Abstract
Animals balance nutrient storage and mobilization to maintain metabolic homeostasis, a process that is disrupted in metabolic diseases like obesity and diabetes. Here, we show that DHR38, the single fly ortholog of the mammalian nuclear receptor 4A family of nuclear receptors, regulates glycogen storage during the larval stages of Drosophila melanogaster. DHR38 is expressed and active in the gut and body wall of larvae, and its expression levels change in response to nutritional status. DHR38 null mutants have normal levels of glucose, trehalose (the major circulating form of sugar), and triacylglycerol but display reduced levels of glycogen in the body wall muscles, which constitute the primary storage site for carbohydrates. Microarray analysis reveals that many metabolic genes are mis-regulated in DHR38 mutants. These include phosphoglucomutase, which is required for glycogen synthesis, and the two genes that encode the digestive enzyme amylase, accounting for the reduced amylase enzyme activity seen in DHR38 mutant larvae. These studies demonstrate that a critical role of nuclear receptor 4A receptors in carbohydrate metabolism has been conserved through evolution and that nutritional regulation of DHR38 expression maintains the proper uptake and storage of glycogen during the growing larval stage of development.
The single Drosophila ortholog of the mammalian NR4A family of nuclear receptors, DHR38, regulates glycogen storage during larval stages.
Nuclear receptors are ligand-regulated transcription factors that are defined by a conserved zinc finger DNA-binding domain (DBD) and a C-terminal ligand-binding domain (LBD). Functional studies have shown that many of these factors play a central role in maintaining metabolic homeostasis, often by acting as metabolic sensors that drive feedback or feed-forward transcriptional programs that act on the compound bound by the receptor (1). A number of ligand-independent orphan nuclear receptors have also been implicated in metabolic control, although their mechanisms of action remain more poorly understood.
In this study, we focus on the nuclear receptor 4A (NR4A) family of orphan nuclear receptors, a subclass that is represented by three paralogs in mammals: Nur77 (NR4A1), Nur-related protein 1 (Nurr1; NR4A2), and neuron-derived orphan receptor 1 (NOR-1; NR4A3). Crystallographic studies have shown that the ligand-binding pocket of NR4A receptors is filled with bulky hydrophobic amino acid side chains and that the LBD adopts a canonical protein fold characteristic of the agonist-bound, transcriptionally active state (2,3). As a result, the activity of NR4A receptors is determined largely by their expression level and posttranslational modifications rather than a specific ligand (4,5). A number of recent studies have revealed pleiotropic metabolic functions for these receptors (6). Nur77 and NOR-1 promote glucose utilization and oxidative metabolism, respectively, after β-adrenergic stimulation in skeletal muscle (7,8). Nur77, Nurr1, and NOR-1 also participate in the up-regulation of hepatic gluconeogenesis in response to glucagon signaling and can increase blood glucose levels (9). In contrast to this diabetes-promoting role, Nur77 and NOR-1 have been shown to increase insulin sensitivity in adipocytes (10). These studies thus reveal distinct organ-specific functions for NR4A receptors in modulating glucose homeostasis and diabetes progression and raise the question of their role in controlling the systemic physiology of the animal. One study to date has explored this topic, showing that mice lacking Nur77 develop hepatic steatosis and increased insulin resistance when challenged with a high-fat diet (11). This phenotype, however, is difficult to interpret due to the compensatory overexpression of Nurr1 and NOR-1 that is observed in Nur77 mutants. It is not possible to investigate systemic functions in animals that are completely deprived of NR4A signaling because of the early lethality associated with double mutant combinations (12).
We are using Drosophila as a simple model system to investigate the metabolic functions of the NR4A nuclear receptors. The fly genome encodes a single NR4A ortholog, DHR38, with 48% overall amino acid identity to Nur77 and 64% identity in the DBD (13). DHR38 null mutants die during metamorphosis, with developmental defects in the formation of the adult cuticle (14). This late lethality provides an opportunity to define the metabolic dysfunction associated with a complete loss of NR4A function. Here, we show that, like its mammalian counterparts, DHR38 expression in larvae is modulated by the nutritional status of the animal. DHR38 is expressed in the larval gut and body wall, the major tissues involved in nutrient uptake and energy expenditure. DHR38 null mutants have normal levels of glucose, trehalose, and triacylglycerol, but display significantly reduced levels of glycogen. Consistent with this, microarray studies identify a number of key genes involved in carbohydrate metabolism that are mis-regulated in DHR38 mutants. These include significant down-regulation of the two genes that encode amylase, the enzyme responsible for digesting complex carbohydrates, consistent with the reduced amylase enzyme activity seen in DHR38 mutant larvae. Pgm, which encodes phosphoglucomutase, a key enzyme in glycogenesis, is also significantly underexpressed in DHR38 mutants. We conclude that increased levels of DHR38 expression in feeding larvae promote the appropriate uptake and storage of carbohydrates. In addition, our results demonstrate that carbohydrate metabolism represents an evolutionarily ancestral function for the NR4A subclass of nuclear receptors.
Results
DHR38 is expressed in the larval gut and body wall
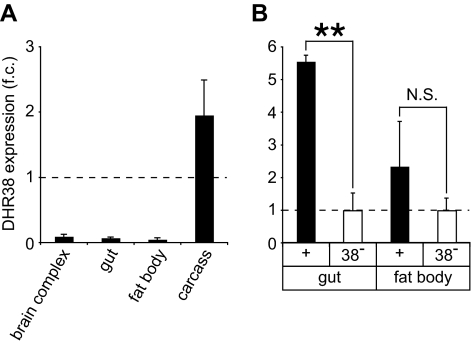
As a first step toward functional characterization of DHR38, we determined its spatial pattern of expression. Although earlier studies have shown that DHR38 is expressed throughout embryonic and larval stages, these efforts were confounded by its low level of expression, which is near background levels of detection (13,14). To circumvent this difficulty, we performed quantitative RT-PCR (qRT-PCR) on dissected organs, including the brain complex, gut, fat body, and carcass from fed third instar larvae, and measured levels of DHR38 mRNA relative to an internal rp49 mRNA control. We found that DHR38 transcripts are highly enriched in the larval carcass, which includes the epidermis and body wall muscle (Fig. 1A). To determine whether the low levels of DHR38 expression detected in the gut and the fat body are above background levels, we measured DHR38 mRNA in these two tissues dissected from either control or DHR38 null mutant larvae, which carry the molecularly defined DHR38Y214 null allele in combination with a deficiency that removes the DHR38 locus (15). This revealed that DHR38 is expressed at low levels in the gut but not in the larval fat body (Fig. 1B).
Figure 1.
DHR38 expression profile. A, DHR38 is most abundantly expressed in the larval carcass. DHR38 transcript levels were measured by qRT-PCR using RNA samples isolated from dissected third instar larval brain complexes, gut, fat body, or carcass. The fold-change (f.c.) in these mRNA levels relative to the level in the whole animal is depicted. B, DHR38 is expressed at low levels in the larval gut but not in the fat body. Levels of DHR38 mRNA were determined by qRT-PCR using RNA isolated from dissected guts or fat bodies of control or DHR38Y214/Df(2)KetelRX32 mutant third instar larvae (38−). The amount of DHR38 mRNA in wild-type tissue is presented relative to the level detected in null mutant animals. The region amplified by PCR is fully deleted in the DHR38 mutant. Error bars represent sem, n ≥ 3 independent samples of 3–10 animals each; **, P < 0.01. N.S., Not significant, Student’s t test.
DHR38 expression is regulated by the nutritional status of the animal
Nuclear receptors of the NR4A subgroup are orphan receptors, the activity of which is determined largely by their expression level (4,5). In addition, the expression of all three mammalian NR4A receptors is responsive to the feeding status of the animal (9). To determine whether a similar mode of regulation exists in Drosophila, DHR38 mRNA levels were measured in larvae that were maintained for 24 h either in the absence of food or on a rich diet consisting of yeast paste. This study revealed that DHR38 expression is significantly higher in fed larvae (Fig. 2A). These results are consistent with published microarray data, which indicate that DHR38 mRNA levels are reduced upon starvation (16).
Figure 2.
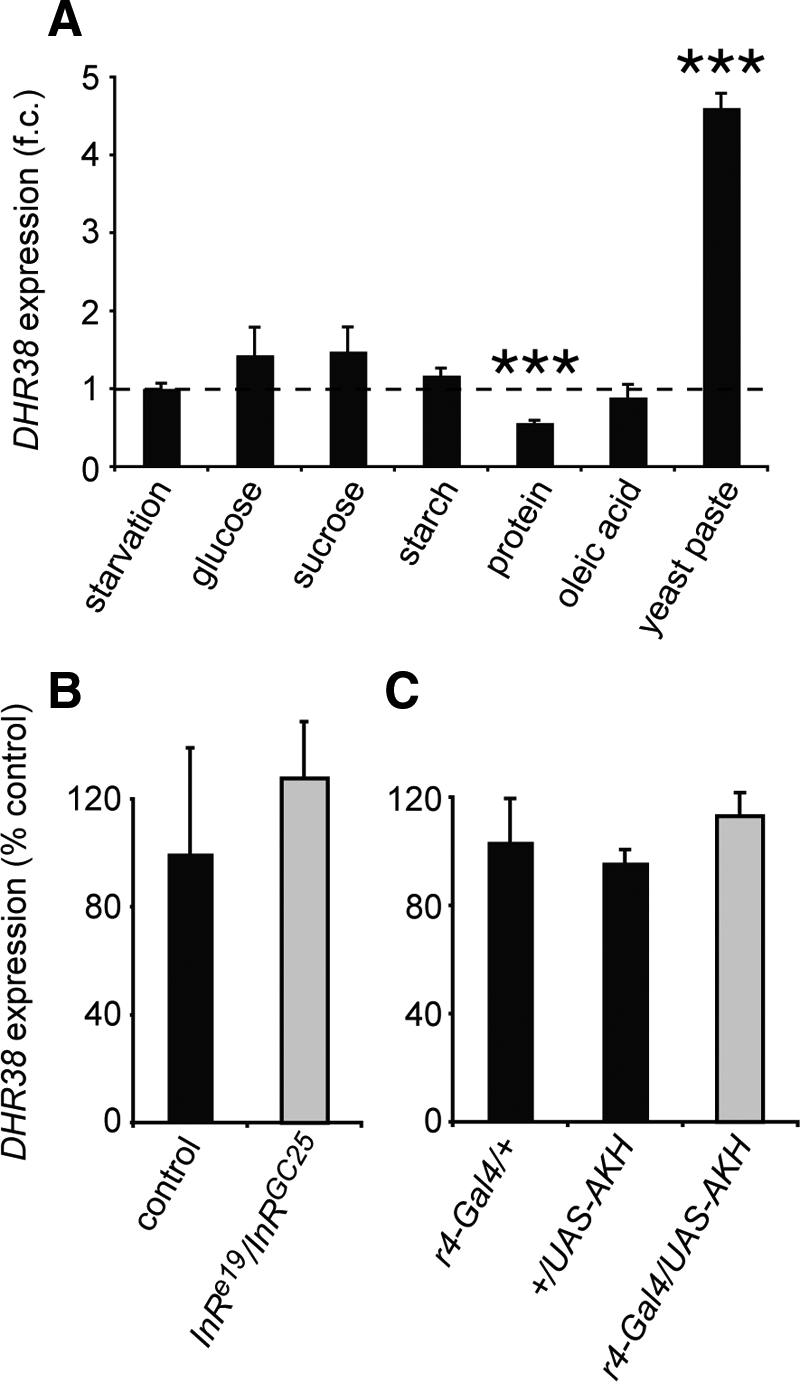
DHR38 expression is regulated by the nutritional status of the animal. A, DHR38 expression is regulated by dietary factors. Late second instar larvae were either starved or maintained on different food sources, as listed, for 24 h. RNA was then extracted, and the levels of DHR38 and rp49 mRNA were determined by qRT-PCR. Levels of DHR38 mRNA normalized to the levels of rp49 mRNA are presented as a fold-change (f.c.) between the amount in fed animals and the amount in starved animals. The qRT-PCR signal detected in starved DHR38 null mutant larvae is less than 0.05% that seen in starved control animals, indicating that it is significantly above background levels (data not shown). B and C, DHR38 expression is not affected by changes in insulin or AKH signaling. Total RNA from control, InR mutant, or AKH-overexpressing larvae was analyzed by qRT-PCR for changes in DHR38 mRNA levels relative to the levels of an internal rp49 mRNA control. B, Second instar InRe19/InRGC25 larvae raised at 25 C that display a strong loss of InR function (gray bar) (38) have the same levels of DHR38 expression as those in w1118 control larvae (black bar). InR inactivation was verified by the small size of InR mutant larvae relative to controls (data not shown). C, Control third instar larvae that carry either the fat body-specific r4-GAL4 driver or the UAS-AKH transgene (black bars) have the same level of DHR38 mRNA as larvae that carry both transgenes and thus overexpress AKH in the fat body (gray bar). The efficiency of AKH overexpression was confirmed by measuring circulating trehalose levels, which are higher in AKH-overexpressing larvae when compared with the controls (20). Error bars represent sem, n ≥ 3 independent samples of 3–10 animals each; ***, P < 0.001, Student’s t test.
In an effort to identify specific nutritional cues that might be responsible for this regulation, larvae were transferred to media that contained either sugars, starch, fatty acids, or proteins. None of these dietary conditions, however, was sufficient to up-regulate DHR38 expression, although mRNA levels are significantly reduced in animals maintained on a pure protein diet (Fig. 2A). These results are consistent with the inability of these individual nutrients to support larval growth and suggest that changes in DHR38 expression are linked to the availability of a complete diet (17). These observations also raise the interesting possibility that DHR38 might transduce nutritional signals in Drosophila larvae.
Several pathways have been identified in Drosophila that signal changes in nutritional status, including a conserved insulin/IGF system counterbalanced by the glucagon analog adipokinetic hormone (AKH) (18,19,20). In vertebrates, NR4A expression is induced by insulin and glucagon in a tissue-specific manner, with insulin stimulating NOR-1 and Nur77 expression in adipocytes and glucagon inducing the three paralogs in hepatocytes (9,10). To test whether a similar form of regulation might exist in Drosophila, we assayed DHR38 mRNA levels in larvae that carry a strong loss of function mutation in the single insulin/IGF receptor homolog InR (Fig. 2B). DHR38 mRNA levels were also measured in larvae that specifically overexpress AKH in the fat body, which shares functions with the mammalian liver (Fig. 2C). This expression is sufficient to promote hyperglycemia and decrease levels of fat body triacylglycerol (20). DHR38 expression, however, is not significantly altered under either of these conditions, suggesting that it is controlled independently of these metabolic sensors.
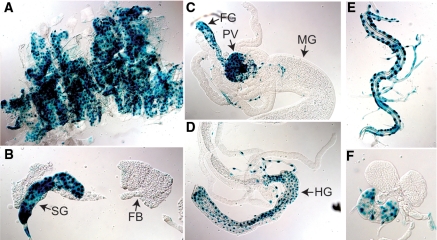
The activity of mammalian NR4A receptors is known to be modulated by a number of factors, including posttranslational modification and dimerization with other nuclear receptors (4). We therefore examined whether we could detect changes in the activity of the DHR38 LBD in response to the nutritional status of the animal. For this purpose, we used transgenic animals that carry a heat-inducible construct that encodes the yeast GAL4 DNA-binding domain fused to the DHR38 LBD (hsp70-GAL4-DHR38) in combination with a UAS-nlacZ reporter gene that directs the synthesis of nuclear-localized β-galactosidase (2,21). This system provides a faithful means of following LBD activity within the animal. No changes in DHR38 LBD activity were detected upon comparing fed and starved larvae at several developmental stages (data not shown). Interestingly, however, the activity of the DHR38 LBD is restricted to specific tissues, with high levels of activation in the body wall, salivary glands, regions of the gut, trachea, and prothoratic gland (Fig. 3). Two of these, the body wall and gut, correspond to tissues in which the receptor is normally expressed (Fig. 1, A and B). This pattern of LBD activation suggests that, like its mammalian counterparts, DHR38 activity is controlled posttranslationally. In addition, it indicates that changes in DHR38 expression level provide the primary means for its regulatory response to the nutritional status of the animal.
Figure 3.
Activation of the DHR38 LBD is spatially restricted. Fed second instar larvae that carry both the hsp70-GAL4-DHR38 and UAS-nlacZ transgenes were heat treated and allowed to recover for approximately 3 h, after which organs were dissected and stained with X-gal to detect β-galactosidase activity (21). High levels of DHR38 LBD activity are restricted to the carcass (A), the salivary glands (SG) (B), the foregut (FG) and proventriculus (PV) (C), and hindgut (HG) (D), as well as the tracheae (E) and prothoracic gland (F). In contrast, no activity is detected in the fat body (FB) (B) or larval midgut (MG) (C). Control experiments have shown that the UAS-nlacZ reporter is capable of being expressed in all tissues at this stage in development (39).
DHR38 mutants display reduced levels of glycogen
Many of the basic metabolic pathways that maintain homeostasis in vertebrates are conserved in the fly (reviewed in Refs. 2,22). This includes the use of glycogen and triacylglycerol as the major intracellular stored forms of energy, whereas the disaccharide trehalose, and to a lesser extent glucose, function as circulating sugars. To uncover possible metabolic functions for DHR38, we measured the levels of these compounds under both feeding and starved conditions in control and DHR38 mutant animals. Animals that carry the DHR38Y214 mutation in combination with Df(2)Ketel, a deficiency that removes the DHR38 locus, were used to provide a complete loss of DHR38 function (15).
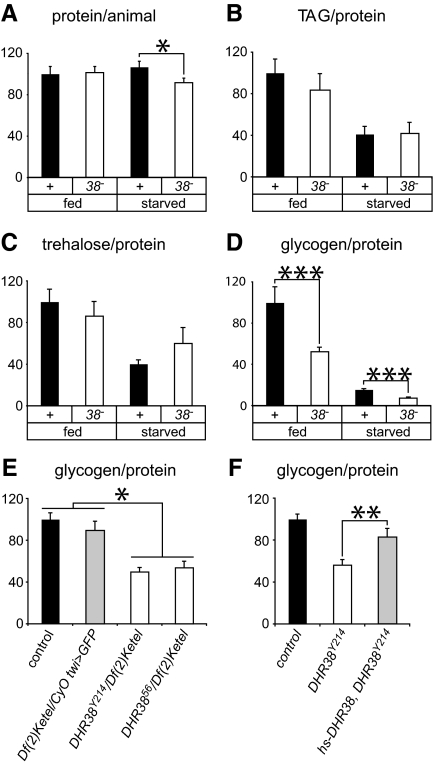
Control and DHR38 mutant larvae were subjected to complete starvation, and the animals were followed for several days. No detectable difference was observed in the starvation response of control and mutant animals, with both strains surviving 2 d in the absence of food (Supplemental Fig. 1A, published on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). We went on to determine whether we could detect defects in the levels of basic metabolites in fed or starved DHR38 mutants. The soluble protein content of fed control and mutant larvae was similar, although DHR38 mutant larvae had slightly reduced amounts of soluble protein upon starvation (Fig. 4A). In contrast, the levels of triacylglycerol are unaffected in the mutant, under either fed or starved conditions (Fig. 4B). Glucose and trehalose levels are also unaffected in either fed or starved DHR38 mutants (Fig. 4C and Supplemental Fig. 1B). Glycogen levels, however, are significantly reduced in both fed and starved mutants (Fig. 4D). This effect was confirmed by measuring glycogen levels in animals that carry two other DHR38 mutant allele combinations, DHR3856/Df(2)Ketel and DHR38Y214 homozygotes (Fig. 4, E and F). In addition, the reduced glycogen levels observed in DHR38Y214 mutants can be efficiently rescued by a hs-DHR38 transgene that expresses the wild-type receptor (Fig. 4F). DHR38 is thus required to promote appropriate glycogen storage in Drosophila larvae. In spite of this defect, however, the DHR38 mutants are able to maintain relatively normal trehalose levels under both fed and starved conditions, although the mechanisms that underlie this response remain unclear (Fig. 4C).
Figure 4.
DHR38 mutants display reduced levels of glycogen. A–D, control and DHR38Y214/Df(2)Ketel (38−) late second instar larvae were either collected or starved for 24 h, and homogenates were assayed for protein (A), triacylglycerol (TAG) (B), trehalose (C), or glycogen (D). A, The amount of protein per larva is presented relative to a control level of 100% and is slightly reduced in starved DHR38 mutants. B–D, Triacylglycerol, trehalose, and glycogen levels were normalized to the amount of protein and are presented relative to the level in fed controls. The differences in trehalose levels between control and DHR38 mutant animals are not significant. E, Reduced glycogen levels are present in two different DHR38 null mutant combinations. Glycogen levels were measured in starved control larvae, larvae that are heterozygous for the Df(2)Ketel deficiency, or null mutant larvae that carry either the DHR38Y214 or DHR3856 allele in combination with the Df(2)Ketel deficiency. F, The reduced glycogen levels in homozygous DHR38Y214 mutant larvae are rescued by constitutive expression of a hs-DHR38 transgene. Error bars represent sem, n ≥ 6 independent samples of 5–40 animals each; *, P < 0.05; **, P < 0.01; ***, P < 0.001, Student’s t test.
Glycogen is stored in the larval muscle
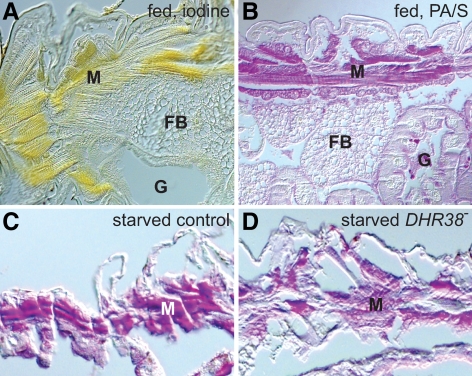
The liver and skeletal muscle constitute the two major sites for glycogen storage in vertebrates. An analogous distribution of glycogen depots has been observed in adult Drosophila, with the highest levels of glycogen present in the fat body and lower levels in the halteres, flight muscle, and gut (23). Little work, however, has been done to examine the glycogen distribution in Drosophila larvae, with only a few reports describing low levels of glycogen in the fat body and salivary glands (24,25). We therefore used iodine and periodic acid/Schiff (PA/S) staining on paraffin sections of fixed larvae to determine the spatial distribution of glycogen stores. This study revealed that glycogen is most abundant in the body wall muscles, the primary muscles used for larval movement, with much lower levels in the fat body and gut (Fig. 5, A and B). These observations are consistent with the spatial expression pattern of many genes involved in glycogen metabolism, which are most abundantly expressed in the larval carcass (26). In addition, glycogen levels are detectably lower in the muscles of DHR38 mutant larvae, as revealed by PA/S staining (Fig. 5, C and D). Taken together, those results indicate that glycogen is primarily stored in the larval muscle and that this process depends on DHR38 function.
Figure 5.
DHR38 mutants display reduced levels of glycogen in the muscle, which is the primary tissue for glycogen storage in larvae. A and B, Fed control third instar larvae were dissected and stained with either iodine vapor (A) or PA/S reagent (B), revealing high levels of glycogen in the muscle (M) but not the fat body (FB) or gut (G). C and D, PA/S staining of starved control or DHR38Y214/Df(2)Ketel third instar larvae reveals that glycogen levels are reduced in the muscles of DHR38 mutant larvae.
DHR38 regulates carbohydrate metabolism gene expression
Microarray studies were conducted in an effort to determine the molecular mechanisms by which DHR38 regulates glycogen levels. RNA was extracted from fed control and DHR38 mutant larvae, labeled, and hybridized to Agilent Drosophila 44K microarrays (Agilent Technologies, Santa Clara, CA). All experiments were conducted in triplicate to facilitate statistical analysis. The raw data were quantile-normalized, and gene expression changes were determined using GeneSifter (1.75-fold cutoff, adjusted P ≤ 0.02). This study identified 437 genes that are down-regulated in DHR38 mutants and 431 genes that are up-regulated (Supplemental Table 1).
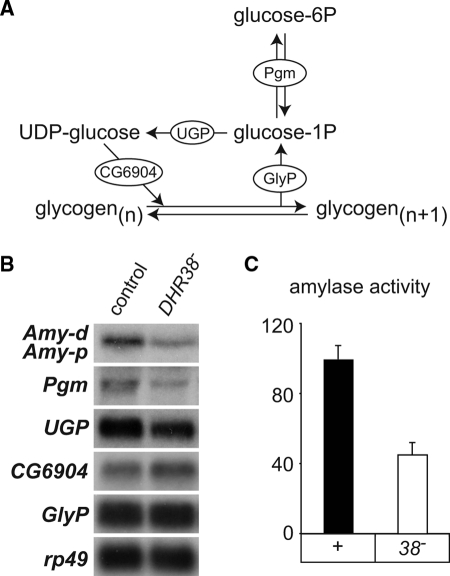
Comparison of the DHR38-regulated genes with a list of genes that comprise the major metabolic pathways in Drosophila revealed a significant overlap in these data sets (χ2 test, P = 6 × 10−5) (Supplemental Table 2). These include genes that act in a number of processes, including amino acid catabolism and lipid metabolism. We focused our attention on those genes that might impact carbohydrate uptake or glycogen synthesis, because these are the primary pathways in feeding larvae that could contribute to the changes in glycogen content seen in DHR38 mutant animals. Two genes that encode amylases are among the most highly down-regulated genes in DHR38 mutants: Amy-p (−11.4-fold) and Amy-d (−11.0-fold). These genes encode enzymes that are predicted to perform the first step in the breakdown of complex dietary carbohydrates (26). One other gene encodes an amylase in Drosophila, Amyrel (27). It is, however, less abundantly expressed than the Amy genes and is unaffected in DHR38 mutants. We also identified one gene that is mis-regulated in DHR38 mutants and that plays a role in glycogen metabolism, Pgm (−1.8-fold). This gene encodes phosphoglucomutase, which catalyzes the interconversion of glucose-1-phosphate, the precursor for glycogen synthesis, and glucose-6-phosphate, the primary form of intracellular sugar (Fig. 6A). Northern blot hybridization confirmed the results of our microarray study, showing markedly reduced expression of the Amy genes and Pgm (Fig. 6B). A similar effect is seen in animals that carry other DHR38 mutant allele combinations (Supplemental Fig. 2). We also examined the expression of three other genes that play a critical role in glycogen metabolism: UGP (CG4347), which encodes uridine diphosphate-glucose pyrophosphorylase; CG6904, which encodes glycogen synthase (the key enzyme in glycogen synthesis); and GlyP (CG7254), which encodes glycogen phosphorylase, the rate-limiting enzyme in glycogenolysis (Fig. 6A). None of these genes, however, are significantly or reproducibly mis-regulated in DHR38 mutant larvae (Fig. 6B and Supplemental Table 1). Taken together, these data indicate that DHR38 regulates a subset of genes involved in carbohydrate metabolism.
Figure 6.
DHR38 regulates the expression of genes involved in carbohydrate metabolism. A, Schematic representation of glycogen metabolism in Drosophila. Glycogen synthesis involves the conversion of glucose-6-phosphate to glucose-1-phosphate through the action of phosphoglucomutase (Pgm). The production of uridine diphosphate-glucose (UDP) by UGP (CG4347) provides the building blocks for glycogen synthesis, mediated by glycogen synthase (CG6904). Glycogen phosphorylase (GlyP) is the rate-limiting enzyme that controls glycogen breakdown. B, Amy and Pgm expression are markedly reduced in DHR38 mutants. RNA isolated from fed control and DHR38Y214/Df(2)Ketel (38−) larvae was analyzed by Northern blot hybridization to detect Amy-p and Amy-d, Pgm, UGP, CG6904, and GlyP expression. Blots were hybridized with rp49 as a control for loading and transfer. The Amy-p and Amy-d genes are highly similar in sequence and length and thus cannot be distinguished by Northern blot analysis. C, Amylase activity is reduced in DHR38 mutant larvae. Amylase activity was measured in dissected guts from third instar feeding larvae using a fluorescent substrate.
We next tested the possibility that the reduced expression of either Amy-p/Amy-d or Pgm in DHR38 mutant larvae contributes to their reduced levels of glycogen. Measuring amylase enzyme activity in both control and DHR38 mutants revealed that amylase levels are reduced approximately 2-fold, reflecting the reduced levels of Amy mRNA seen in these animals (Fig. 6, B and C). Efficient disruption of Amy expression by RNAi, however, has no effect on whole-animal glycogen levels (Supplemental Fig. 3A). Similarly, inactivation of Pgm expression by RNAi does not impact the glycogen content of these animals (Supplemental Fig. 3B). Our results suggest that no single DHR38 target gene is sufficient to explain the reduced levels of glycogen in these animals. Rather, it is likely that DHR38 coordinates the expression of multiple genes that act together to maintain glycogen homeostasis.
Discussion
The physiological functions of NR4A nuclear receptors have remained unclear due to the presence of multiple redundant vertebrate paralogs that perform divergent organ-specific roles in regulating glucose homeostasis and diabetes progression (9,10). Here, we characterize the metabolic functions of DHR38, the single ancestral NR4A nuclear receptor in Drosophila, and demonstrate that it is required for proper glycogen storage in the feeding larva. These results add DHR38 to the growing list of invertebrate nuclear receptors controlling energy homeostasis and confirm that a key role for NR4A receptors in carbohydrate metabolism has been conserved during evolution.
Most nuclear receptors involved in metabolic regulation function as sensors for small metabolites, which modulate their transcriptional activity (1). In contrast, structural studies have demonstrated that NR4A family members do not possess a ligand-binding cavity and function as constitutively active receptors (2,3). The activity of NR4A receptors is thus primarily determined by their expression level. In line with this observation, DHR38 expression is induced by favorable nutritional conditions, whereas the activity of its LBD, as determined using the GAL4-DHR38 ligand sensor system (21), is unaffected by the nutritional status of the animals (data not shown). Therefore, similar to its vertebrate counterparts, DHR38 activity appears to be primarily regulated at the transcriptional level. This response is most likely indirect and could represent a novel signaling pathway for favorable nutritional conditions because it is not affected by the two major nutrient-sensing systems in Drosophila, the insulin and AKH pathways (Fig. 2, B and C).
DHR38 is essential for efficient glycogen storage in the muscle, which we identified as the primary storage site for glycogen in larvae. Similarly, skeletal muscle glycogen represents 90% of the total stored complex carbohydrates in vertebrates. Glycogen is used in situ in glycolytic muscle fibers to provide sugar precursors for glycolysis during exercise, a function conserved in Drosophila where it provides the primary source of energy for adult muscle function (23,28). Glycogen stored in the larval muscle may therefore perform a similar role, providing the primary energy source to support muscle function during starvation. Indeed, larval movement is critical for the hyperactivity and dispersal behavior that is seen upon nutrient deprivation (29). This is thought to be an adaptive behavior that allows animals to explore their environment in search of new food sources. A specific function for glycogen in adaptive muscle physiology fits with the metabolic profile of DHR38. Levels of glycogen, triacylglycerol, and trehalose all drop in starved larvae, consistent with their use as an energy source to maintain viability (Fig. 4, B–D). The reduced levels of glycogen in DHR38 mutants, however, do not impair their ability to survive a period of starvation (Supplemental Fig. 1A), consistent with a more specific function of glycogen in larval muscle physiology. Finally, it is interesting to note that Nur77 is preferentially expressed in fast-twitch glycolytic muscles, where it acts to promote glucose utilization and glycogenolysis, hallmarks of high muscle glycogen content (7). This result raises the possibility that NR4A receptors exert an evolutionarily conserved role in maintaining muscle physiology through their effects on carbohydrate metabolism and glycogen homeostasis.
Our microarray analysis identified candidate metabolic genes that could mediate DHR38 function in regulating glycogen storage, including two amylases (Amy-d and Amy-p) and the converting enzyme phosphoglucomutase (Pgm). Amy-d and Amy-p are most abundantly expressed in the larval gut, whereas Pgm is highly expressed in the larval carcass, reflecting the patterns of DHR38 expression and suggesting that they may be direct regulatory targets of the receptor (26). This possibility is consistent with the identification of potential NR4A binding sites in sequences adjacent to Amy-d and Amy-p (Supplemental Fig. 4). This binding site has been well defined and can be recognized by DHR38 in vitro (13,30). A related sequence located downstream from Amy-p can also be bound by DHR38 protein, although at lower affinity (data not shown), suggesting that DHR38 could directly regulate Amy-d and Amy-p transcription.
Individually inactivating these putative target genes, however, was not sufficient to phenocopy the low glycogen accumulation observed in DHR38 mutants (Supplemental Fig. 3). For the phosphoglucomutase gene Pgm, this finding was unexpected, because changes in Pgm activity have been correlated with glycogen content in natural Drosophila populations (31). It is possible that undetected variation at additional loci participates in controlling glycogen levels in those animals. Alternatively, another study reached the opposite conclusion based on analysis of the two common electrophoretic variants at the Pgm locus (32). We conclude that DHR38 coordinates the expression of multiple genes that act together to maintain glycogen homeostasis. Similarly, vertebrate nuclear receptors of the NR4A family coregulate a number of genes to control common metabolic processes. In the liver, for example, NR4As stimulate the expression of the glucose-6-phosphatase G6pc, the fructose bisphosphatases Fbp1 and Fbp2, the enolase Eno3, and the glucose transporter Glut2 (Slc2a2) to promote gluconeogenesis (9).
The identification of a critical metabolic role for DHR38 is consistent with our overall understanding of NR4A function. Mammalian NR4A receptors contribute to a wide range of biological pathways, including apoptosis, neurological disease, inflammation, carcinogenesis, and atherogenesis (5). Similarly, in addition to its role in muscle carbohydrate homeostasis described here, DHR38 has an essential developmental function in late pupae, when it is required for the proper integrity of the adult cuticle. This function may be related to the ability of the DHR38 LBD to be activated by ecdysteroids, which control cuticle deposition (2). DHR38 null mutants die at the end of metamorphosis with rupturing of the cuticle at leg joints and consequent hemolymph leakage (15). Several adult cuticle genes are expressed at reduced levels in DHR38 mutant pupae, one of which, Acp65A, appears to be directly regulated by the receptor and specifically expressed at the joints (33). No changes in glycogen content or ATP levels, however, are detected in these pupae (Supplemental Fig. 5), suggesting that this represents a specific developmental function for the receptor. In addition, DHR38 is expressed at high levels in the adult brain and regulates DOPA (3,4-dihydroxyphenylalanine) decarboxylase expression in several tissues, suggesting that roles for NR4A receptors in dopaminergic neuron function have been conserved through evolution (26,34). Characterization of the tissue-specific functions of DHR38, as well as its roles in physiology and metabolism during adult stages, should provide further insights into its regulatory activities and provide a framework for understanding how NR4A receptors integrate their widespread developmental and metabolic functions in the animal.
Materials and Methods
Fly stocks and culture conditions
The w1118 strain was used as a control in all experiments. Two DHR38 mutant alleles were used in this study: DHR38Y214 (15) and DHR3856 (14). DHR38Y214 is an excision mutant that deletes the coding region for both the DBD and LBD, whereas DHR3856 is a mutant that introduces a premature stop codon in the three DHR38 transcripts. Mutant phenotypes were examined in animals carrying a DHR38 mutation in combination with Df (2)KetelRX32 (35), a small deficiency that removes the DHR38 locus. The heat-inducible P[w+; hs-DHR38] transgene was used for rescue experiments (14). Fly stocks were maintained at 18–25 C on standard cornmeal-agar-yeast food. For metabolic experiments, larvae were grown on agar/molasses and/or agar egg caps supplemented with fresh yeast paste at low density (50–100 larvae per egg cap) at 25 C, unless noted otherwise. For controlled feeding experiments, larvae were placed on filter paper humidified with distilled water (starvation), 20% glucose, 20% sucrose, 20% starch (S9765; Sigma, St. Louis, MO), 20% casein, or 5 mg/ml oleic acid.
Quantitative RT-PCR and Northern blot analysis
Animal and tissue samples were dissected and/or collected in TriPure isolation reagent (Roche, Basel, Switzerland) and frozen in liquid nitrogen. Total RNA was isolated following the manufacturer’s instruction. cDNA was prepared from 0.5 μg of RNA in a 25 μl reaction using the Protoscript cDNA kit (New England Biolabs, Ipswich, MA). PCRs of 25 μl were prepared in 96-well plates using SYBR Green master mix (Bio-Rad, Hercules, CA), with a primer concentration of 3 mm (DHR38f-GCAACATAACTACAACTCGCACA, DHR38r-AGCTTCGACAGCAGCAGTG, rp49f-ATGCTAAGCTGTCGCACAAA, and rp49r-CGATGTTGGGCATCAGATACT). Quantitative PCR was performed using a Bio-Rad iCycler (MyiQ Single Color). Data were collected from at least three independent samples. To determine the relationship between mRNA abundance and PCR cycle number, all primer sets were calibrated using serial dilutions of cDNA preparations. Relative abundance is reported as DHR38 mRNA levels relative to rp49 mRNA levels. The data were analyzed using efficiency-corrected comparative quantitation. For Northern blot analysis, total RNA samples were fractionated by formaldehyde agarose gel electrophoresis, transferred to a nylon membrane, and cross-linked by UV irradiation, as described (36). Blot hybridization and washing was performed as described (36).
Metabolic assays
For trehalose assays, larvae were homogenized in 100 μl ice-cold trehalase buffer [5 mm Tris (pH 6.6), 137 mm NaCl, and 2.7 mm KCl] (see Ref. 37), centrifuged at maximum speed (15,000 × g) for 3 min, and the resulting supernatant was immediately incubated at 70 C for 5 min; 30 μl of 1/10 diluted samples were treated with or without trehalase (11 mU/μl, T8778; Sigma) overnight at 37 C. Resulting glucose levels were assayed (GAGO-20; Sigma). Trehalose levels were obtained by subtracting the amount of free glucose in the untreated sample from the total glucose present in the sample treated with trehalase and were normalized to protein amounts in each homogenate using a Bradford assay (Bio-Rad). Glucose and glycogen assays were performed as described (16).
Glycogen staining
Larvae were pinned down, fixed in Carnoy’s fixative for 2 h at room temperature, and transferred to 4 C overnight. Animals were then dehydrated in an ethanol series followed by xylene, embedded in paraplast, and 6-μm sections were collected. After deparaffinization and hydration, sections were either exposed to iodine vapor for 3 min and mounted in glycerol or stained using a PA/S stain (395B-1KT; Sigma) following the manufacturer’s instruction.
Amylase activity measurement
Three dissected guts were homogenized in 100 μl ice-cold 10 mm Tris (pH 7.4) and centrifuged at maximum speed for 3 min. A 50-μl sample of supernatant was used to measure amylase activity using the EnzCheck kit (E33651; Molecular Probes, Eugene, OR), following the manufacturer’s instructions. Bacillus sp. α-Amylase (A6380; Sigma) was used to establish the standard curve.
Microarrays
Control w1118 and DHR3856/Df(2)Ketel mutant larvae were collected 18 h after the second to third instar molt. RNA was isolated from these animals using TriPure isolation reagent (Roche) and purified on RNeasy columns (QIAGEN, Valencia, CA). Samples were prepared in triplicate to facilitate subsequent statistical analysis. Probe labeling, hybridization to two-color Agilent Drosophila 44K arrays, and scanning were performed by the University of Utah Microarray Core Facility. The data were quantile-normalized using R, and the fold changes in gene expression and t statistics were determined using GeneSifter (VizX Labs, Seattle, WA). P values were calculated using the Benjamimi and Hochberg correction for false-discovery rate. Comparison between microarray datasets was performed with Microsoft Access. Microarray data from this study can be accessed at National Center for Biotechnology Gene Expression Omnibus (accession no. GSE23047).
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center, the National Institute of Genetics Fly Stock Center, and the Vienna Drosophila RNAi Center for fly stocks. We also thank the University of Utah Microarray Core Facility and B. Milash for help with analyzing the microarray data, J. Evans for technical help, A. Schmid for help with sectioning, and M. Horner for critical reading of the manuscript.
Footnotes
Present address for A.F.R.: Max Planck Institute for the Biology of Aging, Köln 50931, Germany.
This work was supported by a Fondation pour la Recherche Médicale postdoctoral fellowship (A.F.R.) and by the National Institute of Health Grant 1R01DK075607.
Disclosure Summary: The authors have nothing to disclose.
Abbreviations: AKH, Adipokinetic hormone; DBD, DNA-binding domain; LBD, ligand-binding domain; NOR-1, neuron-derived orphan receptor 1; NR4A, nuclear receptor 4A; Nurr1, Nur-related protein 1; PA/S, periodic acid/Schiff; qRT-PCR, quantitative RT-PCR.
First Published Online November 17, 2010
References
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ 2001 Nuclear receptors and lipid physiology: opening the X-files. Science 294:1866–1870 [DOI] [PubMed] [Google Scholar]
- Baker KD, Shewchuk LM, Kozlova T, Makishima M, Hassell A, Wisely B, Caravella JA, Lambert MH, Reinking JL, Krause H, Thummel CS, Willson TM, Mangelsdorf DJ 2003 The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell 113:731–742 [DOI] [PubMed] [Google Scholar]
- Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker NP, Perlmann T 2003 Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 423:555–560 [DOI] [PubMed] [Google Scholar]
- Benoit G, Malewicz M, Perlmann T 2004 Digging deep into the pockets of orphan nuclear receptors: insights from structural studies. Trends Cell Biol 14:369–376 [DOI] [PubMed] [Google Scholar]
- Maxwell MA, Muscat GE 2006 The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal 4:e002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummasti S, Tontonoz P 2008 Adopting new orphans into the family of metabolic regulators. Mol Endocrinol 22:1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao LC, Zhang Z, Pei L, Saito T, Tontonoz P, Pilch PF 2007 Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol Endocrinol 21:2152–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearen MA, Myers SA, Raichur S, Ryall JG, Lynch GS, Muscat GE 2008 The orphan nuclear receptor, NOR-1, a target of β-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology 149:2853–2865 [DOI] [PubMed] [Google Scholar]
- Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P 2006 NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med 12:1048–1055 [DOI] [PubMed] [Google Scholar]
- Fu Y, Luo L, Luo N, Zhu X, Garvey WT 2007 NR4A orphan nuclear receptors modulate insulin action and the glucose transport system: potential role in insulin resistance. J Biol Chem 282:31525–31533 [DOI] [PubMed] [Google Scholar]
- Chao LC, Wroblewski K, Zhang Z, Pei L, Vergnes L, Ilkayeva OR, Ding S, Reue K, Watt MJ, Newgard CB, Pilch PF, Hevener AL, Tontonoz P 2009 Insulin resistance and altered systemic glucose metabolism in mice lacking Nur77. Diabetes 58:2788–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, Conneely OM 2007 Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med 13:730–735 [DOI] [PubMed] [Google Scholar]
- Fisk GJ, Thummel CS 1995 Isolation, regulation, and DNA-binding properties of three Drosophila nuclear hormone receptor superfamily members. Proc Natl Acad Sci USA 92:10604–10608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlova T, Pokholkova GV, Tzertzinis G, Sutherland JD, Zhimulev IF, Kafatos FC 1998 Drosophila hormone receptor 38 functions in metamorphosis: a role in adult cuticle formation. Genetics 149:1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlova T, Lam G, Thummel CS 2009 Drosophila DHR38 nuclear receptor is required for adult cuticle integrity at eclosion. Dev Dyn 238:701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS 2009 Drosophila HNF4 regulates lipid mobilization and β-oxidation. Cell Metab 9:228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang J 1978 The nutritional requirements of Drosophila. In: Ashburner M, Wright T, eds. The genetics and biology of Drosophila. New York: Academic Press; 159–192 [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R 2002 Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296:1118–1120 [DOI] [PubMed] [Google Scholar]
- Kim SK, Rulifson EJ 2004 Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature 431:316–320 [DOI] [PubMed] [Google Scholar]
- Lee G, Park JH 2004 Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics 167:311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L, Necakov AS, Sampson HM, Ni R, Hu C, Thummel CS, Krause HM 2006 Dynamic regulation of Drosophila nuclear receptor activity in vivo. Development 133:3549–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold P, Perrimon N 2007 Drosophila and the genetics of the internal milieu. Nature 450:186–188 [DOI] [PubMed] [Google Scholar]
- Wigglesworth VB 1949 The utilization of reserve substances in Drosophila during flight. J Exp Biol 26:150–163 [DOI] [PubMed] [Google Scholar]
- Butterworth FM, Bodenstein D, King RC 1965 Adipose tissue of Drosophila melanogaster. I. An experimental study of larval fat body. J Exp Zool 158:141–153 [DOI] [PubMed] [Google Scholar]
- Kosta A, Dimopoulou K, Drosou V, Thomopoulos GN 2000 Glycogen distribution in the larval salivary gland cells during the development of Drosophila melanogaster and Drosophila auraria: an ultrastructural cytochemical study. J Zool 251:61–69 [Google Scholar]
- Chintapalli VR, Wang J, Dow JA 2007 Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39:715–720 [DOI] [PubMed] [Google Scholar]
- Da Lage JL, Renard E, Chartois F, Lemeunier F, Cariou ML 1998 Amyrel, a paralogous gene of the amylase gene family in Drosophila melanogaster and the Sophophora subgenus. Proc Natl Acad Sci USA 95:6848–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg CC, Jurczak MJ, Danos AM, Brady MJ 2006 Glycogen branches out: new perspectives on the role of glycogen metabolism in the integration of metabolic pathways. Am J Physiol Endocrinol Metab 291:E1–E8 [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang Y, Xu J, Shen P 2005 Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc Natl Acad Sci USA 102:13289–13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TE, Fahrner TJ, Johnston M, Milbrandt J 1991 Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science 252:1296–1300 [DOI] [PubMed] [Google Scholar]
- Verrelli BC, Eanes WF 2001 The functional impact of Pgm amino acid polymorphism on glycogen content in Drosophila melanogaster. Genetics 159:201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fucci L, Gaudio L, Rao R, Spano A, Carfagna M 1979 Properties of the two common electrophoretic variants of phosphoglucomutase in Drosophila melanogaster. Biochem Genet 17:825–836 [DOI] [PubMed] [Google Scholar]
- Bruey-Sedano N, Alabouvette J, Lestradet M, Hong L, Girard A, Gervasio E, Quennedey B, Charles JP 2005 The Drosophila ACP65A cuticle gene: deletion scanning analysis of cis-regulatory sequences and regulation by DHR38. Genesis 43:17–27 [DOI] [PubMed] [Google Scholar]
- Davis MM, Yang P, Chen L, O'Keefe SL, Hodgetts RB 2007 The orphan nuclear receptor DHR38 influences transcription of the DOPA decarboxylase gene in epidermal and neural tissues of Drosophila melanogaster. Genome 50:1049–1060 [DOI] [PubMed] [Google Scholar]
- Erdelyi M, Mathe E, Szabad J 1997 Genetic and developmental analysis of mutant Ketel alleles that identify the Drosophila importin-β homologue. Acta Biol Hung 48:323–338 [PubMed] [Google Scholar]
- Karim FD, Thummel CS 1991 Ecdysone coordinates the timing and amounts of E74A and E74B transcription in Drosophila. Genes Dev 5:1067–1079 [DOI] [PubMed] [Google Scholar]
- Teleman AA, Chen YW, Cohen SM 2005 Drosophila melted modulates FOXO and TOR activity. Dev Cell 9:271–281 [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS 2001 A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292:107–110 [DOI] [PubMed] [Google Scholar]
- Kozlova T, Thummel CS 2002 Spatial patterns of ecdysteroid receptor activation during the onset of Drosophila metamorphosis. Development 129:1739–1750 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.