Abstract
Activin, a member of the TGF-β superfamily, is an important modulator of FSH synthesis and secretion and is involved in reproductive dysfunctions and cancers. It also regulates ovarian follicle development. To understand the mechanisms and pathways by which activin regulates follicle function, we performed a microarray study and identified 240 activin regulated genes in mouse granulosa cells. The gene most strongly inhibited by activin was Cyp26b1, which encodes a P450 cytochrome enzyme that degrades retinoic acid (RA). Cyp26b1 has been shown to play an important role in male germ cell meiosis, but its expression is largely lost in the ovary around embryonic d 12.5. This study demonstrated that Cyp26b1 mRNA was expressed in granulosa cells of follicles at all postnatal developmental stages. A striking inverse spatial and temporal correlation between Cyp26b1 and activin-βA mRNA expression was observed. Cyp26b1 expression was also elevated in a transgenic mouse model that has decreased activin expression. The Cyp26 inhibitor R115866 stimulated the proliferation of primary cultured mouse granulosa cells, and a similar effect was observed with RA and activin. A pan-RA receptor inhibitor, AGN194310, abolished the stimulatory effect of either RA or activin on granulosa cell proliferation, indicating an involvement of RA receptor-mediated signaling. Overall, this study provides new insights into the mechanisms of activin action in the ovary. We conclude that Cyp26b1 is expressed in the postnatal mouse ovary, regulated by activin, and involved in the control of granulosa cell proliferation.
A novel activin regulated gene, Cyp26b1, was identified in the mouse ovary; the roles of Cyp26b1 and retinoic acid in the ovary are explored.
Mammalian ovarian follicle formation and development involves establishment of the initial follicle pool, follicle growth, proper maturation of eggs, and timely production and release of hormones (1,2). This process is critical for propagation of the species as well as for development and homeostasis. Regulation of follicle formation and development requires intrinsic and endocrine factors as well as interactions between multiple cell types within the ovary (1,2,3,4,5). Among the many intraovarian factors, substantial evidence indicates that activin and inhibin, members of the TGF-β superfamily, play important autocrine/paracrine roles in this process (6,7,8,9). Activin and inhibin were first isolated from gonadal sources as endocrine factors that either stimulate (activin) or suppress (inhibin) the synthesis and secretion of FSH by the pituitary gland (10,11,12,13,14,15,16). Later studies indicated that activin acts predominantly as a local paracrine and autocrine factor (17,18). Effects of activin on many biological processes, including wound repair (19,20), inflammation (20,21,22), renal tubule morphogenesis (23), hair follicle development (24), neuroprotection (19), glucose metabolism (25), and stem cell growth and differentiation (25,26), have been reported. In the ovary, activin can regulate granulosa cell proliferation and differentiation (6,27,28,29,30), increase FSH receptor expression in undifferentiated granulosa cells (31,32), and stimulate oocyte maturation (33). However, the genes regulated by activin in the ovary, and how the products of these genes may affect ovarian follicle formation and development, remain largely unknown.
Like other members of the TGF-β superfamily, activin signals through a receptor serine-threonine kinase/Smad protein pathway (34,35). The βA- and βB-subunits of activin as well as the α-subunit of the activin antagonist inhibin are all expressed in the ovary and localized predominantly to granulosa cells (36,37,38,39,40). Components of the activin signaling system, including activin receptors, signaling Smad proteins, and activin binding protein and antagonist follistatin, are also expressed within the granulosa cells (6,41,42,43). Expression of activin subunits and signaling proteins within the follicle further supports an autocrine/paracrine role of activin in this cell type and suggests granulosa cells are targets of activin action in the ovary.
Through a microarray study of activin-treated primary cultured mouse granulosa cells, we obtained a global view of activin effects on ovarian gene expression/pathways and identified Cyp26b1 (cytochrome P450, family 26, subfamily b, polypeptide 1) as a novel activin down-regulated gene that was expressed in the postnatal ovary. Because Cyp26b1 encodes an enzyme that degrades the potent morphogen retinoic acid (RA) (44,45), we further discovered that RA and a Cyp26 inhibitor stimulated granulosa cell proliferation. We show that RA receptor (RAR)-mediated signaling is involved in both RA- and activin-induced granulosa cell proliferation. Our findings provide new insights into the mechanisms of activin action in the ovary and suggest Cyp26b1 and RA to be novel candidates for regulating postnatal follicle formation and development.
Materials and Methods
Animals
Wild-type or MT-α inhibin transgenic mice on a CD-1 background (Harlan, Indianapolis, IN) (46) were maintained on a 12-h light, 12-h dark cycle (lights off at 1700 h) with food and water available ad libitum. Animals were cared for in accordance with all federal and institutional guidelines.
Primary granulosa cell collection, culture, and treatment
Granulosa cells were collected from ovaries from 22- to 23-d-old immature mice through follicle puncture and cultured as described previously (47). Oocytes were filtered out with a 40-μm cell strainer (BD Falcon, Bedford, MA). Granulosa cells were either used directly for RNA isolation or cultured in a humidified incubator at 37 C and 5% CO2 in a DMEM/F-12 medium (Invitrogen, Grand Island, NY) supplemented with 2 μg/ml insulin, 5 nm sodium selenite, 5 μg/ml transferrin, 0.04 μg/ml hydrocortisone, 50 μg/ml sodium pyruvate, and 10% fetal bovine serum (Invitrogen) (47). Cultured cells were given treatments at the following concentrations: activin A, 2 nm (R&D Systems, Minneapolis, MN); follistatin, 3 nm (provided by Dr. Teresa Woodruff, Northwestern University, Chicago, IL); TGF-β1, 100 pm (R&D Systems); all-trans RA, 0.007, 0.07, or 0.7 μm (Sigma, St. Louis, MO); R115866, 0.007, 0.07,or 0.7 μm (provided by Johnson and Johnson, Beerse, Belgium); or AGN193109, 1 μm (Santa Cruz Biotechnology, Santa Cruz, CA). Treatments were given for 24 h for microarray or real-time PCR studies and 72 h for cell proliferation assays. The doses of treatments were based on reported studies (45,47).
RNA isolation
Total RNA was isolated either from primary granulosa cells or from whole ovaries or testes using a QIAGEN RNeasy minikit (QIAGEN, Valencia, CA). RNA was subjected to on column deoxyribonuclease digestion and analyzed for quality using a bioanalyzer. Before microarray assays, a small portion of the samples was used for real-time RT-PCR analysis to confirm a consistent effect of treatments across replicate experiments on marker genes, including estrogen receptors (ERs) (47) and inhibin-α (43).
Microarray and data analysis
An Illumina BeadArray (Illumina, Inc., San Diego, CA) was used for gene expression profiling (48,49). RNA from PBS- (P, control), activin A- (A), and activin A plus its antagonist follistatin (A+F)-treated groups was isolated from four independent experiments. About 100 ng of each RNA sample was amplified using a modified Eberwine T7-based amplification protocol and hybridized to the Illumina Sentrix Mouse-6 Expression BeadChip (Illumina) that contains 48,000 sets of oligonucleotides (48,49). Among these probes, 14,641 gave positive signals in the granulosa cells, and the genes with expression values less than the background levels were removed from further analysis. A Bioconductor lumi package with the variance-stabilizing transform + robust spline normalization scheme was used to preprocess the data (50,51). Because each treatment condition was repeated four times, samples from experiment 1 were assigned as P1, A1, and A+F1, samples from experiment 2 as P2, A2, and A+F2, and so on. For each comparison, i.e. A-P, the package computed A1-P1, A2-P2, A3-P3, and A4-P4 first. Next, to evaluate the statistical significance of these differences, the empirical Bayes shrinkage moderated t statistics in lumi Bioconductor package was applied (52). To estimate the false discovery rate, the Bonferroni method was used. Only the genes with a false discovery rate less than 0.05 and with a 1.5- or more-fold change in gene expression were considered as significant.
Real-time PCR
Total RNA from granulosa cells or whole tissues was reverse transcribed with avian myeloblastosis virus-reverse transcriptase (Fisher Scientific, Pittsburgh, PA). Real-time PCR was then performed on a 7300 real-time PCR system using SYBR Green PCR master mix (Applied Biosystems, Warrington, UK). Primers were designed according to the complete mouse cDNA sequences of the above genes. A list of the primers used is shown in Table 1. Ribosomal protein L19 was used as an internal control for all measurements. Specificity of all the real-time PCR reactions was also confirmed by a single peak in the melt curves and a single band of the predicted size after agarose gel electrophoresis of the PCR products (data not shown).
Table 1.
List of primers used in this study
| Genes | PCR primers (5′–3′) | Product size (bp) |
|---|---|---|
| Rpl19 | CTG AAG GTC AAA GGG AAT GTG | 195 |
| GGA CAG AGT CTT GAT GAT CTC | ||
| Nap1l5 | AAG ATC GAG GCC AAG TTT GA | 143 |
| TCA TCA TCC TCT CCC TCC AG | ||
| Mycl1 | ACG GCA CTC CTA GTC TGG AA | 136 |
| ACG GTC ACC ACG TCA ATC TC | ||
| Fst | GAG TGA CTT ACT CCA GCG CC | 266 |
| AGC TTC CTT CAT GGC ACA CT | ||
| Mid1ip1 | GGC CTG AGT CAC TTG GAG AG | 149 |
| AAG CCG ATC TCC TGC TTG TA | ||
| Ihh | GAG CTC ACC CCC AAC TAC AA | 118 |
| TGA CAG AGA TGG CCA GTG AG | ||
| Rgs2 | GAG GAG AAG CGG GAG AAA AT | 150 |
| TTC CTC AGG AGA AGG CTT GA | ||
| Nr1h4 | AAG GGG ATG AGC TGT GTG TT | 120 |
| TGT ACA CGG CGT TCT TGG TA | ||
| Figf | GTA TGG ACT CAC GCT CAG CA | 137 |
| GGC GAC TTC TAC GCA TGT CT | ||
| Slit2 | CGT TTG GAA AAT GTT CAG CA | 103 |
| TGA AAC TGT CGT TCC CAA CA | ||
| Itgb1 | GTC AGC AAC GCA TAT CTG GA | 100 |
| ACA TTC CTC CAG CCA ATC AG | ||
| Cyp26b1 | AGC TAG TGA GCA CCG AGT GG | 146 |
| GGG CAG GTA GCT CTC AAG TG- | ||
| Inha | CTC CCA GGC TAT CCT TTT CC | 112 |
| TGG CCG GAA TAC ATA AGT GA | ||
| Esr1 | ACC ATT GAC AAG AAC CGG AG | 170 |
| CCT GAA GCA CCC ATT TCA TT | ||
| Esr2 | TGT GTG TGA AGG CCA TGA TT | 138 |
| TCT TCG AAA TCA CCC AGA CC | ||
| Cyp19a1 | GAC ACA TCA TGC TGG ACA CC | 100 |
| TGC CAG GCG TTA AAG TAA CC | ||
| Inhba | GAT CAT CAC CTT TGC CGA GT | 143 |
| TGG TCC TGG TTC TGT TAG CC |
Cell proliferation assay
Granulosa cells were plated at 4–5 × 104 cells/well, and HeLa cells were plated at 3 × 103 cells/well into 96-well plates. After treatments, cell proliferation assays were performed using a CellTiter96 AQueous One Solution cell proliferation assay reagent (Promega, Madison, WI) following the manufacturer’s instructions. Absorbance at 490 nm was obtained using a 96-well plate reader. Cell numbers were calculated according to readings from standards plated in the same plates.
In situ hybridization
Ovaries collected from CD-1 mice at different ages were fixed in 4% paraformaldehyde and embedded in paraffin. Five-micrometer serial sections were obtained and mounted on Superfrost-Plus slides (Fisher Scientific). Digoxigenin-labeled RNA antisense and sense probes for Cyp26b1 were kindly provided by Dr. David Page (Massachusetts Institute of Technology, Boston, MA). Digoxigenin-labeled RNA antisense and sense probes for activin-βA (Inhba) were generated by cloning of a 143-bp PCR product (see primers in Table 1) into the SP6/T7 dual-promoter pCR II vector using a TA cloning kit (Invitrogen) and labeling of the transcript with a digoxigenin RNA labeling kit (Roche, Indianapolis, IN). The probes were hybridized to the sections at 50 C overnight and visualized with an alkaline phosphatase-conjugated antidigoxigenin antibody (Santa Cruz) and 4-nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indoyl-phosphate, 4-toluidine salt chromogen (Sigma). Sections were counterstained with nuclear Fast Red (Aldrich Chemical Corp., Milwaukee, WI).
Western blot
Protein homogenates were collected from primary cultured granulosa cells or ovaries or testes of 20-d-old mice. Protein homogenates were prepared in a buffer containing 50 mm Tris-HCl, 120 mm NaCl, 5 mm KCl, 1 mm MgSO4, 1 mm CaCl2, 10% glycerol, 0.5 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride (Roche Molecular Biochemicals, Indianapolis, IN) and 0.1 mm bacitracin (Sigma) (pH 7.4) at 4 C. Proteins were electrophoresed and transferred to nitrocellulose membranes. Blots were incubated overnight at 4 C with goat anti-Cyp26b1 antibody (Everest Biotech, Oxfordshire, UK; 1:1000 dilution), followed by a 1-h incubation at room temperature with horseradish peroxidase-labeled antigoat secondary antibody (Zymed Laboratories, South San Francisco, CA; 1:8000 dilution). Proteins were then visualized by chemiluminescence. The blots were scanned by densitometry. The intensities of the protein bands were analyzed using the public domain NIH Image program (National Institutes of Health; rsb.info.nih.gov/nih-image). The pixel intensity of each protein band was normalized against that of the corresponding loading control, which was actin. The relative intensity of the protein band was then obtained from the ratio of the experimental group over the control.
Statistics
Data are presented as the means ± se. For comparisons between two groups, a Student’s two-tailed t test was used. For comparisons among multiple groups, one-way ANOVA followed by a Tukey’s test was used. P < 0.05 was considered significant.
Results
Gene expression profiles
To obtain gene expression profiles of activin targets, primary granulosa cells were collected from d 22–23 immature mice and treated with activin A (2 nm), activin A (2 nm) plus its antagonist follistatin (3 nm), or PBS (control) for 24 h. RNA from each treatment group was then isolated for microarray analysis. The results revealed that a total of 240 genes were significantly regulated by activin A compared with control (A-P). Among those 240 activin-regulated genes, 236 were specific because their regulation was abolished or reversed in the activin A plus follistatin treatment group (Supplemental Fig. 1, published on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org). Among these 236 genes, 127 genes were stimulated (range 1.5–18 fold) and 109 genes were inhibited (range 1.5–11 fold) by activin (Fig. 1). Cotreatment with follistatin almost totally reversed activin A’s effects on the expression of these genes (Fig. 1). In most cases, the combination of activin A and follistatin resulted in a change opposite to that of activin A alone, suggesting that follistatin (which was present in access) was acting to suppress endogenous activin action. This is consistent with the fact that these cells are known to produce activin A in culture (53). Among the activin-stimulated genes, follistatin was induced 5.72-fold and inhibin-α was induced 3.17-fold, consistent with previous reports and therefore serving as validation for the microarray results (54,55).
Figure 1.
A heat map overview of all the genes that are significantly regulated by activin A as identified by Illumina BeadArray (n = 4). Red, Up-regulation; green, down-regulation. Act, Activin A; Fst, follistatin.
Using Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com), we found that activin regulated genes in granulosa cells are strongly associated with functions including cell signaling, cell death, cellular growth and proliferation, cancer, cell movement, development, cell cycle, and tissue morphology (Supplemental Fig. 2). This is consistent with the known direct actions of other TGF-β superfamily ligands (56,57,58,59,60). To further understand mechanisms of activin action and validate our data, we also performed signaling pathway analysis according to the Kyoto Encyclopedia of Genes and Genomes pathway definitions. The MAPK signaling pathway was the pathway that was most significantly regulated by activin A (Supplemental Fig. 3A). The MAPK signaling pathway is critical for cell proliferation, differentiation, apoptosis, and development (for a review see Ref. 61). Consistent with this finding, we observed that activin A stimulated mouse granulosa cell proliferation in primary culture, and this stimulatory effect was reversed by follistatin (Supplemental Fig. 3B). Follistatin alone suppressed granulosa cell proliferation (Supplemental Fig. 3B).
Quantitative PCR verification
To confirm the results from the microarray assay, seven genes induced by 2- to 15-fold and four repressed by 2- to 6-fold on the array were selected for validation by quantitative RT-PCR. These genes were selected based on a combination of criteria to include genes expected to be regulated by activin (Fst and Inha) and to include genes that were regulated over the dynamic range observed in the array study. Real-time PCR was performed, and these results were consistent with the microarray measurements, although there was some variance in the magnitude of the induction or repression (Fig. 2A). All of these genes also showed rapid activin responses in experiments in which granulosa cells were treated with activin A for only 4 h. The activin responses after 4 h were generally similar to the corresponding longer-term activin responses after 24 h, although, as expected, some differences were observed between the two time points with several genes further increasing or decreasing by 24 h (Fig. 2B).
Figure 2.
Validation of microarray results and time-course study of activin effect on selected genes. A, Validation of select activin up- and down-regulated genes obtained from microarray results with real-time RT-PCR (P < 0.05 for all; microarray, n = 4; real-time RT-PCR, n = 3–5). B, Time-course study of activin effect on selected genes. The same data for 24 h from A are plotted here as part of the time-course comparison. Primary cultured granulosa cells were treated with activin A or PBS (control) for 4 or 24 h, and mRNA levels were measured with real-time RT-PCR (P < 0.05 for all, n = 3–5).
A novel activin-regulated gene: Cyp26b1
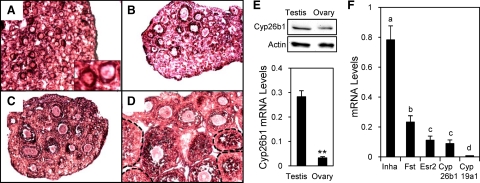
Cyp26b1 is the gene that was most significantly down-regulated by activin (−11-fold), suggesting that it may be an important factor mediating the intraovarian functions of activin. It has been shown to play an important role in male germ cell meiosis and survival, but its expression is reduced in the ovary around embryonic d 12.5–13.5 (44,45). To confirm and further localize the expression of Cyp26b1 in the postnatal ovary, we performed in situ hybridization and showed that Cyp26b1 mRNA was localized to pregranulosa cells of primordial follicles in the d 1 ovary and to granulosa cells of follicles at all developmental stages in d 6, 10, and 20 ovaries (Fig. 3, A–D). No staining was observed in ovaries probed with a control sense probe for Cyp26b1 (Supplemental Fig. 4). At postnatal d 1, the mRNA levels of Cyp26b1 were substantially higher in the testis than in the ovary (Fig. 3E, bottom panel); similar results were observed in d 6, 10, and 20 ovaries. The protein levels of Cyp26b1 were also higher in the testis than in the ovaries (relative intensity 1.51:1), although the difference was not as large as observed for mRNA levels (Fig. 3E, top panel). The relative mRNA levels of Cyp26b1 were compared with select well-characterized ovarian genes in mouse granulosa cells. Cyp26b1 mRNA was expressed at a comparable level with the ERβ mRNA, the predominant form of ER in the mouse ovary (Fig. 3F). As expected, inhibin-α and follistatin mRNAs showed high levels of expression, while Cyp19A1 (aromatase) mRNA was almost undetectable in the granulosa cells from immature animals due to lack of FSH stimulation (Fig. 3F), consistent with reports by others (62,63).
Figure 3.
Localization and relative expression levels of Cyp26b1 in the mouse ovary. A–D, In situ hybridization of Cyp26b1 in postnatal d 1 (A), 6 (B), 10 (C), and 20 (D) ovaries. Insert (A), Enlarged picture of two primordial follicles. E, top panel, Western blot showing the Cyp26b1 protein levels in d 1 testes and ovaries. Actin was detected as a loading control. Bottom panel, Comparison of Cyp26b1 mRNA levels in d 1 ovaries and testes as measured by real-time PCR. **, P < 0.005 (n = 4). F, Comparison of Cyp26b1 mRNA levels with inhibin-α (Inha), follistatin (Fst), ERβ (Esr2), and Cyp19a1 in d 22 mouse granulosa cells as measured by real-time PCR. Consistent amplification efficiency was observed for the primers for Inha, Fst, and Esr2 in the d 22 granulosa cell samples and for Cyp19a1 in adult ovaries (efficiency = 100 ± 3%, R2 = 0.990–0.999). Different letters indicate statistically significant differences, according to ANOVA followed by Tukey’s test (P < 0.05, n = 3–8).
Activin regulation of Cyp26b1 mRNA levels
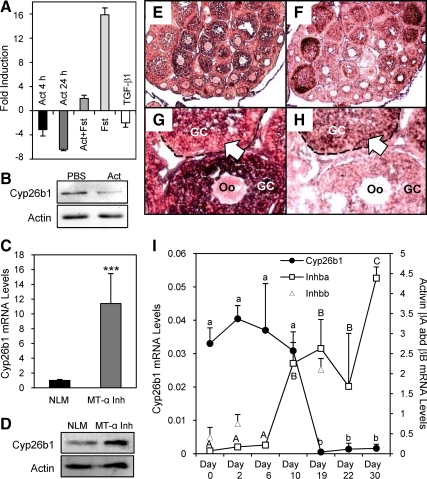
Activin suppression of Cyp26b1 mRNA levels was confirmed with real-time PCR analysis of RNA collected from primary cultured granulosa cells at both 4 and 24 h of treatments. This suppression was reversed when an excess amount of follistatin was given together with activin A, and follistatin alone significantly increased Cyp26b1 mRNA levels because these cells produce endogenous activin (36,37,38,64). TGF-β1 also decreased Cyp26b1 mRNA levels but to a lesser extent (Fig. 4A), suggesting a common effect among related TGF-β superfamily members. The suppressive effect of activin on Cyp26b1 expression was also observed at the protein level (the relative intensity of Cyp26b1 protein band in activin vs. PBS-treated samples is 1:0.44) (Fig. 4B). To confirm the importance of activin in regulating ovarian Cyp26b1 levels in vivo, we examined Cyp26b1 expression in a transgenic mouse model that overexpresses the inhibin-α subunit gene from a metallothionein-I promoter (MT-α inhibin) and as a consequence has decreased activin levels (65). These mice show decreased fertility and exhibit a variety of ovarian pathologies (46,66). The mRNA and protein levels of Cyp26b1 were significantly increased in the MT-α inhibin transgenic mouse ovary (the relative intensity of Cyp26b1 protein band in the normal litter mate (NLM) vs. MT-α inhibin transgenic mouse ovary is 1:1.64) (Fig. 4, C and D), consistent with a suppressive effect of activin on Cyp26b1 expression.
Figure 4.
Activin regulation of Cyp26b1 gene expression and the inverse correlation of Cyp26b1 and activin βA mRNA expression. A, Effects of activin A (Act), activin A+follistatin (Act+Fst), follistatin (Fst) alone, and TGF-β1 on Cyp26b1 mRNA levels in primary cultured granulosa cells (P < 0.05 for all, n = 3). B, Effect of activin A (Act) on Cyp26b1 protein levels. Actin was detected as a loading control. C, Comparison of Cyp26b1 mRNA levels in MT-α inhibin transgenic (MT-α Inh) mouse ovaries with those in the NLM ovaries. ***, P < 0.0001 (n = 7). D, Western blot pictures showing Cyp26b1 protein levels in NLM and MT-α Inh mouse ovaries. Actin was detected as a loading control. E–H, In situ hybridization of Cyp26b1 (E and G) and activin-βA (F and H) in postnatal d 20 ovaries. E and F show lower magnification; G and H show higher magnification. Complementary expression patterns of Cyp26b1 and activin-βA mRNA within one follicle is indicated by open arrows. GC, Granulosa cells. Oo, oocytes. I, Comparison of mRNA levels of Cyp26b1, activin-βA (Inhba), and activin-βB (Inhbb) in developing ovaries as measured by real-time PCR. Different letters indicate statistically significant differences, according to ANOVA followed by Tukey’s test (lowercase letters for Cyp26b1; uppercase letters for activin βA). P < 0.05 (n = 3–5).
In situ hybridization studies revealed that Cyp26b1 was lost in more mature follicles that express high levels of activin-βA subunit mRNA (Fig. 4, E and F) and that even within a single follicle, the expression of Cyp26b1 was decreased in regions in which activin-βA mRNA was highly expressed (Fig. 4, G and H, indicated by open arrows). This pattern was more pronounced in d 20 ovaries than in early ovaries, possibly due to low activin production at earlier times (see data below). No staining was observed in ovarian sections probed with a control sense probe for Cyp26b1 (Supplemental Fig. 5). A striking inverse correlation between Cyp26b1 expression and activin-βA expression was also observed during postnatal ovary development. Cyp26b1 mRNA levels remained high from d 0 to d 10 and decreased in d 19–30 ovaries, whereas activin-βA mRNA levels were low from d 0 to d 6 and increased starting at d 10 (Fig. 4I). We have also examined activin-βB mRNA levels in d 0, 2, and 19 mouse ovaries. The results revealed that activin-βB mRNA levels were lower at postnatal d 0 and 2 and increased at d 19, suggesting an inverse correlation between Cyp26b1 mRNA and activin-βB mRNA levels as well (Fig. 4I). Future studies will also examine the effect of activin B on Cyp26b1 expression.
RA and the Cyp26 inhibitor R115866 increase mouse granulosa cell proliferation
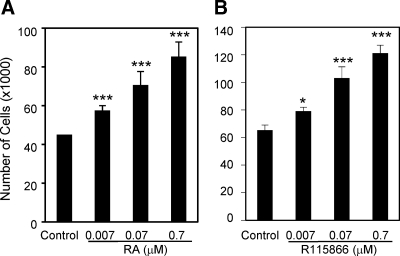
Cyp26b1 encodes an enzyme that degrades RA, and changes in Cyp26b1 expression can alter RA levels and result in significant biological consequences (44,45,67). To investigate functions of Cyp26b1 and RA in the ovary, we examined effect of RA on the proliferation of primary cultured mouse granulosa cells. RA significantly stimulated granulosa cell proliferation in a dose-response manner (Fig. 5A). As a control, we also examined the effects of RA on HeLa cell proliferation. Consistent with reports by others (68,69), RA moderately suppressed HeLa cell proliferation at high concentrations, although the effect was not significant (Supplemental Fig. 6). These results demonstrate that RA is a proliferation-stimulating factor for mouse granulosa cells and suggest that Cy26b1 may act to suppress granulosa cell proliferation. To examine this hypothesis, we treated primary cultured granulosa cells with the specific Cyp26 inhibitor R115866 (45) and found that R115866 increased granulosa cell numbers in a dose-response manner (Fig. 5B).
Figure 5.
Effects of RA (A) and the Cyp26 inhibitor, R115866 (B) on proliferation of primary cultured mouse granulosa cells. *, P < 0.05; ***, P < 0.005 (n = 3–7).
A pan-RAR inhibitor attenuates RA and activin-induced granulosa cell proliferation
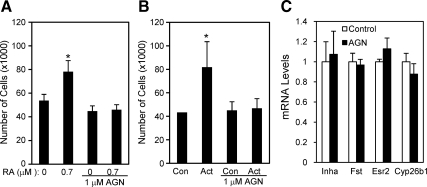
To examine signaling mechanisms involved in the stimulatory effect of RA, we treated mouse granulosa cells with RA in the presence of the pan-RAR inhibitor AGN193109 (70,71). AGN193109 completely blocked the stimulatory effect of RA on granulosa cell proliferation (Fig. 6A), whereas no cell toxicity was observed (76 vs. 75% cell viability in control and AGN193109 treated groups, respectively), suggesting that the effect was mediated through RARs. Because activin stimulates mouse granulosa cell proliferation and also suppresses Cyp26b1 expression, some of the proliferative effects of activin may be mediated by decreased Cyp26b1, leading to increased RA levels. We tested this and found that AGN194310 also abolished the stimulatory effect of activin on granulosa cell proliferation (Fig. 6B), indicating an involvement of RAR-mediated signaling in activin-induced granulosa cell proliferation. To examine the specificity of the effect of AGN194310 on activin-induced cell proliferation, we measured the mRNA levels of select activin regulated genes including inhibin-α (55), follistatin (54), ERα (Esr1) (47), and Cyp26b1 after AGN194310 treatment. The mRNA levels of these genes were not altered by AGN194310, suggesting that the drug is not acting as a general inhibitor of activin signaling (Fig. 6C).
Figure 6.
Effects of the pan-RAR inhibitor, AGN193109, on RA- (A) or activin A (B)-induced proliferation of primary cultured mouse granulosa cells and lack of effect of AGN193109 on select activin target genes (C). Con, Control; Act, activin A. *, P < 0.05 (n = 3–5).
Discussion
To understand the intraovarian functions of activin, we used global gene expression profiling to identify networks of genes that are either stimulated or inhibited by activin.
By treating primary granulosa cells with activin and activin plus follistatin, we were able to identify a group of genes whose expression is significantly regulated by activin compared with PBS and whose regulation is abolished or reversed when activin is given together with excess follistatin (by comparing activin plus follistatin vs. activin). Because excess follistatin was used in the study, it is likely that endogenous activin signaling was suppressed. Treatment with follistatin alone would help to further distinguish the extent to which endogenous activin signaling contributes to follistatin regulation of the activin regulated genes that we have identified.
Stimulatory effects of activin on granulosa cell proliferation have been reported previously in rat (30) and cow (72), although it has also been reported that proliferation and differentiation effects of activin in rat are not apparent unless FSH is present (73). Our study revealed a stimulatory effect of activin on proliferation of primary cultured mouse granulosa cells. We did not detect any changes in most of the key proteins involved in the execution of apoptosis, the caspases, with the exception of a decrease in caspase-6 expression (−1.75 fold) after activin treatment, suggesting suppression of apoptosis in the mouse granulosa cells. It has been reported that activin suppresses apoptosis in granulosa cells of cultured preantral follicles (74), although it induces apoptosis in liver cells (75), B cells (76,77), and decidual cells (78).
The microarray study revealed that Cyp26b1 is expressed in postnatal granulosa cells and is the gene most significantly suppressed by activin. It has been reported that Cyp26b1 expression is largely lost in female embryonic gonads around embryonic d 12.5, and this is believed to be critical for maintaining proper RA levels and ultimately allowing meiosis to progress (44,45). Expression of Cyp26b1 in the postnatal ovary has not been previously investigated. Although Cyp26b1 mRNA is expressed in much less abundance in the ovary than in the testis, it can be detected in pregranulosa cells at d 1 and is maintained at relatively high levels in the ovary during the first 10 d after birth before decreasing by d 19. The higher expression levels of Cyp26b1 during the first 10 d after birth coincide with active germ cell nest breakdown, massive oocyte apoptosis, somatic cell proliferation, and establishment of the initial follicle pool (3). Activin has been shown to be involved in early follicle formation and development (39,65), and here we demonstrate that it is a regulator of Cyp26b1, suggesting that Cyp26b1 may mediate some of activin’s actions in the ovary.
The suppressive effect of activin on Cyp26b1 expression was detected not only at 24 h treatment but also at 4 h treatment, suggesting a rapid regulation. Activin signals predominantly through Smad proteins, and Smads 3 and 4 can directly bind to Smad binding elements to regulate target gene expression (79). We have identified multiple potential Smad binding elements in the promoter region of the Cyp26b1 gene. However, activin can also function through other signaling pathways. Related to our finding that activin regulates the MAPK pathway, the MAPK pathway can in turn regulate Smad proteins. The nuclear factor-κB or phosphatidylinositol 3-kinase/AKT pathways can modulate or be activated by Smad pathways as well (80). Whether activin regulation of Cyp26b1 represents a direct transcriptional effect or an indirect effect through other signaling pathways requires further study.
In Cyp26b1 knockout mice, male germ cells undergo massive apoptosis when meiosis takes place prematurely at embryonic d 13.5 in the testis, whereas female germ cell numbers appear to be normal at birth in the ovary (67). This observation is not surprising because Cyp26b1 expression is already absent in the normal embryonic ovary at embryonic d 12.5 and meiosis can be initiated normally. No further analysis of postnatal ovary development was carried out in the Cyp26b1 knockout mice because they die shortly after birth. We observed that the Cyp26 inhibitor R115866 stimulated granulosa cell proliferation, suggesting that Cyp26b1 inhibits granulosa cell proliferation in the ovary. The stimulatory effect of R115866 on granulosa cell proliferation is consistent with the moderate stimulatory effect of activin on granulosa cell proliferation, suggesting that activin may induce cell proliferation in part through suppressing Cyp26b1 expression. These observations suggest that in addition to its recently reported roles in determining male germ cell fate and survival in embryos, Cyp26b1 may be a novel factor that mediates the autocrine/paracrine functions of activin in regulating ovarian follicle formation and development in the postnatal ovaries.
The importance of RA in ovary development has been suggested by several studies. Knockout of stimulated by retinoic acid gene 8 (Stra8), a RA-inducible gene whose expression is negatively regulated by Cyp26b1, results in depletion of oocytes at 8 wk after birth (81). Vitamin A and 9-cis-RA stimulate oocyte development and maturation (82,83). In this study, we discovered that RA is a potent stimulator of granulosa cell proliferation. The pan-RAR inhibitor, AGN194310, abolished the stimulatory effect of either RA or activin on granulosa cell proliferation, indicating an involvement of RAR-mediated signaling in both RA- and activin-induced granulosa cell proliferation.
Overall, this study provides new insights into activin functions in the female reproductive system. It demonstrates that Cyp26b1 is expressed in the postnatal mouse ovary and is regulated by activin and suggests that the activin may regulate RA pathways to modulate mouse granulosa cell proliferation and ovarian functions.
Supplementary Material
Acknowledgments
We thank Tyler Wellington for tissue processing and Jacob Avraham for help with primary cell cultures. We appreciate the generosity of Dr. David Page for providing Cyp26b1 probe constructs and Johnson and Johnson at Beerse, Belgium, for providing R115866. We also thank the Northwestern University microarray core and Dr. Pan Du and Dr. Simon Lin from Robert H. Lurie Comprehensive Cancer Center Bioinformatics Core for helping with the microarray assay and data analysis. The microarray data reported in this paper have been deposited in the Gene Expression Ominbus database (accession no. GSE20583).
Footnotes
This work was supported by Grant HD21921 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (to K.E.M.), a DePaul University Faculty Summer Research Grant (to J.L.K.), a DePaul University Undergraduate Research Assistantship (to A.G.), Elizabeth Morse Genius Charitable Trust Fund (to G.R.), and Louis Stokes Alliance for Minority Participation Program from the National Science Foundation (to G.R.).
Disclosure Summary: The authors have nothing to disclose.
Abbreviations: ER, Estrogen receptor; MT, metallothionein-I promoter; NLM, normal litter mate; RA, retinoic acid; RAR, RA receptor.
First Published Online November 17, 2010
References
- McGee EA, Hsueh AJ 2000 Initial and cyclic recruitment of ovarian follicles. Endocr Rev 21:200–214 [DOI] [PubMed] [Google Scholar]
- Edson MA, Nagaraja AK, Matzuk MM 2009 The mammalian ovary from genesis to revelation. Endocr Rev 30:624–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epifano O, Dean J 2002 Genetic control of early folliculogenesis in mice. Trends Endocrinol Metab 13:169–173 [DOI] [PubMed] [Google Scholar]
- Barnett KR, Schilling C, Greenfeld CR, Tomic D, Flaws JA 2006 Ovarian follicle development and transgenic mouse models. Hum Reprod Update 12:537–555 [DOI] [PubMed] [Google Scholar]
- Richards JS, Pangas SA 2010 The ovary: basic biology and clinical implications. J Clin Invest 120:963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight PG, Glister C 2001 Potential local regulatory functions of inhibins, activins and follistatin in the ovary. Reproduction 121:503–512 [DOI] [PubMed] [Google Scholar]
- Pangas SA, Rademaker AW, Fishman DA, Woodruff TK 2002 Localization of the activin signal transduction components in normal human ovarian follicles: implications for autocrine and paracrine signaling in the ovary. J Clin Endocrinol Metab 87:2644–2657 [DOI] [PubMed] [Google Scholar]
- Welt C, Sidis Y, Keutmann H, Schneyer A 2002 Activins, inhibins, and follistatins: from endocrinology to signaling. A paradigm for the new millennium. Exp Biol Med (Maywood, NJ) 227:724–752 [DOI] [PubMed] [Google Scholar]
- de Kretser DM, Hedger MP, Loveland KL, Phillips DJ 2002 Inhibins, activins and follistatin in reproduction. Human Reproduction Update 8:529–541 [DOI] [PubMed] [Google Scholar]
- Schwartz NB, Channing CP 1977 Evidence for ovarian “inhibin”: suppression of the secondary rise in serum follicle stimulating hormone levels in proestrous rats by injection of porcine follicular fluid. Proc Natl Acad Sci U S A 74:5721–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi M 1988 [Control mechanism of FSH secretion from the pituitary]. Nippon Sanka Fujinka Gakkai Zasshi 40:973–978 [PubMed] [Google Scholar]
- Ying SY 1988 Inhibins, activins, and follistatins: gonadal proteins modulating the secretion of follicle-stimulating hormone. Endocr Rev 9:267–293 [DOI] [PubMed] [Google Scholar]
- de Kretser DM, Robertson DM 1989 The isolation and physiology of inhibin and related proteins. Biol Reprod 40:33–47 [DOI] [PubMed] [Google Scholar]
- Vale W HA, Rivier C, Yu J 1990 The inhibin/activin family of hormones and growth factors. In: Sporn M RA ed. Growth factors and their receptors. Berlin: Springer-Verlag; 211–248 [Google Scholar]
- Woodruff TK, Mayo KE 1990 Regulation of inhibin synthesis in the rat ovary. Annu Rev Physiol 52:807–821 [DOI] [PubMed] [Google Scholar]
- Carroll RS, Kowash PM, Lofgren JA, Schwall RH, Chin WW 1991 In vivo regulation of FSH synthesis by inhibin and activin. Endocrinology 129:3299–3304 [DOI] [PubMed] [Google Scholar]
- DePaolo LV 1997 Inhibins, activins, and follistatins: the saga continues. Proc Soc Exp Biol Med 214:328–339 [DOI] [PubMed] [Google Scholar]
- Mather JP, Moore A, Li RH 1997 Activins, inhibins, and follistatins: further thoughts on a growing family of regulators. Proc Soc Exp Biol Med 215:209–222 [DOI] [PubMed] [Google Scholar]
- Sulyok S, Wankell M, Alzheimer C, Werner S 2004 Activin: an important regulator of wound repair, fibrosis, and neuroprotection. Mol Cell Endocrinol 225:127–132 [DOI] [PubMed] [Google Scholar]
- Werner S, Alzheimer C 2006 Roles of activin in tissue repair, fibrosis, and inflammatory disease. Cytokine, growth factor reviews 17:157–171 [DOI] [PubMed] [Google Scholar]
- Phillips DJ, de Kretser DM, Hedger MP 2009 Activin and related proteins in inflammation: not just interested bystanders. Cytokine, growth factor reviews 20:153–164 [DOI] [PubMed] [Google Scholar]
- Jones KL, de Kretser DM, Patella S, Phillips DJ 2004 Activin A and follistatin in systemic inflammation. Mol Cell Endocrinol 225:119–125 [DOI] [PubMed] [Google Scholar]
- Kojima I, Maeshima A, Zhang YQ 2001 Role of the activin-follistatin system in the morphogenesis and regeneration of the renal tubules. Mol Cell Endocrinol 180:179–182 [DOI] [PubMed] [Google Scholar]
- McDowall M, Edwards NM, Jahoda CA, Hynd PI 2008 The role of activins and follistatins in skin and hair follicle development and function. Cytokine, growth factor reviews 19:415–426 [DOI] [PubMed] [Google Scholar]
- Xia Y, Schneyer AL 2009 The biology of activin: recent advances in structure, regulation and function. J Endocrinol 202:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe T, Miyazono K 2009 Roles of TGF-beta family signaling in stem cell renewal and differentiation. Cell research 19:103–115 [DOI] [PubMed] [Google Scholar]
- Findlay JK 1993 An update on the roles of inhibin, activin, and follistatin as local regulators of folliculogenesis. Biol Reprod 48:15–23 [DOI] [PubMed] [Google Scholar]
- Findlay JK, Drummond AE, Dyson M, Baillie AJ, Robertson DM, Ethier JF 2001 Production and actions of inhibin and activin during folliculogenesis in the rat. Mol Cell Endocrinol 180:139–144 [DOI] [PubMed] [Google Scholar]
- Rabinovici J, Spencer SJ, Jaffe RB 1990 Recombinant human activin-A promotes proliferation of human luteinized preovulatory granulosa cells in vitro. J Clin Endocrinol Metab 71:1396–1398 [DOI] [PubMed] [Google Scholar]
- Miró F, Hillier SG 1996 Modulation of granulosa cell deoxyribonucleic acid synthesis and differentiation by activin. Endocrinology 137:464–468 [DOI] [PubMed] [Google Scholar]
- Xiao S, Robertson DM, Findlay JK 1992 Effects of activin and follicle-stimulating hormone (FSH)-suppressing protein/follistatin on FSH receptors and differentiation of cultured rat granulosa cells. Endocrinology 131:1009–1016 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Nakamura K, Igarashi S, Tano M, Miyamoto K, Ibuki Y, Minegishi T 1995 Interaction between activin A and cAMP in the induction of FSH receptor in cultured rat granulosa cells. J Endocrinol 147:103–110 [DOI] [PubMed] [Google Scholar]
- Sadatsuki M, Tsutsumi O, Yamada R, Muramatsu M, Taketani Y 1993 Local regulatory effects of activin A and follistatin on meiotic maturation of rat oocytes. Biochem Biophys Res Commun 196:388–395 [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL 2002 Signal transduction by the TGF-β superfamily. Science 296:1646–1647 [DOI] [PubMed] [Google Scholar]
- Datto M, Wang XF 2000 The Smads: transcriptional regulation and mouse models. Cytokine Growth Factor Rev 11:37–48 [DOI] [PubMed] [Google Scholar]
- Woodruff TK, D'Agostino J, Schwartz NB, Mayo KE 1988 Dynamic changes in inhibin messenger RNAs in rat ovarian follicles during the reproductive cycle. Science 239:1296–1299 [DOI] [PubMed] [Google Scholar]
- Meunier H, Rivier C, Evans RM, Vale W 1988 Gonadal and extragonadal expression of inhibin α, βA, and βB subunits in various tissues predicts diverse functions. Proc Natl Acad Sci USA 85:247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykema JC, Rahal JO, Mayo KE 1991 Regulation of inhibin and activin genes in the rat ovary. In: Gibori G, ed. Signaling mechanisms and gene expression in the ovary. New York: Springer-Verlag; 99–111 [Google Scholar]
- Kipp JL, Kilen SM, Bristol-Gould S, Woodruff TK, Mayo KE 2007 Neonatal exposure to estrogens suppresses activin expression and signaling in the mouse ovary. Endocrinology 148:1968–1976 [DOI] [PubMed] [Google Scholar]
- Sidis Y, Fujiwara T, Leykin L, Isaacson K, Toth T, Schneyer AL 1998 Characterization of inhibin/activin subunit, activin receptor, and follistatin messenger ribonucleic acid in human and mouse oocytes: evidence for activin’s paracrine signaling from granulosa cells to oocytes. Biol Reprod 59:807–812 [DOI] [PubMed] [Google Scholar]
- MacConell LA, Leal AM, Vale WW 2002 The distribution of β-glycan protein and mRNA in rat brain, pituitary, and gonads: implications for a role for β-glycan in inhibin-mediated reproductive functions. Endocrinology 143:1066–1075 [DOI] [PubMed] [Google Scholar]
- Drummond AE, Le MT, Ethier JF, Dyson M, Findlay JK 2002 Expression and localization of activin receptors, Smads, and β-glycan to the postnatal rat ovary. Endocrinology 143:1423–1433 [DOI] [PubMed] [Google Scholar]
- Shimasaki S, Koga M, Buscaglia ML, Simmons DM, Bicsak TA, Ling N 1989 Follistatin gene expression in the ovary and extragonadal tissues. Mol Endocrinol 3:651–659 [DOI] [PubMed] [Google Scholar]
- Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J, Hamada H, Koopman P 2006 Retinoid signaling determines germ cell fate in mice. Science 312:596–600 [DOI] [PubMed] [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC 2006 Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci USA 103:2474–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BN, McMullen ML, Pei L, Yates CJ, Mayo KE 2001 Reproductive deficiencies in transgenic mice expressing the rat inhibin α-subunit gene. Endocrinology 142:4994–5004 [DOI] [PubMed] [Google Scholar]
- Kipp JL, Kilen SM, Woodruff TK, Mayo KE 2007 Activin regulates estrogen receptor gene expression in the mouse ovary. J Biol Chem 282:36755–36765 [DOI] [PubMed] [Google Scholar]
- Kuhn K, Baker SC, Chudin E, Lieu MH, Oeser S, Bennett H, Rigault P, Barker D, McDaniel TK, Chee MS 2004 A novel, high-performance random array platform for quantitative gene expression profiling. Genome Res 14:2347–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes M, Freudenberg J, Thompson S, Aronow B, Pavlidis P 2005 Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res 33:5914–5923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM 2008 lumi: a pipeline for processing Illumina microarray. Bioinformatics 24:1547–1548 [DOI] [PubMed] [Google Scholar]
- Lin SM, Du P, Huber W, Kibbe WA 2008 Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res 36:e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK 2004 Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3 [DOI] [PubMed] [Google Scholar]
- Shikone T, Matzuk MM, Perlas E, Finegold MJ, Lewis KA, Vale W, Bradley A, Hsueh AJ 1994 Characterization of gonadal sex cord-stromal tumor cell lines from inhibin-α and p53-deficient mice: the role of activin as an autocrine growth factor. Mol Endocrinol 8:983–995 [DOI] [PubMed] [Google Scholar]
- Dalkin AC, Haisenleder DJ, Yasin M, Gilrain JT, Marshall JC 1996 Pituitary activin receptor subtypes and follistatin gene expression in female rats: differential regulation by activin and follistatin. Endocrinology 137:548–554 [DOI] [PubMed] [Google Scholar]
- LaPolt PS, Soto D, Su JG, Campen CA, Vaughan J, Vale W, Hsueh AJ 1989 Activin stimulation of inhibin secretion and messenger RNA levels in cultured granulosa cells. Mol Endocrinol 3:1666–1673 [DOI] [PubMed] [Google Scholar]
- Shi Y, Massagué J 2003 Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113:685–700 [DOI] [PubMed] [Google Scholar]
- von Bubnoff A, Cho KW 2001 Intracellular BMP signaling regulation in vertebrates: pathway or network? Dev Biol 239:1–14 [DOI] [PubMed] [Google Scholar]
- Hill CS 2001 TGF-β signalling pathways in early Xenopus development. Curr Opin Genet Dev 11:533–540 [DOI] [PubMed] [Google Scholar]
- Gordon KJ, Blobe GC 2008 Role of transforming growth factor-β superfamily signaling pathways in human disease. Biochim Biophys Acta 1782:197–228 [DOI] [PubMed] [Google Scholar]
- Siegel PM, Massagué J 2003 Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat Rev 3:807–821 [DOI] [PubMed] [Google Scholar]
- Zhang W, Liu HT 2002 MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res 12:9–18 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Robayna IJ, Alliston TN, Buse P, Firestone GL, Richards JS 1999 Functional and subcellular changes in the A-kinase-signaling pathway: relation to aromatase and Sgk expression during the transition of granulosa cells to luteal cells. Mol Endocrinol 13:1318–1337 [DOI] [PubMed] [Google Scholar]
- Kwintkiewicz J, Cai Z, Stocco C 2007 Follicle-stimulating hormone-induced activation of Gata4 contributes in the up-regulation of Cyp19 expression in rat granulosa cells. Mol Endocrinol 21:933–947 [DOI] [PubMed] [Google Scholar]
- Meunier H, Cajander SB, Roberts VJ, Rivier C, Sawchenko PE, Hsueh AJ, Vale W 1988 Rapid changes in the expression of inhibin α-, βA-, and βB-subunits in ovarian cell types during the rat estrous cycle. Mol Endocrinol 2:1352–1363 [DOI] [PubMed] [Google Scholar]
- McMullen ML, Cho BN, Yates CJ, Mayo KE 2001 Gonadal pathologies in transgenic mice expressing the rat inhibin α-subunit. Endocrinology 142:5005–5014 [DOI] [PubMed] [Google Scholar]
- Bristol-Gould SK, Hutten CG, Sturgis C, Kilen SM, Mayo KE, Woodruff TK 2005 The development of a mouse model of ovarian endosalpingiosis. Endocrinology 146:5228–5236 [DOI] [PubMed] [Google Scholar]
- Maclean G, Li H, Metzger D, Chambon P, Petkovich M 2007 Apoptotic extinction of germ cells in testes of Cyp26b1 knockout mice. Endocrinology 148:4560–4567 [DOI] [PubMed] [Google Scholar]
- Oridate N, Lotan D, Mitchell MF, Hong WK, Lotan R 1995 Inhibition of proliferation and induction of apoptosis in cervical carcinoma cells by retinoids: implications for chemoprevention. J Cell Biochem 23:80–86 [DOI] [PubMed] [Google Scholar]
- Massad LS, Turyk ME, Bitterman P, Wilbanks GD 1996 Interferon-α and all-trans-retinoic acid reversibly inhibit the in vitro proliferation of cell lines derived from cervical cancers. Gynecol Oncol 60:428–434 [DOI] [PubMed] [Google Scholar]
- Johnson AT, Klein ES, Gillett SJ, Wang L, Song TK, Pino ME, Chandraratna RA 1995 Synthesis and characterization of a highly potent and effective antagonist of retinoic acid receptors. J Med Chem 38:4764–4767 [DOI] [PubMed] [Google Scholar]
- Johnson AT, Wang L, Gillett SJ, Chandraratna RA 1999 High affinity retinoic acid receptor antagonists: analogs of AGN 193109. Bioorg Med Chem Lett 9:573–576 [DOI] [PubMed] [Google Scholar]
- Hulshof SC, Figueiredo JR, Beckers JF, Bevers MM, Vanderstichele H, van den Hurk R 1997 Bovine preantral follicles and activin: immunohistochemistry for activin and activin receptor and the effect of bovine activin A in vitro. Theriogenology 48:133–142 [DOI] [PubMed] [Google Scholar]
- Park Y, Maizels ET, Feiger ZJ, Alam H, Peters CA, Woodruff TK, Unterman TG, Lee EJ, Jameson JL, Hunzicker-Dunn M 2005 Induction of cyclin D2 in rat granulosa cells requires FSH-dependent relief from FOXO1 repression coupled with positive signals from Smad. J Biol Chem 280:9135–9148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun SY, Eisenhauer KM, Minami S, Billig H, Perlas E, Hsueh AJ 1996 Hormonal regulation of apoptosis in early antral follicles: follicle-stimulating hormone as a major survival factor. Endocrinology 137:1447–1456 [DOI] [PubMed] [Google Scholar]
- Chen W, Woodruff TK, Mayo KE 2000 Activin A-induced HepG2 liver cell apoptosis: involvement of activin receptors and smad proteins. Endocrinology 141:1263–1272 [DOI] [PubMed] [Google Scholar]
- Nishihara T, Okahashi N, Ueda N 1993 Activin A induces apoptotic cell death. Biochem Biophys Res Commun 197:985–991 [DOI] [PubMed] [Google Scholar]
- Nishihara T, Ohsaki Y, Ueda N, Koseki T, Eto Y 1995 Induction of apoptosis in B lineage cells by activin A derived from macrophages. J Interferon Cytokine Res 15:509–516 [DOI] [PubMed] [Google Scholar]
- Tessier C, Prigent-Tessier A, Bao L, Telleria CM, Ferguson-Gottschall S, Gibori GB, Gu Y, Bowen-Shauver JM, Horseman ND, Gibori G 2003 Decidual activin: its role in the apoptotic process and its regulation by prolactin. Biol Reprod 68:1687–1694 [DOI] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE 1998 Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell 1:611–617 [DOI] [PubMed] [Google Scholar]
- Javelaud D, Mauviel A 2005 Crosstalk mechanisms between the mitogen-activated protein kinase pathways and Smad signaling downstream of TGF-β: implications for carcinogenesis. Oncogene 24:5742–5750 [DOI] [PubMed] [Google Scholar]
- Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC 2006 In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 38:1430–1434 [DOI] [PubMed] [Google Scholar]
- Gómez E, Royo LJ, Duque P, Carneiro G, Hidalgo C, Goyache F, Lorenzo PL, Alvarez I, Facal N, Diez C 2003 9-Cis-retinoic acid during in vitro maturation improves development of the bovine oocyte and increases midkine but not IGF-I expression in cumulus-granulosa cells. Mol Reprod Dev 66:247–255 [DOI] [PubMed] [Google Scholar]
- Whaley SL, Hedgpeth VS, Britt JH 1997 Evidence that injection of vitamin A before mating may improve embryo survival in gilts fed normal or high-energy diets. J Anim Sci 75:1071–1077 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.