Abstract
Capillary isoelectric focusing and capillary zone electrophoresis are coupled with laser-induced fluorescence detection to create an ultrasensitive two-dimensional separation method for proteins. In this method, two capillaries are joined through a buffer filled interface. Separate power supplies control the potential at the injection end of the first capillary and at the interface; the detector is held at ground potential. Proteins are labeled with the fluorogenic reagent Chromeo P503, which preserves the isoelectric point of the labeled protein. The labeled proteins were mixed with ampholytes and injected into the first dimension capillary. A focusing step was performed with the injection end of the capillary at high pH and the interface at low pH. To mobilize components, the interface was filled with a high pH buffer, which was compatible with the second dimension separation. A fraction was transferred to the second dimension capillary for separation. The process of fraction transfer and second dimension separation was repeated two dozen times. The separation produced a spot capacity of 125.
1 Introduction
Multidimensional separations can provide exquisite resolution of complex mixtures. O’Farrell reported the archetypical example in 1975, in which a complex protein sample was first subjected to separation by isoelectric focusing followed by a second dimension separation using polyacrylamide gel electrophoresis [1]. This two-dimensional separation was performed on a rectangular electrophoresis plate, and resulted in the formation of a large number of spots, whose position was related to the isoelectric point and molecular weight of the proteins within the sample.
Giddings recognized that the spot capacity of a two-dimensional separation equals the product of the peak capacity of the individual separations for orthogonal separation mechanisms [2]. The combination of two high efficiency separation methods can result in very high spot capacity; isoelectric focusing-polyacrylamide gel electrophoresis can provide a spot capacity of over 10,000 [3].
Multidimensional chromatographic separations have also been developed for analysis of complex mixtures of peptides produced by the proteolytic digestion of cellular homogenates [4,5].
Detection remains an issue in protein analysis. Classic gel electrophoresis typically employs staining technology to reveal proteins with a detection limit in the sub-picomole range. Mass spectrometry can detect tryptic peptides in the high attomole range, although protein analysis tends to have lower sensitivity [6]. There are cases where higher sensitivity detection would be useful. As an extreme example, there is much interest in characterizing the protein content of a single cell, where the average protein abundance can be in the mid-zeptomole range [7–9].
Capillary electrophoresis coupled with laser-induced fluorescence is an attractive alternative to traditional proteomic techniques because it allows for analysis of small sample volumes, has high sensitivity, and can produce six or more orders of magnitude dynamic range [10–11]. We have reported multidimensional separation systems using capillary electrophoresis for sensitive and reproducible analysis of biological samples, including single-cell analyses [12–14]. In our first example, capillary zone electrophoresis at pH 7.5 was coupled with capillary zone electrophoresis at pH 11.1 for the two-dimensional separation of proteins [15]. We also reported the first coupling of capillary sieving electrophoresis, which is the capillary equivalent of polyacrylamide gel electrophoresis, with capillary zone electrophoresis [16]. That system was employed for the high-resolution separation of a cellular homogenate prepared from D. radiodurans. Kraly characterized the reproducibility of that system for the analysis of proteins obtained from human biopsies [17]. The within-day migration time precision was better than 1% in both dimensions and the limit of detection was in the high yoctomole (10−24 mol) range.
There have been a few efforts to couple capillary isoelectric focusing with capillary sieving electrophoresis. Sheng and Pawliszyn reported a two-dimensional capillary electrophoresis separation coupling micellar electrokinetic capillary chromatography in the first dimension to capillary isoelectric focusing using a 10-port valve interface with two loops [18]. Detection occurred on-column with whole-column absorbance imaging. There have been several reports of two-dimensional separations based on capillary isoelectric focusing/capillary zone electrophoresis system that featured an etched porous interface to join two separations on a single capillary [19–20]. Hydrodynamic mobilization was used to transfer plugs across the interface, and the analytes were detected on-column with a UV detector.
Capillary isoelectric focusing has had relatively limited application because of the poor sensitivity produced by absorbance detection across the narrow capillary diameter. Improved detection limits result from the use of labeling chemistry and laser-induced fluorescence detection. Unfortunately, most fluorescent labels are incompatible with capillary isoelectric focusing; those labels convert the cationic lysine residue into a neutral or anionic product, which generates very poor performance during isoelectric focusing. Fortunately, a set of fluorogenic reagents has been developed by Wolfbeis that convert cationic lysine residues into cationic fluorescent products [21–22]. These reagents, the Chromeo dyes, preserve the isoelectric focusing properties of proteins while providing outstanding sensitivity. We have reported three ultrasensitive capillary isoelectric focusing separation systems coupled with laser-induced fluorescence detection. In the first system, attomole detection limit of fluorescently labeled proteins were reported [23]. The sensitivity was limited by the background fluorescence generated by impurities found in the ampholytes. Several modifications were reported in the second system to reduce the background and improve sensitivity by an order of magnitude with concentration detection limits in the femtomolar range and mass detection limits in the zeptomole range [24]. These modifications included photobleaching the ampholytes to reduce impurities and replacing blue excitation with green, since the fluorescence generated by ampholyte impurities is more intense in the blue region of the spectrum. The most recent work employs an array of 32-capillaries for high-throughput analysis [25].
2 Materials and Methods
2.1 Chemicals and materials
Unless stated, all reagents were purchased from Sigma-Aldrich (St. Louis, MO USA). Solutions were made with distilled deionized water (Barnstead Nanopure, Boston, MA USA) and vacuum filtered through a 0.22 µm filter (Millipore, Billerica, MA USA). Biolyte ampholytes 5–8 and Biolyte ampholytes 7–9 where purchased from Bio-Rad (Hercules, CA USA). P503 was purchased from Active Motif (California). Uncoated and LPA coated fused-silica capillaries were purchased from Polymicro Technologies (Phoenix, AZ USA). Drummond glass capillaries used in the interface were from Drummond Scientific Co. (Broomall, PA).
2.2 Sample and ampholyte preparation
Protein samples were originally dissolved in water at a concentration of 10 mg/mL, aliquoted, and stored at −20 °C. Each day, a new sample was taken from the freezer and thawed at room temperature. Proteins were then labeled with Chromeo P503 [21–22, 26–28]. To label proteins, 5 µL of protein solution was added to 15 µL of Na2B4O7 buffer (10 mM, pH 9.2) and 5 µL of a 1 mM solution of Chromeo P503 dye dissolved in methanol. The labeling reaction took 15 min at room temperature and was observed to be complete when the solution color changed from blue to pink; exposure to room light was minimized to avoid photobleaching. Once labeled, 175 µL of ddH2O was added to quench the reaction. Just before an experiment, the sample was further diluted in 5% ampholyte solution containing 3% Pharmalytes 3–10, 1% each of Pharmalytes 5–8 and Pharmalytes 7–9 in 1% Triton X-100, 0.1% Tween 20.
2.3 Capillary electrophoresis instrumentation
Our laser-induced fluorescence detectors have been described in detail [17, 29–30]. Briefly, analytes were detected using a postcolumn sheath-flow cuvette. Fluorescence was excited by a 473-nm solid-state diode-pumped laser (Lasermate Group, Ponoma, CA), collected with an M-PLAN 60x, 0.7 NA microscope objective (Universe Kogaku, Oyster Bay, NY), and filtered with a 580LP long-pass filter (Omega Optical, Brattleboro, VT). Light was detected by an avalanche photodiode single-photon counting module (EG&G Canada, Vaudreuil, Canada). Voltage programming and fluorescence detection were controlled by home-built LabView software. The signal was corrected to account for the dead-time response of the photodetector.
We have previously described the instrumentation and interface for 2-D CE [15–16]. A few modifications were made to couple capillary isoelectric focusing with capillary zone electrophoresis. The two separation capillaries, with an i.d. of 50 µm, were aligned at a buffer-filled interface. Both capillaries were 20 cm in length and coated with either polymerized N-acryloylaminopropanol using the Grignard reaction or linear polyacrylamide provided by Polymicro.
Capillaries were coated using a device similar to that reported by Gao and Liu [31], using a slightly modified procedure from that reported by Gelfi [32]. Thionyl chloride was pumped through 3–4 m capillaries for 4 hours at 65 °C, and the capillary was then purged with N2 for 10 minutes (until bubbling could be seen at the outlet). The capillary was then filled with 1 M vinylmagnesium bromide in THF, which was allowed to react at 50 °C for another 4 hr. The capillary was then washed for 1 hr with THF at 30 psi, followed by a rinse with H2O for another hour. A solution of 0.1% TEMED, 0.1% ammonium persulfate, and 1.5% acryloylaminopropanol was prepared by mixing degassed solutions of each component (prepared immediately prior to this step), and pumped through the capillaries at 15 psi overnight. Capillaries were then washed with H2O for 1 hour, and stored filled with water.
2.4 One dimensional separation
One-dimensional separations were performed on 30 cm long, 50 µm i.d fused-silica capillary coated with polyacrylamide as described earlier [23–24]. The buffer for the capillary zone electrophoresis was 20 mM ammonium hydroxide with 3.5 mM SDS, pH 9. Sample was electrokinetically injected at −5 kV and separated at −15 kV. Anode detection capillary isoelectric focusing was performed in the same capillary. The capillary was cleaned before each run with 3 M HCl for 2 min and ddH2O for 5 min. The capillary was filled with ampholyte and sample mixture. The injection end of the capillary was placed in the catholyte, 40 mM NaOH, and the detection end of the capillary was placed in the cuvette where the sheath flow was the anolyte, 10 mM H3PO4. After focusing was complete, the sheath flow was switched from phosphoric acid to the mobilizing agent, 20 mM NH4OH – 3.5 mM sodium dodecyl sulfate (SDS). Focusing and mobilization voltage remained constant at −15 kV.
2.5 Two-dimensional separation: capillary isoelectric focusing/capillary zone electrophoresis
In two-dimensional separations, the cathode end of the capillary was placed in sodium hydroxide (40 mM, pH 12) and the anode end was in the interface filled with phosphoric acid (10 mM, pH 2). Capillary 2 was filled with the zone separation buffer, ammonium hydroxide and SDS (20 mM and 3.5 mM, pH 9). Before each experiment, the capillaries were rinsed with citric acid (100 mM) or 1% Triton X-100 for 4 min by using a syringe. The capillaries were then rinsed with ddH2O for 5 min. Capillary 1 was filled with a mixture of 5% ampholyte solution and sample by purging the solution through the capillary for 2 min at 5 psi. While filling capillary 1 with the ampholytes, a syringe was used to manually pump the anode into the interface, which flushed the matrix from capillary 1 through the interface and to waste. Focusing voltage was held constant at 660 V/cm for 7 min, or until the current stabilized. Chemical mobilization at the anode (interface) was used for migration of the proteins by changing the interface buffer from phosphoric acid to the second dimension buffer, ammonium hydroxide with SDS.
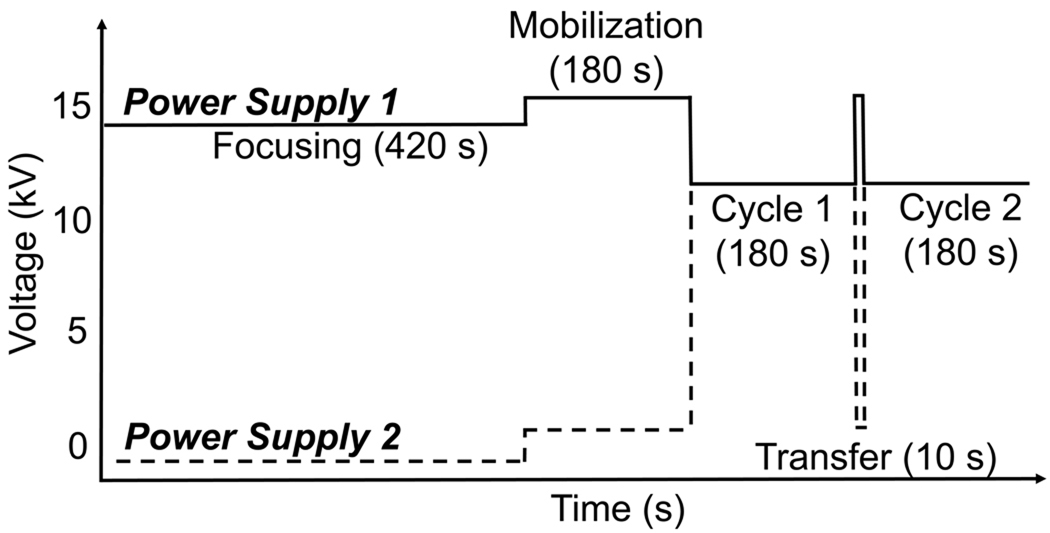
After focusing, fractions were electrokinetically transferred to capillary 2 by the application of 15 kV for 10s. Sample was separated in capillary 2 by applying field strength of 600 V/cm for 180 s. During this time, the net potential across capillary 1 was held at 0 V, preventing migration from capillary 1. These transfer and capillary zone electrophoresis separation cycles were repeated until all components from capillary 1 had been analyzed. The voltage program is illustrated in Figure 1.
Figure 1.
Voltage program for two-dimensional capillary electrophoresis. Analytes were focused in the first capillary during the 420 s focusing step. After focusing, a series of mobilization and second dimension separation cycles were preformed. In these cycles, sample as transferred for 10 s and then subjected to a 180 s duration second-dimension separation.
2.6 Data processing
Data were first corrected for the non-linear response of the avalanche photodiode photon counting modules [11, 33]. The corrected data were then treated with a five-point median filter to remove noise spikes from particles and convoluted with a Gaussian filter that had a 0.1 s standard deviation. The reconstructed two-dimensional electropherogram was convoluted with a two-dimensional Gaussian filter with a 0.4 transfer standard deviation in the isoelectric focusing dimension and a 40-ms standard deviation in the zone electrophoresis dimension.
A nonlinear least-squares routine was used to fit a Gaussian surface to the spots in the two-dimensional electropherogram. The function used for the fit is
where amplitude is the peak maximum, tcIEF is the migration time in the cIEF dimension in units of fractions transferred, is the peak center in the isoelectric focusing dimension, σcIEF is the standard deviation in the cIEF dimension, tcZE is the migration time in the CZE dimension in units of seconds, is the peak center in the CZE dimension, and σCZE is the standard deviation in the CZE dimension. The standard deviations of the Gaussian surface in the two dimensions were used to estimate spot capacity.
3 Results and discussion
3.1 One-dimensional separations of P503 labeled proteins
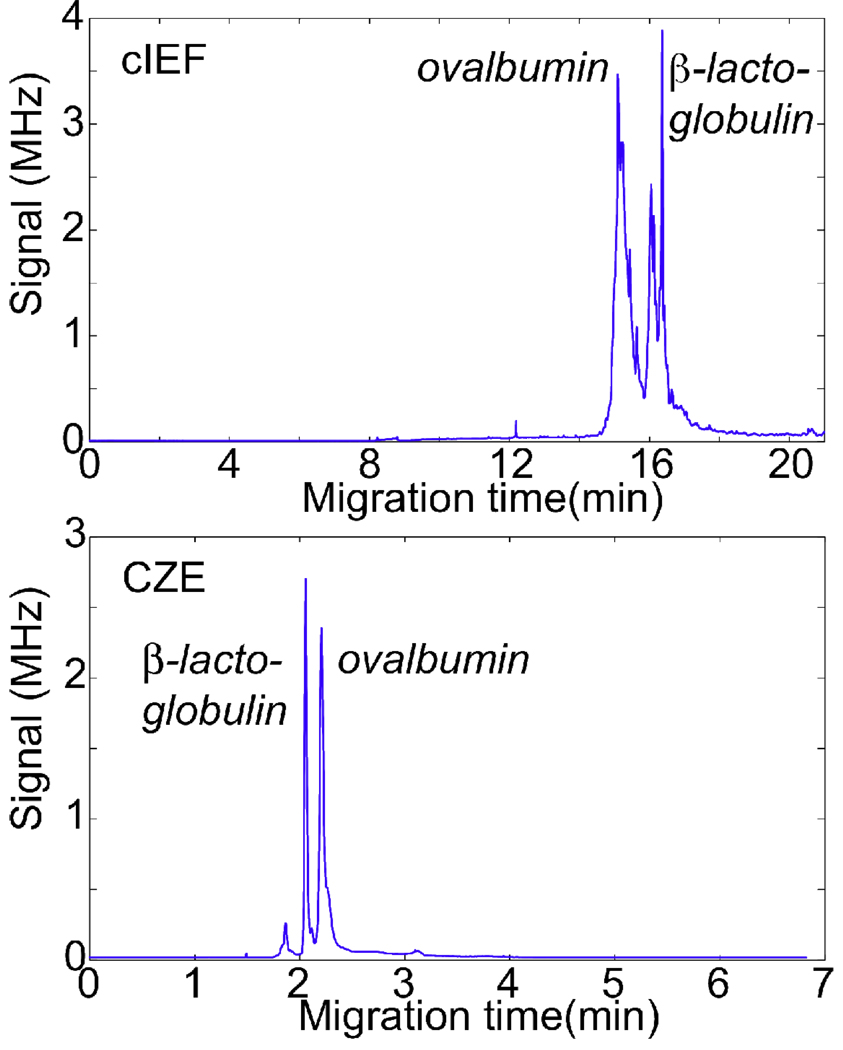
Two standard proteins (β-lactoglobulin (pI = 5.1) and ovalbumin (pI = 4.7)) were labeled with P503 and separated by capillary zone electrophoresis and capillary isoelectric focusing (Fig. 2). The electropherograms for both separations are shown in Figure 2. Zone electrophoresis shows two relatively sharp peaks while the isoelectric focusing separation presents a much more complex electropherogram.
Figure 2.
The top trace shows the capillary isoelectric focusing separation of ovalbumin (7 nM) and β-lactoglobulin (1 nM). Bottom trace shows capillary zone electrophoresis separation of ovalbumin (83 nM) and β-lactoglobulin (34 nM), injected at −5 kV for 2s.
3.2 Two-dimensional capillary isoelectric focusing / capillary zone electrophoresis separation of standard proteins spiked with a pI standard
Chemical mobilization of the components from capillary 1 across the interface to the capillary 2 requires compatibility with all the buffers involved, including the catholyte, anolyte, mobilization buffer, and the 2nd dimension buffer. Chemical mobilization in traditional capillary isoelectric focusing involves displacing the catholyte with a salt solution or acidic solution that matches the anolyte [34, 35]. The ions disrupt the pH gradient and allow migration of the focused zones. Coupling capillary isoelectric focusing with another separation mode requires either the physical separation of the two dimensions through a dialysis loop or that the mobilization buffer matches the 2nd dimension buffer. To simplify the instrument, we employed a basic mobilization buffer, which also served as the separation buffer in capillary zone electrophoresis mode. SDS was added to the capillary zone electrophoresis buffer to improve the separation by complexing with the proteins.
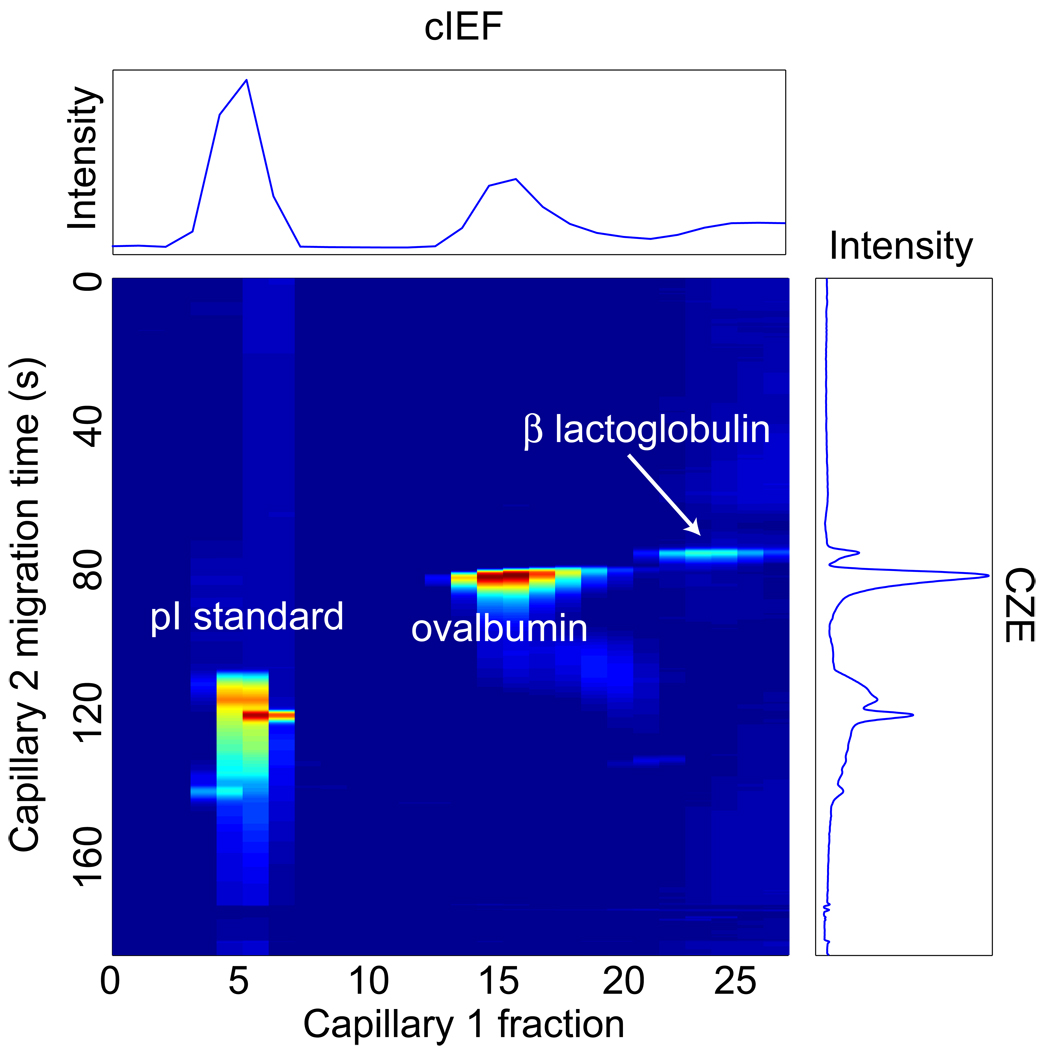
The two standard proteins were labeled with Chromeo 503 and separated by two-dimensional capillary isoelectric focusing / capillary zone electrophoresis. The sample was spiked with a fluorescent pI standard (4.0) synthesized and donated by Šlais [36]. Figure 3 presents the two-dimensional electropherogram. The separation in both dimensions was reasonably good. We used a nonlinear least-squares regression routine to fit a Gaussian surface to the three spots. The results of this fit are summarized in Table 1.
Figure 3.
Two-dimensional capillary isoelectric focusing/capillary zone electrophoresis separation of a 4.0 pI standard, ovalbumin (pI = 4.7), and β-lactoglobulin (pI = 5.2). The false-color heat-map uses red for the most intense and blue for the least-intense signals. The pI trace at the top is the summed electropherogram along that axis. The trace on the right is the summed capillary zone electrophoresis signal. The pI standard was present at 1 ng/mL, the labeled ovalbumin at 5 nM and β-lactoglobulin at 2 nM.
Table 1.
Least squares fit of a Gaussian surface to the spots of Figure 3
| spot | σcIEF (transfer) | σCZE (s) |
|---|---|---|
| β-lactoglobulin | 1.4 | 1.0 |
| ovalbumin | 1.2 | 1.7 |
| standard | 0.9 | 1.6 |
The standard deviation of the spots was 1.2 ± 0.2 transfers in the cIEF dimension and 1.4 ± 0.4 s in the CZE dimension. Note that these are the standard deviation of the spots; the full width at half height is a factor of 2.3 larger and the width at baseline is a factor of 4 larger. The measured peak capacity in the cIEF dimension is 4 and in the CZE dimension is 33, for an overall spot capacity of approximately 125.
The isoelectric focusing dimension produces poorer resolution in the two-dimensional separation compared with the one-dimensional separation. It is clear that the resolution is degraded by the relatively large volume of the fraction transferred to the second capillary. Decreasing this transfer volume will improve resolution, albeit at the expense of longer analysis time.
The laser-induced fluorescence detector worked well in this experiment. After filtering, the noise in the baseline was extremely low, generating a standard deviation of ~600 Hz. The maximum signal was 3 MHz, corresponding to a signal-to-noise ratio of over 5,000 and a concentration detection limit (3 σ) of ~5 pM for ovalbumin. The detection limit was poorer than our previous report [24]; that report employed photobleached ampholytes and green laser excitation, which dramatically improved detection limits.
Acknowledgements
This work was supported by grants from the National Institutes of Health (P50HG002360, R33CA122900). We gratefully acknowledge the donation of fluorescent pI standards by Karel Šlais from the Institute of Analytical Chemistry, Academy of Sciences of the Czech Republic, Brno.
References
- 1.O'Farrell PH. J. Biol. Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 2.Giddings C. Unified Separation Science. New York: Wiley; 1991. [Google Scholar]
- 3.Wittmann-Liebold B, Graack HR, Pohl T. Proteomics. 2006;6:4688–4703. doi: 10.1002/pmic.200500874. [DOI] [PubMed] [Google Scholar]
- 4.Washburn MP, Wolters D, Yates JR. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 5.Stoll DR, Carr PW. J. Amer. Chem. Soc. 2005;127:5034–5035. doi: 10.1021/ja050145b. [DOI] [PubMed] [Google Scholar]
- 6.Towers MW, McKendrick JE, Cramer R. J. Proteome Res. 2010 doi: 10.1021/pr901089j. in press. [DOI] [PubMed] [Google Scholar]
- 7.Krylov SN, Zhang Z, Chan NW, Arriaga E, Palcic MM, Dovichi NJ. Cytometry. 1999;37:14–20. doi: 10.1002/(sici)1097-0320(19990901)37:1<14::aid-cyto2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 8.Dovichi NJ, Hu S. Curr. Opin. Chem. Biol. 2003;7:603–608. doi: 10.1016/j.cbpa.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Wu H, Wheeler A, Zare RN. Proc. Natl. Acad. Sci. USA. 2004;101:12809–12813. doi: 10.1073/pnas.0405299101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu S, Zhang L, Cook LM, Dovichi NJ. Electrophoresis. 2001;22:3677–3682. doi: 10.1002/1522-2683(200109)22:17<3677::AID-ELPS3677>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 11.Whitmore CD, Essaka D, Dovichi NJ. Talanta. 2009;80:744–748. doi: 10.1016/j.talanta.2009.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Krylov S, Arriaga EA, Polakowski R, Dovichi NJ. Anal. Chem. 2000;72:318–322. doi: 10.1021/ac990694y. [DOI] [PubMed] [Google Scholar]
- 13.Hu S, Le Z, Krylov S, Dovichi NJ. Anal. Chem. 2003;75:3495–3501. doi: 10.1021/ac034153r. [DOI] [PubMed] [Google Scholar]
- 14.Hu S, Michels DA, Fazal MA, Ratisoontorn C, Cunningham ML, Dovichi NJ. Anal. Chem. 2004;76:4044–4049. doi: 10.1021/ac0498314. [DOI] [PubMed] [Google Scholar]
- 15.Michels DA, Hu S, Schoenherr RM, Eggertson MJ, Dovichi NJ. Mol. Cell. Proteomics. 2002;1:69–74. doi: 10.1074/mcp.t100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 16.Michels DA, Hu S, Dambrowitz KA, Eggertson MJ, Lauterbach K, Dovichi NJ. Electrophoresis. 2004;25:3098–3105. doi: 10.1002/elps.200405939. [DOI] [PubMed] [Google Scholar]
- 17.Kraly JR, Jones MR, Gomez DG, Dickerson JA, Harwood MM, Eggertson M, Paulson TG, Sanchez CA, Odze R, Feng Z, Reid BJ, Dovichi NJ. Anal. Chem. 2006;78:5977–5986. doi: 10.1021/ac061029+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheng L, Pawliszyn J. Analyst. 2002;127:1159–1163. doi: 10.1039/b206438c. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, El Rassi Z. J. Proteome Res. 2006;5:2001–2008. doi: 10.1021/pr060185u. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Zhang L, Zhu G, Zhang W, Zhang Y. Anal. Chem. 2004;76:6506–6512. doi: 10.1021/ac0493267. [DOI] [PubMed] [Google Scholar]
- 21.Wetzl BK, Yarmoluk SM, Craig DB, Wolfbeis OS. Angew. Chem. Int. Ed. Engl. 2004;43 doi: 10.1002/anie.200460508. 5400-2402. [DOI] [PubMed] [Google Scholar]
- 22.Craig DB, Wetzl BK, Duerkop A, Wolfbeis OS. Electrophoresis. 2005;26:2208–2213. doi: 10.1002/elps.200410332. [DOI] [PubMed] [Google Scholar]
- 23.Ramsay LM, Dickerson JA, Dovichi NJ. Electrophoresis. 2009;30:297–302. doi: 10.1002/elps.200800498. [DOI] [PubMed] [Google Scholar]
- 24.Ramsay LM, Dickerson JA, Dada O, Dovichi NJ. Anal. Chem. 2009;81:1741–1746. doi: 10.1021/ac8025948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dada OO, Ramsay LM, Dickerson JA, Cermak N, Jiang R, Zhu CR, Dovichi NJ. Anal. Bioanal. Chem. 2010 doi: 10.1007/s00216-010-3595-x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wojcik R, Swearingen KE, Dickerson JA, Turner EH, Ramsay LM, Dovichi NJ. J. Chromatogr A. 2008;1194:243–248. doi: 10.1016/j.chroma.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swearingen KE, Dickerson JA, Turner EH, Ramsay LM, Wojcik R, Dovichi NJ. J. Chromatogr A. 2008;1194:249–252. doi: 10.1016/j.chroma.2008.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner EH, Dickerson JA, Ramsay LM, Swearingen KE, Wojcik R, Dovichi NJ. J. Chromatogr A. 2008;1194:253–256. doi: 10.1016/j.chroma.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng YF, Dovichi NJ. Science. 1988;242:562–564. doi: 10.1126/science.3140381. [DOI] [PubMed] [Google Scholar]
- 30.Wu S, Dovichi NJ. J. Chromatogr. 1989;480:141–145. doi: 10.1016/s0021-9673(01)84284-9. [DOI] [PubMed] [Google Scholar]
- 31.Gao L, Liu S. Anal. Chem. 2004;76:7179–7186. doi: 10.1021/ac049353x. [DOI] [PubMed] [Google Scholar]
- 32.Gelfi C, Curcio M, Righetti PG, Sebastiano R, Citterio A, Ahmadzadeh H, Dovichi NJ. Electrophoresis. 1998;19:1677–1682. doi: 10.1002/elps.1150191026. [DOI] [PubMed] [Google Scholar]
- 33.Turner EH, Lauterbach K, Pugsley HR, Palmer VR, Dovichi NJ. Anal. Chem. 2007;79:778–781. doi: 10.1021/ac061778r. [DOI] [PubMed] [Google Scholar]
- 34.Hjerten S, Elenbring K, Kilar F, Liao JL, Chen AJC, Siebert CJ, Zhu MD. J. Chromatogr. 1987;403:47–61. doi: 10.1016/s0021-9673(00)96340-4. [DOI] [PubMed] [Google Scholar]
- 35.Hjerten S, Liao JL, Yao KQ. J. Chromatogr. 1987;387:127–138. doi: 10.1016/s0021-9673(01)94519-4. [DOI] [PubMed] [Google Scholar]
- 36.Šlais K, Horká M, Novácková J, Friedl Z. Electrophoresis. 2002;23:1682–1688. doi: 10.1002/1522-2683(200206)23:11<1682::AID-ELPS1682>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]