Abstract
Alternative splicing (AS) plays a crucial role in the diversification of gene function and regulation. Consequently, the systematic identification and characterization of temporally regulated splice variants is of critical importance to understanding animal development. We have used high-throughput RNA sequencing and microarray profiling to analyze AS in C. elegans across various stages of development. This analysis identified thousands of novel splicing events, including hundreds of developmentally regulated AS events. To make these data easily accessible and informative, we constructed the C. elegans Splice Browser, a web resource in which researchers can mine AS events of interest and retrieve information about their relative levels and regulation across development. The data presented in this study, along with the Splice Browser, provide the most comprehensive set of annotated splice variants in C. elegans to date, and are therefore expected to facilitate focused, high resolution in vivo functional assays of AS function.
Alternative splicing (AS) is the process by which multiple mRNA transcripts are produced from a single precursor transcript through the differential utilization of splice sites. Alternative splicing is one of the key mechanisms that have evolved in metazoans to generate increased transcriptome complexity and recent studies estimate that greater than 95% of human multi-exon genes express multiple splice isoforms (Pan et al. 2008; Wang et al. 2008). Moreover, alternatively spliced exons are often differentially regulated across tissues and during development, suggesting that individual isoforms may serve specific spatial or temporal roles (Hartmann and Valcarcel 2009; Licatalosi and Darnell 2010; Nilsen and Graveley 2010).
The importance of proper regulation of AS during development has been demonstrated in many different instances; one particularly well-studied example is that of the sex determination pathway in Drosophila. In this pathway, the female-specific expression of a splicing regulator transformer stimulates the inclusion of exons in transcripts of the doublesex and fruitless transcription factor genes (Lopez 1998; Forch and Valcarcel 2003). The female-specific isoforms of these transcription factors subsequently activate the expression of genes required for female development, while the male-specific variants induce a gene expression program important for male differentiation (Dulac 2005; Shirangi and McKeown 2007). Similar spatio-temporally regulated AS networks are likely to exist in metazoans. The characterization of these AS networks, and their integration with other layers of gene regulation, will be necessary for a more complete understanding of development (Blencowe 2006).
The nematode C. elegans, with its amenability for genetic manipulations, defined cell lineage, simple but extensively differentiated nervous system, conserved splicing machinery, and established molecular and cell biological resources, provides an attractive yet relatively under-explored model organism with which to study the regulation and functions of AS in vivo. Tissue or developmental stage-specific AS events have been identified in approximately 70 C. elegans genes which, like those identified in vertebrates, are controlled by combinatorial interactions between splicing factors (Kuroyanagi et al. 2007; Barberan-Soler and Zahler 2008; Ohno et al. 2008). These studies, while informative, were limited to the analysis of isoforms represented in expressed sequence tag (EST)/cDNA libraries, which currently do not provide a full coverage of the transcriptome.
The advent of deep sequencing technologies has now enabled transcriptome analyses at unprecedented levels of sensitivity and precision, and these have recently been applied in C. elegans (Hillier et al. 2009; Ramani et al. 2009; Mangone et al. 2010). However, due in part to limited depth of coverage of existing data sets and the ongoing development of analytical software, it has remained a major challenge to systematically categorize and quantify the relative usage of isoforms across development for both known and novel AS events.
In this study, we combined high-throughput sequencing of mRNA (RNA-seq) and quantitative AS microarray profiling to provide a high resolution survey of splicing across C. elegans development. We have identified thousands of new AS events and hundreds of isoforms that are differentially expressed during development. Additionally, we identified candidate cis-elements that likely play a role in regulating these events. To facilitate the use of these data, we have created the “C. elegans Splice Browser,” a web resource in which all of the data in the present study are made accessible. This resource, which represents the most complete set of splice variants reported to date, should serve as a valuable basis for focused analyses of the functional importance of individual splice isoforms in vivo.
Results
Large-scale identification of splice variants in C. elegans by RNA-seq and microarray profiling
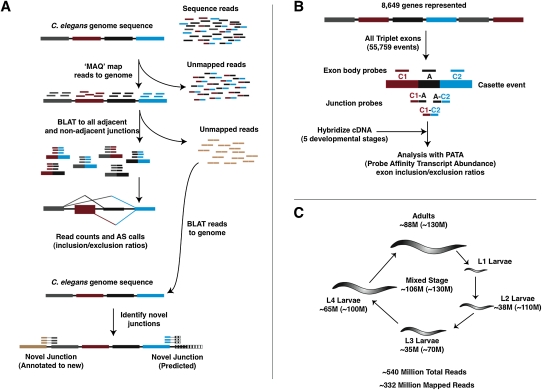
We used two distinct but complementary approaches to identify and quantify splice variant usage in C. elegans (Fig. 1). In the first approach, we performed RNA-seq of polyA+ RNA and identified splice junctions using a computational approach that does not require prior knowledge of splice junction connectivity (Fig. 1A). In parallel, we developed a custom microarray and associated computational tool in which combinations of exon body and splice junction probes were used to monitor single exon skipping or “cassette” type splicing events (Fig. 1B; see Supplemental Methods). Our overall strategy, described in more detail below, was to use RNA-seq profiling to identify and measure as comprehensively as possible alternative splice site usage in the wild-type transcriptome across development, and then to cross-validate and augment this data set with information from quantitative AS microarray profiling data.
Figure 1.
Outline of RNA-seq and microarray approaches for profiling AS in C. elegans. (A) For RNA-seq analysis, short sequence reads from the transcriptomes of animals isolated at multiple or specific developmental stages are first aligned to the genome and those that uniquely map in their entirety are used to identify exonic regions of transcripts. The remaining unmapped reads are then mapped against an exon junction database containing all possible combinations of splice junctions between annotated exons to identify and quantitatively measure AS events. Finally, any remaining unmapped reads are aligned to novel splice junctions that are not annotated or predicted. (B) Our microarray platform uses exon body (labeled C1, A, and C2 in diagram) and exon junction (labeled C1:A, A:C2, and C1:C2) probes to monitor cassette-type AS events, where the internal exon in a triplet of exons can be skipped in spliced mRNAs. We have included probes to monitor a total of 55,759 exon triplets in 8649 genes (∼50% of annotated genes), in addition to a set of 499 previously annotated cassette AS events. Probe sets corresponding to exon triplets with evidence of AS by our RNA-seq analysis are then combined with the PATA algorithm to generate quantitative predictions of relative isoform usage across the developmental stages analyzed. (C) Number of sequence reads uniquely mapping to the C. elegans genome by stage and sample. The values in parentheses indicate the total number of reads generated for each sample.
Our RNA-seq data set was generated from polyA+ RNA isolated either from mixed populations of wild-type hermaphrodite animals of various developmental stages, or from synchronized L4 larvae or adult stage animals (Ramani et al. 2009). These samples yielded 130M, 54M, and 69M reads respectively, over 80% of which could be aligned to the genome. To maximize coverage, we incorporated recently published RNA-seq data sets from four developmental stages (Fig. 1C; Hillier et al. 2009). In total, we analyzed approximately 540 million reads, of which approximately 332 million alignable reads (62%) could be mapped to 19,809 of the 20,534 genes (96.5%) annotated in WormBase (Harris et al. 2010). In contrast, <1.5% of the reads mapped to intergenic regions. The extent of coverage and specific enrichment of reads mapping to annotated genes demonstrates that the RNA-seq data sets in this study afford a high degree of sensitivity and specificity in the detection of expressed genes.
To generate the AS microarray data set, we hybridized cDNAs derived from polyA+ RNA isolated from synchronized wild-type hermaphrodite animals harvested at five developmental stages (larval stages L1 to L4 and young adults). Alternative exon inclusion level measurements were estimated by using a new method capable of accurately modeling the behavior of splice junction probes (see Supplemental Methods; S Mavandadi, JA Calarco, X Wang, Q Pan, BJ Blencowe, and Q Morris, in prep.). Of 8649 microarray profiled genes 7031 (81%) were detected above background in one or more of the developmental stages, suggesting that a large proportion of genes can also be profiled using this complementary method.
To identify splice junctions with our RNA-seq data, we created an exon junction database (EJDB) by joining pairwise combinations of predicted exons from all annotated multi-exon genes from WormBase (version WS200). We also created a control exon junction database by joining randomly shuffled pairs of exons from the EJDB. We identified reads mapping to 104,246 of the 126,628 adjacent junctions (∼82%) in 18,555 (90%) annotated genes (see Supplemental Methods). In contrast, at all depths of coverage surveyed, only a small fraction of reads could be mapped to our control junction database (∼2% relative to the annotated adjacent junctions detected; Supplemental Fig. S1). These results indicate that our splice junction mapping procedure also achieves high specificity and sensitivity.
To assess the quality of our RNA-seq data, we examined how well we could identify a set of previously annotated splice junctions. A total of 4049 nonadjacent splice events are annotated in WormBase and 2842 (∼71%) of these were detected with at least one supporting read and 2257 (∼56%) junctions were detected by five or more reads (Fig. 2A; see Supplemental Methods). At read depths of greater than 200 million reads, the number of detected adjacent and nonadjacent junctions did not substantially increase (Supplemental Fig. S1), suggesting that at our current read depth we have detected the vast majority of AS events in the samples we have analyzed. It is likely that many of the undetected AS events occur in other developmental states (such as males or dauers) or in response to other environmental conditions that are not profiled in our samples. We note that ∼20% of the known AS events that we detected have only a single sequence read spanning the alternatively spliced junctions. However, >90% of single read AS events can be confirmed as coverage is increased (Supplemental Fig. S1). These results provide evidence that the majority of single-read AS events reported here are true events. We therefore report these AS events, and all other single-read junction data in the sections to follow.
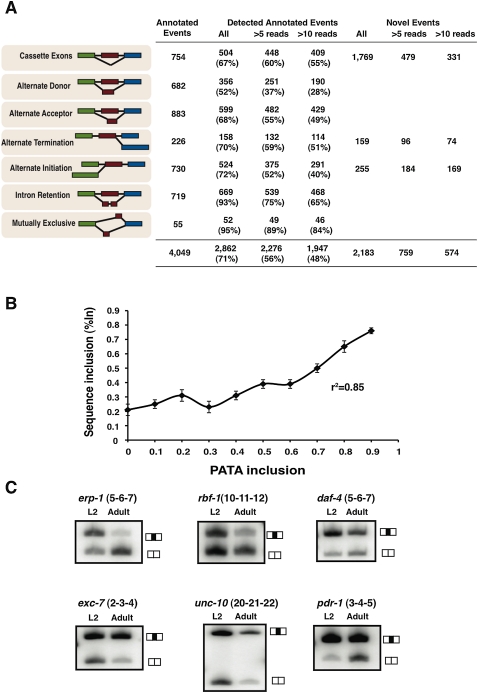
Figure 2.
Identification of known and novel AS events. (A) Table displaying the proportion of different classes of AS events annotated in WormBase that are identified by our RNA-seq analysis with varying degrees of junction count support. Also listed in the last three columns are the number of unannotated AS events identified by our analysis and the number of reads supporting these splice junctions. (B) We compared the %In values for a set of 317 annotated cassette AS events from the true positive set that were detected both by RNA-seq analysis and by our microarray platform. For these AS events, RNA-seq derived %In values profiled across four developmental stages (L2, L3, L4, and adults) were compared to the PATA derived %In values in the same stage. The plot shows a high correlation between the two platforms indicating that both methods are reliable estimates of relative isoform usage. (C) RT-PCR validation of a subset of unannotated AS events in the larval (L2) and adult developmental stages. Primers were designed to anneal to sequences corresponding to neighboring constitutively spliced exons, amplifying two products: one representing isoforms including the alternative exon in transcripts (top bands in gel images) and the other one skipping the alternative exon (bottom bands).
We next cross-validated our RNA-seq data with data generated by AS microarray profiling. Our microarray analysis provides quantitative predictions of the degree to which an alternative exon is included in a mature mRNA transcript, referred to as a percent inclusion (%In) value. %In values can also be generated from the RNA-seq data using ratios of reads mapping to splice junctions formed by exon inclusion and skipping. Focusing on a set of 317 annotated cassette AS events from the “true positive” (cDNA/EST-validated) set that can be monitored both by RNA-seq analysis and AS microarray profiling, we observe a strong correlation (r2 = 0.85) between the %In values measured by these two technologies (Fig. 2B; see Methods).
Collectively, our results demonstrate that the majority of previously annotated AS events in the C. elegans transcriptome are present in our RNA-seq data and that RNA-seq yields accurate measures of relative splice site usage.
Identification of novel splicing events
We also identified novel junctions in our RNA-seq analysis. These were derived from two classes of splicing events: (1) AS events involving annotated splice sites (Novel-class A events), in which the exons and splice sites are correctly annotated, but the splicing event joining the exons has not been previously detected, and (2) splicing events involving unannotated splice sites (Novel-class B events), in which new junctions are created from a predicted or novel exon, or from an unannotated internal splice site.
Our analysis detected 2183 Novel-class A events from 1354 genes. Out of these events, 1769 could be classified as putative cassette AS events, while the remaining 414 were grouped as potential alternative initiation or termination events (Fig. 2A; see Supplemental Methods). We performed semiquantitative RT-PCR validations for 13 of the RNA-seq predicted novel AS events in L2 larvae and adult stage RNA samples, and found that 12 (92%) events had detectable expression of both isoforms (Fig. 2C; Supplemental Fig. S2). A large proportion of the previously unannotated AS events also share key characteristics of known AS events, further supporting the conclusion that they are truly expressed. For example, annotated cassette AS events are significantly enriched for frame-preserving exons relative to the total set of annotated exons (68% vs. 40%, respectively, P = 1 × 10−4, χ2 test). Similarly, our set of novel cassette AS events are also enriched for frame preserving exons (54%, P = 4 × 10−3, χ2 test). In addition, the level of conservation of both annotated and novel exons affected by AS is markedly higher between C. elegans and C. briggsae relative to the total set of annotated exons (64% for annotated and 65% for novel AS events, compared with 55% for all exons, P < 0.05, χ2 test). These analyses, together with our RT-PCR validation experiments strongly suggest that the majority of the previously unannotated splicing events are bona fide AS events.
We further identified 6468 putative Novel-class B events by analyzing reads that had not been mapped to either the genome, annotated splice junctions, or to the novel-type A events (see Supplemental Methods). Of the 6468 junctions, ∼17% connect two novel splice sites while the remaining events involved junctions combining both novel and annotated splice sites. Of 25 putative novel splicing events randomly selected, all were validated by RT-PCR assays (Supplemental Table S1). These results indicate that our method for detecting novel splicing events is specific. We further analyzed each of the 6468 novel splice junctions for evidence of alternative usage of one of their two splice sites, and found that 256 of these novel junctions represent potentially novel alternative splicing events.
In summary, our RNA-seq analysis has identified 8651 putative novel splicing events in more than 2682 genes. Based on our analyses of known and novel junctions, we estimate that transcripts from more than 5000 genes (∼25% of all protein coding genes) of the C. elegans genome undergo AS, with each gene on average generating 2.2 isoforms (see Supplemental Methods). This estimate represents a 2.5-fold increase over what is suggested by EST/cDNA evidence, and a twofold increase over previous estimates for the proportion of C. elegans genes containing one or more AS event (Zahler 2005).
Genome-wide analysis of temporally regulated alternative exon usage
We next determined whether any of the known or novel alternative splice junctions were differentially-regulated during development. Notably, from the RNA-seq data set, we detect 349 AS events that are predicted to have significant differences in junction usage between two or more developmental stages (P < 0.01, Fisher's exact test). These developmentally regulated junctions involved all classes of profiled AS events, and significant quantitative differences were detected between every pair of developmental stages compared (Supplemental Table S2). We performed semiquantitative RT-PCR assays to validate a subset of the RNA-seq derived predictions. Of seven events examined, all were experimentally validated (Fig. 3A; Supplemental Fig. S3). Additionally, of 80 events that could also be monitored by our microarray, 61 (77%) were found to display a %In value difference of 20% or greater between the corresponding stages predicted to undergo differential junction usage by our RNA-seq analysis.
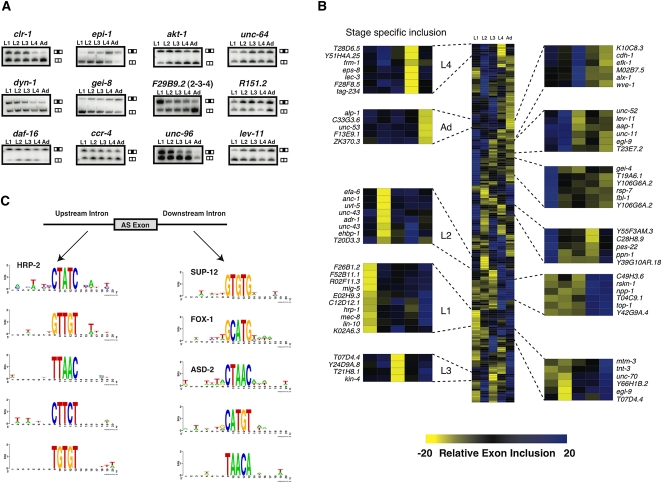
Figure 3.
Identification of temporally regulated AS events. (A) Semiquantitative RT-PCR validation for a subset of regulated AS events across larvae (L1, L2, L3, and L4) to adult (Ad) stages. Primers were designed to amplify both isoforms, and in each case, the two possible isoforms (top bands, included isoform; bottom bands, excluded isoform) show changes in relative intensities across developmental stages. (B) AS exons that were identified to undergo temporal regulation across development of at least 20% were median centered and hierarchically clustered and visualized using Cluster and TreeView (Eisen et al. 1998). Blue boxes indicate higher exon inclusion, while yellow boxes indicate higher exon exclusion. We find exons that are excluded in specific stages (projected panels to the left) while we also notice shared patterns of relative exon inclusion between multiple stages for other subsets of AS events (projected panels to the right). (C) SeedSearcher was used to identify pentamer motifs that are statistically enriched in either the 50 nt upstream of or downstream from splice junctions that are differentially regulated during development. Examples of these enriched motifs are shown either for motifs located upstream of the AS exon (left) or downstream from the AS exon (right). Note that four of these motifs correspond to the published motifs of HRP-2, SUP-12, FOX-1 and ASD-2, and these are indicated.
Although data generated from our RNA-seq analysis are reliable, a number of developmentally regulated AS events are likely to be missed due to an insufficient depth of coverage. However, our microarray platform is sufficiently sensitive to detect some of these missing events. Indeed, 214 additional AS events that lacked sufficient read coverage for reliable quantification were predicted by our microarray to undergo %In value differences of 20% or higher between two or more developmental stages (Fig. 3B; Supplemental Table S3). Out of 21 candidates tested from this set of events, 18 (86%) were confirmed to exhibit significant changes by RT-PCR assays (Fig. 3A; Supplemental Fig. S3; data not shown). These results suggest that our microarray predictions are also accurate and can identify additional regulated AS events that are not detected in our RNA-seq analysis. Intriguingly, the alternative exons and flanking intron sequences of the developmentally regulated splicing events detected in our RNA-seq and microarray analysis are significantly more conserved between C. elegans and C. briggsae than are other AS events (68%, P < 0.025, χ2 test), suggesting that the temporal regulation of this class of AS events is likely to be important for development.
In summary, the combined analysis of the RNA-seq and splicing microarray data sets revealed 574 temporally regulated AS events in 394 genes. When considering the number of events that can be accurately quantified by our methods, and extrapolating to all AS events, we estimate that roughly 30% of AS events are differentially regulated between the C. elegans developmental stages we have profiled, consistent with previous estimates (Barberan-Soler and Zahler 2008). To our knowledge, this study provides the most comprehensive data set and profiling information for developmentally regulated C. elegans AS events to date.
Alternative splicing events are regulated by the interplay between trans-acting splicing regulators and cis-regulatory elements associated with alternative exons and flanking intron and exon sequences (Matlin et al. 2005; Blencowe 2006; Wang and Burge 2008). Identifying these cis-elements and the factors that associate with them is an important step toward better understanding the code governing AS regulation. As a first step toward this goal, we used the SeedSearcher algorithm (Barash et al. 2001) to identify motifs that are enriched in the regulated exons or intron sequences flanking these exons among the set of temporally regulated AS events (see Supplemental Methods). This search identified 12 pentamer motifs enriched in the upstream intronic sequence and 19 pentamer motifs enriched in the downstream intronic region of the temporally-regulated AS events (FDR corrected P-values of <0.01; Fig. 3C; Supplemental Figs. S4, S5). Twelve of the 19 (65%) downstream motifs and seven of the 12 (60%) upstream motifs were previously identified as being enriched in intronic regions flanking AS events by an independent approach (Kabat et al. 2006). The set of motifs we have identified include perfect matches to motifs previously shown to be regulated by the splicing factors ASD-1/FOX-1, ASD-2, SUP-12, or HRP-2 (Fig. 3C; Kuroyanagi et al. 2006, 2007; Ohno et al. 2008; Kabat et al. 2009). Of these factors, only ASD-2 was previously demonstrated to play a role in modulating temporally regulated AS (Ohno et al. 2008). Our results thus suggest that these factors (and as yet uncharacterized RNA binding proteins) and their cognate motifs identified here, play important roles in differential splicing during development.
The C. elegans Splice Browser
In order to make our results easily accessible to the research community, we have created the “C. elegans Splice Browser” (http://splicebrowse.ccbr.utoronto.ca). This web resource includes relevant information from the AS analysis generated in this study. For each of the gene predictions, read counts supporting all detected adjacent, alternative, and novel splice junctions are included. These read counts can be displayed either by developmental stage of interest or as a cumulative count from all stages. Where applicable, microarray-derived %In value measurements are represented as color-coded heat maps that indicate splicing changes throughout development (ways of accessing the data are covered in a tutorial at the website to increase the ease of use). Users can also upload personal data and create customized tracks to compare their data with the current data set. In this regard, the Splice Browser not only serves as a reference database but can also act as a repository for future data sets as they become available. With these features, the Splice Browser should provide biologists with easily accessible, quantitative information regarding alternative splice site usage during C. elegans development. The Splice Browser can thus serve as a basis for driving detailed high-resolution experiments to investigate the spatio-temporal regulation of individual splicing events and to test the functional significance of individual isoforms (JA Calarco, B Raj, BJ Blencowe, and M Zhen, in prep.).
Discussion
Development depends on precisely regulated changes in gene expression. In C. elegans, significant progress has been made to globally map gene expression changes through the use of expression microarrays and through the generation of transgenic reporter lines. However, regulated AS events also contribute to development, yet the spatio-temporal expression patterns as well as the functional specificity of individual isoforms remain poorly understood. Using complementary genome-wide approaches, we have verified the majority of annotated AS events, have identified thousands of novel candidate splice variants, and conclude that AS in C. elegans is more prevalent than previously appreciated. Our data set has also revealed hundreds of splicing events that are differentially regulated during development, providing a comprehensive and quantitative catalog of such events from the samples analyzed. Results from this study complement and extend recent analyses of the C. elegans transcriptome by RNA-seq and microarray profiling (Barberan-Soler and Zahler 2008; Hillier et al. 2009; Ramani et al. 2009; Mangone et al. 2010). We show that ∼25% of C. elegans genes have multiple isoforms, with an average of approximately two isoforms per gene. Furthermore, we estimate that roughly 30% of these alternatively spliced genes show developmental regulation of their isoform; thus, in total we find that 5%–10% of genes display differential splicing across development. These changes are likely to be functionally important since these exons and their surrounding intronic sequences tend to be more conserved than average.
To make these data easily accessible we constructed the “C. elegans Splice Browser” (http://splicebrowse.ccbr.utoronto.ca). This resource allows researchers to assess the relative usage of individual AS events in genes of interest, which will in turn facilitate future downstream in vivo functional analyses. While the set of splice variants already catalogued in the Splice Browser, along with their relative usage at each developmental stage, provide a more complete survey of splicing in C. elegans than previously reported, it is clear that the present data do not represent a comprehensive set of splice variants. In this study, we examined larval development in wild-type hermaphrodites but not in carefully staged embryos, nor in males or dauers, and we only examined one set of environmental conditions. RNA-seq is being used by a wide variety of labs not only to survey these additional stages, but also the transcriptomes of various mutant C. elegans strains. As such, as additional RNA-seq and microarray data generated by either the modENCODE consortium or individual researchers become available, we will integrate these into the Splice Browser, further improving the quality and coverage of C. elegans AS events. In this regard, one of the findings of our current study is the high degree to which splicing events detected by RNA-seq can be validated by an independent platform, in this case AS microarrays. Thus RNA-seq provides an accurate and quantitative view of splicing and we can confidently incorporate future RNA-seq data into the Splice Browser. This will facilitate the systematic investigation of AS in transcriptome regulation, evolution, and in the generation of functional complexity.
Methods
Strains and maintenance
C. elegans N2 bristol wild-type animals were cultured and maintained on NGM agar plates seeded with OP50 E. coli according to standard protocols (Brenner 1974).
RNA isolation, sequencing, and analysis
Poly A+ RNA was isolated and used to synthesize cDNA as described previously (Pan et al. 2004; Ramani et al. 2009). Sequencing reads were mapped to the WS200 version of the C. elegans genome as previously described (Ramani et al. 2009). See Supplemental Methods for details on mapping reads to splice sites, identifying novel splice sites, and other computational analyses.
Microarray design, hybridization, and analysis
Alternative splicing events were identified from alignments of C. elegans mRNA/EST sequences (UniGene Build #26) to C. elegans genomic sequence, essentially as previously described (Pan et al. 2004, 2005). In total, 499 cassette type AS events were identified. For each AS event, three exon probes and three exon junction probes were designed to profile the AS event on the microarray, essentially as previously described (Pan et al. 2004). Microarray hybridizations were performed as described previously (Calarco et al. 2009) and the data were analyzed using a new algorithm (see Supplemental Methods for details).
Database setup
The C. elegans Splice Browser database (http://splicebrowse.ccbr.utoronto.ca) was created using Gbrowse (Stein et al. 2002; Donlin 2009; http://gmod.org) with a mySQL backend. The database was specifically designed to be similar to WormBase (the widely popular C. elegans web resource) and utilized data mirrored from WormBase. Sequence and array data were converted to a format compatible with gbrowse, and the gbrowse admin tutorial was followed extensively (http://gmod.svn.sourceforge.net/viewvc/gmod/Generic-Genome-Browser/trunk/htdocs/tutorial/tutorial.html).
RT-PCR assays
Nonradioactive RT-PCR assays were performed with the OneStep kit (QIAGEN) as recommended by the manufacturer. Radioactive RT-PCR assays were carried out essentially as described previously (Calarco et al. 2009). All primer sequences used in these assays are available in Supplemental Table S4.
Acknowledgments
We thank Chris Wedeles, Bushra Raj, and Valentina Slobodeniuc for experimental assistance, and Nadège Pelte and Arneet Saltzman for helpful comments on the manuscript. We thank The Wellcome Trust Sanger Institute for sequencing our samples. This research was supported in part by a grant from the CIHR (MOP-67011) (B.J.B.), a grant from Genome Canada through the Ontario Genomics Institute (B.J.B. and others), CIHR grants MOP-209314 and MOP-93619 (M.Z.), and CIHR grant 488367 and NSERC grant 487143 (A.G.F.). J.A.C. was supported by an NSERC Alexander Graham Bell Canada Graduate Scholarship, and A.K.R. was supported by a Wellcome Trust fellowship.
Footnotes
[Supplemental material is available for this article. The sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA009279. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE25927.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.114645.110.
References
- Barash Y, Bejerano G, Friedman N 2001. A simple hyper-geometric approach for discovering putative transcription factor binding sites. Lect Notes Comput Sci 2149: 278–293 [Google Scholar]
- Barberan-Soler S, Zahler AM 2008. Alternative splicing regulation during C. elegans development: Splicing factors as regulated targets. PLoS Genet 4: e1000001 doi: 10.1371/journal.pgen.1000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ 2006. Alternative splicing: New insights from global analyses. Cell 126: 37–47 [DOI] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O'Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. 2009. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 138: 898–910 [DOI] [PubMed] [Google Scholar]
- Donlin MJ 2009. Using the Generic Genome Browser (GBrowse). Curr Protoc Bioinformatics Chapter 9: Unit 9.9 doi: 10.1002/0471250953.bi0909s28 [DOI] [PubMed] [Google Scholar]
- Dulac C 2005. Sex and the single splice. Cell 121: 664–666 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forch P, Valcarcel J 2003. Splicing regulation in Drosophila sex determination. Prog Mol Subcell Biol 31: 127–151 [DOI] [PubMed] [Google Scholar]
- Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. 2010. WormBase: A comprehensive resource for nematode research. Nucleic Acids Res 38: D463–D467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann B, Valcarcel J 2009. Decrypting the genome's alternative messages. Curr Opin Cell Biol 21: 377–386 [DOI] [PubMed] [Google Scholar]
- Hillier LW, Reinke V, Green P, Hirst M, Marra MA, Waterston RH 2009. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res 19: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat JL, Barberan-Soler S, McKenna P, Clawson H, Farrer T, Zahler AM 2006. Intronic alternative splicing regulators identified by comparative genomics in nematodes. PLoS Comput Biol 2: e86 doi: 10.1371/journal.pcbi.0020086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat JL, Barberan-Soler S, Zahler AM 2009. HRP-2, the Caenorhabditis elegans homolog of mammalian heterogeneous nuclear ribonucleoproteins Q and R, is an alternative splicing factor that binds to UCUAUC splicing regulatory elements. J Biol Chem 284: 28490–28497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M 2006. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat Methods 3: 909–915 [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Ohno G, Mitani S, Hagiwara M 2007. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol Cell Biol 27: 8612–8621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2010. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AJ 1998. Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu Rev Genet 32: 279–305 [DOI] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW 2005. Understanding alternative splicing: Towards a cellular code. Nat Rev Mol Cell Biol 6: 386–398 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno G, Hagiwara M, Kuroyanagi H 2008. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev 22: 360–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. 2004. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell 16: 929–941 [DOI] [PubMed] [Google Scholar]
- Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ 2005. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet 21: 73–77 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Ramani AK, Nelson AC, Kapranov P, Bell I, Gingeras TR, Fraser AG 2009. High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol 10: R101 doi: 10.1186/gb-2009-10-9-r101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi TR, McKeown M 2007. Sex in flies: What “body–mind” dichotomy? Dev Biol 306: 10–19 [DOI] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. 2002. The generic genome browser: A building block for a model organism system database. Genome Res 12: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM 2005. Alternative splicing in C. elegans. In WormBook (ed. The C. elegans Research Community). doi: 10.1895/wormbook.1.31.1, http://www.wormbook.org [Google Scholar]