Abstract
Thyroid hormone receptors (TRs) are ligand-gated transcription factors with critical roles in development and metabolism. Although x-ray structures of TR ligand-binding domains (LBDs) with agonists are available, comparable structures without ligand (apo-TR) or with antagonists are not. It remains important to understand apo-LBD conformation and the way that it rearranges with ligands to develop better TR pharmaceuticals. In this study, we conducted hydrogen/deuterium exchange on TR LBDs with or without agonist (T3) or antagonist (NH3). Both ligands reduce deuterium incorporation into LBD amide hydrogens, implying tighter overall folding of the domain. As predicted, mass spectroscopic analysis of individual proteolytic peptides after hydrogen/deuterium exchange reveals that ligand increases the degree of solvent protection of regions close to the buried ligand-binding pocket. However, there is also extensive ligand protection of other regions, including the dimer surface at H10–H11, providing evidence for allosteric communication between the ligand-binding pocket and distant interaction surfaces. Surprisingly, C-terminal activation helix H12, which is known to alter position with ligand, remains relatively protected from solvent in all conditions suggesting that it is packed against the LBD irrespective of the presence or type of ligand. T3, but not NH3, increases accessibility of the upper part of H3–H5 to solvent, and we propose that TR H12 interacts with this region in apo-TR and that this interaction is blocked by T3 but not NH3. We present data from site-directed mutagenesis experiments and molecular dynamics simulations that lend support to this structural model of apo-TR and its ligand-dependent conformational changes.
Hydrogen-Deuterium exchange, molecular modelling and site directed mutagenesis provide a structural model for apo-TR LBD, and for its conformational changes triggered by ligand binding in solution.
Thyroid hormone receptors (TRs) are physiologically important transcription factors that belong to the nuclear hormone receptor (NR) family and play roles in regulation of cholesterol levels, metabolism and heart function in adults (1,2,3,4). TRs mediate actions of thyroid hormones, predominantly T3 (3,5,3′ triiodo-l-thyronine) but are transcriptionally active with and without ligands because they bind constitutively to chromatin, often as heterodimers with retinoid X receptors (RXRs). T3 modulates gene transcription by altering the conformation of the TR ligand-binding domain (LBD), which in turn alters the complement of TR-associated coregulators (1,2,3,4).
It is important to understand how ligands influence TR LBD conformation to develop better pharmaceuticals to modulate receptor activity (5,6,7,8). We, and others, obtained x-ray structures of TR LBDs with agonists (4,6,8,9,10,11,12). These structures, with subsequent similar structures of other NRs, reveal that the LBD adopts a canonical three-dimensional fold comprised of 12 conserved α-helices (H) and 4 β-strands (S) (13,14,15,16,17,18). Hormone occupies a buried ligand-binding pocket (LBP) formed by H5–H6 on the top, by H7 and H11 on one side, and along the opposite side by H2, S3 and S4, and H3, being enclosed by a lid formed by the C-terminal part of H11 and H12 (10,11,12,13,14,15,16,17,18). At present, x-ray structures of unliganded (apo-) TRs or TRs with available antagonists have not been reported. Thus, it has not been possible to perform detailed comparisons of TR conformation in active agonist-bound and unliganded and inactive states.
One hormone-dependent change in apo-TR conformation has been inferred from mutational analysis of TR and comparison of liganded TR structures with available x-ray structures of other apo- and antagonist-bound NR LBDs (14,15,16,17); agonist promotes packing of C-terminal H12 into an active position against the LBD. This event completes a coactivator binding surface [activation function 2 (AF-2)] that includes surface-exposed hydrophobic residues from H3 and H5 and partly occluding an overlapping corepressor binding surface that is also comprised of residues from H3 and H5 but extends below the usual position of H12 in the liganded state (15,16,17,18). Although H12 must adopt a distinct position that fully exposes the corepressor binding surface in the absence of ligand, the organization of H12 in apo-TR is unknown (18).
Hormone induces other rearrangements in the LBD. Analysis of dynamics of several NR LBDs suggests that they are disordered without hormone (19,20). Early models suggested that ligand stabilizes the region near the LBP, with the rest of the domain remaining more organized in the absence of ligand (21,22,23). Several specific ligand-dependent changes have been inferred from mutational analysis of TRs. First, in vitro assembly assays indicate that H1, which links the LBD to the receptor DNA-binding domain (DBD), packs tightly against the LBD (H2–H11) with agonists (24,25,26). Second, analysis of temperature (B) factors in x-ray structures of liganded TR-LBDs with human resistance to thyroid hormone syndrome mutations reveals instability in the H1–H3 region (25,26). Because resistance to thyroid hormone mutants retain features of apo-TR LBD conformation with agonists, it is likely that the H1–H3 region is unstructured without ligand. Last, T3-dependent rearrangements in surface salt bridge clusters in the H7–H8 region and H11, near the TR dimer surface, stabilize bound T3 and inhibit apo-TR homodimer formation (24,25,26). Presently, however, the true extent and function of conformational changes that occur upon hormone binding are not known.
It is also important to understand structural alterations that occur in response to TR antagonists to comprehend the molecular basis of their action and to provide information for the structure-directed development of useful antagonist compounds for treatment of thyrotoxicosis and cardiac arrhythmias (18). We identified the first TR antagonists, based on TR LBD x-ray structures and the knowledge that active H12 conformation is required for coactivator binding. We reasoned that ligands that resemble native hormone with appropriately placed extensions could compete for agonist but would dislodge H12, inhibiting coactivator binding and receptor activity (27). Later, x-ray structures of estrogen receptors (ERs) reveal that many antagonists indeed contain bulky extensions that displace H12, which packs over the H3–H5 region of AF-2 (7,14,17,28). Our lead antagonist, NH3, is derived from the synthetic TRβ agonist GC-1 and contains a 5′-nitrophenylethynyl extension predicted to dislodge H12 (27). NH3 binds TRs with nanomolar affinity, blocks TR LBD interactions with coactivators, and antagonizes T3 responses in cell culture, tadpoles, and rats. More surprisingly, NH3 also inhibits TR LBD interactions with corepressors, and we proposed that the ligand repositions H12 so that it occludes the coactivator and the corepressor binding surfaces (27). This idea is not proven, and the extent to which NH3 alters other aspects of LBD conformation is not clear.
Amide hydrogen/deuterium (H/D) exchange can be used to probe protein conformation and dynamics (29,30,31). In this technique, protein samples are incubated in deuterated (D, heavy) water, and exchange of deuterium with protein amide hydrogens (H, light isotope) is detected by mass spectroscopic analysis of complete proteins or individual proteolytic peptides. Because amide hydrogens must contact solution for deuterium exchange to occur, the increase in protein/peptide mass after incubation in heavy water provides a useful index of solvent exposure in different conditions (29,30,31,32,33,34). Several studies confirmed that this technique yields information about ligand-dependent structural perturbations and dynamics of different NR (21,22,23,32,33,34), including the full-length peroxisome proliferator-activated receptor-RXR heterodimer (34). Molecular dynamics (MD) simulations complement this method, and MD has been used to examine other aspects of LBD dynamic behavior (35,36,37,38,39,40,41). Here, we use H/D exchange to probe TR LBD conformation with or without T3 or NH3. This analysis, coupled with TR LBD x-ray structures, prior knowledge of ligand-dependent conformational rearrangements of the LBD, site-directed mutagenesis experiments, and MD simulations, allows us to generate specific hypotheses about apo-LBD organization and effects of TR agonists and antagonists on LBD structure.
Results
TR LBD characterization
Mass-spectroscopic analysis of humanTRβ1 LBD preparations revealed that the protein is intact and composed of dimers and monomers. The average molecular mass of TRβ1 LBD monomer (amino acids 209-461) predicted by MS-digest software is 30,049 Da (28,645 Da from TR plus 1422 Da from His tag and linker). Mass-spectroscopic analysis of apo-TRβ1 LBD preparations reveals peaks of 60,108.1 ± 26.8 Da and 30,039.2 ± 19.3 Da, which correspond to LBD average molecular mass of TR dimers and monomers. The proportion of monomers and dimers determined in apo-TR mass-spectroscopic measurements, based on the absolute intensity of each peak, was approximately 62 and 38% for dimers and monomers, respectively. The monomer/dimer distribution was consistent with previous studies of the protein by size exclusion chromatography and native gel electrophoresis (10,42). Analysis of TRβ1 LBD preparations liganded to T3 and NH3 reveals that they are also comprised of a mix of monomers and dimers in similar conditions (data not shown).
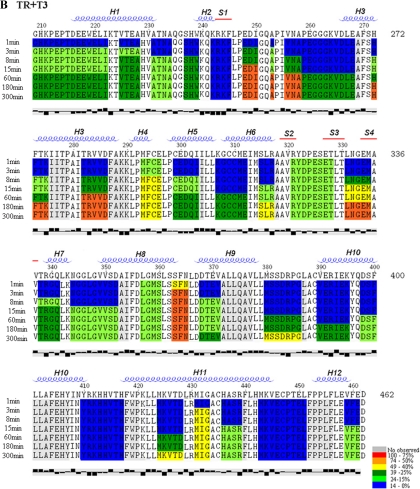
We performed pepsin digestions of apo- and liganded TR LBD preparations and analyzed the mass of each peptide by mass spectroscopy. Proteolysis generated 68 identifiable peptides, covering 92% of the amino acid sequence (Fig. 1). Of these, 31 peptides exhibited better signal to noise ratio (solid underline), covering about 88% of TRβ1 LBD amino acid sequence, and these peptides were used for H/D exchange analysis. The other peptides were found at low intensity in the mass spectrum, and they cover the same region of the protein as the selected peptides. The profile of deuterium incorporation for each peptide of apo-TR, TR+T3, and TR+NH3 is presented in Supplemental Fig. 1 (published on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org).
Figure 1.
Alignment of all peptides generated by pepsin cleavage shows almost complete coverage of the TR sequence (92%). The peptides used in our analysis (88% of sequence coverage) are shown in continuous line.
Ligand reduces LBD solvent exposure
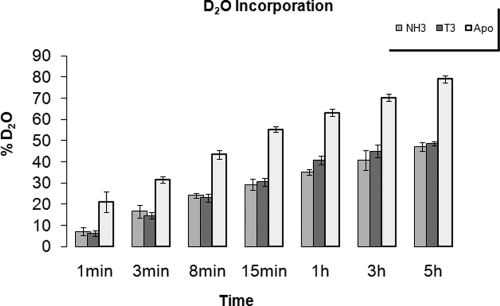
Global mass exchange in apo-TR, TR+T3 and TR+NH3 complexes was analyzed by ESI MS revealing that total deuterium incorporation increased with incubation time in all samples. Deuterium exchange was detected in approximately 80% of amide hydrogen in apo-TR peptides, 48.8% in TR+T3, and 47.2% in TR+NH3 peptides. Thus, the agonist (T3) and antagonist (NH3) both induce tighter folding of the LBD (Fig. 2).
Figure 2.
Ligand-dependent reductions in TR LBD solvent exposure. Deuterium levels at amide bonds in the TR LBD during the total time of the experiment for the receptor in the absence of ligand (apo TR, light gray), in presence of agonist (TR+T3, dark gray) and of antagonist (TR+NH3, medium gray). H/D exchange of the liganded-protein is lower than apo-protein, indicative of protection against deuterium uptake.
Apo-TR LBD conformation
We analyzed deuterium incorporation into individual apo-TR LBD proteolytic peptides as a function of time and projected positions of each peptide onto x-ray structural models of T3-TR LBDs. Analysis of effects of short (1 min) heavy water incubation implies that the apo-LBD is well folded (Fig. 3A). Some apo-LBD peptides were only weakly deuterated, even after long D2O incubation times, as shown by blue color (<15% D incorporation) in the structure projection in Fig. 4A.
Figure 3.
Deuterium incorporation into individual TR-LBD peptides varies with ligand. Deuterium uptake at each time point (t1 to t8 - 1, 3, 8, 15, 60, 180, 300 and 480 min, respectively) for apo-TR, TR+T3 and TR+NH3 is shown, respectively, in A, B and C. The rates of deuterium uptake are color-coded: red shows an uptake over 75%; orange, from 74 to 50%; yellow, from 49 to 40%, dark green, from 39 to 25%, light green, from 24 to 15%; blue, below 14%. The secondary structure was determined by DSSP software program. α-helices are given as α1 - α12, and β-strands are termed as β1 - β4. The last line represents the hydrophobicity of each residue, plotted by Texshade software. The first two residues in each peptide are not colored because they do not participate in H/D exchange (31).
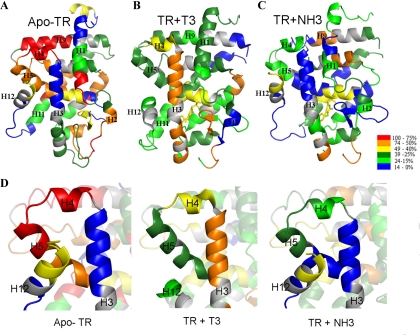
Figure 4.
Projection of deuterium exchange rates onto structural models of TR. The TR LBD structures colored by differences in deuterium uptake, in a gradient color, showing the deuterium incorporation after a 5-h experiment, evidencing the differences among apo-TR (A), TR+T3 (B), andTR+NH3 (C). The scale from low to high D2O uptake is represented by a gradient color, from blue (protected) to red (exposed). D, Close-up view of protected area of H3, H4, H5, and H12 for apo-TR and TR+NH3, which is more exposed in TR+T3. Note that the same area exhibits greatly increased protection from solvent in inactive conformation (apo-TR and TR+NH3, green to blue) when compared with active conformation (TR+T3, orange to red).
Analysis of positions of peptides and the extent of deuteration suggests that apo-LBD conformation resembles agonist-bound LBD (Fig. 3A). Peptides with amino acids that are predicted to form the folded core of the domain (H1, H2, H6, H9, and H10, and β-strands S2 and S3) exhibit low deuterium incorporation (blue). By contrast, peptides with amino acids that lie on the surface of the domain exhibit intermediate or high levels of deuterium incorporation (green and yellow, 15–50%, H3, H5, and the center of H11; orange and red, >50%, the H1 C terminus, S1, H4, and H8). Surprisingly, three parts of the predicted protein surface appear well protected from solvent; the N-terminal H0/H1 region, part of H3 in the AF-2 surface (amino acids 280-286), and the region that encompasses the loop between H11 and C-terminal activation H12 and H12 itself.
Deuterium incorporation into apo-TR peptides generally increased as a function of time (note progression of color from blue through green, yellow and orange to red in Fig. 3A). The most striking changes (<15% deuterium incorporation after 1 min to >85% deuterium incorporation after 5 h) involved the dimer surface (C terminus of H10 and the N terminus of H11) and the H5–H6 region. Thus, both regions of protein are initially protected from solvent but undergo motions that bring them into contact with solution during the incubation. Interestingly, H12 exhibited only moderate deuterium incorporation during the experiment (<15%, blue, to <49%, yellow), implying that it is not highly solvent exposed. Four TR segments remained completely protected (blue) throughout the experiment: the N-terminal part of H1 (amino acids 209-221); β-strands S2 and S3; the C-terminal part of H3 (amino acids 280-286); and a fragment from the C-terminal part of H11 and the loop between H11 and H12 (amino acids 434-450).
Together, our results suggest that apo-TR is well structured and that its fold resembles that of liganded LBD. In addition, the fact that most TR peptides are, at least, partly accessible to deuterium exchange during 5-h incubations implies that apo-LBD is relatively dynamic.
T3-TR conformation
Measurement of deuterium exchange into TR-T3 complex proteolytic peptides supports the conclusion that ligand induces a more compact configuration than apo-LBD (Figs. 3B and 4B). Whereas deuterium incorporation into apo-TR approached 100% after 5 h for some peptides (Table 1), rates of deuterium incorporation into T3-TR peptides reached only 67%, at best, and were usually lower (Table 1 and Fig. 3B). Nearly all peptides from the TR-T3 complex were poorly accessible after 1 min heavy water incubations (<15% incorporation, blue) (Fig. 3B and Table 1). At this time, only H4, the S2/S3 β-sheet region, and H8 exhibited intermediate levels of deuterium incorporation (15–49%). Most regions of TR remained inaccessible (blue) during longer incubations or exhibited only intermediate levels of deuterium incorporation (15 – 40%, green) (Table 1 and Fig. 3B). Exceptions were peptides in the loop between S1 and H3, the upper part of H3, H4, the S3/S4 region, H8, and the center of H11, which all exhibited more than 40% deuterium exchange.
Table 1.
Differences in deuterium uptake after 5 h of deuteration
| Structurea | Sequence | Residuesb
|
Apo-TR (%)c | ± Error (%) | TR-T3 (%)d | ± Error (%) | TR-NH3 (%)e | ± Error (%) | Apo-TR − TR-T3 (%)f | Apo-TR − TR-NH3 (%)g | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beginning | End | ||||||||||
| L0/H1 | GHKPEPTDEEWEL | 209 | 221 | 9 | 0.2 | 27 | 0.3 | 18 | 0.4 | −18 | −9 |
| H1 | KTVTEA | 223 | 228 | 33 | 0.6 | 33 | 0.4 | 17 | 0.4 | 0 | 17 |
| H1/L1 | HVATNA | 229 | 234 | 100 | 1.7 | 17 | 0.2 | 33 | 0.7 | 83 | 67 |
| L1 | TNAQGSHW | 232 | 239 | 25 | 0.4 | 25 | 0.3 | 38 | 0.8 | 0 | −13 |
| H2/B1 | KQKRKF | 240 | 245 | 100 | 1.7 | 0 | 0.0 | 33 | 0.7 | 100 | 67 |
| L2 | LPEDI | 246 | 250 | 100 | 1.7 | 50 | 0.6 | 50 | 1.1 | 50 | 50 |
| L2 | PEDIGQA | 247 | 253 | 50 | 0.9 | 67 | 0.7 | 33 | 0.7 | −17 | 17 |
| L2 | PIVNA | 254 | 258 | 75 | 1.3 | 50 | 0.6 | 50 | 1.1 | 25 | 25 |
| L2/H3 | PEGGKVDLEA | 259 | 268 | 33 | 0.6 | 22 | 0.2 | 22 | 0.5 | 11 | 11 |
| H3 | SHFTKI | 270 | 275 | 50 | 0.9 | 50 | 0.6 | 50 | 1.1 | 0 | 0 |
| H3 | ITRVVDF | 280 | 286 | 14 | 0.2 | 57 | 0.6 | 14 | 0.3 | −43 | 0 |
| H4 | PMFCEL | 291 | 296 | 100 | 1.7 | 40 | 0.4 | 20 | 0.4 | 60 | 80 |
| H5 | PCEDQII | 297 | 303 | 83 | 1.4 | 33 | 0.4 | 33 | 0.7 | 50 | 50 |
| H5/H6 | LKGCCMEI | 305 | 312 | 63 | 1.1 | 25 | 0.3 | 13 | 0.3 | 38 | 50 |
| H6 | MSLRA | 313 | 317 | 40 | 0.7 | 40 | 0.4 | 60 | 1.3 | 0 | −20 |
| B2/B3 | VRYDPESETL | 319 | 328 | 22 | 0.4 | 22 | 0.2 | 11 | 0.2 | 0 | 11 |
| B3/B4 | TLNGEMA | 329 | 335 | 57 | 1.0 | 57 | 0.6 | 29 | 0.6 | 0 | 29 |
| B4/H7 | VTRGQL | 336 | 341 | 67 | 1.1 | 33 | 0.4 | 33 | 0.7 | 33 | 33 |
| H7/H8 | KNGGLGVVSDA | 342 | 352 | 20 | 0.3 | 20 | 0.2 | 30 | 0.6 | 0 | −10 |
| H8 | DLGMSL | 355 | 360 | 100 | 1.7 | 17 | 0.2 | 33 | 0.7 | 83 | 67 |
| H8/L9 | SSFNL | 361 | 365 | 100 | 1.7 | 60 | 0.7 | 40 | 0.8 | 40 | 60 |
| H9 | DDTEVA | 366 | 371 | 83 | 1.4 | 33 | 0.4 | 50 | 1.1 | 50 | 33 |
| H9/L10 | MSSDRPGL | 378 | 386 | 50 | 0.9 | 38 | 0.4 | 38 | 0.8 | 13 | 13 |
| L10H10 | CVERIEKY | 388 | 395 | 75 | 1.3 | 25 | 0.3 | 25 | 0.5 | 50 | 50 |
| H10 | QDSFL | 396 | 400 | 100 | 1.7 | 20 | 0.2 | 60 | 1.3 | 80 | 40 |
| H10H11 | NYRKHHVTHF | 408 | 417 | 50 | 0.9 | 0 | 0.0 | 30 | 0.6 | 50 | 20 |
| H11 | LMKVTDL | 422 | 428 | 86 | 1.5 | 43 | 0.5 | 43 | 0.9 | 43 | 43 |
| H11 | RMIGA | 429 | 433 | 80 | 1.4 | 40 | 0.4 | 40 | 0.8 | 40 | 40 |
| H11 | CHASRF | 434 | 439 | 17 | 0.3 | 17 | 0.2 | 33 | 0.7 | 0 | −17 |
| H11/L12 | HMKVECPTELF | 441 | 451 | 10 | 0.2 | 10 | 0.1 | 10 | 0.2 | 0 | 0 |
| H12 | EVFED | 457 | 461 | 40 | 0.7 | 20 | 0.2 | 40 | 0.8 | 20 | 0 |
Location of the peptide in protein structure.
Number of residues in the beginning and in the end of the peptide.
Percentage of deuterium uptake in Apo-TR.
Percentage of deuterium uptake in TR+T3.
Percentage of deuterium uptake in TR+NH3.
Deuterium uptake difference percentage between Apo-TR and TR+T3.
Deuterium uptake difference percentage between Apo-TR and TR+NH3.
Projection of peptides that exhibit increased solvent protection in the presence of T3 vs. apo-TR onto structural models of the T3-LBD complex suggests that ligand binding altered the conformation and dynamics of the whole domain (Fig. 3B). Several peptides that comprise the region that envelopes the LBP (including the C terminus of H1, H2, H6, and H7–H8) or comprise the hydrophobic core of the domain (part of H4, S2, and parts of H9 and H10) exhibit greatly increased solvent protection with hormone. As predicted from previous mutational studies of TRs (see introductory section), we observed striking increases in ligand protection of the C-terminal part of H1, H2, S1, the loop between H2 and H3, and the H7–H8 region.
Some peptides that comprise part of the LBP that interacts with the outer ring of the ligand, including the N-terminal part of H3, H11, the loop between H11 and H12, and H12 itself, did not exhibit great changes in solvent accessibility with T3. The sole difference was that H12 exhibits a modest increase in solvent protection with hormone [note the transition from yellow (apo) to green (T3) for amino acids 456–461]. Equally surprising, T3 binding led to extensive protection of several regions of the domain that are relatively distant from the LBP, including parts of H4, H9, and the dimer surface at the junction of H10–H11. Thus, solvent accessibility of part of the LBD does not change greatly with hormone and hormone-dependent structural rearrangements affect the entire domain.
Three regions of the domain exhibited increased rates of deuterium exchange after T3 binding, consistent with increased solvent exposure (Fig. 3B). These were S2–S3, implying that ligand binding must promote rearrangements in this region of the LBP that bring this region into contact with solvent. More surprisingly, we observed increased deuterium incorporation at the hinge/H1 region after T3 binding. This is the first evidence that hormone alters the conformation of this region of TR (which links the LBD to the neighboring DBD). Finally, there are striking increases in deuterium incorporation in the upper part of H3 that comprises part of the AF-2 surface along with modest increases in deuterium incorporation in the neighboring H4 region relative to apo-TR. Thus, hormone binding must expose the upper part of AF-2 to solvent (see Discussion).
NH3-TR conformation
As seen with T3-TR, the NH3-TR complex appeared more tightly packed than apo-TR. H/D exchange rates for individual peptides reached only 60% (down from 100% without ligand; Fig. 3C and Table 1). Most peptides exhibited low levels of deuterium incorporation (blue) during short heavy water incubations and reached intermediate levels (green) only after extended incubation, as seen with T3.
Projection of positions of peptides that exhibited increased protection from solvent in the presence of NH3 onto the TR-T3 structural model revealed strong similarities between effects of both ligands on TR conformation. We observed 1) concerted protection in regions of the LBP that envelop the charged ligand carboxylate group and inner ring, especially the region between H1 and H3, the N terminus of H7, and the C terminus of H8; 2) few alterations in solvent accessibility in the region of the LBP that envelops the outer ring and 5′-phenyl ring extension of the ligand, the N terminus of H3, H11, the loop between H11-H12 and H12; 3) marked increases in solvent protection of several regions of protein that are distant from the LBP, including H9 and H11; 4) increased solvent exposure of H0/H1 with ligand.
Several regions of the TR protein did exhibit differences in deuterium exchange rates with NH3 vs. T3 (Table 1). Most strikingly, the C-terminal part of H3 (amino acids 279-285) appeared well protected from solvent in the presence of NH3 but not T3. There were also modest increases in protection in the nearby H4 region (amino acids 290-295) with NH3 vs. T3. These regions were also well protected from solvent in apo-TR, suggesting that NH3-TR conformation resembles apo-TR conformation at this location. NH3 also gave better protection of several regions near to the LBP relative to T3, including the center of H1, the loop between H2 and H3, the N terminus of H6 and S2–S4. Conversely, there was increased solvent exposure of the C terminus of H6 (amino acids 313-317) and the C terminus of H11 with NH3 relative to T3. These variations probably reflect differences in binding mode of the two ligands (see Discussion). Finally, there were differences in regions that are relatively distant from the LBP; NH3 increased solvent protection of H10 relative to apo-TR, but the degree of protection was less than with T3. This implies differences in dimer surface configuration with T3 vs. NH3.
Model of TRβ1 LBD in inactive conformation suggests that H12 docks over H3
Differences between deuteration rates obtained with different TR complexes in the H3–H5 region are shown in Fig. 4 (A–C). Comparison of deuteration in apo-TR and T3-TR complexes or NH3-TR and T3-TR complexes (Fig. 4D) highlight the fact that there is increased protection of the upper part of H3 in the absence of ligand or the presence of the antagonist.
Because NR H12 docks over the H3–H5 region in the presence of some antagonists (14,18,28,42,43), we asked whether similar interactions could account for T3-dependent increases in solvent accessibility of the C-terminal part of H3. We created hybrid structural models consisting of the H0 to H11 of TR LBD+T3 [Protein Data Bank (PDB) ID 3GWS] (10) and H12 from an ERα-antagonist structure with an inactive H12 conformation (the ER-hydroxytamoxifen complex (PDB ID 3ERT) (43) (Fig. 5, A and B). This model shows that H12 binds to the same part of TR H3 that exhibits increased solvent exposure with T3 (Fig. 5, C and D); this corresponds to the upper part of the coactivator/corepressor binding surface. Alignment of TR and ER sequences reveals only 23.7% identity and 58% similarity between LBDs and between the two regions that interact in our apo-TR model, the C-terminal portion of H3, and center of H12 (not shown). Nevertheless, specific residues from TR and ER H3 and H12 that are predicted to interact are conserved (Fig. 5E).
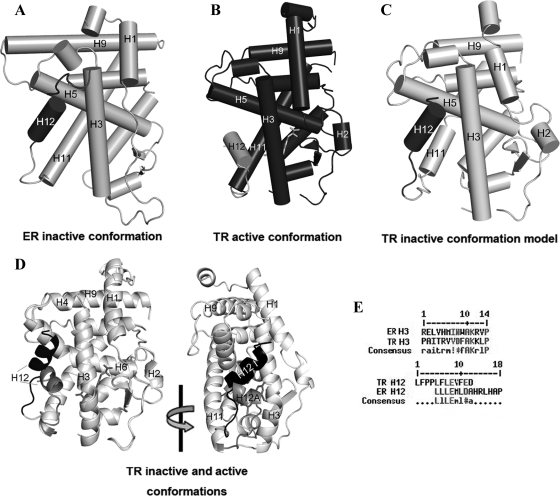
Figure 5.
Model of TRβ1 LBD in inactive (apo) conformation. A, ERα LBD crystal structure used as template for the modeling (gray, PDB ID 3ERT); the H12 (dark gray) is positioning in inactive conformation, docking over H3. B, TR + T3 crystal structure (dark gray, PDB ID 3GWS), also used as template for modeling, with H12 (light gray) in active conformation. C, The proposed model of apo-TR LBD (light gray), with the H12 (dark gray) positioned over H3 in inactive conformation. D, Superposition of holo-TR structure and apo-TR model presenting the two different conformations of H12: active (H12A, gray) and inactive (H12I, dark gray). E, Alignment of ER H3 and TR H3 where the residues that make contacts with H12 are considered similar (gray); alignment of H12 of TR and of ER, where the residues that contacts H3 are similar (gray).
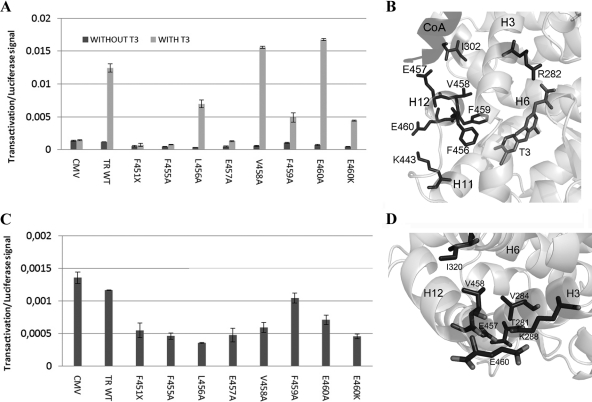
Because the TR-ER hybrid model predicts that TR H12 is required for optimal T3 response and masking of the corepressor binding surface at H3–H5, we mutated TRβ H12 and tested activity of mutant TRs in transactivation assays with or without T3 (Fig. 6 and Table 2). As expected, the deletion mutant F451X (which removes H12) eliminated the T3 response at a standard T3-inducible reporter (Fig. 6A). Ala substitutions at F455, L456, E457, and F459 reduced or eliminated the T3 response, whereas similar mutations at V458 and E460 modestly increased activity. This is consistent with the known structure of the T3-TR complex and previous functional analysis (Fig. 6B); residues that are required for the T3 response contact the hormone (F455 and F459), play a role in packing of H12 against H3–H5 (L456), or contact coactivator (E457), whereas residues that are dispensable for the T3 response (V458 and E460) are partly or completely solvent exposed and make only weak side-chain contacts with nearby residues (I302 on H5 and K443 in H11, respectively).
Figure 6.
TR H12 is needed for activity of T3-TR and unliganded TR. A, Transactivation assay for TR wild-type and TR point mutations in H12 showing responses to T3. B, Structural model of TR H12 interactions in the presence of T3 derived from T3-TR crystal structure showing positions of key residues and interactions with protein or ligand. E457 contacts coactivator. C, Transrepression in the absence of ligand. TR H12 mutations cause increased repression by unliganded TR. D, Structural model of TR H12 interactions in the absence of T3 derived from the ER-TR hybrid model in Fig. 5. Note that V458 and E460 are predicted to form tight interactions with nearby residues on H3.
Table 2.
Mutations, location in protein structure, and effects in transactivation assays
| TR mutant | Interactions in active state | Predicted interactions in inactive state | T3 activation | Basal repression |
|---|---|---|---|---|
| F455A | T3 (binding) | I302 (H5) | − | + |
| L456A | F451 (loop H11–H12) | − | + | |
| E457A | Coactivator | T281 (H3) | − | + |
| V458A | L454 (H12) | V284 (H3), I302 (H5) | + | + |
The same mutations exhibited different effects on unliganded TRs. TRβ451X showed increased transrepression relative to unliganded wild-type TR (Fig. 6C), consistent with the idea that the corepressor binding surface is fully exposed and that TR H12 masks the corepressor binding surface in vivo in the absence of ligand (18,44,45,46). More surprisingly, TR H12 mutations, including TRβV458A and E460A mutations that showed increased T3 response, exhibited a similar phenotype in the absence of ligand. Our TR-ER hybrid model (Fig. 6D) predicts that H12 is buried in the cleft between H3 and H5, with H12 residues in contact with the floor of the cleft (L456, E457, and F459) or side chains of amino acids that surround the cleft (V458 and E460). Thus, multiple H12 amino acids are required to suppress transrepression capacity of unliganded TR, and we suggest that this requirement is consistent with the model in Fig. 5, which predicts that multiple H12 amino acids participate in interactions with the H3–H5 hydrophobic cleft.
MD simulations support the new apo-TR structure model
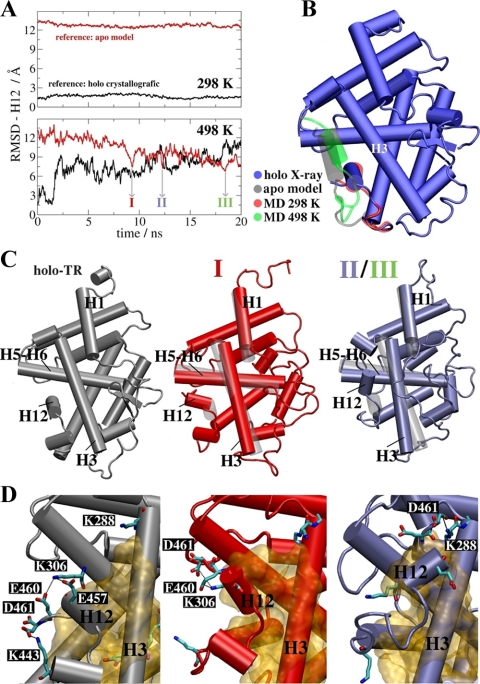
Our structural model for apo-TR illustrates one possible H12 conformation that explains experimental results from H/D exchange. To explore the likelihood that H12 will adopt this position without ligand, we performed a series of MD simulations, starting from the active conformation of the TR LBD, but with ligand removed.
We did not detect H12 repositioning in the apo-TR model under normal MD simulation conditions. Our previous MD studies revealed that the main TR ligand dissociation pathway involves the mobile part of the LBD comprising H3, the loop between H1 and H2, and nearby β-sheets, located opposite to H12 (39,40), but ligand dissociation occurred without appreciable changes in the H12 position. We confirmed that H12 did not change position in three independent simulations at normal temperature (298 K) with or without T3 (Fig. 7). The root mean square deviation (RMSD) between the ligand-free LBD and the crystallographic holo structure remains constant at about 1.5 Å (Fig. 7A, top panel, black line) and at about 12.5 Å relative to the hybrid apo model (red curve). It is likely that displacement of H12 is kinetically suppressed because of extensive packing against the body of LBD and therefore unlikely to occur during relatively short timeframes that are currently feasible with existing MD simulation approaches. A similar result was obtained in simulations of the ER LBD (37).
Figure 7.
Conformational transition from the holo-LBD structure without ligand to apo-TR model via MD. A, RMSD of the H12 Cα atoms positions in two different structural references: the TR+T3 (black line) and apo-TR model conformations (red line). The upper panel shows average RMSD of three independent simulations at 298 K. No conformational change is detected. The lower panel depicts the RMSD of one of the three independent simulations at 498 K. There is an increase in RMSD for holo conformation and a reduction when the reference is the proposed apo model. The arrows indicate the three lowest RMSD structures for the apo model (I–III) selected as starting structures to be annealed to 298 K. B, Structural superposition of the holo-TR LBD bound to T3 (blue), the average LBD structure obtained from simulations at 298 K (red), the hybrid apo-TR LBD model (gray), and the lowest RMSD structure (∼6 Å) for the model obtained in the simulation at 498 K (green). Note similarities in conformations between the former two models and later two models, respectively. C, Comparison between the holo-TR LBD (gray) and the annealed apo-LBD average structures I (red) and II (blue), obtained from the last 5-nsec portions of the annealing runs. Average structures obtained from initial conformations II and III are nearly identical. D, The main polar interactions and hydrophobic contacts (yellow surface) between H12 and the LBD body in the three structures.
To increase the likelihood of observing changes in the H12 position, we carried out three simulations at higher temperature (498 K, Fig. 7A, lower panel) to increase sampling rate of protein motions and overcome energy barriers that prevent large-amplitude movements of H12. Although this temperature is unrealistically high, its overall impact on the LBD structure is not very strong in short simulation times of the order of 10–20 nsec or less, as previously observed under similar simulation conditions (47). At high temperature, H12 RMSD in the holo-TR increases from 1.5 to 10.0 Å (black line, bottom panel, Fig. 7A). This means that the structure diverges from the holo-LBD structure, as expected in high-temperature simulations. However, the RMSD of H12 in the apo model drops from 12.5 to about 6.5 Å. Therefore, behavior of H12 in the apo structure differs from the holo crystallographic structure.
Results outlined above are a consequence of the fact that H12 adopts a new position closer to the one suggested by the apo model than to the holo structure. Comparisons of average apo-LBD structure in simulations at 298 K (red) and the lowest RMSD structure (around 6 Å) obtained from the 498 K simulations (green) reveals H12 in a new position, docked over H3–H5 (Fig. 7B). This is similar to our hybrid apo-TR LBD model (gray) but distinct from the T3-TR LBD complex (blue). Thus, MD simulations performed without ligand predict that H12 displays tendency to dock over the H3–H5 region.
To determine the conformational stability of the new H12 position and to confirm that it is not dependent on structural rearrangements in the LBD that are specific for high temperature, we performed additional simulations that started from different high-temperature apo conformations (labeled I–III, Fig. 7A) and were cooled to 298 K to restore annealed LBD structures. The resulting annealed apo structures aligned closely with the holo-LBD, except for H12, which remained docked over H3–H5 (Fig. 7C, II/III) or trapped in an apparent transition state between conformations (Fig. 7, C and I). Conformational differences were detected in other parts of the apo-LBD, affecting H1, H3, H5, and the lower part of H11, which partly folds into the open LBP as shown in Supplemental Fig. 2. Thus, the novel apo-H12 position was preserved in annealed apo structures, whereas organization of most LBD secondary structure elements was restored to a conformation that resembled holo-TR.
Comparisons of main polar residue interactions that hold H12 in place in the holo (gray) and annealed apo (color) structures were consistent with those predicted by our hybrid TR-ER model (Fig. 7D). In the holo structure, H12 is anchored by salt bridges between residues E457, E460, and D461 with K443 and K288, in addition hydrophobic contacts with protein and the ligand. In structure I, H12 is caught in transition to an inactive position in which interactions between E460/E461 and K306 are prominent. In structures II/III, H12 is found in the suggested apo-structure conformation, where residues D461 and K288 form a salt bridge protecting the hydrophobic region of the C terminus of H3 from hydration (see also Supplemental Fig. 2). Overall, the computed hydration numbers of the C-terminal portion of H3 (comprised of residues 280-285) are markedly smaller in the apo structures II/III relative to TR-T3 (Supplemental Fig. 2), consistent with results of HD exchange.
Discussion
It is important to understand apo-TR conformation and structural alterations that occur with agonist or antagonist binding (see introductory section). Presently, there are no crystal structures of apo-TR or antagonist-bound TRs to compare with agonist-bound TR-LBD. Thus, it has not been possible to perform detailed comparisons of structures of TRs in different activity states.
In this study, we used H/D exchange to learn about apo-TR LBD conformation in solution and structural alterations that occur with different classes of ligands. We find that agonist- and antagonist-liganded TRs are more compact than the equivalent apo-LBD complex. This agrees with previous studies of other NRs that employed a variety of techniques to probe domain organization, including H/D exchange, nuclear magnetic resonance, protease sensitivity, and melting temperature determination (19,20,21,22,23,32,33,34,47). All suggest that the apo-NR LBD is mobile and that ligand induces a tightly packed conformationally restricted state. However, apo-TR is not completely unfolded. Analysis of solvent protection of TR proteolytic peptides, coupled with mapping of their positions onto TR x-ray structures, suggests that apo-TR partly resembles a loosely folded version of agonist-bound TR.
Several investigators have proposed that the NR LBP region (roughly corresponding to the lower part of the domain in Fig. 4) will be completely disordered without ligand (19,20,21,22,23). This is only partly true for TRβ. The portion of the LBP that binds the T3 inner ring and aminopropionate group, including H1, the H1–H3 region, H6, and H7–H8, is indeed highly solvent exposed in apo-TR and better protected with T3 or NH3. Interestingly, some ligand-dependent rearrangements (involving H1, the H1–H3 loop region, and H7–H8) were previously predicted from TR mutational analysis (see introductory section). However, the opposite part of the LBP near the T3 outer ring (H3, H11, H11-H12 loop, and H12) is well protected from solvent, implying that it is relatively ordered with or without ligands. We do not think that this region is completely unaffected by ligand; it is well established that H12 changes location after hormone binding. Rather, we suggest that ligand-dependent conformational rearrangements that affect this region involve transitions between distinct well-ordered states (see below).
Another surprise was the extent to which ligands affect the entire domain, not just the LBP region. We observed increased protection of the dimer surface at the junction of H10 and H11, indicative of allosteric communication between the LBP and dimer surface. T3 is known to inhibit TR LBD homodimer formation in solution (42), and we expected that hormone would increase overall exposure of the dimer surface to solvent as dimers dissociate to form monomers. The fact that we obtain the converse result implies that hormone remodels the dimer surface itself and that tight packing of this region of protein is incompatible with homodimer formation.
Additional ligand-dependent increases in solvent protection involve H4 and H9. We have not uncovered a function for this region of TR, but the analogous region of the androgen receptor is needed for transcriptional activity (48). Ligand also decreased solvent protection of the hinge and N terminus of H1, implying that the TR hinge is packed against the LBD without hormone and exposed with ligands (24,25,26). We have previously suggested (on the basis of low-resolution x-ray structural models of TR dimers and tetramers in solution (10,42) that differences in LBD conformation affect hinge organization. Our H/D exchange data support this idea and raise the possibility that ligand-dependent changes in hinge position could communicate information about LBD conformation to the nearby DBD.
The most striking ligand-dependent alteration in TR conformation involves the C terminus of H3; T3 greatly increases solvent accessibility of this region relative to apo-TR and the NH3-TR complex. Although T3 could directly increase the mobility of the AF-2 surface, we think that the simplest explanation for this result is that H12 packs against H3 without ligand and that T3 increases solvent accessibility by altering H12 position so that it adopts the position similar to that observed in TR-T3 crystal structures. This hypothesis is attractive for several reasons. First, ER H12 adopts a similar position in ER-antagonist x-ray structures (14,17,28,38), as shown in our hybrid TR-ER model in Fig. 5. Second, it would explain why H12 and nearby regions of the TR LBP are protected in the apo state; H12 adopts an ordered and discrete conformation packed against H3. Third, individual H12 residues play different roles with or without hormone, suggesting that H12 adopts different positions in the apo-TR and T3-TR complex. The fact that all C-terminal amino acids are required for suppression of unliganded TR activity implies that they are required for docking of H12 into the cleft and occlusion of the corepressor binding site in vivo. H/D exchange results with NH3 support this notion. Although T3 and NH3 induced a more compact overall LBD structure, NH3 did not increase solvent exposure of this segment of TR H3. Thus, we predict that NH3 changes LBD conformation but does not alter H12 position relative to apo-TR; if H12 is indeed docked over the H3–H5 region in the presence of NH3, this result would explain why NH3 blocks both coactivator and corepressor binding.
Apart from differences in H3 solvent accessibility, few aspects of TR conformation appear different with T3 and NH3. There were subtle differences in solvent protection pattern that can be attributed to differences in ligand-binding mode. Several regions of TR near the LBP exhibited increased solvent exposure with T3 relative to NH3, including the C terminus of H1, part of the loop between S1 and H3, the S2–S4 region, and H8. Conversely, the C terminus of H6 was solvent exposed with NH3 and less so with T3. We cannot explain these effects without detailed TR-NH3 atomic structures, but the fact that these peptides lie near the T3 aminopropionate group suggests that conformational differences are related to the shorter, negatively charged carboxylic acid substituent at this position in NH3. However, other differences may reflect variations in LBD allosteric communication. Unlike T3, NH3 did not induce complete protection of the dimer surface at C terminus of H10 and the H10–H11 loop. This observation has a functional correlate; NH3 does not inhibit TR-TR dimer formation as efficiently as T3 (27). Although we cannot eliminate the possibility that some NH3-specific effects on TR conformation contribute to its antagonist actions, we nevertheless note that the largest difference between T3 and NH3 involves H3 and propose that specific changes in H12 position and AF-2 surface conformation are most important for the ability of NH3 to block TR activity, exactly as predicted by the extension hypothesis (18).
Our model of apo-TR H12 position has implications for current hypotheses about TR/corepressor interactions. The TR LBD corepressor binding surface is composed of hydrophobic residues from H3, H5, and H6 (49). Thus, packing of H12 over H3 should block corepressor binding. This agrees with experimental observations; H12 truncation enhances apo-TR/corepressor interactions in solution (41). Moreover, NH3 prevents TR interactions with corepressors (50). Additionally, point mutations that are predicted to interfere with contacts between H12 and H3–H5 enhance unliganded TR activity, consistent with the idea that inhibit TR H12 interactions with this region of the LBD surface and expose the corepressor binding site in vivo. MD simulations also lend support to hypothesis by revealing spontaneous, noninduced, H12 conformational transitions from ligand-free holo-TR LBD to structures very similar to the proposed apo-TR LBD model, in which the corepressor binding surface will be occluded.
Finally, it is interesting to compare H/D exchange results for TRs with published studies for other NRs. Whereas we observed strong solvent protection of TR H11 and H12 in the absence and presence of different ligands, other groups observed that glucocorticoid receptor H11 and H12 becomes more exposed with antagonist (RU486) relative to agonist and that RXR agonists decreased deuterium incorporation into H11 but not H12 (21,23). These studies suggest that there are considerable differences between NRs in terms of response of H11–H12 to ligands. It will be important to understand these influences to design ligands to control TR and NR activity.
Materials and Methods
Protein expression and purification
Human TRβ LBD (residues 209-461), fused in frame to the C terminus of a poly-histidine (his) tag in a pET dueT plasmid (Novagen, Darmstadt, Germany), was expressed in the Escherichia coli strain BL21 (DE3) as described (51). After purification, protein buffer was changed to 50 mm ammonium acetate (pH 7.0) using a HiTrap desalting column (GE Healthcare, Piscataway, NJ) because we found that this buffer provided best results for mass spectroscopic analysis. To produce liganded TRs, T3 (Sigma Chemical Co., St. Louis, MO) or NH3 was added in a 3-fold molar excess and incubated for 1 h at 4 C. The protein was concentrated up to 12-fold by ultrafiltration (Amicon Ultra 10MWCO; Millipore, Billerica, MA). Protein concentration was determined by Bradford assay, and purity was assessed by Coomassie Blue-stained SDS-PAGE.
Sample preparation and H/D exchange
H/D exchange was initiated with TRβ1 LBD by 3.5-fold dilution of the protein in the same buffer in D2O (pD 7.0) at 25 C with or without ligands (∼70% D2O). The samples were incubated for various times (1, 3, 8, 15, 60, 180, and 300 min), at which point 70-μl TR aliquots were added to 60 μl of 20 mm Na+-phosphate buffer to quench the reaction (pH 2.5). The samples were immediately applied onto a Quattro II triple-quadrupole mass spectrometer (Micromass, Altrincham, UK), equipped with a standard electrospray ionization source, or digested with pepsin (1 mol enzyme per 10 TR protein) at ∼ 0 C for 5 min and then applied to the mass spectrometer as above.
Sequence identification of pepsin-generated peptides
Deuterium level for each peptide was determined from the differences in centroid masses between the deuterated and nondeuterated fragments. The nondigested protein, after deuteration, was used as a control, being compared with the total deuterium incorporated into the peptides, to estimate deuterium loss during the protein digestion. This procedure was applied to both nonliganded and liganded protein.
Data analysis
The MS-Digest software (52) was used to identify the sequence of selected peptide ions and to calculate the protein molecular weight to compare these data with those acquired from deconvolution of each spectra measurement for all samples. Total H/D exchange was calculated as the total number of peptides bound plus one N-terminal hydrogen per peptide minus the number of proline residues. The secondary structure of the protein was calculated by the DSSP program (53). Deuteration rates and hydrophobicity were plotted using TexShade software (54).
Model building
An apo-TRβ1 LBD model was built using both ERα LBD structure complexed with antagonist 4-hydroxytamoxifen (PDB ID 3ERT) (38) and the TRβ1 LBD structure (PDB ID 3GWS) (10) as templates. The sequences were aligned using ClustalW software (55). For the alignment, we used the entire sequence of TRβ1 LBD, part of TRβ1 structure, from helix 0 to helix 11 (PDB ID 3GWS), and the ERα structure in inactive conformation (PDB ID 3ERT) to modify the conformation of helix 12. The model was constructed using MODELLER 9v4 software (56). For a given alignment, 10 model structures were built and evaluated with the PROCHECK software suite (57). All models were similar, and only the best-evaluated model was retained after the analysis.
Mutations and transactivation analyses
The plasmids pCMV-TRβ1, pCMV-TRβ1 mutants, and TRE-F2-2x1 luciferase reporter for mammals transcription assays were described previously (58,59). New TR mutants reported in the present study (pCMX vectors) were created from existing vectors using QuikChange site-directed mutagenesis kits (Stratagene, La Jolla, CA). The mutations were verified by DNA sequencing.
For transactivation assays, HeLa cells were seeded into 24-well plates at a density of 1×105 cells per well and grown in 10% FBS-DMEM under 95% air and 5% CO2 at 37 C overnight with 2 mm glutamine and 50 μg/ml streptomycin. The cells were then cotransfected with 10 ng of pCMV-TRβ1 and with 100 nm TRE (F2) linked with luciferase reporter. The plasmid pRL containing the Renilla luciferase gene was transfected simultaneously and used as a control. TransFectinβ lipid reagent (Bio-Rad, Hercules, CA) was mixed with plasmids in DMEM and incubated at room temperature for 20 min before adding to the culture medium. The ratio of DNA (micrograms) to TransFectin (microliters) was 1:3 (wt/vol). T3 was subsequently added to the culture medium 4 h later and was incubated with the cells overnight. For activation assays, the ligand concentration in the cultures was kept at 10−7 m.
For luciferase assays, the cell monolayer was washed with PBS and harvested with lysis buffer (dual-luciferase reporter assay system; Promega, Madison, WI) and measured in a Safire2 luminescent counter (Tecan, Durham, NC). Renilla luciferase activity was measured in the same lysate to adjust variation caused by transfection efficiencies. Luciferase assays were performed as previously described (58,59).
MD simulations
The complete simulated systems were built with Packmol (60,61), containing the LBD of TRβ, water, and one counterion for each charged residue for electroneutrality. We used a cubic box with 16,600 water molecules with side dimensions of 81 Å. The average thickness of the LBD hydration layer is approximately 25 Å. The initial protein structure was the T3-TRβ LBD complex in the holo conformation (PDB ID 3GWS) (10), from which the ligand was deleted.
All simulations were performed with NAMD (62), applying periodic boundary conditions, a time step of 2.0 fsec, and CHARMM parameters (63). The TIP3P model was used for water (64). All hydrogen-to-heavy-atom bonds were kept rigid. A 14-Å cutoff with smooth switching function starting at 12 Å was used for the van der Waals interactions, whereas electrostatic forces were treated via the particle mesh Ewald method (65). Energy minimization was performed as follows. The energy of the system was minimized by 700 conjugate gradient (CG) steps keeping all protein atoms fixed, except the modeled regions, which were always allowed to move. Fixing only the Cα atoms, another 500 CG steps were performed. Finally, 300 CG steps were carried out without any restrictions.
After this procedure, 22-nsec MD simulations were performed under constant number of particles, temperature, and pressure conditions (NpT ensemble) at 298 K and 1 bar, with velocity rescaling every 2 psec and Langevin barostat with damping coefficient of 5 psec−1. The first 2 nsec were discarded for equilibration of the system. We carried out three independent simulations with this protocol. To capture events of significant conformational changes during the course of the simulated protein motions, we performed a set of three additional simulations of the same system at the canonical ensemble with constant temperature of 498 K and 1 g/cm3 density (see for instance Ref. 47). The initial configurations for these runs, taken from the last step of 298 K simulations, were thermalized at 498 K by rescaling atomic velocities every 0.1 psec with a Berendsen thermostat during 500 psec before starting the set of 20-nsec production runs.
We characterized the helix H12 conformation with the RMSD computed by aligning each frame to two different references (the holo-TR LBD and the proposed ER-TR hybrid model) with the algorithm described by Kearsley (66). The independent structures with lowest RMSD relative to the hybrid model were used as starting configurations for 298 K annealing simulations. These runs lasted for about 20 nsec each. The average apo structures shown in Fig. 7, C and D, and Supplemental Fig. 2 were obtained from the last 5-nsec portion of these trajectories.
Supplementary Material
Acknowledgments
We thank J. R. C. Muniz for helping us with TexShade software.
Footnotes
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) IP (300220/96-0), by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Grants 03/09462-5, 06/00182-8, and 08/00078-1 and National Institutes of Health Grants DK41482 and 51281 to J.D.B.
Disclosure Summary: The authors have nothing to disclose.
First Published Online November 24, 2010
Abbreviations: AF-2, Activation function 2; CG, conjugate gradient; DBD, DNA-binding domain; ER, estrogen receptor; H/D, hydrogen/deuterium; LBD, ligand-binding domain; LBP, ligand-binding pocket; MD, molecular dynamics; NR, nuclear receptor; PDB, Protein Data Bank; RMSD, root mean square deviation; RXR, retinoid X receptor; TR, thyroid hormone receptor.
References
- Laudet V, Gronemeyer H 1995 The nuclear receptors facts book. London: Academic Press; 1–109 [Google Scholar]
- Glass CK, Rosenfeld MG 2000 The corregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14:121–141 [PubMed] [Google Scholar]
- Lazar MA, Chin WW 1990 Nuclear thyroid receptor. J Clin Invest 86:1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro RC, Apriletti JW, Wagner RL, West BL, Feng W, Huber R, Kushner PJ, Nilsson S, Scanlan T, Fletterick RJ, Schaufele F, Baxter JD 1998 Mechanisms of thyroid hormone action: insights from x-ray crystallographic and functional studies. Recent Prog Horm Res 53:351–394 [PubMed] [Google Scholar]
- Swanson EA, Gloss B, Belke DD, Kaneshige M, Cheng SY, Dillmann WH 2003 Cardiac expression and function of thyroid hormone receptor and its mutant. Endocrinology 144:4820–4825 [DOI] [PubMed] [Google Scholar]
- Baxter JD, Dillmann WH, West BL, Huber R, Furlow JD, Fletterick RJ, Webb P, Apriletti JW, Scanlan TS 2001 Selective modulation of thyroid hormone receptor action. J Steroid Biochem Mol Biol 76:31–42 [DOI] [PubMed] [Google Scholar]
- Nagy L, Schwabe JWR 2004 Mechanism of the nuclear receptor molecular switch. Trends Biochem Sci 29:317–324 [DOI] [PubMed] [Google Scholar]
- Wagner RL, Huber BR, Shiau AK, Kelly A, Cunha Lima ST, Scanlan TS, Apriletti JW, Baxter JD, West BL, Fletterick RJ 2001 Hormone selectivity in thyroid hormone receptors. Mol Endocrinol 15:398–410 [DOI] [PubMed] [Google Scholar]
- Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR 1998 Structure and specificity of nuclear receptor-coactivator. Genes Dev 12:3343–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento AS, Dias SM, Nunes FM, Aparício R, Ambrosio AL, Bleicher L, Figueira AC, Santos MA, de Oliveira Neto M, Fischer H, Togashi M, Craievich AF, Garratt RC, Baxter JD, Webb P, Polikarpov I 2006 Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function. J Mol Biol 360:586–598 [DOI] [PubMed] [Google Scholar]
- Togashi M, Borngraeber S, Sandler B, Fletterick RJ, Webb P, Baxter JD 2005 Conformational adaptation of nuclear receptor ligand binding domains to agonists: potential for novel approaches to ligand design. J Steroid Biochem Mol Biol 93:127–137 [DOI] [PubMed] [Google Scholar]
- Ribeiro RC, Feng W, Wagner RL, Costa CH, Pereira AC, Apriletti JW, Fletterick RJ, Baxter JD 2001 Definition of the surface in the thyroid hormone receptor ligand binding domain for association as homodimers and heterodimers with retinoid X receptor. J Biol Chem 276:14987–14995 [DOI] [PubMed] [Google Scholar]
- Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, Redinbo MR 2001 The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science 292:2329–2333 [DOI] [PubMed] [Google Scholar]
- Pike AC, Brzozowski AM, Hubbard RE, Bonn T, Thorsell AG, Engström O, Ljunggren J, Gustafsson JA, Carlquist M 1999 Structure of the ligand-binding domain of oestrogen receptor β in the presence of a partial agonist and a full antagonist. EMBO J 18:4608–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D, Gronemeyer H 1996 A canonical structure for the ligand binding domain of nuclear receptors. Nat Struct Biol 3:87–94 [DOI] [PubMed] [Google Scholar]
- Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D 1995 Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-R. Nature 375:377–382 [DOI] [PubMed] [Google Scholar]
- Heldring N, Pawson T, McDonnell D, Treuter E, Gustafsson JA, Pike AC 2007 Structural insights into corepressor recognition by antagonist-bound estrogen receptors. J Biol Chem 282:10449–10455 [DOI] [PubMed] [Google Scholar]
- Webb P, Nguyen NH, Chiellini G, Yoshihara HA, Cunha Lima ST, Apriletti JW, Ribeiro RC, Marimuthu A, West BL, Goede P, Mellstrom K, Nilsson S, Kushner PJ, Fletterick RJ, Scanlan TS, Baxter JD 2002 Design of thyroid receptor antagonists from first principles. J Steroid Biochem Mol Biol 83:59–73 [DOI] [PubMed] [Google Scholar]
- Kosztin D, Izrailev S, Schulten K 1999 Unbinding of retinoic acid from its receptor studied by steered molecular dynamics. Biophys J 76:188–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Wilson EM, Li Y, Moller DE, Smith RG, Zhou G 2000 Ligand-induced stabilization of PPARγ monitored by NMR spectroscopy: implications for nuclear receptor activation. J Mol Biol 298:187–194 [DOI] [PubMed] [Google Scholar]
- Hamuro Y, Coales SJ, Morrow JA, Molnar KS, Tuske SJ, Southern MR, Griffin PR 2006 Hydrogen/deuterium exchange (H/D-Ex) of PPARγ LBD in the presence of various modulators. Protein Sci 15:1883–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frego L, Davidson W 2006 Conformational changes of the glucocorticoid receptor ligand binding domain induced by ligand and cofactor binding, and the location of cofactor binding sites determined by hydrogen/deuterium exchange mass spectrometry. Protein Sci 15:722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Broderick D, Leid ME, Schimerlik MI, Deinzer ML 2003 Dynamics and ligand-induced solvent accessibility changes in human retinoid X receptor homodimer determined by hydrogen deuterium exchange and mass spectrometry. Biochemistry 43:909–917 [DOI] [PubMed] [Google Scholar]
- Pissios P, Tzameli I, Kushner P, Moore DD 2000 Dynamic stabilization of nuclear receptor ligand binding domains by hormone or corepressor binding. Mol Cell 6:245–253 [DOI] [PubMed] [Google Scholar]
- Huber BR, Sandler B, West BL, Cunha Lima ST, Nguyen HT, Apriletti JW, Baxter JD, Fletterick RJ 2003 Two resistance to thyroid hormone mutants with impaired hormone binding. Mol Endocrinol 17:643–652 [DOI] [PubMed] [Google Scholar]
- Huber BR, Desclozeaux M, West BL, Cunha-Lima ST, Nguyen HT, Baxter JD, Ingraham HA, Fletterick RJ 2003 Thyroid hormone receptor-β mutations conferring hormone resistance and reduced corepressor release exhibit decreased stability in the N-terminal ligand-binding domain. Mol Endocrinol 17:107–116 [DOI] [PubMed] [Google Scholar]
- Shah V, Nguyen P, Nguyen NH, Togashi M, Scanlan TS, Baxter JD, Webb P 2008 Complex actions of thyroid hormone receptor antagonist NH3 on gene promoters in different cell lines. Mol Cell Endocrinol 296:69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engström O, Ohman L, Greene GL, Gustafsson JA, Carlquist M 1997 Molecular basis of agonism and antagonism in the estrogen receptor. Nature 389:753–758 [DOI] [PubMed] [Google Scholar]
- Engen JR, Gmeiner WH, Smithgall TE, Smith DL 1999 Hydrogen exchange shows peptide binding stabilizes motions in Hck SH2. Biochemistry 38:8926–8935 [DOI] [PubMed] [Google Scholar]
- Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR 2006 Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem 78:1005–1014 [DOI] [PubMed] [Google Scholar]
- Englander JJ., Del Mar C, Li W, Englander SW, Kim JS, Stranz DD, Hamuro Y, Woods Jr VL 2003 Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc Natl Acad Sci USA 100:7057–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai SY, Burris TP, Dodge JA, Montrose-Rafizadeh C, Wang Y, Pascal BD, Chalmers MJ, Griffin PR 2009 Unique ligand binding patterns between estrogen receptor α and β revealed by hydrogen-deuterium exchange. Biochemistry 48:9668–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Pérez E, Leid M, Schimerlik MI, de Lera AR, Deinzer ML 2007 Deuterium exchange and mass spectrometry reveal the interaction differences of two synthetic modulators of RXR LBD. Protein Sci 16:2491–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F 2008 Structure of the intact PPAR-γ-RXR-α nuclear receptor complex on DNA. Nature 456:350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhaji YA, Stoica I, Dennis S, Purisima EO, Lumbroso R, Beitel LK, Trifiro MA 2006 Impaired helix 12 dynamics due to proline 892 substitutions in the androgen receptor are associated with complete androgen insensitivity. Hum Mol Genet 15:921–931 [DOI] [PubMed] [Google Scholar]
- McGee TD, Edwards J, Roitberg AE 2008 Preliminary molecular dynamic simulations of estrogen receptor α ligand binding domain from antagonist to apo. Int J Environ Res Public Health 5:111–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik L, Lund JD, Schiøtt B 2007 Conformational dynamics of the estrogen receptor α: molecular dynamics simulations of the influence of binding site structure on protein dynamics. Biochemistry 46:1743–1758 [DOI] [PubMed] [Google Scholar]
- Sonoda MT, Martínez L, Webb P, Skaf MS, Polikarpov I 2008 Ligand dissociation from estrogen receptor is mediated by receptor dimerization: evidence from molecular dynamics simulations. Mol Endocrinol 22:1565–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez L, Webb P, Polikarpov I, Skaf MS 2006 Molecular Dynamics simulations of ligand dissociation from thyroid hormone receptors: evidence of the likeliest escape pathway and its implications for the design of novel ligands. J Med Chem 49:23–26 [DOI] [PubMed] [Google Scholar]
- Martínez L, Sonoda MT, Webb P, Baxter JD, Skaf MS, Polikarpov I 2005 Molecular dynamics simulations reveal multiple pathways of ligand dissociation from thyroid hormone receptors. Biophys J 89:2011–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez L, Nascimento AS, Nunes FM, Phillips K, Aparicio R, Dias SM, Figueira ACM, Lin JH, Nguyen P, Apriletti JW, Neves FA, Baxter JD, Webb P, Skaf MS, Polikarpov I 2009 Gaining ligand selectivity in thyroid hormone receptors via entropy. Proc Natl Acad Sci USA 106:20717–20722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueira AC, Neto Mde O, Bernardes A, Dias SM, Craievich AF, Baxter JD, Webb P, Polikarpov I 2007 Low-resolution structures of thyroid hormone receptor dimers and tetramers in solution. Biochemistry 46:1273–1283 [DOI] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL 1998 The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95:927–937 [DOI] [PubMed] [Google Scholar]
- Zhang J, Hu X, Lazar MA 1999 A novel role for helix 12 of retinoid X receptor in regulating repression. Mol Cell Biol 19:6448–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimuthu A, Feng W, Tagami T, Nguyen H, Jameson JL, Fletterick RJ, Baxter JD, West BL 2002 TR surfaces and conformations required to bind nuclear receptor corepressor. Mol Endocrinol 16:271–286 [DOI] [PubMed] [Google Scholar]
- Togashi M, Nguyen P, Fletterick R, Baxter JD, Webb P 2005 Rearrangements in thyroid hormone receptor charge clusters that stabilize bound 3,5′,5-triiodo-l-thyronine and inhibit homodimer formation. J Biol Chem 280:25665–25673 [DOI] [PubMed] [Google Scholar]
- Martínez L, Souza PC, Garcia W, Batista FA, Portugal RV, Nascimento AS, Nakahira M, Lima LM, Polikarpov I, Skaf MS 2010 On the denaturation mechanisms of the ligand binding domain of thyroid hormone receptors. J Phys Chem B 114:1529–1540 [DOI] [PubMed] [Google Scholar]
- Estébanez-Perpiñá E, Arnold LA, Arnold AA, Nguyen P, Rodrigues ED, Mar E, Bateman R, Pallai P, Shokat KM, Baxter JD, Guy RK, Webb P, Fletterick RJ 2007 A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci USA 104:16074–16079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner RL, Apriletti JW, McGrath ME, West BL, Baxter JD, Fletterick RJ 1995 A structural role for hormone in the thyroid hormone receptor. Nature 378:690–697 [DOI] [PubMed] [Google Scholar]
- Nguyen NH, Apriletti JW, Cunha Lima ST, Webb P, Baxter JD, Scanlan TS 2002 Rational design and synthesis of a novel thyroid hormone antagonist that blocks coactivator recruitment. J Med Chem 45:3310–3320 [DOI] [PubMed] [Google Scholar]
- Figueira AC, Dias SM, Santos MA, Apriletti JW, Baxter JD, Webb P, Neves FA, Simeoni LA, Ribeiro RC, Polikarpov I 2006 Human thyroid receptor forms tetramers in solution, which dissociate into dimers upon ligand binding. Cell Biochem Biophys 44:453–462 [DOI] [PubMed] [Google Scholar]
- Clauser KM, Baker P, Burlingame AL 1999 Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal Chem 71:2871–2882 [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C 1983 Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637 [DOI] [PubMed] [Google Scholar]
- Beitz E 2000 TeXshade: shading and labeling of multiple sequence alignments using LaTeX2e. Bioinformatics 16:135–139 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ 1994 CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL 1993 Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol 234:779–815 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM 1993 PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291 [Google Scholar]
- Feng W, Ribeiro RCJ, Wagner RL, Nguyen H, Apriletti JW, Fletterick RJ, Baxter JD, Kushner PJ, West BL 1998 Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science 5370:1747–1749 [DOI] [PubMed] [Google Scholar]
- Velasco LF, Togashi M, Walfish PG, Pessanha RP, Moura FN, Barra GB, Nguyen P, Rebong R, Yuan C, Simeoni LA, Ribeiro RC, Baxter JD, Webb P, Neves FA 2007 Thyroid hormone response element organization dictates the composition of active receptor. J Biol Chem 282:12458–12466 [DOI] [PubMed] [Google Scholar]
- Martínez JM, Martínez L 2003 Packing optimization for automated generation of complex system’s initial configurations for molecular dynamics and docking. J Comput Chem 24:819–825 [DOI] [PubMed] [Google Scholar]
- Martínez L, Andrade R, Birgin EG, Martínez JM 2009 A package for building initial configurations for molecular dynamics simulations. J Comput Chem 30:2157–2164 [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K 2005 Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M 1998 All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616 [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML 1983 Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935 [Google Scholar]
- Darden T, York D, Pedersen L 1993 Particle mesh Ewald: an Nlog(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092 [Google Scholar]
- Kearsley SK 1989 On the orthogonal transformation used for structural comparisons. Acta Cryst A 45:208–210 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.