Summary
The constitutive androstane receptor (CAR) transactivation can occur in the absence of exogenous ligand and this activity is enhanced by agonists TCPOBOP and meclizine. We use biophysical and cell-based assays to show that increased activity of CAR(TCPOBOP) relative to CAR(meclizine) corresponds to a higher affinity of CAR(TCPOBOP) for the steroid receptor coactivator-1. Additionally, steady-state fluorescence spectra suggest conformational differences between CAR(TCPOBOP):RXR and CAR(meclizine):RXR. Hydrogen/deuterium exchange (HDX) data indicate that the CAR activation function 2 (AF-2) is more stable in CAR(TCPOBOP):RXR and CAR(meclizine):RXR than in CAR:RXR. HDX kinetics also show significant differences between CAR(TCPOBOP):RXR and CAR(meclizine):RXR. Unlike CAR(meclizine):RXR, CAR(TCPOBOP):RXR shows a higher overall stabilization that extends into RXR. We identify residues 339–345 in CAR as an allosteric regulatory site with a greater magnitude reduction in exchange kinetics in CAR(TCPOBOP):RXR than CAR(meclizine):RXR. Accordingly, assays with mutations on CAR at leucine-340 and leucine-343 confirm this region as an important determinant of CAR activity.
Keywords: allosterism, coactivator, H/D exchange, ITC, nuclear receptor, spectroscopy
Introduction
Nuclear receptors (NRs) are multi-domain transcription factors that play a vital role in growth, development and homeostasis (Mangelsdorf et al., 1995). These proteins consist of an N-terminal activation function-1 (AF-1), a central DNA-binding domain (DBD) and a C-terminal ligand-binding domain (LBD) which encompasses the activation function-2 (AF-2). X-ray structures of all NR LBDs determined to date exhibit a common three-dimensional fold. The C-terminal LBD encloses the ligand-binding pocket (LBP) within a twelve α-helix framework (Moras and Gronemeyer, 1998; Wurtz et al., 1996). The ultimate helix H12, also the ligand-dependent activation function 2 (AF-2) of the LBD, plays a critical role in the recruitment of coactivator and corepressor molecules. NRs utilize a common agonist-mediated repositioning of helix H12 to generate the coactivator-binding groove (Glass et al., 1997). However, differences in levels of transcriptional activity in response to various agonists of the same receptor suggest that agonists can fine-tune the conformation of the coactivator-binding groove and consequently the affinity of the activated receptor for coactivator proteins (Bourguet et al., 2000; Gangloff et al., 2001; Oberfield et al., 1999).
CAR plays key roles in the clearance of xenobiotics (Sonoda et al., 2003; Wei et al., 2000) and endogenous toxins such as bilirubin (Huang et al., 2003). In addition, this protein has pharmacological importance due to its role in drug-to-drug interactions (Chang and Waxman, 2006). Also, prolonged CAR activation has been observed to enhance the progression of liver cancer (Huang et al., 2005). For optimal function, CAR heterodimerizes with the retinoid X receptor (RXR) and the CAR:RXR heterodimer can transactivate retinoic acid response elements (RAREs) in an apparent ligand-independent manner (Baes et al., 1994; Choi et al., 1997). Transactivation by murine CAR is further enhanced by agonist ligands such as 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP) (Tzameli et al., 2000) and 1-[(4-chlorophenyl)(phenyl)methyl]-4-(3-methylbenzyl)piperazine (meclizine) (Huang et al., 2004) (Figure 1A). Activated CAR recruits coactivator proteins such as the steroid receptor coactivator-1 (SRC1) by interacting specifically with receptor interacting domain (RID) 2 of SRC1 (Dussault et al., 2002). Androstane metabolites such as androstenol (5α-androst-16-en-3α-ol) have been identified as the physiological inverse agonist ligands of murine CAR (Forman et al., 1998). Androstenol binds to the CAR ligand-binding domain (LBD) where it displaces helix H12 (AF2) from the active-state conformation required to recruit coactivator proteins (Shan et al., 2004). Incidently, meclizine is also a ligand for the human CAR isoform, where it functions as an inverse agonist (Huang et al., 2004).
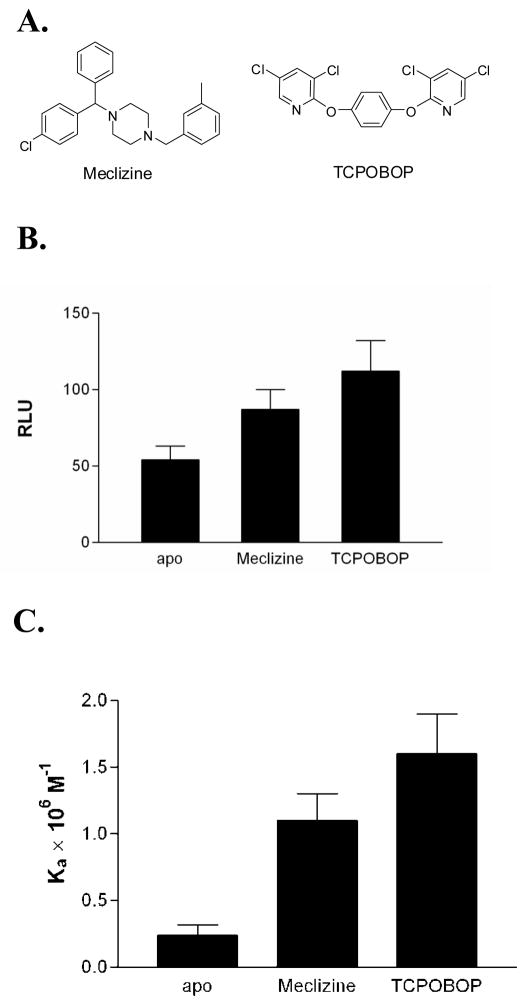
Figure 1. Comparison of the two CAR ligands used in this study.
(A) The chemical structures of CAR and TCPOBOP are shown. (B) TCPOBOP is a more potent agonist in cell-based reporter gene assays using the βRE response element. The data is shown in relative luciferase units (RLU). (C) TCPOBOP promotes higher affinity for coactivator than meclizine in ITC experiments.
Here, we report a comparative study of the role of two agonists of murine CAR. We compare CAR transactivation and coactivator binding affinities in response to the agonists TCPOBOP and meclizine. Further, we utilize fluorescence spectroscopy, HDX and site-specific mutagenesis to identify distinct CAR alterations in conformational dynamics induced upon binding either TCPOBOP or meclizine.
Results
Comparing transactivation and coactivator binding by CAR(TCPOBOP) and CAR(meclizine)
Meclizine has been identified as an mCAR agonist in a high-throughput screen and confirmed by a reporter gene assay using the LXRE response element in HepG2 cells (Huang et al., 2004). Also, in these experiments TCPOBOP is observed to exert a stronger agonist response from CAR than meclizine. Reporter gene assays with a second response element, βRE in CV-1 cells, confirm that TCPOBOP induces higher CAR transactivation than meclizine, independent of the DNA hormone response element or cell-line (Figure 1B). To compare the recruitment of coactivator protein by CAR(TCPOBOP):RXR and CAR(meclizine):RXR, we measured SRC1 binding to ligand-bound CAR using ITC. Quantitative analyses of binding isotherms show that at saturating concentrations of meclizine the affinity for coactivator is less than CAR(TCPOBOP) (Figure 1C). The thermodynamic parameters for recruitment of SRC1 NR2 peptide are presented in Table 1. All three forms of the heterodimer have similar favorable enthalpy change (ΔH). The thermodynamic basis for the difference in affinity is the larger unfavorable entropy change (ΔS) observed in SRC1 binding to CAR(meclizine):RXR compared to SRC1 binding to CAR(TCPOBOP):RXR.
Table 1.
Thermodynamic parameters for SRC1 peptide binding to CAR:RXR in the presence of indicated ligandsa
| Ligand | Kd (μM) | ΔH (kcal/mol) | −TΔS (kcal/mol) | ΔG (kcal/mol) | n |
|---|---|---|---|---|---|
| TCPOBOPb | 0.60 ± 0.10 | −10.0 ± 0.3 | 1.5 | −8.5 | 0.98 ± 0.07 |
| meclizine | 0.91 ± 0.14 | −9.9 ± 0.3 | 1.7 | −8.2 | 0.95 ± 0.08 |
| noneb | 4.9 ± 0.7 | −10.0 ± 0.4 | 2.8 | −7.2 | 1.03 ± 0.06 |
Determined at 25°C and pH 7.2 as described in materials and methods. The reported values are the average of at least three experiments and the errors are the standard deviation.
Originally reported (Wright et al., 2007)
Conformational differences within the CAR:RXR, CAR(TCPOBOP):RXR, and CAR(meclizine):RXR complexes
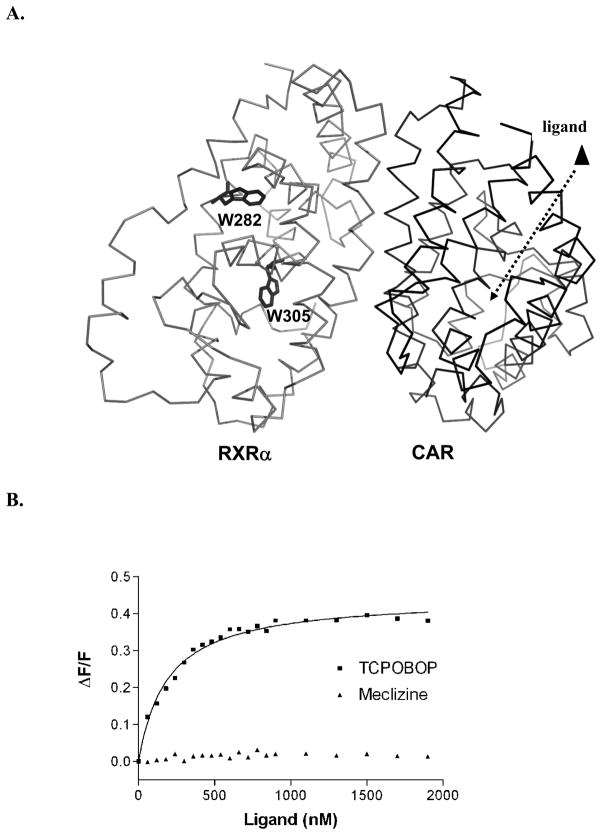
Comparative binding data shows that CAR(TCPOBOP):RXR exhibits a stronger affinity for SRC1 coactivator peptide than the CAR(meclizine):RXR complex. To determine a structural basis for these results, we utilized steady-state quenching of intrinsic tryptophan fluorescence of CAR:RXR to monitor changes in conformation upon ligand binding. Upon titrating TCPOBOP into the CAR:RXR heterodimeric LBD complex, there is a dose-responsive decrease in fluorescence of this protein complex. Since the CAR LBD sequence does not contain any tryptophan residues and an excitation wavelength of 295 nm is chosen to minimize the contribution from tyrosine residues, the TCPOBOP-induced conformational changes are transmitted from CAR across the CAR:RXR interface to the two tryptophan residues on RXR (Figure 2A & B). However, these conformational changes do not appear to significantly affect 9-cis-retinoic acid (9c) binding to CAR:RXR (Supplemental Figure S1). In addition, we determined the effect of ligand binding on each tryptophan on RXR by titrating TCPOBOP into CAR:(W282F)RXR and CAR:(W305F)RXR (Supplemental Figure S2). There is a strong dose-dependent increase in tryptophan quenching induced by TCPOBOP on the CAR:(W282F)RXR mutant but no significant response to the ligand by the CAR:(W305F)RXR mutant (Supplemental Figure S2A). This suggests that while W305 in RXR is within the allosteric pathway that is regulated by TCPOBOP, the environment of W282 does not undergo significant changes in the presence of TCPOBOP.
Figure 2. TCPOBOP binding to CAR results in a conformational change in RXR.
(A) Change in intrinsic tryptophan fluorescence of RXR is monitored in response to increasing levels of CAR ligand. (B) TCPOBOP elicits a dose-responsive change in RXR fluorescence while meclizine does not. See also Figures S1 and S2.
Curiously, titrations of meclizine into CAR:RXR result in no significant changes in the fluorescence spectra (Figure 2B). Therefore, although both TCPOBOP and meclizine function as mCAR agonists, the molecular mechanisms of CAR(TCPOBOP) and CAR(meclizine) transactivation are not identical.
To identify specific structural features in CAR:RXR that rearrange upon ligand binding and to distinguish between CAR(TCPOBOP):RXR and CAR(meclizine):RXR, we performed comprehensive differential, solution phase amide HDX MS experiments on the CAR:RXR, CAR(TCPOBOP):RXR and CAR(meclizine):RXR LBD complexes. For each receptor(ligand) complex, differential HDX kinetics are determined by comparing the kinetics of individual peptic peptides derived from the complex to the corresponding peptic peptides generated from the unliganded receptor.
Amide exchange kinetics of peptides representative of 76 regions of the CAR LBD within CAR:RXR and CAR(TCPOBOP):RXR were compared for the two protein complexes (Figure 3A, Supplemental Figure S3A). From the same data sets, HDX kinetics were determined and compared for peptides representative of 84 segments of RXR in the CAR:RXR and CAR(TCPOBOP):RXR complexes (Figure 3B, Supplemental Figure S3B). We observe statistically significant changes in HDX kinetics in three specific subdomains of CAR. Peptides encompassing helices H2′ and H2″ as well as parts of helix H3 (residues 143–178) show significant protection from exchange in CAR(TCPOBOP):RXR compared to CAR:RXR. In addition, there are substantial reductions in exchange kinetics within the sequences that comprise helix H4 and parts of the β-sheet, helix H6 and helix H7 (residues 199–210 and 228–249). The most significant reductions in HDX behavior of the CAR(TCPOBOP) complex are seen within the region encompassing helices H10, H11 and H12 (residues 327–358). These results reflect the extensive network of hydrophobic interactions between TCPOBOP and CAR that stabilize AF-2 in the active conformation as observed in the structure of CAR(TCPOBOP):RXR(9c) (Suino et al., 2004). In this structure, the TCPOBOP A-ring is sandwiched between CAR Y234 and CAR F227 and the C-ring is sandwiched between CAR Y336 and F244. The C-ring of TCPOBOP also interacts directly with AF-2 (L353) and the linker helix (L346 and T350). These results show a strong correlation between the HDX data from the CAR(TCPOBOP) complex displaying regions with significantly reduced exchange kinetics and the CAR ligand-binding pocket in the CAR(TCPOBOP):RXR(9c) structure.
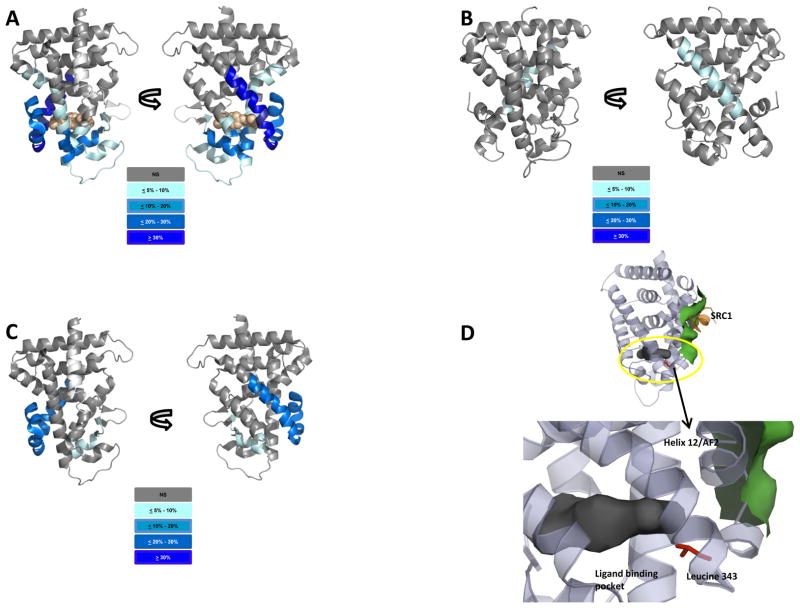
Figure 3. Perturbations in HDX data for CAR and RXR LBDs.
Cartoon rendition of percent reduction in HDX of ligand bound CAR:RXR heterodimer complex relative to unliganded receptor. Increasing protections from deuterium exchange are represented as color gradient from light to dark blue as shown. TCPOBOP is shown as an orange space-filled model. A. CAR portion of CAR(TCPOBOP):RXR (76 peptides). B. RXR portion of CAR(TCPOBOP):RXR (84 peptides). C. CAR portion of the CAR(meclizine):RXR (76 peptides). D. Residues within the encircled region in the unliganded CAR LBD model are mutated and tested for activity. See also Figure S3.
To examine the impact of TCPOBOP binding on the conformational dynamics of RXR within the CAR:RXR heterodimer, we compare the exchange kinetics of all 84 RXR peptides between the CAR:RXR and CAR(TCPOBOP):RXR. Interestingly, we observe that RXR peptides in the region 416–432 display a significant reduction in HDX kinetics in CAR(TCPOBOP):RXR (Figure 3B, Supplemental Figure S3B). These amino acids represent a large segment of helix H10 of RXR which forms the dimerization interface between CAR and RXR (Suino et al., 2004). In summary, residues from both the CAR and RXR dimerization interface display differential exchange kinetics when comparing CAR:RXR with the CAR(TCPOBOP):RXR complex. Taken together, data from HDX and fluorescence titrations suggest that TCPOBOP-binding results in a perturbation of the conformational dynamics within the heterodimer interface as well as conformational changes near the RXR ligand-binding pocket.
We analyzed exchange kinetics for 76 regions of CAR within the CAR(meclizine):RXR and CAR:RXR complexes (Figure 3C, Supplemental Figure S3A). Additionally, we compare HDX behavior for 84 regions of RXR within the CAR:RXR and CAR(meclizine):RXR complexes (Supplemental Figure S3B). The first of two regions within CAR(meclizine):RXR that are significantly protected to exchange when compared to unliganded CAR:RXR includes a segment of the β-sheet and all of helix H6 (residues 228–245). The second protected region of CAR(meclizine):RXR includes helices H10/11 and H12 (residues 327–358) (Figure 3C). As mentioned previously, helix H10/11 is the major contributor to the dimer interface in CAR:RXR and H12 forms the AF-2 subdomain that is critical to its interactions with SRC1. As predicted by fluorescence quenching experiments (Figure 2), and in contrast to CAR(TCPOBOP):RXR, there are no significant differences in exchange kinetics in RXR within the CAR(meclizine):RXR and CAR:RXR complexes (Supplemental Figure S3B).
Stabilization of CAR H10/11 is proportional to transactivation and coactivatior recruitment
Subtle differences in H/D exchange patterns within helices H10-H12, which contains the AF-2 region, are evident between the two ligand complexes. For instance, the C-terminal region of helix H10 (residues 330–338) indicate lesser mobility of CAR(TCPOBOP):RXR (exchange level=−32%) than CAR(meclizine):RXR (exchange level=−10%) (Supplemental Figure S3A). At the AF2, the most prominent differences between the two liganded complexes are in residues 338–343 preceding the AF2 (helix H12). These residues form a short α-helix (H11) in the inverse agonist bound structure of CAR (Moore, 2005) but are part of the longer H10 observed in the CAR(TCPOBOP):RXR structure.
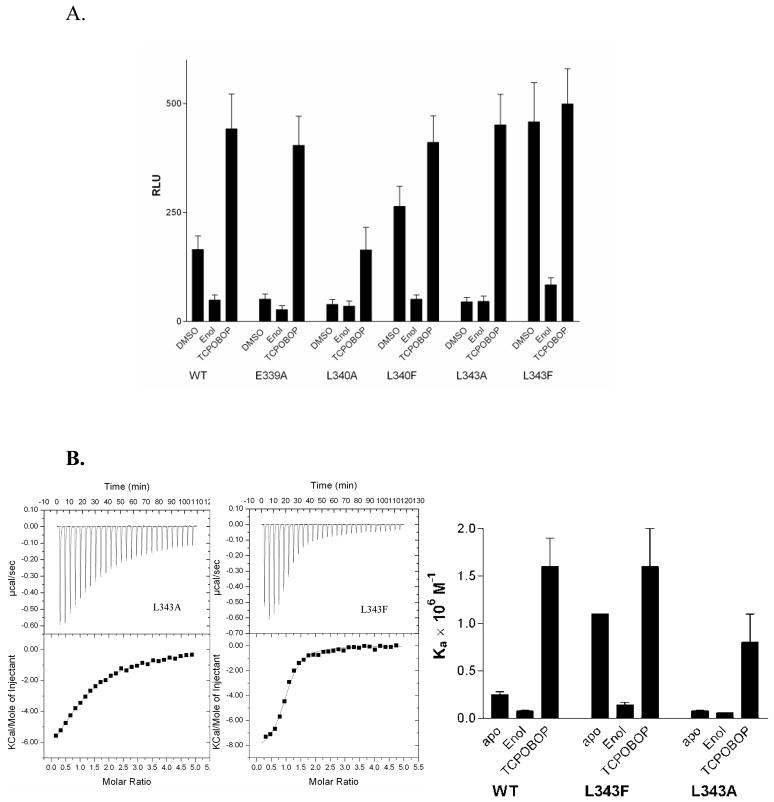
We introduced site-specific mutations within helix H11 to investigate the role of of this subdomain on the transactivation of CAR. The CAR Glu339 forms a hydrogen bond with the backbone amide of Gln245, presumably contributing to the stability of the active-state conformation of CAR (Shan et al., 2004). Also, Leu340 and Leu343 extend into the apolar ligand-binding pocket and can form hydrophobic interactions with residues within the interior of the protein. Thus, alanine substitutions of Glu339, Leu340 and Leu343 should eliminate these interactions resulting in a less constrained helix H11. As predicted, we observe that E339A (Shan et al., 2004), L340A and L343A each substantially decreases CAR transactivation in transfected CV-1 cell-based reporter gene assays (Figure 4A). Transactivation by these CAR mutants is rescued by exogenously added TCPOBOP to wild-type agonist-bound levels. We also reason that the substitution of Leu340 and Leu343 with residues capable of stronger apolar interactions with the CAR apolar ligand-binding pocket would constrain helix H11 to a greater extent than wild-type CAR. To test the hypothesis that the mobility of helix H11 can regulate activity, we designed L340F and L343F mutants based on the two CAR structures (Shan et al., 2004; Suino et al., 2004). Once again as predicted, the L340F and L343F CAR mutants exhibit enhanced transactivation when compared to wild-type CAR, presumably due to increased interactions of the bulkier phenylalanine sidechain with the apolar CAR ligand-binding pocket. Therefore, stronger interactions between the L340F and L343F on helix H11 and the main body of CAR can reduce the dynamics of this helix, and this corresponds to higher transactivation by these mutants.
Figure 4. Transactivation and coactivator binding assays of CAR:RXR.
A. b) Luciferase reporter assay showing ligand (TCPOBOP) mediated CAR:RXR transactivation of the LXRE-TK-Luc reporter plasmid in CV-1 cells. Results are reported in relative luciferase units (RLU). (B) Binding isotherms from titrations of SRC1 peptide into unliganded (L343A)CAR:RXR and (L343F)CAR:RXR and (C) summary of the SRC1 binding assay.
To relate the conformational mobility of helix H11 observed by HDX to the transactivation of the CAR helix H11 mutants, we characterized coactivator binding by these CAR mutants. The affinity of (L343A)CAR:RXR for the SRC1 NR box 2 peptide is lower than wild-type CAR, as suggested by the transactivation profile for this mutant (Figure 4B & C). Additionally, this mutant displays appreciably higher unfavorable entropy (TΔS) (Table 2). In the presence of TCPOBOP, the affinity of (L343A)CAR:RXR for the 13-mer SRC1 peptide is comparable to wild-type CAR(TCPOBOP):RXR (Table 2). Again, in close agreement with the transactivation assay, (L343F)CAR:RXR displays significantly greater affinity for coactivator and lower unfavorable entropy compared to the wild-type complex (Table 2). Also, the affinity of (L343F)CAR(androstenol):RXR for the SRC1 peptide is similar to wild-type CAR(androstenol):RXR (Table 2). These results provide further support for the hypothesis that the local dynamics of H11 plays a critical role in regulating CAR transactivation. It is likely that TCPOBOP imparts greater stability to this region and that this provides the structural basis for the higher levels of transactivation and coactivator binding by CAR(TCPOBOP):RXR than by the unliganded CAR:RXR or CAR(meclizine):RXR complexes.
Table 2.
Thermodynamic parameters for SRC1 peptide binding to the indicated CAR:RXR complexesa
| Complex | Kd (μM) | ΔH (kcal/mol) | −TΔS (kcal/mol) | ΔG (kcal/mol) | n |
|---|---|---|---|---|---|
| L343A (no ligand) | 14 ± 1.7 | −10.6 ± 0.5 | 4.0 | −6.6 | 1.0 ± 0.13 |
| L343A (TCPOBOP) | 2.0 ± 0.24 | −11.5 ± 0.8 | 3.7 | −7.8 | 0.92 ± 0.14 |
| L343F (no ligand) | 0.92 ± 0.05 | −8.5 ± 0.2 | 0.30 | −8.2 | 0.97 ± 0.05 |
| L343F (androstenol) | 4.2 ± 0.29 | −10.0 ± 0.2 | 2.7 | −7.3 | 1.0 ± 0.04 |
Determined at 25°C and pH 7.2 as described in materials and methods. The reported values are the average of at least three experiments and the errors are the standard deviation.
Discussion
Since CAR is active in the unliganded state, displays increased transactivation in the presence of meclizine and highest transactivation in the presence of TCPOBOP, it is an ideal protein to study the relationship of conformational dynamics and activity in NRs. We have compared transcriptional activation, coactivator recruitment, and conformational dynamics of CAR:RXR in the presence of two functionally distinct agonists. Cell-based reporter gene assays and ITC experiments show that TCPOBOP elicits greater transactivation and that this increase is proportional to the difference in affinity for SRC1. Comprehensive analysis of differential HDX behavior of the two ligand-bound forms clearly shows that the impact of TCPOBOP on the conformational dynamics of the CAR structure is broader and of greater magnitude than that of meclizine. This result is surprising since the two ligands are very similar in molecular size and chemical structure (Figure 1A). It is possible that meclizine does not make as many efficient contacts within the CAR ligand-binding pocket as compared to TCPOBOP or that the CAR(meclizine) complex does not establish the intricate layers of interactions observed in the CAR(TCPOBOP) structure. Additionally, there are more rotatable bonds within the meclizine structure which can allow it to adapt more readily to the CAR ligand-binding pocket unlike the relatively rigid TCPOBOP which can bind CAR more strongly but can also restrict the dynamics of the protein.
The global HDX kinetics observed for these three complexes shows that the CAR:RXR complex is more dynamic with the rank order of CAR:RXR ≫ CAR(meclizine):RXR > CAR(TCPOBOP):RXR, and that it follows the same pattern as the values for TΔS observed upon binding the LXXLL coactivator peptide (Table 1). This parallel suggests that, globally, the higher affinity for SRC1 results from a reduction in unfavorable entropy. The binding of the ligand, TCPOBOP (and to a lesser extent meclizine) restricts the conformational dynamics of the protein. Thus, coactivator binding is accompanied by a smaller net change in dynamics of CAR and a smaller positive TΔS component to the interaction. Since ΔS values depend on other factors in addition to degrees of freedom of the polypeptide chain (e.g. solvent reorganization), any correlation between ΔS and dynamics must include these factors. Nonetheless, the coactivator peptide binding and HDX data when taken together suggest that the overall reduction in protein motion is correlated with higher affinity for SRC-1 by CAR:RXR.
Besides the global properties observed, there are two specific areas of difference in the HDX data that distinguish TCPOBOP from meclizine. First, the binding of TCPOBOP to CAR also impacts the dynamics of RXR in CAR:RXR. This is not surprising given the large conformational changes in the CAR helix H10 which forms a significant portion of the heterodimer interface. Second, the greatly reduced HDX kinetics within helix H10/11 in CAR(TCPOBOP):RXR and the data from mutagenesis studies of residues within this region show that reducing mobility of this helix is a key component of the increase in activity by CAR agonist. Observations that regions within the LBD other than AF-2 can be major determinants of coactivator affinity and transactivation levels have been reported previously in the androgen receptor (Estebanez-Perpina et al., 2007), estrogen receptor (Dai et al., 2008) and PPARγ (Bruning et al., 2007; Choi et al., 2010).
The HDX and H11 mutagenesis data also provide insight into the structure of unliganded CAR:RXR (which has not been reported) as well as dynamic and conformational changes associated with the transitions between the active and very active states of CAR. The relatively shorter H12 in CAR compared to other NRs and the resulting interaction between the C-terminal carbonyl interacting with the side chain of K205 (Dussault et al., 2002) is understood to position helix H12 in the active position in the unliganded protein resulting in its constitutive activity. This helix H12 conformation is retained in the CAR(TCPOBOP) complex (Suino et al., 2004). In the presence of the inverse agonist, androstenol, helix H10 dissociated into two helices with the C-terminal residues (338–343) forming the short H11 (Shan et al., 2004). The significant differences in mobility observed in the H/D exchange data for residues 338–343 suggest that in the unliganded state H10 may more closely resemble the CAR(androstenol) structure with a shorter, more mobile, helix H11. Thus, unliganded, active CAR may have a conformation that contains a short H11 similar to the inactive CAR(androstenol) structure but with helix H12 in an orientation similar to the very active CAR(TCPOBOP) conformation (Figure 5). In the CAR(TCPOBOP):RXR complex helices H10 and H11 fuse into a single helix H10. The L340F and L343F CAR mutants appear to mimic TCPOBOP in stabilizing helix H11 which leads to enhanced coactivator recruitment and increases the levels of CAR transactivation.
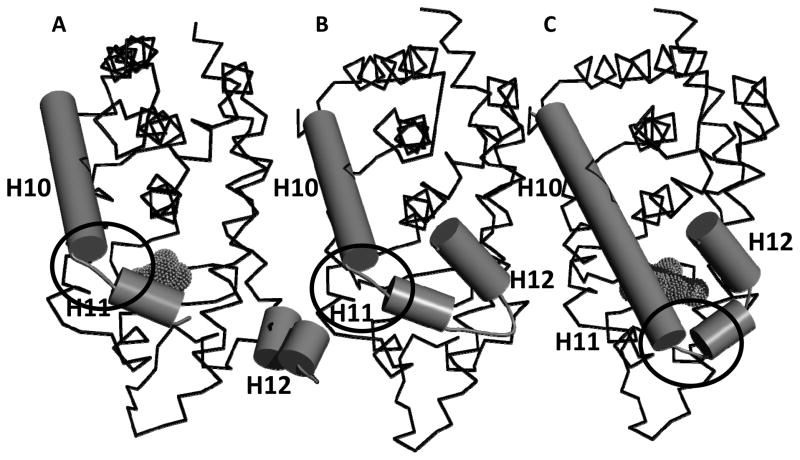
Figure 5. Comparison of CAR LBD structures.
(A). Structure of the CAR(androstenol) complex (REF); (B) Proposed model for the structure of unliganded CAR based on the HDX data and site-directed mutagenesis studies for residues 338–343; (C). Structure of the CAR(TCPOBOP) complex (REF). The circled regions have different conformations in the CAR(androstenol) and CAR(TCPOBOP) structures. Ligands are shown in mesh format.
In summary, these results provide insight into the mechanism of graded modulation of CAR activity for structurally similar agonists. It remains to be determined whether these mechanisms are common to other NRs or unique to a subset of NR family members. For instance, studies on the estrogen receptor have indicated that allosteric communication between dimeric partners can be mediated via helix H11 (Nettles et al., 2004). More importantly, these results provide the foundation for designing compounds that target CAR with varying degrees of agonist activity.
Experimental Procedures
Protein Expression and Purification
Protein was prepared as described previously (Wright et al., 2007). Briefly, the mCAR LBD (residues 117–358) was subcloned into pET15b (Novagen) with an N-terminal hexahistidine tag from mCAR cDNA provided by B. M. Forman. The human RXR LBD (residues 225–462) in pACYC184 was a gift from B. Wisely (Glaxo Smith-Kline, Inc.). The plasmids of CAR and RXR were cotransformed into Escherichia coli BL21(DE3) Gold cells (Novagen) and coexpressed for 20 hours at 20°C after induction with 0.5 mM IPTG. The cells were lysed by French press. The CAR:RXR heterodimer was purified by affinity chromatography using Ni-NTA (Qiagen) followed by size-exclusion chromatography on a Superdex S75 column (GE Healthcare). The mobile phase for size-exclusion chromatography consisted of 20mM Tris-HCl buffer at pH 8.0, 250 mM NaCl, 2.5 mM EDTA, 5.0 mM DTT for fluorescence and HDX experiments and 50 mM HEPES pH 7.5, 125mM KCl, and 2 mM TCEP for ITC experiments.
Reporter gene assays
CV-1 cells were maintained in DMEM/F-12 media containing 10% fetal bovine serum and 1000 U/ml penicillin and 1 mg/ml streptomycin. Immediately prior to the assay, the media was changed to DMEM/F-12 with 10% charcoal-dextran treated FBS and no antibiotics. Lipofectamine® (Invitrogen) was used to transfect cells with 50 ng/well pCMX mCAR, 100 ng/well pCMV-TK-luc containing three copies of either retinoid acid receptor β2 response element (βRE) or liver X receptor response element (LXRE) and 25 ng of pRL CMV expressing renilla luciferase as an internal control. The cells were dispensed on 96-well plates and ligands were added four hours after transfection. The ligand concentrations used were TCPOBOP (1.0 μM), androstenol (10 μM) and meclizine (20 μM). After 36 hours, cells were lysed. Activity was determined using the dual luciferase assay kit (Promega) following the manufacturer’s instructions. The reported results are the average from three separate experiments and the error bars represent the standard deviation.
Isothermal Titration Calorimetry
Experiments were performed as described previously using the Microcal VP-ITC instrument (Wright et al., 2007). Briefly, 100–400 μM SRC1 box 2 peptide was titrated into 8–36 μM CAR:RXR LBDs in 50 mM HEPES pH 7.4, 125 mM KCl and 1 mM TCEP. A four-fold excess of TCPOBOP or a ten-fold excess of meclizine was included where indicated. Data were analyzed using the one-site binding model in the Origin 7.0 software provided with the instrument. The reported results are the average from four separate experiments and the error bars represent the standard deviation.
Hydrogen/Deuterium Exchange (HDX) Analysis
Solution phase amide HDX was performed on liganded and unliganded CAR:RXR LBD heterodimers using a fully automated system that is described elsewhere (Busby et al., 2007; Chalmers et al., 2006). The changes reported in HDX are from direct comparisons between CAR(ligand):RXR to the corresponding regions within the unliganded CAR:RXR. Briefly, 4 μL of a 10 μM protein solution containing either the co-purified, preformed CAR:RXR heterodimer or CAR:RXR bound to either meclizine or TCPOBOP (20 mM Tris-Cl, pH 8, 250 mM NaCl, 5 mM DTT) was diluted up to 20 μL with D2O dilution buffer (20 mM Tris-Cl, pH 8, 250 mM NaCl, 5 mM DTT) and incubated at 4°C for the following periods of time: 10, 30, 60, 300, 900, and 3600 seconds. Following deuterium on-exchange, unwanted forward and back exchange was minimized and the protein was denatured by dilution to 50μL with 1% TFA in 3 M Urea (held at 1°C). The protein sample was then passed across an immobilized pepsin column (prepared in house) at 200 ul/min (0.1% TFA, 1°C) and the resulting peptides were trapped onto a C18 trap column (Microm Bioresources). Peptides were gradient eluted (4% CH3CN to 40% CH3CN, 0.3% formic acid over 15 min at 2°C) over a 2.1 mm × 50 mm C18 reverse phase HPLC column (Hypersil Gold, Thermo Electron) and electrosprayed directly into a high resolution orbitrap mass spectrometer (LTQ-Exactive, ThermoElectron, San Jose CA). Data were processed using in-house software (Pascal et al., 2006; Pascal et al., 2009; Pascal et al., 2007) and Microsoft Excel followed by visualization with pyMol (DeLano Scientific, South San Francisco CA). For the differential analysis between the unliganded and meclizine bound forms of the CAR:RXR, average percent deuterium incorporation was calculated for 76 regions of the CAR portion and 84 regions of the RXR portion of the CAR:RXR heterodimer in either its unliganded or meclizine bound state following 10, 30, 60, 300, 900, and 3600 seconds of on-exchange with deuterium. In addition, for the differential analysis between the unliganded and TCPOBOP bound forms of the CAR:RXR, average percent deuterium incorporation was calculated for 76 regions of the CAR and 84 regions of RXR in either its liganded or TCPOBOP bound state following 10, 30, 60, 300, 900, and 3600 seconds of on-exchange with deuterium. To determine differences in exchange between the CAR portions of the unliganded and the ligand bound CAR:RXR, the average percent deuterium values for all regions of the CAR from the unliganded CAR:RXR heterodimer were subtracted from the average percent deuterium values of the identical CAR regions from the CAR:RXR heterodimer bound to either meclizine or TCPOBOP. The same procedure was used to determine differences in exchange between the RXR portions of the heterodimer in either its unliganded or ligand bound states.
Ligand binding assays
Binding was measured by monitoring the decrease in intrinsic tryptophan fluorescence upon addition of ligand. For binding of CAR ligands, protein was diluted to a concentration of 2.0 μM with fluorescence buffer (3% DMSO, 20 mM Tris-HCl pH 8.0, 150 mM NaCl). Protein was then degassed for 15 minutes. Titrations were performed on the Perkin Elmer LS55 spectrofluorometer. Scans were performed using an excitation wavelength of 295 nm and emission scan of 305–400nm. The excitation and emission slit widths were 8.0 nm. Either DMSO or ligand was added in 2 μL increments into a 2.5 mL protein solution. Titrations of 9-cis-retinoic acid (9c) into CAR:RXR were performed in a similar manner except a protein concentration of 0.5 μM was used and the excitation and emission slit widths were 4.0 nm. Control experiment titrating TCPOBOP and meclizine into 2.0 μM molecular tryptophan showed no effect on fluorescence.
Supplementary Material
Supplemental Figure S1: CAR ligands do not significantly affect 9c (9cRA) binding to CAR:RXR. Steady-state fluorescence quenching measured at 340 nm upon titrating 9c into 250 nM CAR:RXR (■), CAR(TCPOBOP):RXR (▴) and CAR(meclizine):RXR.
Supplemental Figure S2: Emission spectra of 1.0 μM CAR:RXR, CAR:(W282F)RXR and CAR:(W305F)RXR. (A). The majority steady-state fluorescence quenching within the CAR:RXR heterodimer arises from W305. Mutation of W305 results in an almost complete loss of fluorescence signal. The W282F mutant still retains approximately 75% of its fluorescence at the emission maximum of 340 nm. (B). Titration of TCPOBOP into CAR:(W282F)RXR shows that there is a dose-dependent quenching of RXR W305 fluorescence.
Supplemental Figure S3- Comparison of changes in HDX rates in regions of ligandedCAR and RXR vs. unligandedCAR:RXR. (A). Comparison of differences in deuterium exchange for CAR from CAR(TCPOBOP):RXR and CAR(meclizine)RXR compared to unliganded receptor. (B). Comparison of differences in deuterium exchange for RXR from CAR(TCPOBOP):RXR and CAR(meclizine)RXR compared to unliganded receptor. Increasing protections from deuterium exchange compared to unliganded receptor are represented as color gradient from light to dark blue. Gray indicates no significant change in exchange between ligand bound and unliganded receptor.
Acknowledgments
This work is supported by National Institutes of Health Grant DK066394 (E.J.F.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–1551. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000;21:381–388. doi: 10.1016/s0165-6147(00)01548-0. [DOI] [PubMed] [Google Scholar]
- Bruning JB, Chalmers MJ, Prasad S, Busby SA, Kamenecka TM, He Y, Nettles KW, Griffin PR. Partial agonists activate PPARgamma using a helix 12 independent mechanism. Structure. 2007;15:1258–1271. doi: 10.1016/j.str.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Busby SA, Chalmers MJ, Griffin PR. Improving digestion efficiency under H/D exchange conditions with activated pepsinogen coupled columns. International Journal of Mass Spectrometry. 2007;259:130–139. [Google Scholar]
- Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- Chang TK, Waxman DJ. Synthetic drugs and natural products as modulators of constitutive androstane receptor (CAR) and pregnane X receptor (PXR) Drug Metab Rev. 2006;38:51–73. doi: 10.1080/03602530600569828. [DOI] [PubMed] [Google Scholar]
- Choi HS, Chung M, Tzameli I, Simha D, Lee YK, Seol W, Moore DD. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J Biol Chem. 1997;272:23565–23571. doi: 10.1074/jbc.272.38.23565. [DOI] [PubMed] [Google Scholar]
- Choi JH, Banks AS, Estall JL, Kajimura S, Bostrom P, Laznik D, Ruas JL, Chalmers MJ, Kamenecka TM, Bluher M, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466:451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai SY, Chalmers MJ, Bruning J, Bramlett KS, Osborne HE, Montrose-Rafizadeh C, Barr RJ, Wang Y, Wang M, Burris TP, et al. Prediction of the tissue-specificity of selective estrogen receptor modulators by using a single biochemical method. Proc Natl Acad Sci U S A. 2008;105:7171–7176. doi: 10.1073/pnas.0710802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussault I, Lin M, Hollister K, Fan M, Termini J, Sherman MA, Forman BM. A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol Cell Biol. 2002;22:5270–5280. doi: 10.1128/MCB.22.15.5270-5280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estebanez-Perpina E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, Pallai P, Shokat KM, Baxter JD, Guy RK, et al. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci U S A. 2007;104:16074–16079. doi: 10.1073/pnas.0708036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, Moore DD. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Gangloff M, Ruff M, Eiler S, Duclaud S, Wurtz JM, Moras D. Crystal structure of a mutant hERalpha ligand-binding domain reveals key structural features for the mechanism of partial agonism. J Biol Chem. 2001;276:15059–15065. doi: 10.1074/jbc.M009870200. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rose DW, Rosenfeld MG. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Chua SS, Qatanani M, Han Y, Granata R, Moore DD. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc Natl Acad Sci U S A. 2003;100:4156–4161. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, Moore DD. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol Endocrinol. 2005;19:1646–1653. doi: 10.1210/me.2004-0520. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Wei P, Schrader WT, Moore DD. Meclizine is an agonist ligand for mouse constitutive androstane receptor (CAR) and an inverse agonist for human CAR. Mol Endocrinol. 2004;18:2402–2408. doi: 10.1210/me.2004-0046. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DD. CAR: three new models for a problem child. Cell Metab. 2005;1:6–8. doi: 10.1016/j.cmet.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- Nettles KW, Sun J, Radek JT, Sheng S, Rodriguez AL, Katzenellenbogen JA, Katzenellenbogen BS, Greene GL. Allosteric control of ligand selectivity between estrogen receptors alpha and beta: implications for other nuclear receptors. Mol Cell. 2004;13:317–327. doi: 10.1016/s1097-2765(04)00054-1. [DOI] [PubMed] [Google Scholar]
- Oberfield JL, Collins JL, Holmes CP, Goreham DM, Cooper JP, Cobb JE, Lenhard JM, Hull-Ryde EA, Mohr CP, Blanchard SG, et al. A peroxisome proliferator-activated receptor gamma ligand inhibits adipocyte differentiation. Proc Natl Acad Sci U S A. 1999;96:6102–6106. doi: 10.1073/pnas.96.11.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal B, Chalmers MJ, Busby SA, Southern M, Mader C, Griffin P, Tsinoremas N. Software for the calculation of backbone amide deuterium levels from H/D exchange MS data. Mol Cell Proteomics. 2006;5:S90–S90. doi: 10.1186/1471-2105-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal BD, Chalmers MJ, Busby SA, Griffin PR. HD desktop: an integrated platform for the analysis and visualization of H/D exchange data. J Am Soc Mass Spectrom. 2009;20:601–610. doi: 10.1016/j.jasms.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal BD, Chalmers MJ, Busby SA, Mader CC, Southern MR, Tsinoremas NF, Griffin PR. The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data. BMC Bioinformatics. 2007;8:156. doi: 10.1186/1471-2105-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Vincent J, Brunzelle JS, Dussault I, Lin M, Ianculescu I, Sherman MA, Forman BM, Fernandez EJ. Structure of the murine constitutive androstane receptor complexed to androstenol: a molecular basis for inverse agonism. Mol Cell. 2004;16:907–917. doi: 10.1016/j.molcel.2004.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Rosenfeld JM, Xu L, Evans RM, Xie W. A nuclear receptor-mediated xenobiotic response and its implication in drug metabolism and host protection. Curr Drug Metab. 2003;4:59–72. doi: 10.2174/1389200033336739. [DOI] [PubMed] [Google Scholar]
- Suino K, Peng L, Reynolds R, Li Y, Cha JY, Repa JJ, Kliewer SA, Xu HE. The Nuclear Xenobiotic Receptor CAR; Structural Determinants of Constitutive Activation and Heterodimerization. Mol Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20:2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Wright E, Vincent J, Fernandez EJ. Thermodynamic characterization of the interaction between CAR-RXR and SRC-1 peptide by isothermal titration calorimetry. Biochemistry. 2007;46:862–870. doi: 10.1021/bi061627i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1: CAR ligands do not significantly affect 9c (9cRA) binding to CAR:RXR. Steady-state fluorescence quenching measured at 340 nm upon titrating 9c into 250 nM CAR:RXR (■), CAR(TCPOBOP):RXR (▴) and CAR(meclizine):RXR.
Supplemental Figure S2: Emission spectra of 1.0 μM CAR:RXR, CAR:(W282F)RXR and CAR:(W305F)RXR. (A). The majority steady-state fluorescence quenching within the CAR:RXR heterodimer arises from W305. Mutation of W305 results in an almost complete loss of fluorescence signal. The W282F mutant still retains approximately 75% of its fluorescence at the emission maximum of 340 nm. (B). Titration of TCPOBOP into CAR:(W282F)RXR shows that there is a dose-dependent quenching of RXR W305 fluorescence.
Supplemental Figure S3- Comparison of changes in HDX rates in regions of ligandedCAR and RXR vs. unligandedCAR:RXR. (A). Comparison of differences in deuterium exchange for CAR from CAR(TCPOBOP):RXR and CAR(meclizine)RXR compared to unliganded receptor. (B). Comparison of differences in deuterium exchange for RXR from CAR(TCPOBOP):RXR and CAR(meclizine)RXR compared to unliganded receptor. Increasing protections from deuterium exchange compared to unliganded receptor are represented as color gradient from light to dark blue. Gray indicates no significant change in exchange between ligand bound and unliganded receptor.