Summary
The E2F family of transcription factors is critical for the control of cell cycle progression. We now show that the specific inactivation of E2F3 in mouse embryo fibroblasts (MEFs) results in a disruption of the centrosome duplication cycle. Loss of E2F3, but not E2F1, E2F2, E2F4, or E2F5 results in unregulated cyclin E-dependent kinase activity, defects in nucleophosmin B association with centrosomes, and premature centriole separation and duplication. Consequently, this defect leads to centrosome amplification, mitotic spindle defects, and aneuploidy. Our findings implicate the E2F3 transcription factor as an important link that orchestrates DNA and centrosome duplication cycles, ensuring the faithful transmission of genetic material to daughter cells.
Introduction
The centrosome directs the polarity of the mitotic spindle and is essential for the fidelity of chromosome segregation and genomic stability (Pihan and Doxsey, 1999). Normal cells have one centrosome composed of two tightly associated centrioles, surrounded by an amorphous pericentriolar material. During transit through G1 to S phase of the cell cycle, the single centriole pair separates and each centriole begins to duplicate. During S and G2, the two newly formed centriole pairs mature and move toward opposite poles of the nuclear material in preparation for mitosis (Hinchcliffe and Sluder, 2001). Loss of centrosome duplication control, leading to centrosome amplification, has been associated with most human tumors and is thought to be a key event in tumor progression of the breast, prostate, and colon (Ghadimi et al., 2000; Lingle et al., 1998, 2002; Lingle and Salisbury, 1999; Pihan et al., 1998). Indeed, centrosome amplification and aneuploidy are common events triggered by the loss of tumor suppressors such as p53, p21CIP1, GADD45, BRCA1, BRCA2, and APC and by the expression of oncogenes such as Ras, STK15, MAPKK, and Mos (Fodde et al., 2001; Fukasawa et al., 1996; Fukasawa and Vande Woude, 1997; Hollander et al., 1999; Kaplan et al., 2001; Mantel et al., 1999; Saavedra et al., 1999, 2000; Tutt et al., 1999; Xu et al., 1999).
In order to accurately segregate chromosomes to each of the daughter cells during mitosis, centrosomes must be duplicated once and only once during the cell cycle. While the many steps involved in centrosome duplication have been elucidated, little is known about the nature of the activities that normally coordinate this process with the cell cycle. Recent work from multiple laboratories has demonstrated that the Rb/E2F transcriptional program is important for the timely expression of a plethora of genes involved in DNA replication, centrosome duplication, DNA repair, and mitosis, consistent with the finding that loss of E2F1, E2F2, and E2F3 arrest cells at multiple stages of the cell cycle (Ishida et al., 2001; Muller et al., 2001; Ren et al., 2002; Wu et al., 2001). These results raise the possibility that the E2F program may serve to coordinate a variety of essential processes required for the timely and accurate progression of cells through the cell cycle. We have taken a genetic approach to dissect the roles played by E2F transcription factors in the regulation of the centrosome cycle and find that the E2F3 family member plays a specific function in the control of this important process.
Results
Loss of E2F3 leads to centrosome amplification
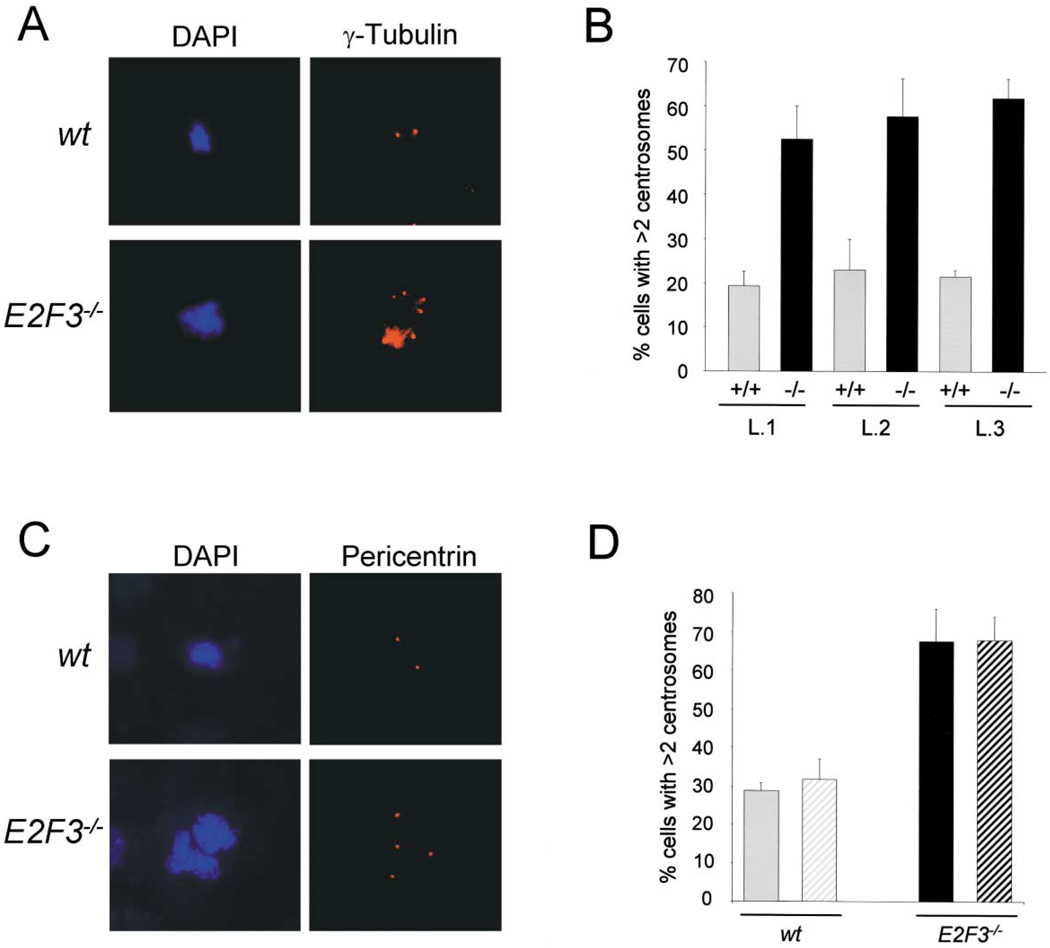
To explore the potential involvement of the E2F transcriptional program in centrosome biology, we derived MEFs from E2F1, E2F2, E2F3, E2F4, and E2F5 knockout mice (Leone et al., 2001; Wu et al., 2001) and assessed the status of centrosomes in early passage cell populations. The E2F3 locus encodes two gene products, E2F3a and E2F3b, whose expression is driven by two different promoters (Leone et al., 2000). Since E2F3a and E2F3b appear to play a dominant role in the control of cell cycle progression (Leone et al., 1998; Humbert et al., 2000; Wu et al., 2001), we initially analyzed wild-type and E2F3−/− MEFs lacking both E2F3 products. Centrosomes numbers were determined and quantified by immunohistochemistry using antibodies specific for γ-tubulin, a core component of the centrosome that is central to the proper nucleation of the α- and β-tubulin subunits of the mitotic spindle (Archer and Solomon, 1994; Dictenberg et al., 1998; Doxsey et al., 1994; Merdes and Cleveland, 1997; Zheng et al., 1998). Analysis of E2F3−/− MEFs derived from multiple litters revealed that a high proportion of cells (approximately 60%) in proliferating populations have more than two centrosomes (Figure 1B), with many cells containing greater than four centrosomes (Figure 1A). In contrast, less than 20% of wild-type cells contained more than two centrosomes (Figures 1A and 1B). This level of centrosome amplification is typical of wild-type cells and is likely a consequence of cytokinesis defects that arise upon culturing cells in vitro (Borel et al., 2002; Meraldi et al., 2002). The severity of the centrosome defect in E2F3−/− cells is similar to that previously found in p53−/− cells (Fukasawa et al., 1996; data not shown). Immunohistochemistry using antibodies against pericentrin, another core component of centrosomes (Archer and Solomon, 1994; Dictenberg et al., 1998; Doxsey et al., 1994; Merdes and Cleveland, 1997; Zheng et al., 1998), confirmed the high frequencies of centrosome amplification in E2F3−/− cells detected by γ-tubulin staining (Figures 1C and 1D).
Figure 1. Targeted disruption of E2F3 results in centrosome amplification.
A: Immunohistochemical analysis of centrosomes from wild-type and E2F3−/− cells using a primary γ-tubulin antibody and a rhodamine-conjugated secondary antibody (right panel); the left panel shows the same cells stained with DAPI.
B: Percentage of centrosome amplification in early passage (p 2) MEFs of three independent litters (L.1–L.3) of wild-type ( ) and E2F3−/− cells (■). The first and second litters include three wild-type and four E2F3−/− embryos each; the third litter includes one wild-type and one E2F3−/− embryo. Centrosomes were stained with a monoclonal antibody against γ-tubulin and a rhodamine-conjugated secondary antibody; the results represent the percentage of cells with three or more centrosomes.
) and E2F3−/− cells (■). The first and second litters include three wild-type and four E2F3−/− embryos each; the third litter includes one wild-type and one E2F3−/− embryo. Centrosomes were stained with a monoclonal antibody against γ-tubulin and a rhodamine-conjugated secondary antibody; the results represent the percentage of cells with three or more centrosomes.
C: Immunohistochemical analysis of the centrosome status of wild-type and E2F3−/− cells using a primary antibody against pericentrin and a rhodamine-conjugated secondary antibody (right panel); the left panel shows the DAPI-stained mitoses.
D: Percentage of cells with three or more centrosomes from three wild-type and three E2F3−/− embryos from two independent litters. Shown are the percentages resulting from staining wild-type cells ( ) or E2F3−/− embryos (■) with γ-tubulin (solid bar) or pericentrin (striped bar). For all the graphs presented in this section, we counted >200 cells per embryo from two different experiments.
) or E2F3−/− embryos (■) with γ-tubulin (solid bar) or pericentrin (striped bar). For all the graphs presented in this section, we counted >200 cells per embryo from two different experiments.
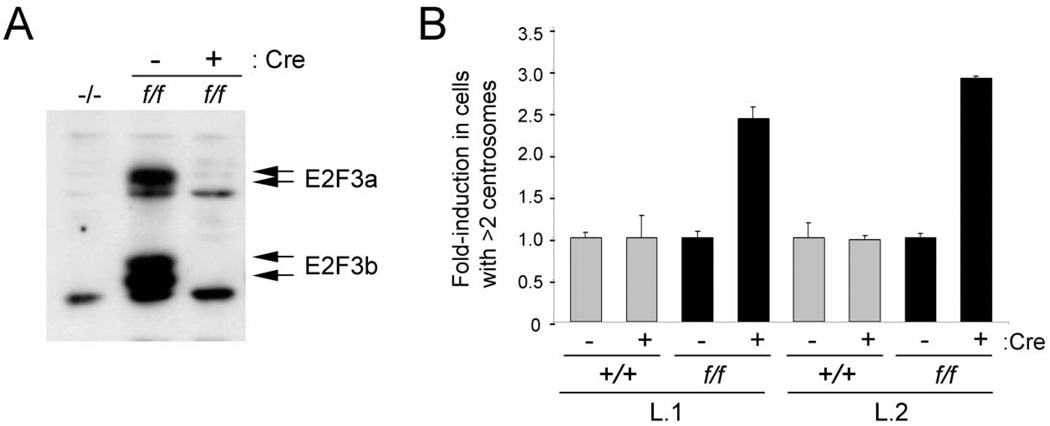
To determine whether centrosome amplification is a direct consequence resulting from the inactivation of E2F3, we ablated E2F3 function from MEFs harboring a conditional allele of E2F3 (E2F3f/f) and assessed the immediate effects on centrosome numbers. In E2F3f/f MEFs, exon 3, which encodes the DNA binding domain common to both E2F3a and E2F3b, is flanked by loxP sites. Introduction of the Cre recombinase into these cells via retroviral vectors results in deletion of exon 3 and ablation of E2F3a and E2F3b proteins (Wu et al., 2001) (Figure 2A). While expression of Cre in wild-type cells did not result in centrosome amplification, expression of Cre in E2F3f/f cells led to a similar level of centrosome amplification as that observed in E2F3−/− MEFs (Figure 2B), suggesting that E2F3 plays a cell-autonomous role in control of the centrosome cycle.
Figure 2. Conditional ablation of E2F3 results in centrosome amplification.
A: Western blot analysis of E2F3−/− (−/−) and E2F3f/f (f/f) MEFs infected with control (−) or Cre-expressing (+) retroviruses. Arrows indicate endogenous E2F3a and E2F3b proteins.
B: Fold induction in centrosome amplification in two independent litters (L.1 and L.2) of wild-type ( ) or E2F3f/f (■) MEFs. Cells were infected with control (−) or Cre-expressing (+) retroviruses as indicated, and centrosomes were stained with a primary antibody against γ-tubulin and a rhodamine-conjugated secondary antibody.
) or E2F3f/f (■) MEFs. Cells were infected with control (−) or Cre-expressing (+) retroviruses as indicated, and centrosomes were stained with a primary antibody against γ-tubulin and a rhodamine-conjugated secondary antibody.
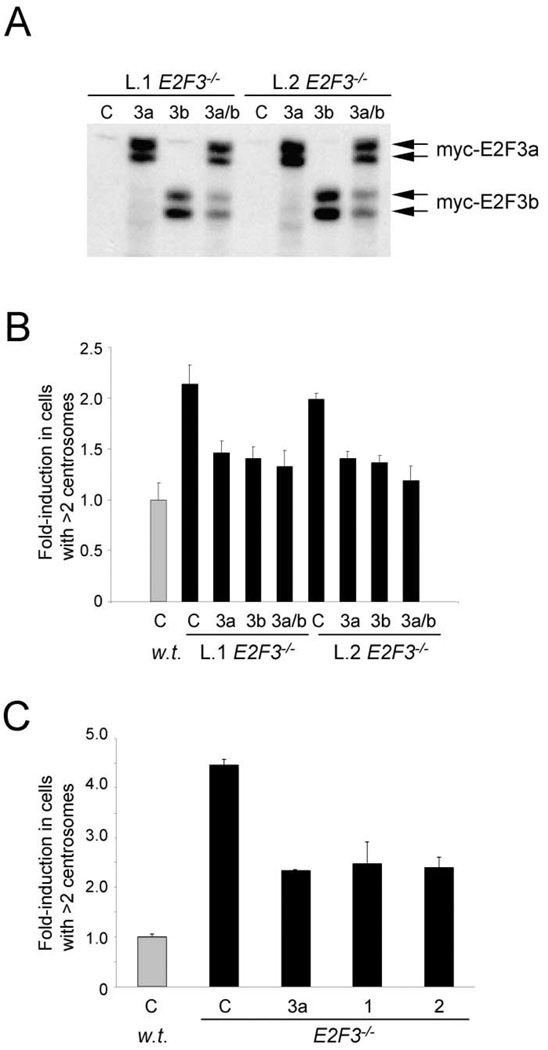
To confirm that loss of E2F3 is the primary cause for centrosome amplification in E2F3−/− MEFs, myc-tagged versions of E2F3 were reintroduced into E2F3−/− MEFs and the centrosome status was determined as before. As shown in Figures 3A and 3B, exogenous expression of E2F3a and E2F3b in cells derived from two different E2F3−/− embryos significantly suppressed, in each case, the number of cells harboring more than two centrosomes. These results demonstrate that centrosome amplification is a direct consequence stemming from the loss of E2F3 and not from a secondary genetic change that might have occurred in the generation of E2F3−/− cells. Rescue experiments with exogenous E2F3a or E2F3b alone did not yield any insight as to which E2F3 family member is responsible for preventing centrosome amplification since overexpression of either E2F3a or E2F3b protein could rescue to the same extent (Figures 3A and 3B). Generation of E2F3a and E2F3b knockout mice should clarify the roles played by each of these proteins in the control of the centrosome cycle.
Figure 3. Ectopic expression of E2F3a and E2F3b suppresses centrosome amplification in E2F3−/− MEFs.
A: Western blot analysis of protein lysates from two litters (L.1, L.2) of wild-type or E2F3−/− cells infected with control (C), myc-E2F3a-expressing (3a), myc-E2F3b-expressing (3b), or with both myc-E2F3a- and myc-E2F3b-expressing (3a/b) retroviruses. Cell lysates were subjected to SDS-PAGE, transferred unto PDVF membranes, and probed with an antibody that recognizes the myc epitope of the myc-tagged E2F3a and E2F3b proteins.
B: Fold induction of centrosome amplification in wild-type ( ) or E2F3−/− (■) ells. MEFs were infected with retroviruses as in (A), and centrosomes were stained with a primary antibody against γ-tubulin and a rhodamine-conjugated secondary antibody.
) or E2F3−/− (■) ells. MEFs were infected with retroviruses as in (A), and centrosomes were stained with a primary antibody against γ-tubulin and a rhodamine-conjugated secondary antibody.
C: Fold induction of centrosome amplification in wild-type ( ) or E2F3−/− (■) cells infected as in (A), except that parallel plates were infected with retroviruses expressing HA-hE2F1 (1) or HA-hE2F2 (2).
) or E2F3−/− (■) cells infected as in (A), except that parallel plates were infected with retroviruses expressing HA-hE2F1 (1) or HA-hE2F2 (2).
Deregulation of the centrosome cycle is specific to loss of E2F3
While E2F3 shares many structural and functional properties with E2F1 and E2F2, two E2F family members also thought to act as transcriptional activators important for promoting cellular proliferation (Leone et al., 1998; Trimarchi and Lees, 2002), other experiments indicate that these three family members may have unique functions in cells (Cloud et al., 2002; DeGregori et al., 1997; Field et al., 1996; Humbert et al., 2000; Leone et al., 1998, 2001; Saavedra et al., 2002; Wu et al., 2001; Yamasaki et al., 1996, 1998; Ziebold et al., 2001). To address whether appropriate control of the centrosome cycle could also be mediated by E2F1 or E2F2, we initially overexpressed these activities and assessed their ability to alleviate the centrosome amplification defect in E2F3−/− cells. As shown in Figure 3C, overexpression of E2F1 or E2F2 also rescued the centrosome amplification phenotype to the same extent as E2F3a, indicating that these E2F activities, when overexpressed, are sufficient to significantly suppress centrosome amplification in E2F3−/− MEFs.
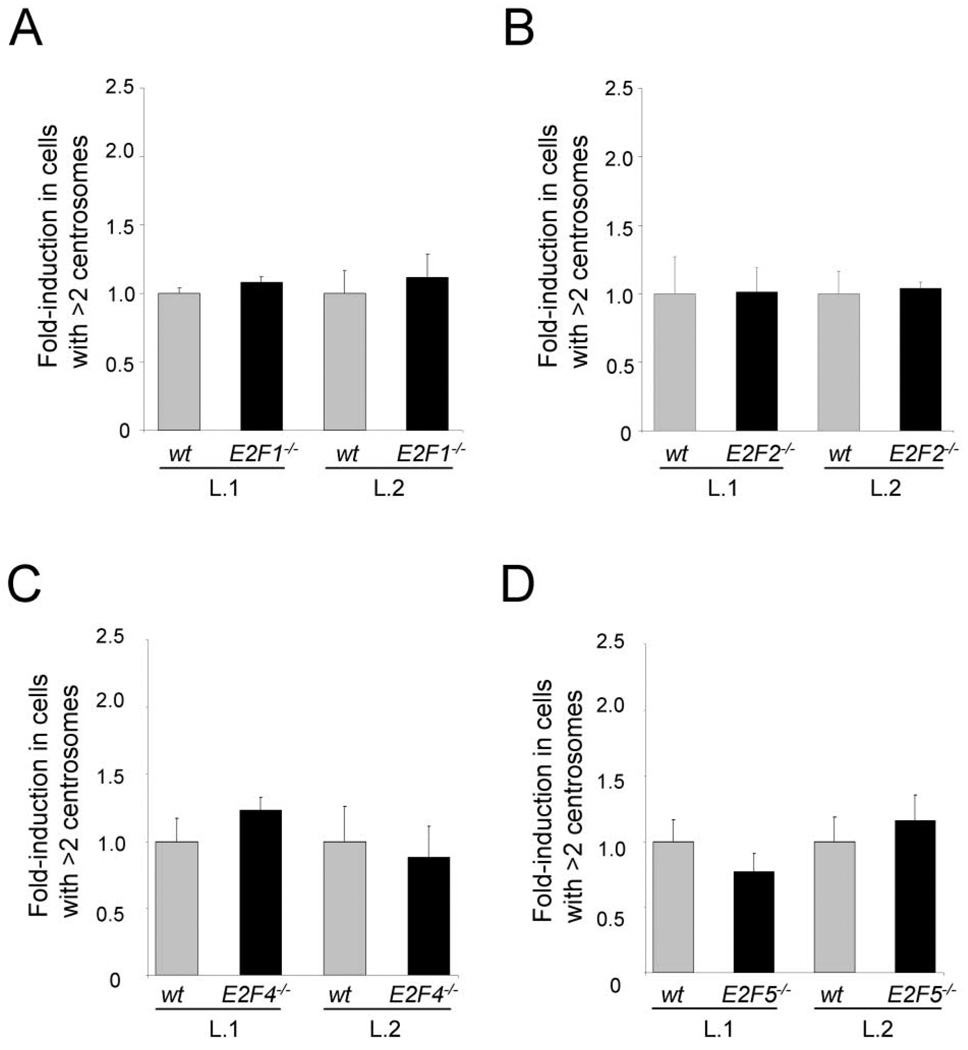
Because overexpression studies do not accurately reflect the normal function of proteins, we used anti-γ-tubulin antibodies to perform a similar analysis in E2F1−/− and E2F2−/− MEFs as that described above for E2F3−/− cells. Analysis of MEFs from multiple different litters revealed no difference in centrosome numbers between E2F1−/−, E2F2−/−, and their wild-type counterparts (Figures 4A and 4B). We also extended our analysis to determine a potential role for members of the repressor subclass of E2F factors, comprised of E2F4 and E2F5, in the control of centrosome duplication. As shown in Figures 4C and 4D, E2F4−/− and E2F5−/− MEFs had a similar frequency of centrosome amplification as wild-type littermate cells. Taken together, our findings suggest that centrosome amplification is a defect specific to the loss of the E2F3 family member.
Figure 4. Centrosome amplification is specific to loss of E2F3.
A: Percentage of centrosome amplification in wild-type ( ) or E2F1−/− (■) MEFs from two independent litters (L.1 and L.2).
) or E2F1−/− (■) MEFs from two independent litters (L.1 and L.2).
B: Percentage of centrosome amplification in wild-type ( ) or E2F2−/− (■) MEFs from two independent litters (L.1 and L.2).
) or E2F2−/− (■) MEFs from two independent litters (L.1 and L.2).
C: Percentage of centrosome amplification in wild-type ( ) or E2F4−/− (■) MEFs from two independent litters (L.1 and L.2).
) or E2F4−/− (■) MEFs from two independent litters (L.1 and L.2).
D: Percentage of centrosome amplification in wild-type ( ) or E2F5−/− (■) MEFs from two independent litters (L.1 and L.2). Centrosomes from early passage (p 2) MEFs were stained with a primary γ-tubulin antibody and a rhodamine-conjugated secondary antibody.
) or E2F5−/− (■) MEFs from two independent litters (L.1 and L.2). Centrosomes from early passage (p 2) MEFs were stained with a primary γ-tubulin antibody and a rhodamine-conjugated secondary antibody.
Loss of E2F3 leads to aneuploidy
Since centrosome amplification can lead to spindle defects and aneuploidy (Fodde et al., 2001; Fukasawa et al., 1996; Fukasawa and Vande Woude, 1997; Hollander et al., 1999; Kaplan et al., 2001; Mantel et al., 1999; Saavedra et al., 1999, 2000; Tutt et al., 1999; Xu et al., 1999), we first investigated whether centrosome defects in E2F3−/− cells were associated with defects in the assembly of the mitotic spindle. To this end, the microtubule-nucleation function of centrosomes was determined by immunofluorescence using antibodies specific to γ-tubulin and α-tubulin. As shown in Figures 5A and 5B, centrosomes in E2F3-deficient cells colocalized with active sites of microtubule nucleation, often leading to aberrant mitoses.
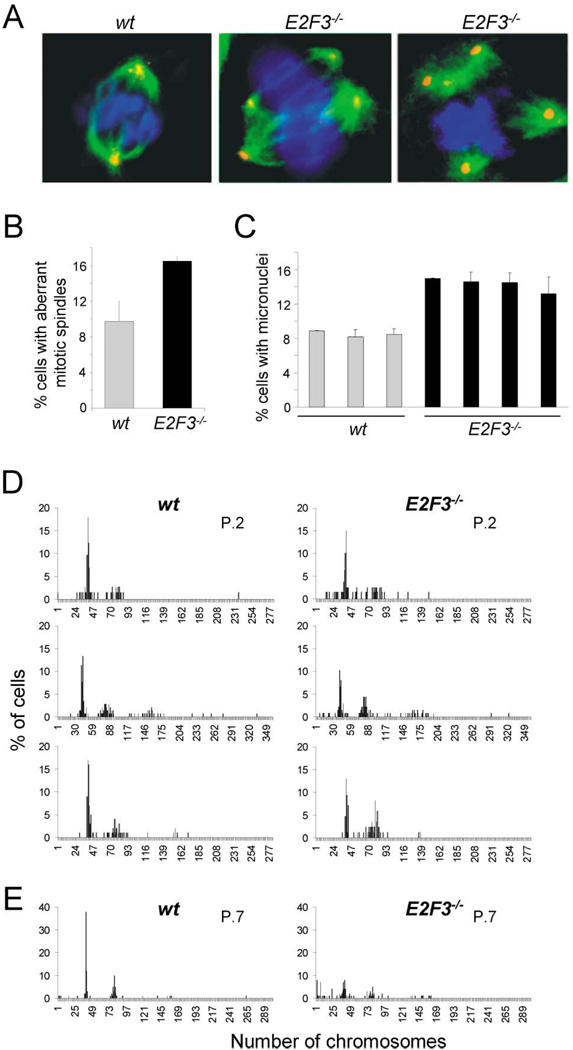
Figure 5. Loss of E2F3 results in aneuploidy.
A: Spindle abnormalities were detected by coimmunostaining the mitotic asters with a primary antibody against α-tubulin and an ALEXA-488-conjugated secondary antibody (shown in green), and the centrosomes with a primary antibody against γ-tubulin and a rhodamine-conjugated secondary antibody (shown in red). The left panel shows a wild-type cell in metaphase with two centrosomes and a normal, bipolar mitotic spindle; the middle and right panel show two E2F3−/− cells with multipolar mitotic spindles.
B: Percentage of wild-type ( ) or E2F3−/− (■) mitotic cells with abnormal multipolar spindles.
) or E2F3−/− (■) mitotic cells with abnormal multipolar spindles.
C: Percentage of wild-type ( ) and E2F3−/− (■) cells containing micronuclei. MEFs derived from three wild-type and four E2F3−/− embryos from one litter are presented. The results were derived from counting 2000 cells in interphase per group (from duplicate plates).
) and E2F3−/− (■) cells containing micronuclei. MEFs derived from three wild-type and four E2F3−/− embryos from one litter are presented. The results were derived from counting 2000 cells in interphase per group (from duplicate plates).
D: Analysis of ploidy in early passage (p 2) wild-type (left panels) and E2F3−/− (right panels) MEFs.
E: Analysis of ploidy in late passage (p 7) wild-type (left panel) or E2F3−/− (right panel) MEFs. At least 50 chromosome spreads were counted per group. Each pair of panels represents wild-type and E2F3−/− MEFs from the same litter. The four pairs presented are from independent litters.
Aneuploidy, a cellular consequence often associated with centrosome amplification and mitotic spindle defects, was measured by two independent methods, the micronucleus assay and direct chromosome counts. A micronucleus is a chromosome or chromosome fragment that is missegregated during mitosis as a consequence of spindle damage and/or a chromosome break, and it appears in the cytoplasm as a DNA-containing sphere surrounded by nuclear membrane. Thus the extent of micronucleus formation reflects the frequency of cells in a population that is losing chromosomes (Saavedra et al., 1999, 2000). Quantification of cells derived from multiple E2F3-deficient embryos revealed a significant increase in micronucleus formation relative to cells derived from wild-type embryos (Figure 5C), suggesting that E2F3−/− MEFs suffer a higher rate of chromosome loss. To more directly assess whether loss of E2F3 can lead to aneuploidy, chromosome numbers were counted in cells from early and late passage populations. As expected, analysis of early passage wild-type cells derived from different litters revealed that the majority of cells contain a 2N content of DNA (Figures 5D and 6). However, the number of chromosomes in E2F3−/− cells varied among MEF preparations derived from different litters, with most preparations containing a near-normal 2N complement of DNA (Figure 5D, top two panels), but some preparations exhibiting clear polyploidy (bottom panel). At later passages, the distribution of chromosomes in E2F3−/− cells was broadened to include many cells containing fewer than 40 chromosomes, while wild-type cells retained mostly a 2N complement of DNA, with a smaller subset of cells having a 4N complement (Figure 5E, bottom panel). We conclude from these findings that loss of E2F3 is associated with increased genetic instability and aneuploidy.
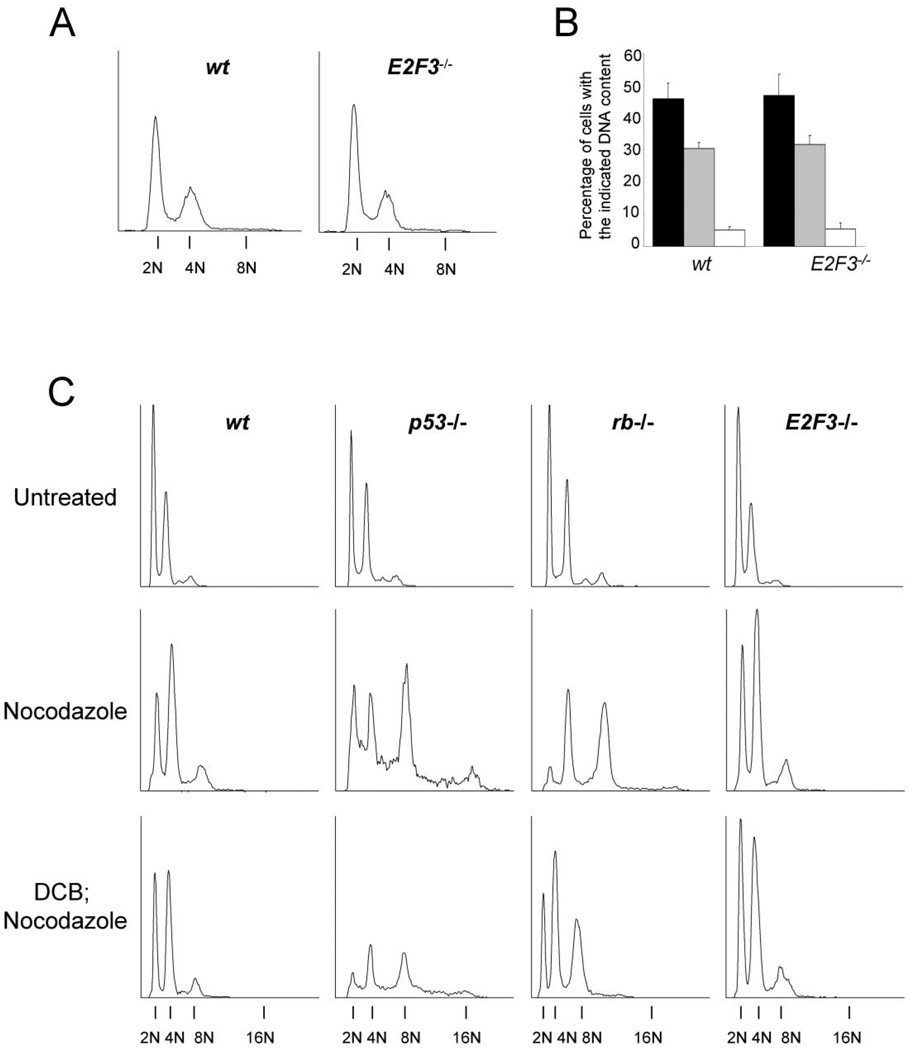
Figure 6. Loss of E2F3 does not result in multinucleation or failure of the tetraploidy checkpoint.
A: Flow cytometric analysis of various early passage mouse embryonic fibroblasts (MEFs). Passage 2 wt or E2F3−/− MEFs were processed as described in Experimental Procedures and analyzed by flow cytometry. Shown are the flow cytometric analyses of cells, showing their respective DNA contents (2N, 4N, or 8N).
B: Quantitative analysis of the percentage of passage 2 wt or E2F3−/− MEF cells with the indicated DNA contents. Values indicate cells with 2N (■), 4N ( ), or 8N (□) DNA contents ± the standard deviation between four embryos from independent litters.
), or 8N (□) DNA contents ± the standard deviation between four embryos from independent litters.
C: Flow cytometric analysis of control, nocodazole, or DCB and nocodazole-treated MEFs. Passage 5 wt, p53−/−, rb−/−, or E2F3−/− MEFs were left untreated or were treated with nocodazole for 72 hr, or with DCB for 36 hr followed by nocodazole treatment for 36 hr. Shown are cells with a 2N, 4N, 8N, or 16N DNA content.
E2F3−/− cells have a functional G1/S tetraploidy checkpoint
Recent observations suggest that more than one model may explain how centrosome amplification occurs (Borel et al., 2002; Meraldi et al., 2002; Tarapore et al., 2001). One of the models proposes that direct deregulation of the centrosome duplication cycle can lead to centrosome amplification (Tarapore et al., 2001). The other model proposes that defective cytokinesis can result in binucleation and the inheritance of two pairs of centrosomes, leading to the amplification of centrosomes during subsequent cell cycles (Borel et al., 2002; Meraldi et al., 2002). To determine whether this latter mechanism may be responsible for centrosome amplification in E2F3−/− cells, we assessed gross ploidy status in these cells by flow cytometry. This analysis revealed aDNA content in E2F3−/− cells that was almost identical to that of wild-type cells (Figures 6A and 6B), suggesting that loss of E2F3 does not lead to increased multinucleation in the population.
To explore whether G1/S checkpoint function was intact in E2F3−/− MEFs, cells were treated with nocodazole, dihydrocytochalasin B (DCB; data not shown), or DCB followed by nocodazole as described previously (Borel et al., 2002; Lanni and Jacks, 1998). Nocodazole prevents polymerization of the α- and β̃-tubulin subunits of the mitotic spindle, whereas DCB disrupts the normal formation of a contractile actin ring necessary for proper cytokinesis (Aubin et al., 1981; Martineau et al., 1995). Since loss of either Rb or p53 is known to abrogate G1/S checkpoint function (Di Leonardo et al., 1997, Lanni and Jacks, 1998; Niculescu et al., 1998), Rb−/− and p53−/− fibroblasts were utilized as controls for this assay. Treatment of wild-type or E2F3−/− cells with nocodazole, or DCB followed by nocodazole, resulted in the accumulation of cells with a 4N content of DNA (Figure 6C). As expected, similar treatment of Rb−/− or p53−/− cells led to an accumulation of cells with DNA contents higher than 4N. Taken together, these experiments suggest that E2F3−/− cells are not multinucleated and have an intact G1/S tetraploidy checkpoint. Therefore, defects in cytokinesis or checkpoint function are unlikely to account for the centrosome amplification defects observed in E2F3−/− cells.
E2F3 controls cyclin E expression and centriole separation
A number of other reasons could account for the abnormally high number of centrosomes observed in E2F3-deficient cells, including premature centriole separation or centrosome reduplication. One of the first steps required before the onset of centrosome duplication is the phosphorylation, by cyclin E-dependent kinase activity, of nucleophosmin B at threonine 199 (T199). Phosphorylation at T199 leads to the release of nucleophosmin B from centrosomes and the separation of paired centrioles (Okuda et al., 2000). In its unphosphorylated state, nucleophosmin B remains associated with centrosomes and is thought to prevent the separation of paired centrioles and thus preclude centrosome duplication. In previous studies, we demonstrated that the concerted ablation of E2F1, E2F2, and E2F3 proteins from MEFs led to an increase in cyclin E mRNA levels (Wu et al., 2001); however, the identity of the specific E2F family member responsible for regulating cyclin E expression was not investigated. Nonetheless, these observations raised the possibility that deregulated cyclin E expression could be a contributing factor toward the centrosome amplification observed in E2F3−/− MEFs.
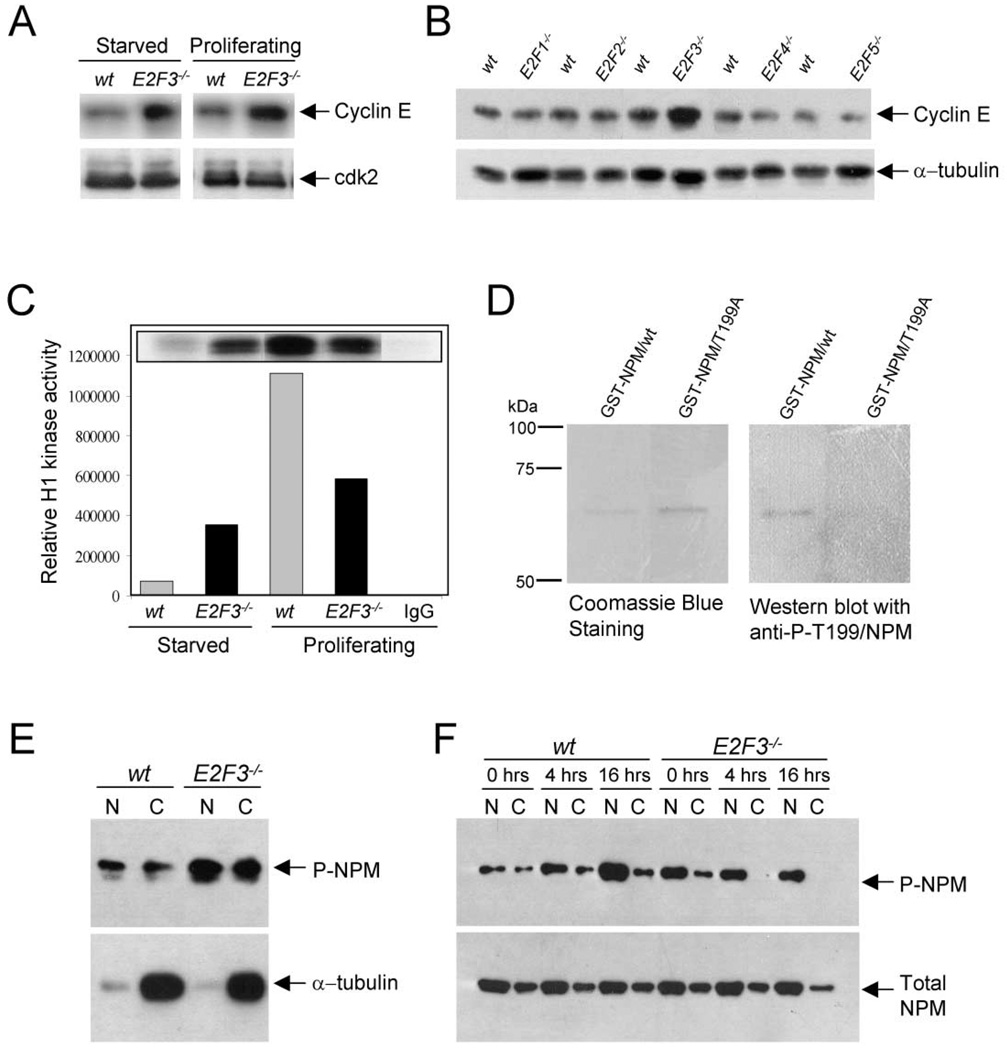
To first determine whether the loss of E2F3 may lead to deregulated cyclin E expression, we analyzed cyclin E protein levels in wild-type and E2F3-deficient MEFs by Western blot assays. As shown in Figure 7A, cyclin E but not cdk2 protein levels were consistently higher in both quiescent and proliferating E2F3−/− cells than in wild-type control cells. This observed increase was specific to the loss of E2F3 since disruption of each of the other E2Fs had no effect on cyclin E protein levels (Figure 7B). Concordant with an increase in cyclin E protein, cyclin E-dependent kinase activity was modestly, but reproducibly elevated in quiescent E2F3−/− cells (Figure 7C). Moreover, while cyclin E-dependent kinase activity in wild-type cells could be significantly induced by serum-mediated growth stimulation, further activation in E2F3−/− cells was minimal.
Figure 7. Loss of E2F3 leads to premature cyclin E-dependent kinase activation.
A: Western blot analysis showing Cyclin E (top panel) or Cdk2 (as a loading control, bottom panel) protein levels in proliferating and starved wild-type and E2F3−/− cells.
B: Western blot analysis showing Cyclin E protein levels in serum-starved wild-type, E2F1−/−, E2F2−/−, E2F3−/−, E2F4−/−, and E2F5−/− cells. The bottom panel shows α-tubulin levels, used as loading controls. The lysate from E2F3−/− MEFs presented were derived from a different litter than the ones shown in (B).
C: Relative activity of cyclin E-dependent kinase in protein lysates derived from starved and proliferating wild-type and E2F3−/− cells, using histone-1 (H1) as a substrate.
D: Western blot analysis of isolated GST-tagged wt nucleophosmin B (GST-NPM/wt) or a GST-tagged mutant nucleophosmin, in which the threonine 199 site was changed to alanine (GST-NPM/T199A). The left panel shows the coomassie-stained gel to show the equal loading of protein, and the right panel a Western blot probed with an antibody against phospho-T199 nucleophosmin B.
E: Western blot analysis of nuclear and cytoplasmic protein fractions from serum-starved wt or E2F3−/− MEFs. The Western blot was probed with the antibody described in (D). To ensure that equal loading was achieved, the same membrane was probed with α-tubulin.
F: wt and E2F3−/− cells were cultured under serum starvation conditions and released from the G1 block by serum addition. Nuclear or cytoplasmic fractions were obtained 0, 4, and 16 hr after serum addition. The Western blot was probed with the same antibody used in (D) and (E). As a loading control, we probed the same membrane with an antibody against total nucleophosmin (NPM).
Considering the established link between cyclin E-dependent phosphorylation of nucleophosmin and centrosome duplication, our results raised the possibility that deregulated cyclin E activity in E2F3-deficient cells could result in the untimely phosphorylation and release of nucleophosmin B from centrosomes, leading to premature centriole separation and duplication. To explore this possibility, we used an antibody specific for the T199-phosphorylated form of nucleophosmin to assess its phosphorylation status in E2F3−/− cells. To test the specificity of this antibody, purified GST-tagged wild-type or mutant nucleophosmin, in which threonine 199 was changed to alanine (T199A), was preincubated with purified cyclin E-cdk2 complexes and then subjected to SDS-PAGE. Commassie blue staining of blots demonstrated equal loading of proteins into gels (Figure 7D, left panel). Importantly, the anti-phospho-T199-specific nucleophosmin antibody recognized the wild-type but not the mutant form of nucleophosmin by Western blot analysis (Figure 7D, right panel), demonstrating the specificity of these antibodies for the T199-phosphorylated form of nucleophosmin.
Using these specific antibodies, we next explored the phosphorylation status of nucleophosmin in E2F3−/− cells synchronized by serum deprivation. As shown in Figure 7E, quiescent E2F3−/− cells had increased levels of phospho-T199 nucleophosmin relative to wild-type cells. Serum stimulation of quiescent wild-type cells into the cell cycle resulted in an accumulation of phospho-T199 nucleophosmin that peaked by 16 hr post induction. In contrast, further increases in phospho-T199 nucleophosmin were not observed in E2F3−/− serum-stimulated cells. The T199 phosphorylation status of nucleophosmin during the cell cycle of wild-type and mutant cells was consistent with the observed cyclin E-dependent kinase activity in these cells (compare with Figure 7C, and data not shown). While in all cases the total levels of nucleophosmin protein did not vary significantly, we were unable to detect any phospho-nucleophosmin in cytoplasmic fractions of serum-stimulated E2F3−/− cells, the significance of which remains to be examined.
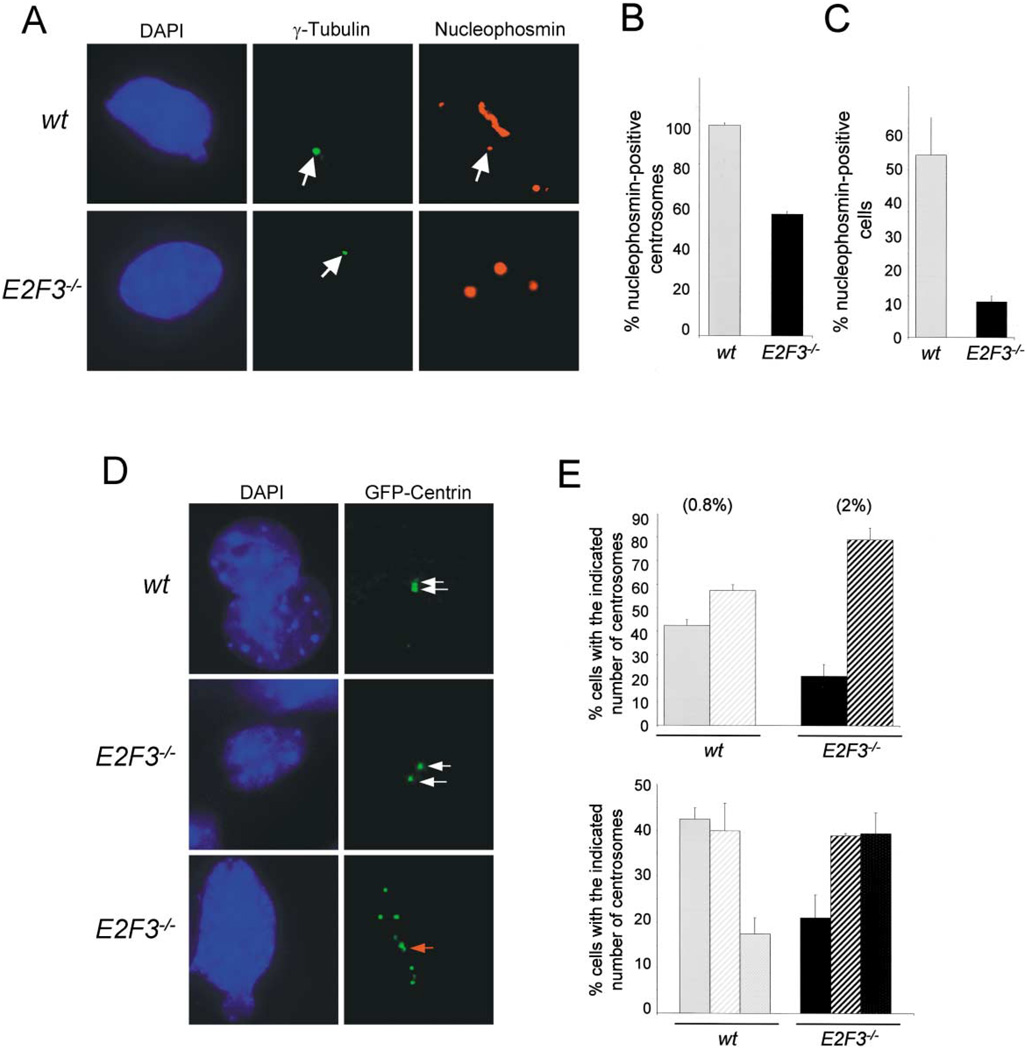
We then compared the association of nucleophosmin B with centrosomes in quiescent wild-type and E2F3−/− cells by immunostaining cells with nucleophosmin B-specific and γ-tubulin-specific antibodies (Okuda et al., 2000). As reported previously, nucleophosmin localized to essentially all the unduplicated centrosomes in wild-type cells (Okuda et al., 2000) (>99%; Figures 8A and 8B). In contrast, nucleophosmin localized to only half of the unduplicated centrosomes in E2F3−/− cells (~55%). Since quiescent E2F3−/− cells have a significantly lower proportion of unduplicated centrosomes relative to wild-type cells (20% versus 55%, respectively), the actual fraction of cells containing centrosome-associated nucleophosmin is significantly lower in E2F3−/− than that in wild-type cells (Figure 8C). These data demonstrate that inactivation of E2F3 results in defects in nucleophosmin association with centrosomes. We propose that inappropriate cyclin E-dependent kinase activation and phosphorylation of nucleophosmin in E2F3−/− cells may lead to the premature release of nucleophosmin from unduplicated centrosomes or, alternatively, to the improper loading of nucleophosmin on centrosomes during the previous mitosis.
Figure 8. Deregulation of cyclin E-dependent kinase activity in E2F3−/− cells leads to premature centriole separation.
A: Coimmunostaining of γ-tubulin (middle panels) and nucleophosmin B (right panels) in cells cultured under serum-starvation conditions.
B: Percentage of nucleophosmin B-positive unduplicated centrosomes in wild-type ( ) or E2F3−/− (■) cells under serum starvation conditions.
) or E2F3−/− (■) cells under serum starvation conditions.
C: Percentage of cells with nucleophosmin B-positive centrosomes in the total population of wild-type ( ) or E2F3−/− (■) cells cultured under serum starvation.
) or E2F3−/− (■) cells cultured under serum starvation.
D: Wild-type and E2F3−/− cells expressing GFP-centrin-2. Shown from top to bottom: a wild-type cell with an unduplicated centrosome, prior to the separation of centrioles; an E2F3−/− cell with a separated centrosome; an E2F3−/− cell with multiple, separated centrioles. The left panels show the corresponding DAPI-stained nuclei. White arrows indicate individual centrioles while the red arrow indicates a separated centriole associated with a nascent centriole.
E: Top panel, percentage of wild-type ( ) or E2F3−/− cells (■) containing one (solid bars) or ≥2 separated centrosomes. Bottom panel, percentage of wild-type (
) or E2F3−/− cells (■) containing one (solid bars) or ≥2 separated centrosomes. Bottom panel, percentage of wild-type ( ) or E2F3−/− cells (■) containing unduplicated (solid bars), ≥2 separated and unduplicated (stripes), or ≥ 2 separated and duplicated centrosomes (dotted) in density-arrested conditions. Centrioles were visualized using GFP-centrin-2, and the calculated percentages are based on the results from counting 100 cells. Shown in parenthesis are the percentage of BrdU-positive cells in the population.
) or E2F3−/− cells (■) containing unduplicated (solid bars), ≥2 separated and unduplicated (stripes), or ≥ 2 separated and duplicated centrosomes (dotted) in density-arrested conditions. Centrioles were visualized using GFP-centrin-2, and the calculated percentages are based on the results from counting 100 cells. Shown in parenthesis are the percentage of BrdU-positive cells in the population.
Finally, centrosomes in wild-type and E2F3−/− cells were also visualized by the introduction of a GFP-tagged version of centrin-2 (GFP-centrin), an essential component of centrioles. There are two main advantages in analyzing centrosomes in cells expressing GFP-centrin-2. One advantage is that centrosomes can be visualized and quantified during interphase. A second advantage is that the resolution of its visualization by direct immunofluorescent microscopy is sufficient to discriminate between centriole pairs, single separated centrioles, and separated centrioles with an associated nascent centriole as they begin to duplicate (White et al., 2000). Wild-type and E2F3−/− cells were thus infected with a retrovirus vector expressing GFP-centrin-2, brought to quiescence by density arrest, fixed and visualized by fluorescent microscopy. Cells were classified as having one centrosome if GFP-positive centrioles were tightly paired (Figure 8D; top right panel) and as having two or more centrosomes if the centrioles were separated or amplified (Figure 8D; middle and bottom right panels, respectively). Under density-arrested conditions, approximately 45% of wild-type and 20% of E2F3−/− MEFs contained one unduplicated centrosome (Figure 8E, top panel). Interestingly, a high proportion of the quiescent E2F3−/− MEFs containing separated centrioles were in the process of duplicating their centrosomes, as indicated by the presence of a growing nascent daughter centriole adjacent to the larger original centriole (Figures 8D [see arrow in bottom panel] and 8E). These observations suggest that separated centrosomes in E2F3−/− MEFs can undergo centrosome duplication prematurely. The premature separation of centrosomes in E2F3−/− MEFs was not due to the inability of E2F3 null cells to undergo growth-arrest under these conditions, as very few of these cells continued to replicate their DNA (2% of cells were BrdU positive; Figure 8E). Together, these findings suggest that inappropriately high levels of cyclin E-dependent kinase activity in quiescent E2F3−/− cells may result in the phosphorylation and release of nucleophosmin from centrosomes, leading to centriole separation and premature centrosome duplication, and eventually to centrosome amplification.
Discussion
Genome-wide analysis of gene expression suggests that E2F regulates the expression of genes involved in G1/S progression and DNA replication, as well as the expression of genes involved in apoptosis, DNA repair, and mitosis (Ishida et al., 2001; Muller et al., 2001; Ren et al., 2002). Recent observations have revealed extensive complexity among the E2F family of transcription factors. While E2F4 and E2F5 are required in mediating a p16INK4a-induced cell growth arrest, the E2F1, E2F2 and E2F3 members are essential for cell cycle progression and cellular proliferation (Gaubatz et al., 2000; Wu et al., 2001). Moreover, the expression and activities of the various E2F family members can be regulated by each other (Trimarchi and Lees, 2002). Our present work identifies and describes a function that is unique to the E2F3 gene in regulating the centrosome duplication cycle. We find that inactivation of E2F3 leads to an increase in cyclin E expression, inappropriate cyclin E-dependent kinase activation, and a defect in nucleophosmin B association with unduplicated centrosomes. These events appear to result in premature centriole separation and duplication, centrosome amplification, and genetic instability. While E2F3 has been shown to be critical for DNA replication and normal cellular proliferation, and thus its loss may be expected to decrease cellular proliferation rates of cancer cells (Leone et al.; 1998, 2001; Humbert et al., 2000; Wu et al., 2001), its loss would also promote genetic instability and, paradoxically, may enhance the metastatic potential of tumor cells.
Previous observations indicated a role for the E2F transcriptional program in the control of centrosome biology. Initial clues from studies using hydroxyurea-arrested tumor cells suggested that centrosome reduplication could be inhibited by a nonphosphorylatable form of Rb or by p16INK4a (Meraldi et al., 1999). In addition, introduction of the papilloma virus E7 oncogene into primary human fibroblasts was shown to induce centrosome amplification (Duensing et al., 2000). More recently, genome-wide microarray analysis revealed that loss of the p107 and p130 pocket proteins can lead to the upregulation of nek2 expression (Ren et al., 2002), and that overexpression of E2Fs can lead to the activation of RanBP (Muller et al., 2001); nek2 and RanBP are two genes whose products localize to centrosomes and affect centriole cohesion and microtubule nucleation, respectively (Carazo-Salas et al., 2001; Meraldi et al., 1999; Wilde and Zheng, 1999). Whether these and other centrosome-related genes are specific transcriptional targets regulated by E2F3 remains to be determined.
Recent publications (Borel et al., 2002; Meraldi et al., 2002; Tarapore et al., 2001) have proposed two models to explain how centrosome amplification may occur. The first model suggests that loss of tumor suppressor function, such as p53, leads to centrosome amplification by the uncoupling of the centrosome duplication cycle and the cell cycle (Tarapore et al., 2001). Recent observations have revealed that defects in cytokinesis may also lead to polyploidy and the acquisition of more than one centrosome per daughter cell, leading to centrosome amplification (Borel et al., 2002; Meraldi et al., 2002). The fact that early passage E2F3−/− cells have similar DNA contents as wild-type cells suggests that loss of E2F3 does not induce major changes in ploidy. Moreover, E2F3-deficient cells have a competent G1/S tetraploidy checkpoint since they arrest normally when challenged with agents that disrupt spindle formation (nocodazole) or cytokinesis (DCB). Together with the observation that E2F3−/− cells have increased frequencies of separated centrosomes that are often associated with nascent daughter centrioles, these findings suggest that loss of E2F3 leads to increased centrosome amplification by inducing a defective centrosome duplication cycle, rather than indirectly by affecting ploidy or cytokinesis. Even though we believe that the micronucleus formation in E2F3−/− cells is due to increases in spindle abnormalities, we have not yet ruled out whether the micronuclei formation is due to other possible mechanisms such as defects in the spindle assembly checkpoint machinery (Amon, 1999) or the induction of chromosome breaks, amechanism commonly associated with the expression of certain oncogenes such as Ras (Denko et al., 1994; Saavedra et al., 1999, 2000).
How might E2F3 regulate the centrosome duplication cycle? The E2F3 locus encodes for two gene products, E2F3a and E2F3b (Leone et al., 2000), which differ in their expression patterns during the cell cycle. E2F3a protein levels oscillate during the cell cycle, reaching peak levels late in G1 and early S phase, a time when E2F target genes are maximally activated. In contrast, E2F3b protein is constant throughout the cell cycle and can be found in association with Rb during G0 (Leone et al., 1998, 2000). These properties of the E2F3s have led to the suggestion that E2F3a may function as an activator, and E2F3b as a repressor of transcription, and thus these two E2F3 isoforms may have opposing roles in the control of cell proliferation. Consistent with this notion, the targeted inactivation of both E2F3a and E2F3b may lead to a decrease in expression of many E2F target genes and an increase in others (such as cyclin E) (Wu et al., 2001). We speculate that the E2F3 locus regulates centrosome duplication in two ways. First, E2F3b, by mediating the repression of cyclin E, may prevent the premature activation of cyclin E/cdk2, phosphorylation of nucleophosmin B, and separation of centrioles. Second, E2F3a, by activating the expression of G1/S-specific genes involved in the inhibition of centrosome rereplication (such as BRCA1, Wang et al., 2000; Xu et al., 1999) may prevent the inappropriate reduplication of centrosomes. Thus, loss of E2F3a and E2F3b function may disrupt the coordinated expression of activities required for the timely duplication of centrosomes, leading to premature centrosome duplication and amplification. While overexpression of E2Fs can partly restore normal centrosome numbers in E2F3−/− cells, overexpression studies are difficult to interpret, and the analysis of E2F-deficient cells clearly demonstrates that E2F3 is unique in its ability to impact on the centrosome duplication cycle. At least in part, functional specificity among E2Fs in the control of centrosome duplication may reflect their ability to regulate distinct subsets of target genes (DeGregori et al., 1997; Ishida et al., 2001; Moroni et al., 2001; Muller et al., 2001). The generation of mice and cells deficient for the individual E2F3s should provide important insights on how this transcriptional program regulates centrosome duplication.
The observed alterations in the centrosome cycle of E2F3−/− cells are consistent with previous roles proposed for cyclin E-dependent kinase activity in the regulation of centrosome duplication (Hinchcliffe et al., 1999; Lacey et al., 1999; Matsumoto et al., 1999; Meraldi et al., 1999). Cyclin E/cdk2 has been shown to phosphorylate nucleophosmin B on threonine 199, an event that promotes nucleophosmin B dissociation from centrosomes late in G1, allowing centriole separation and duplication to occur as cells progress through G1/S and S phase, respectively (Okuda et al., 2000). Cyclin E/cdk2 also phosphorylates and stabilizes mMsp1p (esk2), a protein kinase required subsequent to centriole separation for the formation of nascent centrioles (Fisk and Winey, 2001). Additionally, cyclin E/cdk2 can phosphorylate CP110, a protein that plays a critical role in promoting duplication of centrosomes (Chen et al., 2002). While our data support a role for deregulated cyclin E activity in mediating centrosome amplification in E2F3−/− cells, the premature induction of cyclin E/cdk2 activity is not likely the sole factor responsible for this phenomenon, as overexpression of cyclin E has been shown to result in only modest increases in centrosome amplification (Mussman et al., 2000; Spruck et al., 1999). Moreover, the inactivation of Rb, which also results in the premature activation of cyclin E/cdk2, does not lead to centrosome amplification to any appreciable extent and does not increase the frequency of centrosome amplification induced by loss of E2F3 (Fukasawa et al., 1996; and data not shown). Nonetheless, the finding that the specific loss of E2F3 function perturbs both the centrosome cycle (this study) and cell cycle kinetics (Humbert et al., 2000; Wu et al., 2001) suggests that this E2F family member is unique in its ability to impact on two cellular processes whose timing of execution is crucial for the maintenance of genomic stability: centrosome duplication and DNA replication.
In summary, our results identify and define a specific role for E2F3 in the control of the centrosome duplication cycle. E2F3 may thus be viewed as an activity important for coordinating two critical processes during the cell cycle, DNA replication and centrosome duplication, and as such represents a key activity essential for the appropriate segregation of genetic material to each daughter cell.
Experimental procedures
Generation of mouse embryonic fibroblasts
The generation of the conventional (E2F3−/−) and the conditional (E2F3f/f) E2F3 knockout mice has been described previously (Leone et al., 2001; Wu et al., 2001). The E2F1−/−, E2F2−/−, E2F4(+/−), and E2F5(+/−) mice were a generous gift from Dr. Michael Greenberg and Drs. Stuart Orkin, Joseph Nevins, and David Livingston, respectively. All the mouse embryonic fibroblasts (MEFs) utilized in this study were generated from 13.5 day embryos of a mixed 129/Sv and C57Bl6 background, where the individual contributions of both genetic backgrounds were similar. Unless otherwise specified, MEFs were cultured in 15% FBS/DMEM in an atmosphere of 5.5%–6% CO2.
Immunohistochemistry
MEFs were plated at 4 × 104 cells per well into 2-well tissue culture chamber slides and grown for 2–3 days. Cells were fixed in cold 4% parafolmaldehyde, washed in PBS, permeabilized in 1% NP40/PBS solution, and blocked in 5% BSA in PBS. Centrosomes were stained overnight at 4°C with monoclonal antibodies against pericentrin (Transduction Laboratories) or γ-tubulin (Sigma) and an anti-mouse rhodamine-conjugated secondary antibody (Pierce). The mitotic spindle was stained with α-tubulin (Sigma) and an Alexa 488-conjugated secondary antibody (Molecular Probes). The nucleophosmin B and γ-tubulin colocalization was achieved by fixing the cells with methanol:acetone (1:1), blocking with 15% goat serum, and staining with rhodamine-conjugated anti-mouse and Alexa 488-conjugated anti-rabbit secondary antibodies, respectively. Cells were washed in PBS and counterstained with 4,6-diamidino-2-phenylindole (DAPI). For each experiment involving calculations of the frequencies of centrosome or spindle defects, at least 100 cells from each of two or more chambers were counted per group.
Proliferation assays
For the serum arrest experiments, 8 × 104 cells were plated into 60 mm tissue culture plates, allowed to attach for 16 hr, and then incubated in medium containing 0.2% fetal bovine serum (FBS) for 60 hr. For the density arrest experiments, cells were allowed to remain contact inhibited for 48 hr. BrdU was added to the medium in order to determine the efficacy of the density and serum arrest, and BrdU incorporation was determined as described previously (Leone et al., 1998). Briefly, cells were fixed in a 1:1 methanol:acetone solution, rinsed twice with PBS, and treated with 1.5 N HCl. BrdU incorporation was detected using a primary anti-BrdU antibody and a rhodamine-conjugated secondary antibody; nuclei were counterstained with DAPI. The percentage of BrdU-positive cells was obtained from the analysis of at least 500 nuclei per time point.
Retroviral infections
The retroviral constructs utilized in this study were generated by standard cloning techniques. We utilized the following constructs in our study: pBABE-puro, pBABEhygro, pBABEpuro-CRE, pBABEhygro-E2F3a (myc-tagged), pBABEhygro-E2F3b (myc-tagged), pBABEpuro-hE2F1 (HA-tagged), pBA-BEpuro-hE2F2 (HA-tagged), and pBABEhygro-GFP-centrin-2. Constructs were transfected into ecotropic Phoenix cells by the CaCl2 procedure and high-titer viruses were harvested 48 and 60 hr after transfection. Primary MEFs were incubated with medium from the transfected Phoenix cells containing the indicated retroviruses and 4 µg/ml polybrene. Twenty-four hours after infection, cells were selected using hygromycin (150 µg/ml) or puromycin (2.5 µg/ml) and cultured in selection medium for 60 hr. Cells were cultured in the absence of selection for 24 hr and plated as described in the previous section. Cells infected with pBABEhygro-GFP-centrin-2 were not selected with hygromycin because the cells expressing GFP-centrin-2 can be directly visualized by microscopy.
Western blot and kinase assays
Protein lysates were obtained by incubating cells in gel shift lysis buffer (50 mM HEPES; pH 7.9; 250 mM KCl; 0.1 mM EDTA; 0.1 mM EGTA; 0.1% NP40 and 10% glycerol; 0.5 mM NaF; 0.1 mM NaVO4; 0.1 mM PMSF; 10 mM β-glycerophosphate; 0.1 mMDTT; 0.1 mg/ml aprotinin; 0.1 mg/ml leupeptin) for 20 min at 4°C. Proteins were separated in SDS polyacrylamide gels and transferred to polyvinylidene fluoride membranes. Blots were blocked with Tris-buffered saline (TBS) containing 5% skim milk for 2 hr and then incubated overnight (4°C) in TBS containing 0.5 µg/ml of antibodies against E2F3 (C-18, Santa Cruz), Cdk2 (M-2, Santa Cruz), Cyclin E (M-20, Santa Cruz), and α-tubulin (Sigma), phosphoT199 nucleophosmin (Cell Signaling Technology), CDK4 (C-22, Santa Cruz), or total nucleophosmin B (Zymed) as indicated. Western blots for detection of phosphorylated nucleophosmin were done similarly, except that the serine/threonine phosphatase inhibitor calyculin A (Cell Signaling Technology) was included in the culture medium 5 min prior to harvest at a concentration of 100 nM and microcystin LR (Calbiochem) was included in the lysis solutions at a concentration of 10 µM. Cytoplasmic extracts were obtained by incubating cell pellets in hypotonic lysis buffer (10 mM HEPES; pH 7.5; 10 mM KCl; 3 mM MgCl2; 0.05%; 1mM EDTA; 0.5 mM NaF; 0.1 mM NaVO4; 0.1 mM PMSF; 10 mM β-glycerophosphate; 0.1 mM DTT; 0.1 mg/ml aprotinin; 0.1 mg/ml leupeptin) for 20 min at 4°C. Nuclear lysates were obtained by incubating the pellets remaining from the cytoplasmic extractions in gel lysis buffer for 20 min at 4°C. Primary antibodies were detected using a horseradish peroxidase-conjugated secondary antibody and the ECL reagent (Amersham). For determining the specificity of the Cdk2-dependent phosphorylation of nucleophosmin, GST-NPM/wt and GST-NPM/T199A (thr199 is replaced by ala) were subjected to in vitro kinase reaction with Cdk2/cyclin E purified from baculovirus, as described previously (Okuda et al., 2000; Tokuyama et al., 2001). The reaction mixtures were run on SDS-PAGE, transferred onto a membrane, and immunoblotted with anti-phosphoT199/NPM antibody. Kinase assays using histone 1 (H1) were performed as described previously (Leone et al., 1998).
Analysis of ploidy
The micronucleus assay was done as described previously (Saavedra et al., 1999, 2000). Briefly, 4 × 104 cells were plated into each well of a 2-well chamber slide (Falcon). After 48 hr in culture, cells were fixed in 4% paraformaldehyde and stained with DAPI. Micronuclei appear as spherical structures with a similar morphology to the nucleus except that their sizes range from 1/10th to 1/100th the size of a nucleus; 1000 cells were counted for each genotype analyzed. To calculate ploidy, we obtained chromosomes by standard procedures. Briefly, asynchronously growing cells were treated with colcemid to inhibit the formation of the mitotic spindle, collected, and placed in a hypotonic solution. After fixation in methanol:acetic acid (3:1 ratio), cells were dropped into glass coverslips and stained with DAPI. At least 100 chromosome spreads per group were analyzed.
Flow cytometric analysis
Flow cytometry was done as previously described (Leone et al., 1998). Briefly, cells were collected, fixed in 70% ethanol, resuspended in PBS, treated with 2N HCl containing 0.2 mg/ml pepsin, and neutralized with 0.1 M sodium tetraborate. Following washes with PBS, cells were incubated in RNase A (0.5 mg/ml) and propidium iodide (50 µg/ml), diluted to an equal concentration, and analyzed by flow cytometry. For the endoreduplication assays, cells were treated with nocodazole alone at a concentration of 0.125 µg/ml, as described in Lanni and Jacks (1998), or medium, for 72 hr. Parallel plates were treated with DCB (10 µM) for 36 hr or with DCB for 36 hr followed by nocodazole treatment for 36 hr (Borel et al., 2002). Cells were then analyzed for flow cytometry.
SIGNIFICANCE
E2F3 has been shown to play a critical role in the control of S phase entry. Our current finding that the specific loss of E2F3 leads to aberrant centrosome duplication demonstrates that the E2F3 family member has the unique role of coordinating two important processes during the cell cycle: DNA and centrosome duplication. Thus, by integrating these two processes during the cell cycle, E2F3 ensures the accurate transmission of genetic material to each daughter cell. In cancer, however, inactivation of E2F3 may be a two-edged sword: its loss may decrease the rate of cellular proliferation, but may also lead to genetic instability and thus increase the metastatic potential of cancer cells.
Acknowledgments
We thank Lizhao Wu and Juan Wu for the preparation of mouse embryo fibroblasts and Jana Opavska for technical assistance. We thank Dr. Jeffrey Salisbury for providing the centrin-GFP construct. This work was supported by grants from the NIH/NCI (to G.L.). H.I.S. was supported by an NCI award; C.T. was supported by an Up on the Roof Human Cancer Genetics Postdoctoral Fellowship and an American Cancer Society Fellowship. G.L. is a V-Foundation and PEW Scholar.
References
- Amon A. The spindle checkpoint. Curr. Opin. Genet. Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- Archer J, Solomon F. Deconstructing the microtubule-organizing center. Cell. 1994;76:589–591. doi: 10.1016/0092-8674(94)90496-0. [DOI] [PubMed] [Google Scholar]
- Aubin JE, Osborn M, Weber K. Inhibition of cytokinesis and altered contractile ring morphology induced by cytochalasins in synchronized PtK2 cells. Exp. Cell Res. 1981;136:63–79. doi: 10.1016/0014-4827(81)90038-0. [DOI] [PubMed] [Google Scholar]
- Borel F, Lohez OD, Lacroix FB, Margolis RL. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc. Natl. Acad. Sci. USA. 2002;99:9819–9824. doi: 10.1073/pnas.152205299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas RE, Gruss OJ, Mattaj IW, Karsenti E. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 2001;3:228–234. doi: 10.1038/35060009. [DOI] [PubMed] [Google Scholar]
- Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Cloud JE, Rogers C, Reza TL, Ziebold U, Stone JR, Picard MH, Caron AM, Bronson RT, Lees JA. Mutant mouse models reveal the relative roles of E2F1 and E2F3 in vivo. Mol. Cell. Biol. 2002;22:2663–2672. doi: 10.1128/MCB.22.8.2663-2672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denko NC, Giaccia AJ, Stringer JR, Stambrook PJ. The human Ha-ras oncogene induces genomic instability in murine fibroblasts within one cell cycle. Proc. Natl. Acad. Sci. USA. 1994;24:5124–5128. doi: 10.1073/pnas.91.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leonardo A, Khan SH, Linke SP, Greco V, Seidita G, Wahl GM. DNA replication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 pr pRB function. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Duensing S, Lee LY, Duensing A, Basile J, Piboonniyom S, Gonzalez S, Crum CP, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA. 2000;97:10002–10007. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG, Jr, Livingston DM, Orkin SH, Greenberg ME. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, van Es JH, Breukel C, Wiegant J, Giles RH, Clevers H. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat. Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- Fukasawa K, Vande Woude GF. Synergy between the Mos/mitogen-activated protein kinase pathway and loss of p53 function in transformation and chromosome instability. Mol. Cell. Biol. 1997;17:506–518. doi: 10.1128/mcb.17.1.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- Gaubatz S, Lindeman GJ, Ishida S, Jakoi L, Nevins JR, Livingston DM, Rempel RE. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell. 2000;6:729–735. doi: 10.1016/s1097-2765(00)00071-x. [DOI] [PubMed] [Google Scholar]
- Ghadimi BM, Sackett DL, Difilippantonio MJ, Schrock E, Neumann T, Jauho A, Auer G, Ried T. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer. 2000;27:183–190. [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001;15:1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Hollander MC, Sheikh MS, Bulavin DV, Lundgren K, Augeri-Henmueller L, Shehee R, Molinaro TA, Kim KE, Tolosa E, Ashwell JD, et al. Genomic instability in Gadd45a-deficient mice. Nat. Genet. 1999;23:176–184. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. E2f3 is critical for normal cellular proliferation. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan KB, Burds AA, Swedlow JR, Bekir SS, Sorger PK, Nathke IS. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat. Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, Nevins JR. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, Miron A, Nevins JR. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol. Cell. Biol. 2000;20:3626–3632. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park C-H, Giangrande P, Wu L, Saavedra HI, Field SJ, et al. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell. 2001;8:105–113. doi: 10.1016/s1097-2765(01)00275-1. [DOI] [PubMed] [Google Scholar]
- Lingle WL, Salisbury JL. Altered centrosome structure is associated with abnormal mitoses in human breast tumors. Am. J. Pathol. 1999;155:1941–1951. doi: 10.1016/S0002-9440(10)65513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. USA. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proc. Natl. Acad. Sci. USA. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel C, Braun SE, Reid S, Henegariu O, Liu L, Hangoc G, Broxmeyer HE. p21(cip-1/waf-1) deficiency causes deformed nuclear architecture, centriole overduplication, polyploidy, and relaxed microtubule damage checkpoints in human hematopoietic cells. Blood. 1999;93:1390–1398. [PubMed] [Google Scholar]
- Martineau SN, Andreassen PR, Margolis RL. Delay of HeLa cell cleavage into interphase using dihydrocytochalasin B: retention of a postmitotic spindle and telophase disc correlates with synchronous cleavage recovery. J. Cell Biol. 1995;131:191–205. doi: 10.1083/jcb.131.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr. Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Cleveland DW. Pathways of spindle pole formation: different mechanisms; conserved components. J. Cell Biol. 1997;138:953–956. doi: 10.1083/jcb.138.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni MC, Hickman ES, Denchi EL, Caprara G, Colli E, Cecconi F, Muller H, Helin K. Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussman JG, Horn HF, Carroll PE, Okuda M, Tarapore P, Donehower LA, Fukasawa K. Synergistic induction of centrosome hyperamplification by loss of p53 and cyclin E overexpression. Oncogene. 2000;19:1635–1646. doi: 10.1038/sj.onc.1203460. [DOI] [PubMed] [Google Scholar]
- Niculescu AB, 3rd, Chen X, Smeets M, Hengst L, Prives C, Reed SI. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol. Cell. Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Pihan GA, Doxsey SJ. The mitotic machinery as a source of genetic instability in cancer. Semin. Cancer Biol. 1999;9:289–302. doi: 10.1006/scbi.1999.0131. [DOI] [PubMed] [Google Scholar]
- Pihan GA, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey SJ. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra HI, Fukasawa K, Conn CW, Stambrook PJ. MAPK mediates RAS-induced chromosome instability. J. Biol. Chem. 1999;274:38083–38090. doi: 10.1074/jbc.274.53.38083. [DOI] [PubMed] [Google Scholar]
- Saavedra HI, Knauf JA, Shirokawa JM, Wang J, Ouyang B, Elisei R, Stambrook PJ, Fagin JA. The RAS oncogene induces genomic instability in thyroid PCCL3 cells via the MAPK pathway. Oncogene. 2000;19:3948–3954. doi: 10.1038/sj.onc.1203723. [DOI] [PubMed] [Google Scholar]
- Saavedra HI, Wu L, de Bruin A, Timmers C, Rosol TJ, Weinstein M, Robinson ML, Leone G. Specificity of E2F1, E2F2 and E2F3 in mediating Rb function. Cell Growth Differ. 2002;13:215–225. [PubMed] [Google Scholar]
- Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- Tarapore P, Horn HF, Tokuyama Y, Fukasawa K. Direct regulation of the centrosome duplication cycle by the p53-p21Waf1/Cip1 pathway. Oncogene. 2001;20:3173–3184. doi: 10.1038/sj.onc.1204424. [DOI] [PubMed] [Google Scholar]
- Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. Specific phosphorylation of nucleophosmin on Thr(199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J. Biol. Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- Tutt A, Gabriel A, Bertwistle D, Connor F, Paterson H, Peacock J, Ross G, Ashworth A. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol. 1999;9:1107–1110. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- Wang A, Schneider-Broussard R, Kumar AP, MacLeod MC, Johnson DG. Regulation of BRCA1 expression by the Rb-E2F pathway. J. Biol. Chem. 2000;275:4532–4536. doi: 10.1074/jbc.275.6.4532. [DOI] [PubMed] [Google Scholar]
- White RA, Pan Z, Salisbury JL. GFP-centrin as a marker for centriole dynamics in living cells. Microsc. Res. Tech. 2000;49:451–457. doi: 10.1002/(SICI)1097-0029(20000601)49:5<451::AID-JEMT7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, et al. The E2F1, E2F2, and E2F3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/−) mice. Nat. Genet. 1998;18:360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Purification and assay of gamma tubulin ring complex. Methods Enzymol. 1998;298:218–228. doi: 10.1016/s0076-6879(98)98021-1. [DOI] [PubMed] [Google Scholar]
- Ziebold U, Reza T, Caron A, Lees JA. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 2001;15:386–391. doi: 10.1101/gad.858801. [DOI] [PMC free article] [PubMed] [Google Scholar]