DC, Wu JC, Sadler PL, Laprade K, Moore LL, Noritake A, Chu DS 2009. Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet 5: e1000611 doi: 10.1371/journal.pgen.1000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Watson JD, Spencer WC, O'Brien T, Cha B, Albeg A, Treinin M, Miller DM 3rd 2010. Time-lapse imaging and cell-specific expression profiling reveal dynamic branching and molecular determinants of a multi-dendritic nociceptor in C. elegans. Dev Biol 345: 18–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. doi: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Strome S, Wood WB 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35: 15–25 [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G 1999. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126: 4861–4871 [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56: 110–156 [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100: 64–119 [DOI] [PubMed] [Google Scholar]
- Thellmann M, Hatzold J, Conradt B 2003. The Snail-like CES-1 protein of C. elegans can block the expression of the BH3-only cell-death activator gene egl-1 by antagonizing the function of bHLH proteins. Development 130: 4057–4071 [DOI] [PubMed] [Google Scholar]
- Touroutine D, Fox RM, Von Stetina SE, Burdina A, Miller DM 3rd, Richmond JE 2005. acr-16 encodes an essential subunit of the levamisole-resistant nicotinic receptor at the Caenorhabditis elegans neuromuscular junction. J Biol Chem 280: 27013–27021 [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI 1995. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83: 207–218 [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol 8: e1000371 doi: 10.1371/journal.pbio.1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina SE, Watson JD, Fox RM, Olszewski KL, Spencer WC, Roy PJ, Miller DM 3rd 2007. Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol 8: R135 doi: 10.1186/gb-2007-8-7-r135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao Y, Wong K, Ehlers P, Kohara Y, Jones SJ, Marra MA, Holt RA, Moerman DG, Hansen D 2009. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics 10: 213 doi: 10.1186/1471-2164-10-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL 2009. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev 23: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xu S, Xia L, Wang J, Wen S, Jin P, Chen D 2009. The bantam microRNA is associated with Drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet 5: e1000444 doi: 10.1371/journal.pgen.1000444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You FM, Huo N, Gu YQ, Luo MC, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD 2008. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinformatics 9: 253 doi: 10.1186/1471-2105-9-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller G, Henz SR, Laubinger S, Weigel D, Rätsch G 2008. Transcript normalization and segmentation of tiling array data. Pac Symp Biocomput 2008:527–538 [PubMed] [Google Scholar]
- Zeller G, Henz SR, Widmer CK, Sachsenberg T, Rätsch G, Weigel D, Laubinger S 2009. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J 58:1068–1082 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfie M 2002. Identification of genes expressed in C. elegans touch receptor neurons. Nature 418: 331–335 [DOI] [PubMed] [Google Scholar]